Abstract
Human DNA polymerase η (hPolη) is one of the newly identified Y-family of DNA polymerases. These polymerases synthesize past template lesions that are postulated to block replication fork progression. hPolη accurately bypasses UV-associated cis–syn cyclobutane thymine dimers in vitro and contributes to normal resistance to sunlight-induced skin cancer. We describe here mutational analysis of motif II, a highly conserved sequence, recently reported to reside in the fingers domain and to form part of the active site in Y-family DNA polymerases. We used a yeast-based complementation system to isolate biologically active mutants created by random sequence mutagenesis, synthesized the mutant proteins in vitro and assessed their ability to bypass thymine dimers. The mutability of motif II in 210 active mutants has parallels with natural evolution and identifies Tyr52 and Ala54 as prime candidates for involvement in catalytic activity or bypass. We describe the ability of hPolη S62G, a mutant polymerase with enhanced activity, to bypass five other site-specific lesions. Our results may serve as a prototype for studying other members of the Y-family DNA polymerases.
Keywords: bypass polymerases/DNA replication/Polη/Rad30A/Y-family
Introduction
DNA in cells is damaged by numerous endogenous and exogenous agents. The majority of altered nucleotides in DNA are removed by DNA repair processes that restore the original DNA sequence. However, some of the damage may escape repair and many of the unrepaired lesions block DNA synthesis by replicative DNA polymerases. As a result, the replication fork stalls and the cell must then employ additional mechanisms to complete replication in the presence of the unrepaired lesion (Lindahl and Wood, 1999).
In the last two years, a new family of DNA polymerases has been identified and are referred to as the Y-family, translesion synthesis (TLS) or bypass DNA polymerases (Bridges, 1999; Johnson et al., 1999c; Goodman and Tippin, 2000; Livneh, 2001; McDonald et al., 2001; Ohmori et al., 2001; Wang, 2001). These polymerases traverse blocking lesions in template DNA in vitro and it is hypothesized that they function in vivo to facilitate replication across damaged or missing bases. The Y-family of DNA polymerases includes Escherichia coli Pol IV (dinB; Wagner et al., 1999) and Pol V (umuC; Reuven et al., 1999; Tang et al., 1999) that have been reported to be required for the major mechanism of damage-inducible mutagenesis. The yeast Saccharomyces cerevisiae contains two Y-family DNA polymerases: Rev1, a DNA-dependent dCMP transferase (Nelson et al., 1996) and DNA polymerase η, encoded by the RAD30 gene (McDonald et al., 1997; Johnson et al., 1999b). Homologs of these bypass polymerases have been identified in higher eukaryotes. The mammalian translesion polymerases are similar to those of S.cerevisiae but include at least two other bypass polymerases that are similar to polymerase η. These are DNA polymerase κ, encoded by hDINB1 (Gerlach et al., 2001) and DNA polymerase ι, encoded by hRAD30B (McDonald et al., 2001).
Human DNA polymerase η (hPolη), encoded by hRAD30A, is inactive in the cancer-prone genetic disorder xeroderma pigmentosum variant syndrome (XPV; Johnson et al., 1999a; Masutani et al., 1999b). XPV patients are 1000-times more susceptible to sunlight-induced skin cancer than are normal individuals (Cleaver, 1999). In culture, XPV cells are hypersensitive to the mutagenic effects of UV irradiation (Maher et al., 1976). Purified hPolη synthesizes past cys–syn thymine–thymine (T–T) dimers in vitro with efficiency comparable with that observed for undamaged DNA, inserting primarily complementary adenines opposite the lesions (Johnson et al., 2000; Masutani et al., 2000). The phenotype of XPV cells and patients can be explained by the diminished accuracy of T–T dimer bypass caused by lack of hPolη.
In addition to T–T dimers, hPolη has been shown to replicate across a wide spectrum of damage with efficiency and accuracy that depends on the lesion and sequence context (Haracska et al., 2000a,b, 2001; Masutani et al., 2000; Vaisman et al., 2000; Zhang et al., 2000; Levine et al., 2001; Yu et al., 2001). The enzyme preferentially inserts complementary nucleotides opposite some of these lesions and non-complementary nucleotides opposite others. hPolη, like the other translesion synthesis polymerases, exhibits exceptionally low fidelity when replicating undamaged DNA (Matsuda et al., 2000). The low fidelity of these enzymes suggested they might have functions in addition to bypass of DNA damage. Recently, evidence has been presented that shows hPolη and other translesion synthesis polymerases are involved in somatic hypermutation of the immunoglobulin variable genes (Rogozin et al., 2001; Zeng et al., 2001).
The primary sequences of Y-family polymerases bear no discernible homology to those of other DNA polymerases. However, the recently solved crystal structures reveal a modified right hand-shaped enzyme, similar to the structures of the other DNA polymerases (Ling et al., 2001; Trincao et al., 2001; Zhou et al., 2001). The polymerase domain of hPolη resides in the 513 N-terminal amino acids of the 713 amino acid polypeptide (Masutani et al., 1999a; Trincao et al., 2001), while the 70 C-terminal residues are required for nuclear localization (Kannouche et al., 2001; Kondratick et al., 2001). The N-terminal region of hPolη shares extensive sequence homology with the N-terminal portion of other related translesion polymerases, including five conserved motifs designated I–V (Kulaeva et al., 1996; Johnson et al., 1999c) or A, B, D, E and F (Masutani et al., 1999b).
We present here mutational analysis of the conserved motif II/region B, revealed in the three-dimensional structures of Y-family DNA polymerases to reside in the fingers domain and to form part of the active site. We chose to substitute random nucleotides for all of the codons corresponding to 22 amino acids within motif II. We utilized a yeast-based functional complementation system to isolate active mutants and, by DNA sequencing of >200 such mutants, established a pattern of amino acid substitution that has parallels with evolutionary conservation and identifies residues that are likely to be involved in catalysis. We synthesized 95 of the active mutant proteins in vitro and, by using a primer extension assay, found accord between in vivo selection in our complementation system and the ability of the polymerase variants to bypass T–T dimers in vitro. One of the mutants, hPolη S62G, was found to exhibit higher activity than wild-type hPolη on undamaged DNA and on templates containing diverse lesions. Because motif II is highly conserved, our findings have implications for other bypass polymerases across the evolutionary spectrum.
