Abstract
We characterized a new signaling pathway leading to the activation of cAMP-responsive element-binding protein (CREB) in several cell lines affected by mitochondrial dysfunction. In vitro kinase assays, inhibitors of several kinase pathways and overexpression of a dominant-negative mutant for calcium/calmodulin kinase IV (CaMKIV), which blocks the activation of CREB, showed that CaMKIV is activated by a mitochondrial activity impairment. A high calcium concentration leading to the disruption of the protein interaction with protein phosphatase 2A explains CaMKIV activation in these conditions. Transcrip tionally active phosphorylated CREB was also found in a ρ0 143B human osteosarcoma cell line and in a MERRF cybrid cell line mutated for tRNALys (A8344G). We also showed that phosphorylated CREB is involved in the proliferation defect induced by a mitochondrial dysfunction. Indeed, cell proliferation inhibition can be prevented by CaMKIV inhibition and CREB dominant-negative mutants. Finally, our data suggest that phosphorylated CREB recruits p53 tumor suppressor protein, modifies its transcriptional activity and increases the expression of p21Waf1/Cip1, a p53-regulated cyclin-dependent kinase inhibitor.
Keywords: calcium–calmodulin kinase IV/cell proliferation/cyclicAMP response element-binding protein/mitochondrial dysfunction/retrograde communication
Introduction
Cells can modulate the expression of nuclear genes in response to changes in the activity level of their organelles. For example, mitochondria dysfunction induces the activation of transcription factors in a response called ‘retrograde communication’ (Parikh et al., 1987; Liao and Butow, 1993). Several lines of evidence previously established that alterations in the mitochondrial activity result in changes in nuclear gene expression (Wang and Morais, 1997). In yeast, numerous studies have shown that a retrograde signaling pathway can act as a homeostatic or stress response mechanism to adjust metabolic activities to the alterations of mitochondria activity. It has been particularly well characterized for the CIT2 gene, encoding a peroxisomal isoform of citrate synthase that catalyzes the first step in the glyoxylate cycle. In cells lacking mitochondrial DNA (mtDNA), increased expression of CIT2 is dependent on the activation of RTG1 and RTG3 encoding basic helix–loop–helix leucine zipper (bHLH-Zip) transcription factors (Liao and Butow, 1993). In yeast, detailed mechanisms on the regulation of the mitochondria to nucleus signaling and metabolic control through Rtg proteins in response to mitochondria impairment have been described recently (Komeili et al., 2000; Sekito et al., 2000).
The interorganelle ‘retrograde communication’ pathways have been described not only in yeast (Liao and Butow, 1993; Komeili et al., 2000; Sekito et al., 2000) but also in plant (Mayfield, 1990) and mammalian cells (Biswas et al., 1999). However, the signaling pathway(s) used by mitochondria to communicate the state of activity to the nucleus is still poorly understood in mammalian cells.
Mitochondrial activity plays a crucial role in cellular physiology, as emphasized recently by the discovery of a wide variety of human diseases caused by or associated with mitochondrial alterations such as genetic defects in mtDNA or nuclear DNA that affect the respiratory chain and ADP phosphorylation (Wallace, 1999). Among these diseases, myoclonus epilepsy with ragged-red fibers (MERRF) is a maternally inherited disorder characterized by point mutations in the mtDNA-encoded tRNALys gene which impairs respiration and mitochondrial protein synthesis (Lombes et al., 1989). Cells cultured with ethidium bromide (EtBr) partly or completely lose their mtDNA (mtDNA-depleted or ρ0 cells) and consequently cannot carry out oxidative phosphorylation (OXPHOS) and thus glycolysis is their only source of ATP (King and Attardi, 1989). Recently, these cells have been used to study mitochondrial disorders due to genetic defects such as deletions in mtDNA, depletion of mtDNA or reduction of mtDNA copy number (King and Attardi, 1989; Moraes et al., 1991). These mtDNA-depleted cells usually have a proliferation defect characterized by a longer doubling time, as observed for mtDNA-depleted HeLa S3 and 143B cells (Vaillant and Nagley, 1995; Buchet and Godinot, 1998), but no molecular control of the cell cycle after inhibition of mitochondria has been evidenced so far.
Mitochondrial dysfunction also greatly modifies nuclear gene expression (Epstein et al., 2001). Indeed, increased expression of ANT-1 and ATP synthase β-subunit mRNAs was described in skeletal muscle of patients with respiratory defects resulting from mtDNA point mutations (Heddi et al., 1993), and increased mRNAs encoding cytochrome oxidase (COX IV and COX VIaL) were also observed in ρ0 cells (Li et al., 1995). Biswas et al. (1999) recently described that in mouse C2C12 cells, the decrease of mtDNA content initiates a signaling pathway leading to the activation of JNK and increased transcription of ryanodine receptor-1 Ca2+ release channel and COX Vb genes. While eukaryotic cells can respond to mitochondrial dysfunction by a modulation of nuclear gene expression, evidence for signaling pathways and transcriptional mechanisms are lacking in most instances. Interestingly, several genes encoding mitochondrial proteins contain CRE (cAMP response element) sequences such as some respiratory chain subunits (Scacco et al., 2000), cytochrome c (Gopalakrishnan and Scarpulla, 1994), manganese superoxide dismutase (Kim et al., 1999) or carnitine palmitoyltransferase (Brady et al., 1992).
In this study, we analyzed the effect of mitochondrial activity inhibition on the signaling and transcriptional machinery of mammalian cells. We identified CRE-binding protein (CREB) as a new transcription factor involved in the ‘retrograde communication’ pathways between impaired mitochondria and the nucleus. We used several cellular models: a mtDNA-depleted murine fibrosarcoma cell population (L929), a true ρ0 human osteosarcoma cell line (143B), a cybrid cell line with a nucleotide mutation (A8344G) in the mitochondria-encoded tRNALys gene and an acute inhibition of mitochondria in 293 cells treated with metabolic mitochondrial inhibitors. We found that mitochondrial activity impairment is sufficient to trigger CREB phosphorylation on Ser133 by calcium/calmodulin kinase IV (CaMKIV). In turn, phosphorylated CREB mediates a proliferation defect in mtDNA-depleted L929 cells by increasing p53 transcriptional activity that leads to the up-regulation of p21Waf1/Cip1, a cyclin-dependent kinase inhibitor gene regulated by p53. Our results suggest that the activation of a ‘retrograde communication’ signaling pathway initiated by a mitochondrial dysfunction could be an important modulator of the cell cycle in cells submitted to energetic stress conditions.
Results
Functional and morphological characterization of the mtDNA-depleted L929 cell line
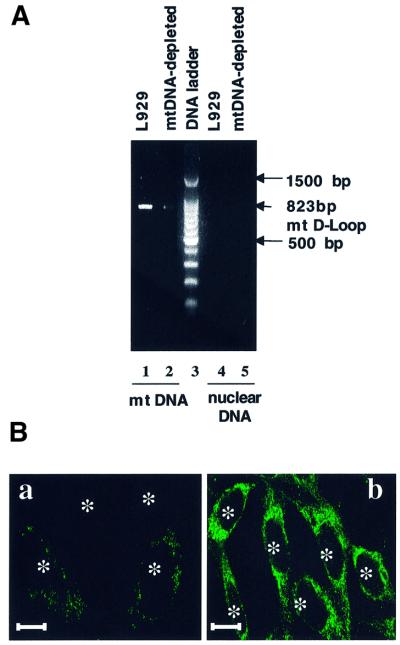
PCR analysis revealed that EtBr-treated L929 cells are dramatically depleted in but not completely devoid of mtDNA. As shown in Figure 1A, using specific primers for the D-loop fragment in mouse mtDNA, we amplified a fragment of the expected size (823 bp) in L929 cells, but only a very weak band was obtained for EtBr-treated cells. No amplification was obtained with nuclear DNA, confirming the specificity for mtDNA. Consistently, the mtDNA-encoded COX I protein was weakly stained in mtDNA-depleted L929 compared with parental cells (Figure 1B). The functional effect of EtBr treatment on oxidative phosphorylation was assessed further on in situ respiration as measured by the respiratory control ratio (RCR). EtBr treatment induces a complete inhibition of the oxidative respiration (RCR = 1) in mtDNA-depleted cells (see Supplementary data available at The EMBO Journal Online). The total cellular ATP measured in mtDNA-depleted L929 cells was reduced by 60% and was no longer sensitive to the uncoupler FCCP, suggesting that the residual ATP content in these cells is supplied by glycolysis (data not shown).