Results
hPolη complements the UV sensitivity of S.cerevisiae rad30rad52
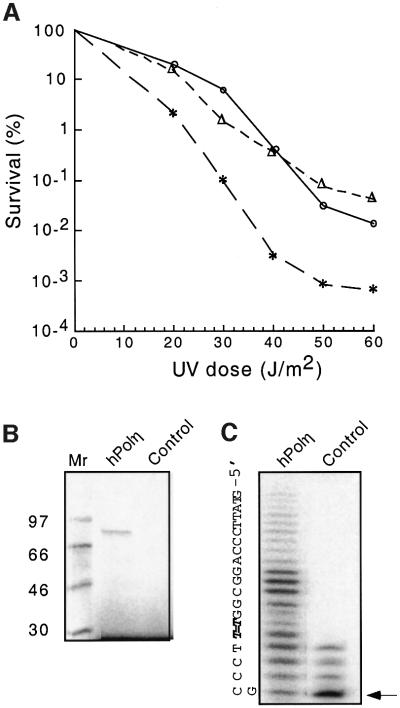
As a first step toward isolating and characterizing active hPolη mutants, we established in vivo and in vitro systems for detecting the function of plasmid-encoded enzymes. To select for mutants that are active in vivo, we used a complementation assay based on the UV sensitivity of the S.cerevisiae rad30rad52 double mutant. Deletion of the yeast hPolη homolog Rad30p and the recombination enzyme Rad52p greatly reduces UV survival (McDonald et al., 1997). As shown in Figure 1A, expression of wild-type hPolη in rad30rad52 cells enhances UV resistance as much as 100-fold at high doses, compensating for loss of endogenous Polη and increasing survival to that observed for the rad52 single mutant. This enhancement in survival provided the basis for selecting active mutants from plasmid libraries harboring hPolη variants.
Fig. 1. Plasmid-encoded wild-type human DNA polymerase η confers UV resistance and catalyzes bypass of cis–syn cyclobutane thymine dimers. (A) hPolη enhances UV resistance of the S.cerevisiae rad30rad52 double mutant that lacks both host Polη (Rad30p) and (Rad52p). Expression of hPolη in rad30rad52 cells increases survival to that observed with rad52 cells. ∗, rad30rad52; O, rad52; Δ, rad30rad52 transformed with pYEX-hPolη. (B) SDS–polyacrylamide gel analysis of the [35S]methionine-labeled products produced by in vitro transcription and translation with either pYEX-hPolη or pYEX-4T (control). Molecular weight markers are shown in the left lane. (C) In vitro bypass of a site-specific cis–syn T–T dimer in a primer extension assay. The template lesion (indicated by T=T) was four nucleotides downstream of the 3′ primer terminus (indicated by an arrow).
To assess the ability of the plasmid-encoded polymerase to bypass DNA lesions, we produced the enzyme in vitro and measured DNA synthesis on oligonucleotide template-primers containing site-specific adducts. A plasmid carrying the wild-type hPolη cDNA, pYEX-hPolη, served as a template for PCR amplification with an upstream primer containing the bacteriophage T7 promoter sequence. The amplified hPolη cDNA, under control of the T7 promoter, was thereafter used as a template for in vitro transcription and translation in the presence of 35S-labeled methionine. As shown in Figure 1B, SDS gel electrophoresis of the 35S-labeled translation product revealed a single major band migrating as expected for hPolη (78 kDa), whereas no labeled products were observed in reactions containing the control plasmid. The ability of in vitro synthesized hPolη to catalyze DNA polymerization and translesion synthesis was confirmed in primer extension assays. As shown in Figure 1C, the enzyme incorporated nucleotides onto a template-primer containing a cis–syn cyclobutane T–T dimer, a major lesion produced by UV damage to DNA. Extension opposite and past the dimer increased with both time of incubation and enzyme concentration (data not shown). The synthesis seen in the control reaction appears to be catalyzed by DNA polymerase activity intrinsic to the reticulocyte lysate used for transcription and translation. The extent of synthesis by the control was uniform among different lysates and in no case was significant bypass observed.
Characterization of translesion synthesis by in vitro synthesized and by purified hPolη
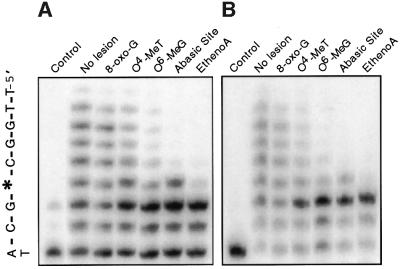
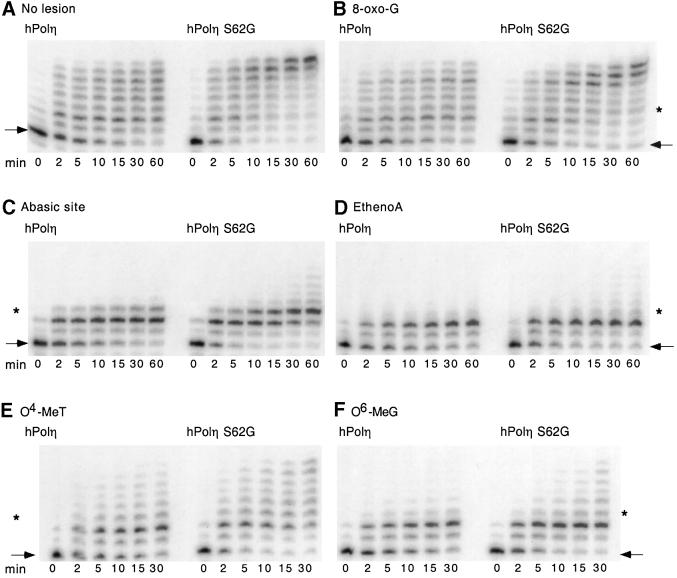
Wild-type hPolη has been shown to bypass a variety of DNA lesions, although differing DNA sequence contexts often complicate assessment of the relative efficiencies of bypass of different lesions. Prior to mutant characterization, we established the relative efficiencies of bypass of five adducts by in vitro synthesized wild-type hPolη, using a single common nucleotide sequence context containing different lesions at the same site. The enzyme bypassed 8-oxo-guanine (8-oxo-G), the major oxygen free radical-induced adduct, with an efficiency (80%) approaching that observed for the template having no lesion (Figure 2A). In our particular sequence context, hPolη bypassed the methylating agent-induced lesions O4-methylthymine (O4-MeT) and O6-methylguanine (O6-MeG) with lower efficiencies i.e. 55 and 20%, respectively, relative to the unmodified template. With an abasic site, hPolη incorporated a single nucleotide opposite the lesion, but failed to significantly extend the incorporated nucleotide. No significant incorporation was observed opposite 1, N6-ethenoadenine (ethenoA), a lesion associated with DNA damage by a metabolite(s) of the human carcinogen vinyl chloride. To demonstrate that this pattern of bypass is intrinsic to hPolη itself, we expressed the polymerase domain of the enzyme in E.coli (Masutani et al., 1999b; Trincao et al., 2001) and measured the activity of the purified protein on the same template primers. As shown in Figure 2B, bypass by the purified enzyme was very similar, if not identical, to that exhibited by the in vitro synthesized polymerase, indicating that the observed pattern of translesion synthesis is characteristic of hPolη. As reported by others for different sequence contexts (Masutani et al., 1999b; Johnson et al., 2000), we observed that hPolη copied a template containing a T–T dimer with efficiency comparable with that found for the same template without the lesion (Figure 1C and data not shown).
Fig. 2. Translesion synthesis by in vitro synthesized and purified hPolη on templates containing site-specific lesions in the same sequence context. Bypass across six different lesions within the same template sequence was measured in primer extension assays containing hPolη synthesized in vitro (A) or hPolη polymerase domain purified from E.coli (B). The template contained either no lesion, 8-oxo-G, O4-MeT, O6-MeG, an abasic site or ethenoA. The sequence of the single-stranded template is indicated with an asterisk marking the position of the lesion. Controls, both without expression vector, were reticulocyte lysate in (A) and purified cell extract in (B).
Creation of a library of hPolη motif II mutants
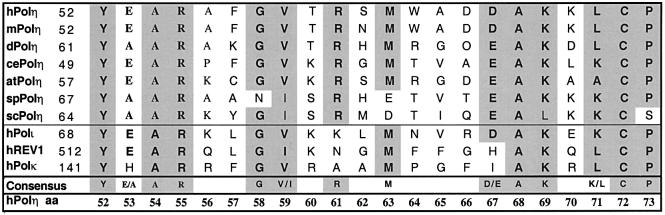
Motif II is highly conserved in natural evolution within the translesion synthesis DNA polymerases, suggesting that it may be important for the biological functions of these enzymes. Figure 3 shows the sequence of 22 amino acids in motif II, 11 of which are invariant, or nearly so, among Polηs across the evolutionary spectrum and among the Y-family DNA polymerases. As an initial approach to ascertaining the mutability of these motif II residues in hPolη and their roles in nucleotide incorporation and lesion bypass, we created a library of mutants containing amino acid replacements at all 22 positions. We substituted an oligonucleotide containing 7.5% random substitutions at each nucleotide encoding the amino acids Tyr52 through Pro73. The mutant library was calculated to contain five nucleotide substitutions per clone (Suzuki et al., 1996) and to encode approximately four amino acid substitutions per clone. The pYEX-hPolη-LibB plasmids were transformed into E.coli to create the hPolη motif II random library encoding ∼5 × 105 mutants.
Fig. 3. Alignment of motif II primary sequences. Amino acid sequence homology of motif II in human Polη, Polη from other species and related human DNA polymerases, were identified using the Blast search engine (NCBI). Conserved residues are indicated in bold type in shaded boxes. The sequences displayed are hPolη, mPolη (mouse), dPolη (Drosophila melanogaster), cePolη (Caenorhabditis elegans), atPolη (Arabidopsis thaliana), spPolη (Schizosaccharomyces pombe), scPolη (S.cerevisiae), hPolι (human), hREV1 (human) and hPolκ (human). The numbers on the left indicate the position of the N-terminal motif II residue in the protein.
Selection of hPolη motif II mutants that enhance UV survival
Active hPolη mutants were selected based on their ability to complement the UV sensitivity of S.cerevisiae rad30rad52, as demonstrated by the wild-type enzyme in Figure 1A. Plasmids comprising the mutant library were isolated from E.coli and transformed into rad30rad52 cells. The resulting yeast library contained approximately 1 × 105 independent hPolη mutants. The yeast library was grown to stationary phase, plated and exposed to UV-C radiation (40 J/m2). After three days, surviving colonies were picked for further analysis. By comparing the surviving fraction of cells transformed with the mutant library to that of cells transformed with plasmids encoding wild-type hPolη, we calculated that ∼2% of the library consists of active mutants.
The ability of selected mutants to rescue cells from lethal UV damage was verified by isolating the mutant plasmids from surviving colonies, retransforming them into the rad30rad52 host and exposing the transformants to increasing amounts of UV radiation. Many of the mutants conferred resistance comparable with that of wild type hPolη, exhibiting survival ∼100-fold greater than that of non-transformed cells (Figure 1A and data not shown).
Characterization of the unselected and selected mutant libraries
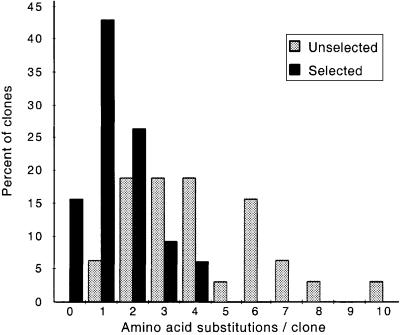
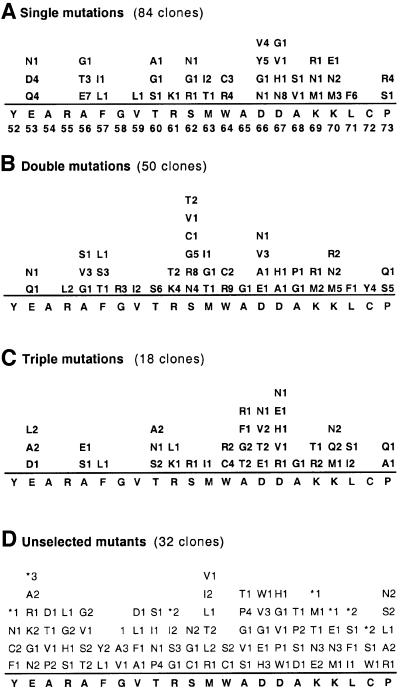
Selection of active hPolη mutants in our complementation system yielded >500 independent clones. We sequenced the DNA-encoding motif II in 210 plasmids from the selected active hPolη library and in 32 plasmids from the unselected library (plasmids from yeast colonies that were not exposed to UV). As anticipated, the number of predicted amino acid substitutions in the two libraries differed (Figure 4). The average number of substitutions in the selected active mutants was 1.2. The range was from zero to four and there was a preference for single substitutions; thus, we found 84 single mutants, 50 double mutants, 18 triple mutants and 12 quadruply-substituted mutants among the selected variants. In contrast, the average number of substitutions per clone in the unselected mutants was 3.8, as expected (see above). The range was from one to 10 substitutions and double, triple and quadruple mutants were equally frequent. The greater number of substitutions per mutant in the unselected library presumably reflects the presence of deleterious mutations that inactivate hPolη or otherwise diminish its ability to promote UV survival. In fact, 30% of the clones in the unselected library contained nonsense codons that would result in truncated proteins (Figure 5D); in contrast, only 6% of clones in the selected library contained nonsense mutations and these proved to be false positives. Consistent with positive genetic selection for UV survival, the wild-type amino acid sequence was found in 31 clones in the active mutant library, 80% of which contained silent mutations at the nucleotide level.
Fig. 4. Amino acid substitutions in selected and unselected motif II libraries. The number of substitutions in 210 clones from the selected library and 32 clones from the unselected library were determined by DNA sequencing.