Fig. 1. Effects of EtBr-treatment on mitochondria in L929 cells. (A) PCR amplification of a specific mtDNA fragment in the mitochondrial D-loop performed on purified mtDNA (lanes 1 and 2) and nuclear DNA (lanes 4 and 5). (B) Immunofluorescence patterns of cells stained with a monoclonal antibody against the COX I subunit: (a) mtDNA-depleted cells; (b) L929 cells. Asterisks indicate the localization of nuclei. Scale bars = 10 µm.
CREB is constitutively activated by phosphorylation on Ser133 in mtDNA-depleted L929 cells
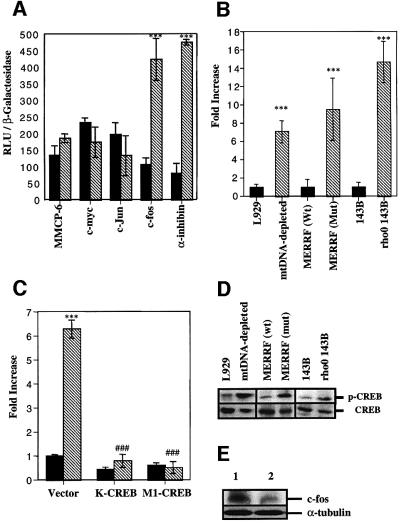
To address the question of whether or not the inhibition of mitochondrial activity modulates the transcriptional activity, L929 or mtDNA-depleted L929 cells were transiently co-transfected with a plasmid encoding β-galactosidase and different luciferase reporter constructs transactivated by various transcription factors. As illustrated in Figure 2A, the luciferase activity was increased specifically for the constructs driven either by the authentic α-inhibin promoter (Pei et al., 1991) or by the authentic c-fos promoter, both containing CRE sites. CREB transactivation is also observed in a ρ0 143B cell line or clonal cybrids obtained from ρ0 143B cells that have been repopulated with mtDNA from a MERRF patient harboring a tRNALys mutation (A8344G) (King and Attardi, 1989). Luciferase activity was compared in L929, mtDNA-depleted L929 cells, cybrids containing either 100% mutated or wild-type mtDNA, and 143B and ρ0 143B cells after transient transfection with the luciferase reporter construct driven by the α-inhibin promoter (Figure 2B). The different cell lines associated with severe defects in respiratory chain activity show a significant increase in CREB-dependent luciferase activity. This effect seems to be specific for mitochondrial inhibition, as glycolysis inhibition with 2-deoxy-d-glucose is without any effect on CREB activation (data not shown). In mtDNA-depleted L929 cells, the overexpression of K-CREB and M1-CREB, two dominant-negative mutants for CREB (Bonni et al., 1999), completely inhibited the luciferase activity driven by the α-inhibin promoter (Figure 2C). Western blotting analysis revealed that CREB phosphorylation on Ser133 is induced in mtDNA-depleted L929 cells, mutated MERRF cybrids and ρ0 143B cells (Figure 2D). Confocal microscopy and subcellular fractionation–western blot analyses show that phosphorylated CREB is found mainly in nuclei of the different cell lines associated with a mitochondrial dysfunction (data not shown). To make sure that phosphorylated CREB is able to modify gene expression, we checked the amount of c-fos expression, a CREB-regulated immediate-early gene (Hall et al., 2001). As can be seen in Figure 2E, c-fos protein is more abundant in mtDNA-depleted cells.
Fig. 2. (A) Luciferase activity in transiently transfected L929 control and mtDNA-depleted cells with luciferase constructs driven by c-myc, c-Jun, authentic MMCP-6, c-fos and α-inhibin promoters. L929 (black) and mtDNA-depleted L929 (hatched) cells, were transiently co-transfected with β-galactosidase and the luciferase constructs. (B) CREB-dependent luciferase activity in L929, mtDNA-depleted L929, wild-type (wt) MERRF cybrid, mutated (mut) MERRF cybrid, 143B and ρ0 143B cells transiently transfected with the luciferase reporter construct driven by the α-inhibin promoter and an expression plasmid encoding β-galactosidase. (C) CREB-dependent luciferase activity in L929 (black) and mtDNA-depleted L929 (hatched) cells transiently transfected with K-CREB or M1-CREB, together with the CREB-sensitive luciferase reporter construct and an expression plasmid encoding β-galactosidase. Luciferase activity was determined 24 h post-transfection. Results are expressed in relative light units (RLU) after normalization for β-galactosidase activity (A) or in fold increase of control cells (B and D) as means ± 1 SD (*** and ###, significantly different from L929 and mtDNA-depleted L929 cells with P <0.001). (D) Western blot analysis of phospho-CREB (p-CREB) and CREB assessed for the different cell lines used in (B). (E) Western blot analysis for c-fos and α-tubulin assessed for L929 (lane 2) and mtDNA-depleted L929 (lane 1).