Fig. 5. Amino acid substitutions in motif II of selected active hPolη mutants and unselected mutants. Mutations observed in selected clones harboring one (A), two (B) or three (C) amino acid changes and mutations (D) in unselected clones. Asterisks indicate nonsense codons. Substitutions at each residue are listed, along with the number of times each substitution was observed. The double mutants are: E53N,A68P; E53Q,K69M; R55L,A56G; R55L,F57L; A56S,S62V; A56V,S62G; F57S,S62G; F57T,D66A; G58R,K70M; V59I,K69R; V59I,K70M; T60S,R61T; T60S,S62T; T60S,D66V; T60S,D66V; R61T,L71F; R61K,C72Y; S62R,W64R; S62R,D66N; S62R,K70N; S62C,P73Q; S62N,P73S; M63G,W64R; M63T,D66E; M63I,K69M; W64R,D67H; W64C,K70R; A65G,A68G; D67A,K70M. The triple mutants are: E53A,T60S,A65T; E53L,R61L,D66T; E53L,W64C,D66N; E53D,W64C,L71F; A56S,F57L,D67R; A56E,D67N,L71I; T60A,A65F,K70N; T60N,A65G,K70M; T60A,D67E,K70N; R61K,D66V,D67H; S62R,W64C,P73A; M63I,W64C,D66E; W64R,D67V,P73Q; W64R,A68G,K70Q; A65G,D66V,K69R; A65R,K69T,L71S; D66T,K69R,K70Q.
Amino acid substitutions in selected, active motif II mutants
The predicted amino acid substitutions in 32 of the unselected mutants appeared to be located at random across motif II residues (Figure 5D). In contrast, amino acid substitutions in the 210 selected active hPolη mutants showed a specific distribution (Figures 5A–C). We found that, despite its evolutionary conservation (Figure 3), the primary sequence of motif II is highly plastic, tolerating replacements at all but two of the 22 positions. At some positions, as many as seven different amino acids could replace the wild-type residue (e.g. Asp66). Remarkably, some of the mutants contained as many as four amino acid substitutions (e.g. V59I, S62G, L71F and C72Y) and yet retained activity comparable with that of wild-type hPolη (data not shown).
No amino acid replacements were found at Tyr52 and Ala54. These two amino acids are strictly conserved in natural evolution (Figure 3) and, together with the lack of replacements in our active mutants, we infer from this preservation that both of these residues may be critical for UV resistance. In addition to Tyr52 and Ala54, four residues were not replaced among the single mutants (Figure 5A). Of these four, Arg55, Gly58 and Cys72 are highly conserved in natural evolution (Figure 3). Only one replacement was observed for each of these amino acids (i.e. R55L, G58R and C72Y) among multiple-substituted mutants (Figures 5A–C). We infer from the lack of single mutants at Arg55, Gly58 and Cys72 and the paucity of replacements in multiply substituted variants that the naturally occurring side chains at these positions may also be important in conferring UV survival. Compensation for loss of such wild-type residues mediated by co-selected replacements is possible in multiple-substituted variants.
In vitro synthesis of hPolη mutant proteins and bypass of cis–syn thymine dimers
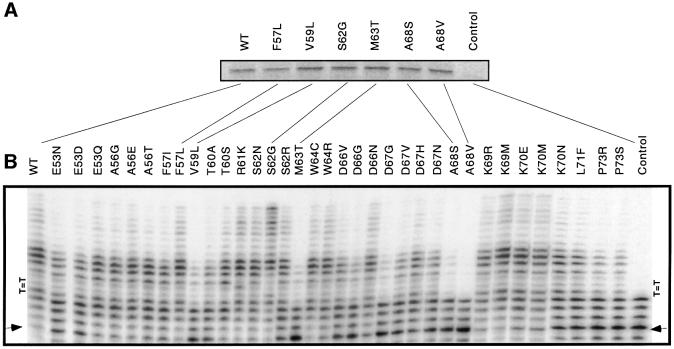
To examine DNA polymerization and translesion synthesis by the selected active hPolη mutants, we established a system for high-throughput in vitro transcription and translation in a 96-well format. Plasmids encoding 95 mutants served as a template for PCR amplification. The amplified cDNAs were then used for coupled in vitro transcription and translation in the presence of [35S]methionine. For each mutant, a single major band with intensity comparable with that of the wild-type enzyme was observed in SDS gels (Figure 6A and data not shown). The equivalent amounts of protein synthesized allowed us to compare relative levels of catalytic activity and bypass frequency within the 95 mutants.
Fig. 6. Bypass of thymine dimers by hPolη motif II single mutants. (A) Mutant hPolη proteins harboring single amino acid substitutions were synthesized in a high-throughput transcription and translation system in the presence of [35S]methionine. Amino acid substitutions in the mutants are indicated along with their position in the protein (e.g. E53A indicates a Glu to Ala substitution at position 53). (B) Bypass of a T–T dimer by the in vitro synthesized mutants was measured in primer extension assays. The position of the lesion (T=T) was four nucleotides downstream from the 3′ primer terminus (arrow), as indicated here and in the template sequence in Figure 1.
Wild-type hPolη efficiently copies past T–T dimers (Figure 1A; Masutani et al., 1999b; Johnson et al., 2000). We used the in vitro synthesized hPolη proteins in a primer extension assay to examine the ability of mutant enzymes to copy an oligonucleotide template containing a site-specific T–T dimer, as illustrated for the wild-type enzyme in Figure 1C. Most of the mutants that were able to rescue UV sensitivity in our complementation system bypassed the dimer to an extent of at least 20% of that observed for wild-type hPolη (Figure 6B). A few of the mutants, including A68S and A68V, although able to complement the UV sensitivity of rad30rad52 host cells, showed a severe reduction in bypass. Importantly, the control vector pYEX-hPolηdum (rightmost lane in Figure 6B) failed to support any detectable bypass. Thus, there is a concordance between the ability of the hPolη mutants to confer UV resistance in vivo and to bypass thymine dimers in vitro and we therefore infer that our complementation assay selects for mutants that can catalyze translesion synthesis. Moreover, we observed a tight correlation between the efficiency in bypassing T–T dimers (Figure 6) and polymerization activity on umodified DNA (data not shown).
hPolη S62G is a more active polymerase than wild-type hPolη
In preliminary studies with undamaged templates, we found that several hPolη mutant proteins catalyzed nucleotide incorporation at rates similar to or greater than that of the wild-type enzyme. One of these mutants contained a single amino acid substitution, Ser62 to Gly. Six different multiple-substituted mutants that contained S62G along with other replacements also showed relatively high activity (Figures 5A–C and data not shown).
To reconfirm that the enhanced synthesis observed for S62G hPolη represents an authentic increase in catalytic activity, we compared the initial rates of nucleotide incorporation in assays containing equal amounts of 35S-labeled enzyme [determined by trichloroacetic acid (TCA) precipitation of the respective lysates used for transcription-translation]. Importantly, both of the freshly prepared lysates showed a single major 35S-labeled band in SDS gels, as illustrated for different preparations in Figures 1 and 6. Based on a linear increase in incorporation on the unmodified template primer during the initial 5 min of reaction, we calculated that the specific activity of the S62G mutant is 3-times that of the wild-type enzyme (data not shown).
As shown in Figure 6, the S62G polymerase copied past T–T dimers to a greater extent than the wild-type enzyme. The ability of the S62G polymerase to copy unmodified DNA and DNA containing other site-specific lesions is illustrated in Figure 7. As shown in Figure 7A, the S62G polymerase accumulated longer copies of the unmodified template than the wild-type enzyme at every time point up to 1 h. The S62G mutant also exhibited more efficient bypass of lesions generated by oxygen damage (8-oxo-G, Figure 7B) and alkylating agents (O4-MeT and O6-MeG; Figure 7E and F); the mutant likewise displayed slightly enhanced synthesis opposite and past abasic sites (Figure 7C) and ethenoA adducts (Figure 7D), though bypass was still very inefficient. These results illustrate the importance of catalytic efficiency in bypassing different DNA lesions.
Fig. 7. Translesion synthesis by in vitro synthesized wild-type and S62G hPolη proteins on an unmodified template and templates containing site-specific lesions. Bypass of five lesions in the same DNA sequence context was compared in primer extension assays. The template contained either no lesion, 8-oxo-G, O4-MeT, O6-MeG, an abasic site or ethenoA. The position of the lesion (∗) was three nucleotides downstream of the 3′ primer terminus (arrow). The assays, containing 2 µl of a 1:20 dilution of in vitro synthesized wild-type or mutant hPolη, were terminated at different times, as indicated, by addition of formamide. The elongated products observed at time 0 probably reflect limited incorporation at 4°C.
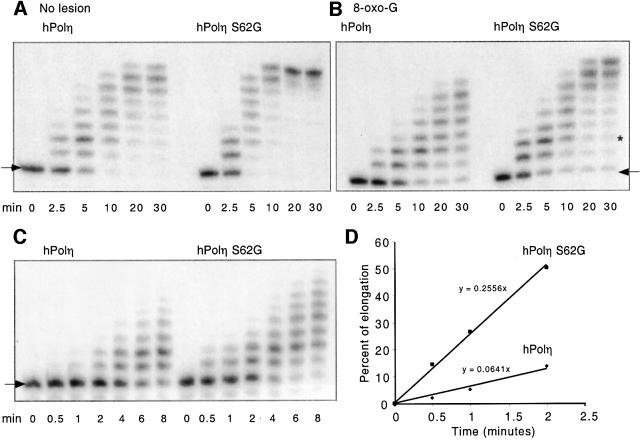
To confirm the enhanced activity and bypass ability of the S62G polymerase, we analyzed the mutant and wild-type polymerase domains purified from E.coli. As shown in Figure 8A and B, in the presence of equal amounts of the proteins, the S62G mutant exhibited greater activity than the wild-type enzyme on both the undamaged and 8-oxo-G-containing template primers. In an additional experiment (Figure 8C and D), we examined initial rates of incorporation in reactions containing an excess of unmodified template primer. Based on a linear increase in incorporation during the initial 2 min of reaction, we calculated that the specific activity of the S62G mutant is 4-times that of the wild-type enzyme. These results are very similar to those obtained with the in vitro synthesized enzymes (see above), corroborating the enhanced incorporation observed for the S62G polymerase. We conclude that the mutant S62G polymerase is a more catalytically active enzyme than wild-type hPol and that it has a higher capacity to bypass DNA lesions.
Fig. 8. Translesion synthesis by purified wild-type and S62G hPolη proteins on an unmodified template and a template containing 8-oxo-G. Primer extension and bypass of a site-specific 8-oxo-G lesion were analyzed in assays containing 0.1 pg/µl (2 nM) of purified wild-type or S62G polymerase domain. The time course of incorporation over 30 min is shown on (A) unmodified and (B) 8-oxo-G-containing template primers. The template lesion (∗) was three nucleotides downstream of the 3′ primer terminus (arrow). Initial rates of incorporation in assays containing a 2-fold higher concentration of undamaged template primer (C) were quantitated by phosphoimage analysis (D).
Discussion
Replication of nucleic acids is essential to the maintenance of living organisms. Until recently, five different families of DNA polymerases were known (Steitz, 1999). Although these polymerases have diverse amino acid sequences and diverse functions in DNA replication and repair, all appear to possess a similar overall architecture and a common catalytic mechanism (Hubscher et al., 2000). The shared three-dimensional structure is conveniently compared to a human right hand, possessing ‘thumb’, ‘palm’ and ‘finger’ domains.
In the last two years, a new sixth family of DNA polymerases has been identified, the Y-family of TLS or bypass DNA polymerases (Bridges, 1999; Johnson et al., 1999c; Goodman and Tippin, 2000; Livneh, 2001; McDonald et al., 2001; Ohmori et al., 2001; Wang, 2001). Members of this growing family of polymerases are capable of bypassing DNA lesions that are believed to block replication fork progression. Sequence alignment among members of the bypass DNA polymerases has revealed five conserved regions at the N-terminus of the proteins (Kulaeva et al., 1996; Johnson et al., 1999c; Masutani et al., 1999b). Interestingly, there is no recognizable sequence homology with the polymerase domains common to other known DNA polymerases. Newly published crystal structures of Y-family DNA polymerases reveal an overall architecture similar to that of other DNA polymerases. The structures are familiar in resembling a human right hand, with ‘thumb’, ‘palm’ and ‘finger’ domains, although certain modifications appear to distinguish the Y-family from previously characterized DNA polymerases (Ling et al., 2001; Trincao et al., 2001; Zhou et al., 2001).
In this work we chose to characterize human DNA polymerase η as a prototype for the other members of the Y-family DNA polymerase family. hPolη polymerizes across the UV-induced cis–syn T–T dimer with high efficiency and relatively high accuracy, inserting primarily adenines opposite the lesion. The ability to bypass these adducts and to incorporate the appropriate nucleotides is presumably the result of a flexible active site that allows polymerization across bulky helix-distorting lesions.
To gain insight into the function of specific amino acids in the activity of hPolη, we used random mutagenesis to study the conserved sequence designated motif II (Johnson et al., 1999c). We had hypothesized, based on its conservation and its position between motifs I and III in the linear sequence, that motif II is directly involved in catalysis. The position of motif II in the ‘fingers’ domain in the three-dimensional structures of hPolη (Trincao et al., 2001) and other Y family DNA polymerases (Ling et al., 2001; Zhou et al., 2001) corroborates this hypothesis.