Identification of the kinase responsible for the phosphorylation of CREB
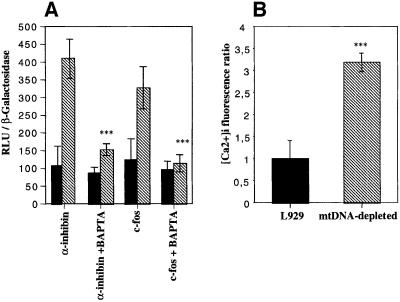
We next examined the activity of several kinase cascades leading to CREB activation in mtDNA-depleted L929 cells. We first measured the in vitro cAMP-dependent protein kinase (PKA) activity in cell homogenates, but no difference was observed between L929 and mtDNA-depleted cells, while PKA is activated in a time-dependent manner in cells treated with 100 µM dibutyryl-cAMP (see Supplementary data). We then tested whether CREB phosphorylation was mediated by a Ca2+-dependent mechanism. Cell treatment with BAPTA, an intracellular Ca2+ chelator, inhibits luciferase activity observed in mtDNA-depleted L929 cells on both CREB reporter constructs (Figure 3A), while BCECF, an unrelated probe tested to rule out the potentially toxic effect, is without any effects (data not shown). We also confirmed that intracellular calcium concentration in cells loaded with Fluo-3, a fluorescent calcium indicator, is higher in mtDNA-depleted L929 cells (Figure 3B).
Fig. 3. (A) Effect of a 6 h treatment with 20 µM BAPTA-AM on the CREB-sensitive luciferase activity after transfection of L929 (black) or mtDNA-depleted L929 (hatched) cells with luciferase reporter genes driven by either CREB-regulated c-fos or α-inhibin promoters. Luciferase activity was determined and expressed as RLU after normalization for β-galactosidase activity (n = 6; ***, significantly different from mtDNA-depleted cells with P <0.001). (B) Intracellular calcium concentration ratio in L929 and mtDNA-depleted cells loaded with 10 µM Fluo-3-AM. Results are expressed in fold increase of control cells, and represent the mean ± SD for n = 3 (***, significantly different from L929 with P <0.001).
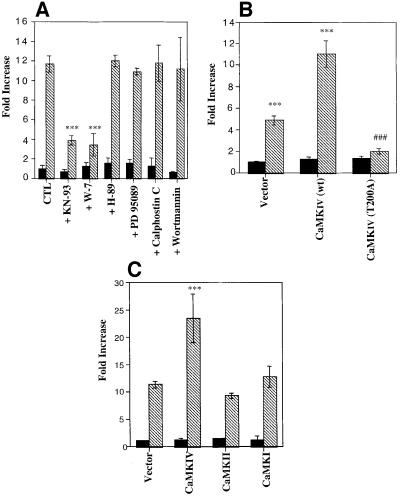
As calcium is able to activate several kinase pathways leading to CREB phosphorylation on Ser133 such as CaMKI, II or IV, p70S6K, rsk2, MSK, PKCs, Ras/Raf/MAPK and MAPKAP-K2, allowing the interaction with the CREB-binding protein (CBP/p300) (De Cesare et al., 1999), we used different inhibitors to modulate the CREB-dependent luciferase activity. As shown in Figure 4A, only KN-93, a CaMK inhibitor, and W-7, a calmodulin antagonist, were able to inhibit CREB-dependent transcription significantly, while inhibitors of the PKA (H-89), PKC (calphostin C), mitogen-activated protein kinase (PD 95089) or phosphatidylinositol 3-kinase (wortmannin) pathway have no effect. We thus investigated the possible role of CaMKI, II, and IV in CREB activation. Expression of a dominant-negative form of CaMKIV (CaMKIV T200A) (Chatila et al., 1996) completely suppressed the activation of the CREB reporter construct in transfected mtDNA-depleted L929 cells, while overexpression of the wild-type enzyme enhanced the luciferase activity only in mtDNA-depleted cells (Figure 4B). When these cells are transiently transfected with plasmids encoding the wild type of different isoforms (CaMKI, II and IV), only CaMKIV overexpression is able to enhance the CREB-dependent luciferase activity by an additional 2-fold increase (Figure 4C).