Motif II contains the consensus sequence YxARxxGVx RxxxxxDAKxxCP (Figure 3). DNA sequencing of hPolη mutants, which confer UV resistance and maintain polymerase activity, revealed that motif II tolerates a wide range of amino acid substitutions (Figure 5). However, no functional mutants were isolated that had replacements at the conserved residues Tyr52 or Ala54 (Figure 5). These amino acids are presumably critical for polymerase or bypass activity and are likely to have a direct function in catalysis by hPolη and other bypass polymerases; they reside in helix D in hPolη, which appears to be involved in interaction with the incoming dNTP (Trincao et al., 2001). The essentiality of Tyr52 and Ala54 for activity and/or bypass, implied by the lack of replacements in this study, needs to be confirmed by saturation mutagenesis and re-selection.
Three other residues were also conserved, judging from the lack of single mutants at positions Arg55, Gly58 and Cys72. The involvement of these amino acids in catalysis and bypass is a significant possibility, since compensatory substitutions in multiple-substituted mutants can counteract the loss of critical wild-type residues (Figure 5A and B). Only conservative substitutions were observed at Val59, implying that hydrophobicity at this position may have functional significance. The substitutability of wild-type residues at these four positions can also be definitively addressed by saturation mutagenesis.
Examination of replication and translesion synthesis activity in vitro by 95 mutant proteins that are active in vivo revealed a positive correlation between their polymerization activity on unmodified DNA (data not shown) and their efficiency in bypassing T–T dimers. This finding highlights the importance of catalytic activity for the bypass capacity of hPolη and indicates that mutations in motif II affect these two functions in parallel. The correlation of activities extends to other template lesions we have examined (Figure 6 and data not shown). In analyzing the ability of the hPolη mutant S62G to incorporate nucleotides on an undamaged oligonucleotide template and to synthesize past diverse types of damage in the same sequence context, we found that the mutant polymerase exhibited greater than wild-type activity on the lesion-free template as well as on all modified templates. Moreover, the mutant enzyme maintained the same relative efficiency of lesion bypass as the wild-type enzyme (no lesion >8-oxo-G >O4-MeT >O6-MeG >abasic site >ethenoA). These findings again underscore the importance of catalytic efficiency in bypassing different DNA modifications and illustrate the parallel effects of mutations in motif II on bypass of diverse lesions.
The finding that inactivation and deletion of Polη decreases UV survival of human (Cordonnier and Fuchs, 1999; Yamada et al., 2000) and yeast cells (McDonald et al., 1997; Roush et al., 1998) provides strong evidence that the enzyme is involved in bypass of UV damage in vivo. It is assumed that the survival deficit reflects direct involvement of the catalytic activity of Polη in translesion synthesis and this assumption is supported by the ability of wild-type Polη to bypass UV dimers in vitro (Washington et al., 2000). Our finding that hPolη mutants, which complement UV sensitivity in yeast, invariably exhibit bypass activity in vitro provides independent evidence that the ability of this enzyme to catalyze translesion synthesis is indeed responsible for conferring UV resistance in cells. This conclusion is reinforced by the isolation of mutants (e.g. S62G) that are more active polymerases than wild-type hPolη and exhibit increased bypass ability. Nonetheless, bypass of alterations in DNA is likely to be a complex process involving multiple proteins, including those required for the restart of DNA replication. It is possible that deletion and inactivation of Polη disrupts protein–protein interactions that are essential for translesion synthesis in vivo.
In conclusion, our results show that a yeast-based complementation system can provide libraries of hPolη variants with altered properties. By examining variant enzymes selected in this system, we were able to demonstrate the importance of motif II for hPolη bypass activity and to provide the first experimental evidence indicating an important role for Tyr52 and Ala54 in maintaining the catalytic activity of the enzyme. Com parable systems for positive genetic selection of variants of other human bypass polymerases are presently lacking. Given the high degree of evolutionary conservation among the bypass DNA polymerases, the ability to readily isolate hPolη mutants foretells a prominent role for hPolη as an informative model for this important new family of polymerases.
Materials and methods
Cloning of the hPolη cDNA
hPolη cDNA was produced by RT–PCR amplification (Access RT–PCR, Promega) of mRNA isolated from HeLa cells using a direct mRNA mini kit (Oligotex, Qiagen). The 2.2 kb DNA product was cloned into the PCR 2.1-TOPO vector (Invitrogen) and subcloned using BamHI and EcoRI into the yeast shuttle vector pYEX-4T (Clontech) to yield pYEX-hPolη. This construct allowed expression of hPolη fused to an N-terminal glutathione S-transferase (GST) peptide. The predicted fusion protein sequence was verified by DNA sequencing.
Construction of the random hPolη mutant library
Site-directed mutagenesis was performed on pYEX-hPolη, introducing the silent mutations T85C, G87C, A88C to create a BspEI site and A228G, G231A to create an NheI site, yielding pYEX-hPolη (BspEI–NheI); the numbers refer to the hPolη open reading frame. To avoid contamination with incompletely cut vectors when preparing the random library, a non-functional stuffer vector, pYEX-hPolηdum, was constructed by replacing the BspEI–NheI 142 bp fragment of hPolη with an oligonucleotide fragment (5′-TCCGGATCGCGATCACGTG-3′, 5′-GCTAGCACGTGATCGCGAT-3′). This ‘dummy’ vector harbors a nonsense codon corresponding to amino acid 36 of hPolη, to generate a truncated product.
The hPolη random library was constructed by annealing two single-stranded DNA oligonucleotides: oligo 1 was a 75-mer corresponding to the sense nucleotides 79–153 and contained a BspEI site for cloning (5′-CCTCATCTCCGGAATAAACCTTGTGCAGTTGTACAGTACAAATCATGGAAGGGTGGTGGAATAATTGCAGTGAGT-3′); oligo 2 was a 100-mer corresponding to the anti-sense strand nucleotides 236–137 and contained an NheI site (5′-TGTGCTAGCAGAAGATCTGGACATAACTTCTTAGCATCATCTGCCCACATACTTCTAGTGACTCCAAATGCACGAGCTTCATAACTCACTGCAATTATTC-3′). The nucleotides in bold type in oligo 2 were designed to contain 92.5% wild-type nucleotides at each position and 2.5% each of the other three nucleotides. The 17 bp complementary regions of hybridization are underlined. Oligo 1 and oligo 2 were annealed at their non-random complementary regions and the partial duplexes were extended by mixing 100 pmol of each primer in 50 µl of PCR mix [1× Taq buffer, 2.5 mM MgCl2, 0.5 mM of all four dNTPs and 5 µl of Taq (Promega)]. The reaction was performed by heating to 95°C for 5 min, cooling to 42°C for 2 min, followed by 60 min at 72°C. The resulting DNA was digested with BspEI and NheI and purified by phenol chloroform and ethanol precipitation. The purified DNA was inserted into pYEX-hPolηdum in place of the stuffer fragment. Plasmids containing the random library were transformed into E.coli XL10-Gold (Stratagene) and the number of transformed cells was determined by plating an aliquot onto LB agar plates containing 100 µg/ml of carbenicillin. The remainder of the library was amplified by growing the transformed E.coli XL10-Gold in 3 liters of LB medium containing 100 µg/ml of carbenicillin for 16 h at 37°C. The random library pYEX-hPolη-LibB plasmids were isolated by maxiprep (Wizard plus maxiprep, Promega).
Genetic selection for active mutants
The yeast strain M59 carrying disruptions of RAD30 and RAD52 (MATα ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 rad30::HIS3 rad52–8::TRP1rad30rad52–8; McDonald et al., 1997) was generously provided by Drs John McDonald and Roger Woodgate. Cells were transformed with pYEX-hPolη-LibB plasmids by using a lithium acetate procedure (Burke et al., 2000) and grown to an OD600 of 2 at 30°C in synthetic complete medium lacking tryptophan, histidine and uracil (Burke et al., 2000). The cells were spread (100 µl/plate) on agar plates containing the same medium and exposed to 40 J/m2 of UV-C radiation (maximum λ 254 nm). The plates were incubated at 37°C in the dark for 72 h and surviving colonies were picked for further analysis.
DNA sequence analysis of motif II
pYEX-Polη-LibB plasmids carrying the mutant hPolη cDNA were isolated from yeast, retransformed into E.coli XL1-Blue (Stratagene) and purified (Wizard SV 96 miniprep kit; Promega). Following purification the 142 bp fragment, BspEI–NheI was sequenced using the primer 5′-AGCGTCGACGGCTGACCTTGTAGCTTTTGTAGTCTC-3′.
High-throughput in vitro synthesis of hPolη mutant proteins
Mutant plasmids were used as templates for PCR amplification in 96-well plates using a hPolη upstream primer that contained the T7 phage promoter sequence (5′-TTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGCTTGGTACCGAGCTCGGATCCAAAATGGCTACTGGACAGGATC-3′) and a hPolη downstream primer that contained the sequence for a hexahistidine tag (5′-CGGAATTCCTAATGGTGATGGTGATGATGAGCGGCCGCATGTGTTAATGGCTTAAAAAATGATTCC-3′). Five microliters of the amplified product (2240 bp), containing hPolη cDNA under control of the T7 promoter, served as a template for in vitro transcription and translation using 20 µl/well of ‘TNT T7 Quick for PCR DNA’ kit (Promega) in a 96-well plate. Protein products labeled with [35S]methionine were run on SDS gels to establish expression levels. For activity assays, the in vitro translated proteins were diluted 1:20 in dilution buffer (buffer D; 25 mM Tris–HCl pH 7.9, 0.5 mM EDTA, 1 mM dithiothreitol, 0.05% NP-40 and 25% glycerol).
DNA polymerase assays
DNA polymerase activity was measured in primer extension assays. Templates were hybridized with 5′-end 32P-labeled primer and 5 nM of template primer was incubated at 37°C for 15 min (or as indicated) in 10 µl reaction mixtures containing 2 µl of a 1:20 dilution of in vitro synthesized enzyme or 0.1 pg/µl (2 nM) of purified enzyme, 125 µM each of the four dNTPs in 40 mM Tris–HCl pH 8.0, 5 mM MgCl2, 60 mM KCl, 10 mM DTT, 250 µg bovine serum albumin (BSA) and 2.5% glycerol. Reactions were terminated by the addition of 2.5 µl of formamide solution; the products were analyzed by electrophoresis through 14% denaturing polyacrylamide gels and quantified by phosphoimager analysis. The oligomers used in assays for T–T dimer bypass were: template 5′-GTATTCCCAGGCGG<TT>TCCCATCCAAGTACTAACCAGGCCCGACCCTGCTTGGCTTCCGAGATCAGA-3′ (DMGC3, generously provided by Dr Michael J.Smerdon) and primer 5′-32P- TCTGATCTCGGAAGCCAAGCAGGGTCGGGCCTGGTTAGTACTTGGATG-3′. The templates used for bypass of other lesions (8-oxo-G, O6-MeG, O4-MeT, abasic site, ethenoA and no lesion) were synthesized by the Midland certified reagent company and contained one of the above adducts in the N position: 5′-TTGGC(N)GCAGAATATTGCTAGCGGGAATTCGGCGCG-3′. The primer was 5′-32P-CGCGCCGAATTCCCGCTAGCAATATTCT-3′.
Cloning of cDNAs for the hPolη polymerase domain
cDNAs corresponding to the polymerase domains of wild type and S62G hPolη (amino acids 1–513) were produced by PCR amplification and cloning of the first 1539 nucleotides of the human Polη cDNA from pYEX-hPolη and pYEX-hPolη S62G into the expression vector pET-15B (Novagen) to yield pET-15B -hPolη 513N and pET-15B -hPolη S62G 513N, respectively. These constructs allow expression of the polymerase domain fused to an N-terminal hexahistidine sequence.
Expression and purification of the hPolη polymerase domain
E.coli BL21(DE3)pLysS (Novagen) cells containing pET-15B-hPolη 513N, pET-15B -hPolη S62G 513N or no plasmid as a control, were grown at 37°C to an OD600 of 0.6, induced with 1 mM IPTG and grown in LB broth containing 5% ethanol for another 12 h at 25°C. Cells were harvested by centrifugation, resuspended and incubated for 20 min in ‘BugBuster’ (Novagen) extraction solution containing 10 U/ml Benzonase (Novagen). The clarified extract was then dialyzed against buffer D. The equilibrated extract was loaded onto a DEAE–cellulose column equilibrated with buffer D and the column was washed with buffer D, followed by buffer D containing 100 mM NaCl. Bound proteins were eluted with buffer D containing 250 mM NaCl, dialyzed against buffer N (20 mM Tris–HCl pH 7.9, 0.5 M NaCl, 20% glycerol) containing 5 mM imidazole and loaded onto a His-Bind Ni2+ column (Novagen) equilibrated with buffer N. The column was washed with buffer N and then with buffer N containing 50 mM imidazole. Bound proteins were eluted in step-wise manner with buffer N containing 80 mM, 120 mM or 200 mM imidazole. His-tagged hPolη 513N and hPolη S62G 513N were resolved by SDS–PAGE and visualized by Coomassie Blue staining and by western blotting with anti-hPolη antibody (sc-5936; Santa Cruz Biotechnology). The majority of the Coomassie Blue-stained material migrated with the molecular weight expected for the 519-residue hexahistidine-tagged hPolη polymerase domain (∼52 kDa) and corresponded to the only immunopositive material in the gel. The concentration of the hPolη polymerase domain used in assays was estimated by densitometric quantitation of Coomassie Blue-stained protein observed in an SDS–polyacrylamide gel with BSA as a standard.