Fig. 4. (A) Effect of kinase pathway inhibitors on CREB-dependent luciferase activity in L929 (black) and mtDNA-depleted (hatched) cells. After transfection, cells were incubated for 16 h before the assay with 50 µM KN-93, 50 µM W-7, 10 µM H-89, 20 µM PD 95089, 1 µM calphostin C or 1 µM wortmannin. (B) L929 (black) and mtDNA-depleted L929 (hatched) cells were transiently co-transfected with a plasmid encoding β-galactosidase, the CREB luciferase reporter construct driven by the α-inhibin promoter and an expression vector encoding wild-type CaMKIV or the dominant-negative form CaMKIV T200A. (C) Effect of overexpression of CaMKI, CaMKII and CaMKIV on the activation of the CREB luciferase reporter construct driven by the α-inhibin promoter. L929 (black) and mtDNA-depleted L929 (hatched) cells were transiently co-transfected as described in (B) with an expression vector encoding wild-type CaMKI, II or IV. Results are normalized for β-galactosidase activity, are expressed in fold increase of luciferase activity of the L929 control cells and represent the mean ± SD for n = 3 (***, significantly different from L929 with P <0.001; ###, significantly different from mtDNA-depleted L929 cells with P <0.001).
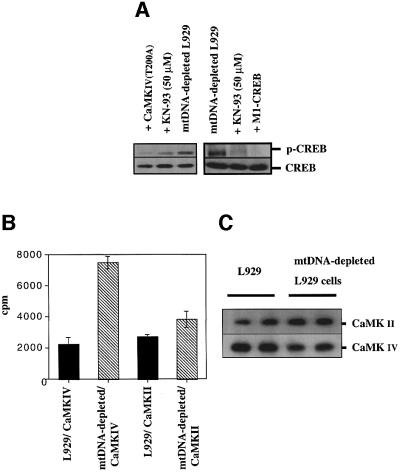
The crucial role of CaMKIV in the phosphorylation of CREB in mtDNA-depleted L929 cells was evidenced further by the fact that overexpression of dominant-negative CaMKIV T200A or KN-93 treatment dramatically inhibited CREB phosphorylation, as observed by western blot analysis (Figure 5A). The overexpression of the dominant-negative M1-CREB also inhibited phosphorylation of endogenous CREB, probably by competing for binding and subsequent phosphorylation (Figure 5A). Finally, endogenous CaMKII and CaMKIV were immunoprecipitated and their activity was determined in vitro. The results shown in Figure 5B and C clearly indicate that the CaMKIV activity is increased by 3- to 4-fold in mtDNA-depleted L929 cells while CaMKII activity seems to be unaffected. The slight increase observed in the CaMKII activity is most likely to be the result of the higher amount of the immunoprecipitated kinase used for the assay (Figure 5C). In conclusion, these results clearly support the hypothesis that CREB phosphorylation in mtDNA-depleted L929 cells is mediated by CaMKIV.
Fig. 5. (A) Effect of dominant-negative CaMKIV T200A or M1-CREB overexpression and the calmodulin kinase inhibitor KN-93 on the CREB phosphorylation pattern in mtDNA-depleted L929 cells. mtDNA-depleted L929 cells were either transfected with a plasmid encoding CaMKIV T200A or M1-CREB or treated with 50 µM KN-93 for 24 h before phospho-CREB and CREB expression were analyzed by western blotting. (B) Endogenous CaMKIV or CaMKII activity in L929 and mtDNA-depleted cells was measured after immuno precipitation. Representative results of one experiment in duplicate are shown and are expressed as c.p.m. (C) The amounts of immunoprecipitated kinases were controlled by western blotting.