Acknowledgments
Acknowledgements
We thank Dr Michael J.Smerdon (Washington State University) for generously providing the T–T dimer oligomer DMGC3, Drs John McDonald and Roger Woodgate (National Institutes of Health) for generously providing the S.cerevisiae strains, Janice Chau for excellent technical assistance, Drs Ashwini Kamath-Loeb and Mickey Fry for enlightening discussions and Ann Blank for critical reading and editing of the manuscript. This work was supported by a Senior Scholar Award from the Ellison Medical Foundation and grants CA78885 and AG01751 from the National Institutes of Health.
References
- Bridges B.A. (1999) DNA repair: polymerases for passing lesions. Curr. Biol., 9, R475–R477. [DOI] [PubMed] [Google Scholar]
- Burke D., Dawson,D. and Stearns,T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Cleaver J.E. (1999) Stopping DNA replication in its tracks. Science, 285, 212–213. [DOI] [PubMed] [Google Scholar]
- Cordonnier A.M. and Fuchs,R.P. (1999) Replication of damaged DNA: molecular defect in xeroderma pigmentosum variant cells. Mutat. Res., 435, 111–119. [DOI] [PubMed] [Google Scholar]
- Gerlach V.L., Feaver,W.J., Fischhaber,P.L. and Friedberg,E.C. (2001) Purification and characterization of polκ, a DNA polymerase encoded by the human DINB1 gene. J. Biol. Chem., 276, 92–98. [DOI] [PubMed] [Google Scholar]
- Goodman M.F. and Tippin,B. (2000) The expanding polymerase universe. Nature Rev. Mol. Cell Biol., 1, 101–109. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash,S. and Prakash,L. (2000a) Replication past O(6)-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol., 20, 8001–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Yu,S.L., Johnson,R.E., Prakash,L. and Prakash,S. (2000b) Efficient and accurate replication in the presence of 7,8-dihydro-8- oxoguanine by DNA polymerase η. Nature Genet., 25, 458–461. [DOI] [PubMed] [Google Scholar]
- Haracska L., Washington,M.T., Prakash,S. and Prakash,L. (2001) Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem., 276, 6861–6866. [DOI] [PubMed] [Google Scholar]
- Hubscher U., Nasheuer,H.P. and Syvaoja,J.E. (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci., 25, 143–147. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999b) Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem., 274, 15975–15977. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (1999c) Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc. Natl Acad. Sci. USA, 96, 12224–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- Kannouche P., Broughton,B.C., Volker,M., Hanaoka,F., Mullenders,L.H. and Lehmann,A.R. (2001) Domain structure, localization and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev., 15, 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratick C.M., Washington,M.T., Prakash,S. and Prakash,L. (2001) Acidic residues critical for the activity and biological function of yeast DNA polymerase η. Mol. Cell. Biol., 21, 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva O.I., Koonin,E.V., McDonald,J.P., Randall,S.K., Rabinovich,N., Connaughton,J.F., Levine,A.S. and Woodgate,R. (1996) Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res., 357, 245–253. [DOI] [PubMed] [Google Scholar]
- Levine R.L., Miller,H., Grollman,A., Ohashi,E., Ohmori,H., Masutani,C., Hanaoka,F. and Moriya,M. (2001) Translesion DNA synthesis catalyzed by human Polη and Polκ across 1,N6-Ethenodeoxy adenosine. J. Biol. Chem., 276, 18717–18721. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Ling H., Boudsocq,F., Woodgate,R. and Yang,W. (2001) Crystal structure of a Y-family DNA polymerase in action. A mechanism for error-prone and lesion-bypass replication. Cell, 107, 91–102. [DOI] [PubMed] [Google Scholar]
- Livneh Z. (2001) DNA damage control by novel DNA polymerases: translesion replication and mutagenesis. J. Biol. Chem., 276, 25639–25642. [DOI] [PubMed] [Google Scholar]
- Maher V.M., Ouellette,L.M., Curren,R.D. and McCormick,J.J. (1976) Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature, 261, 593–595. [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999a) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1999b) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Low fidelity DNA synthesis by human DNA polymerase-η. Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharo myces cerevisiae RAD30 gene, a homologue of Escherichia coli DinB and UmuC, is DNA damage inducible and functions in a novel error-free post-replication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Tissier,A., Frank,E.G., Iwai,S., Hanaoka,F. and Woodgate,R. (2001) DNA polymerase ι and related RAD30-like enzymes. Philos. Trans R. Soc. Lond. B Biol. Sci., 356, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD, RecA and SSB and is specialized for translesion replication. J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Pavlov,Y.I., Bebenek,K., Matsuda,T. and Kunkel,T.A. (2001) Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nature Immunol., 2, 530–536. [DOI] [PubMed] [Google Scholar]
- Roush A.A., Suarez,M., Friedberg,E.C., Radman,M. and Siede,W. (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet., 257, 686–692. [DOI] [PubMed] [Google Scholar]
- Steitz T.A. (1999) DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem., 274, 17395–17398. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Christians,F.C., Kim,B., Skandalis,A., Black,M.E. and Loeb,L.A. (1996) Tolerance of different proteins for amino acid diversity. Mol. Div., 212, 111–118. [DOI] [PubMed] [Google Scholar]
- Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J., Johnson,R.E., Escalante,C.R., Prakash,S., Prakash,L. and Aggarwal,A.K. (2001) Structure of the catalytic core of S.cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell, 8, 417–426. [DOI] [PubMed] [Google Scholar]
- Vaisman A., Masutani,C., Hanaoka,F. and Chaney,S.G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry, 39, 4575–4580. [DOI] [PubMed] [Google Scholar]
- Wagner J., Gruz,P., Kim,S.-R., Yamada,M., Matsui,K., Fuchs,R.P.P. and Nohmi,T. (1999) The dinB gene encodes a novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol. Cell, 4, 281–286. [DOI] [PubMed] [Google Scholar]
- Wang Z. (2001) Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res., 486, 59–70. [DOI] [PubMed] [Google Scholar]
- Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (2000) Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl Acad. Sci. USA, 97, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A., Masutani,C., Iwai,S. and Hanaoka,F. (2000) Comple mentation of defective translesion synthesis and UV light sensitivity in xeroderma pigmentosum variant cells by human and mouse DNA polymerase η. Nucleic Acids Res., 28, 2473–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.L., Johnson,R.E., Prakash,S. and Prakash,L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol., 21, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Winter,D.B., Kasmer,C., Kraemer,K.H., Lehmann,A.R. and Gearhart,P.J. (2001) DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nature Immunol., 2, 537–541. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.L., Pata,J.D. and Steitz,T.A. (2001) Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell, 8, 427–437. [DOI] [PubMed] [Google Scholar]