CREB phosphorylation is induced by inhibitors of mitochondrial activity
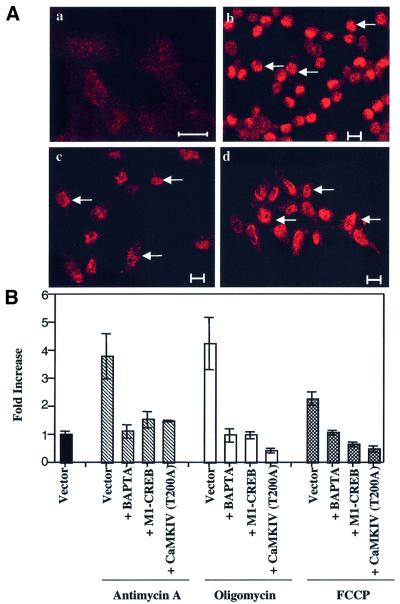
In order to determine whether CREB activation is induced directly by a mitochondrial activity impairment, we tested the effect of metabolic mitochondrial inhibitors on CREB activation. Phospho-CREB localization in cell lines treated with FCCP, oligomycin or antimycin A is almost exclusively nuclear, as shown for L929 cells in Figure 6A. Phosphorylated CREB is also transcriptionally active in cells treated with metabolic inhibitors, as demonstrated by the activation of the CREB reporter gene construct (Figure 6B). Cell incubation with BAPTA and overexpression of dominant-negative form M1-CREB or CaMKIV T200A almost completely inhibited the luciferase activity triggered by the different mitochondrial inhibitors (Figure 6B). Using the Ca2+-sensitive photoprotein apoaequorin as a calcium probe, we confirmed that free cytosolic [Ca2+]cyt is increased in 293 cells treated with mitochondrial inhibitors while mitochondrial [Ca2+]mt is decreased (see Supplementary data). Endo genous CaMKIV activity is also increased (5–7 times) in 293 cells after treatment with oligomycin, antimycin A or FCCP (see Supplementary data). Altogether, these results suggest that mitochondrial inhibitors are also able to trigger CREB phosphorylation through a CaMKIV-dependent mechanism and they demonstrate that CREB activation induced by mitochondria dysfunction is not cell specific as it is observed in two different cell lines.
Fig. 6. (A) Immunolocalization of phospho-CREB on Ser133 in L929 cells treated or not (a) for 6 h with 10 µM FCCP (b), 8 µM oligomycin (c) and 1 µM antimycin A (d) and analyzed the next day by immunofluorescence and confocal microscopy. Bars = 10 µm. Arrows indicate a nuclear localization. (B) CREB-dependent luciferase activity in 293 cells triggered by treatment with 1 µM antimycin A (hatched bars), 8 µM oligomycin (white bars) or 10 µM FCCP (black bars) is inhibited by 20 µM BAPTA for 6 h, or by expression of a dominant-negative form of CREB (M1-CREB) or a dominant-negative mutant of CaMKIV (CaMKIV T200A). Results are expressed as fold increase of control cells (vector) after normalization. The results are representative of two experiments in triplicate and are expressed as means ± 1 SD.
CaMKIV activity is differentially regulated by PP2A in cells affected by mitochondria dysfunction
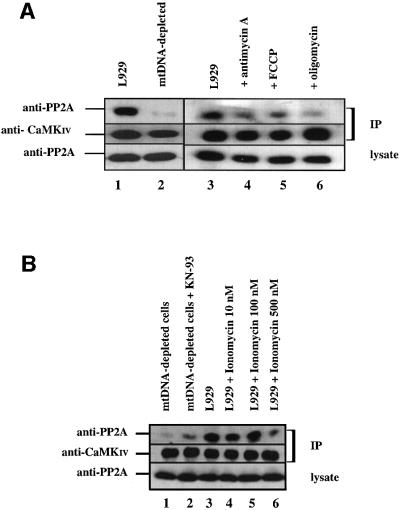
Once activated by phosphorylation on a single threonine residue by the calmodulin-dependent kinase kinase (CaMKK), CaMKIV activity becomes independent of both calcium and calmodulin (Park and Soderling, 1995). However, CaMKIV is known to be inactivated by protein phosphatase 2A (PP2A) (Westphal et al., 1998). To determine whether these enzymes exist as a complex in L929 and mtDNA-depleted L929 cells, we immunoprecipitated endogenous CaMKIV from cell lysates and analyzed the immune complexes by immunoblotting with an antibody specific for the PP2A catalytic C subunit. As shown in Figure 7A, PP2A catalytic C subunit co-immunoprecipitated with CaMKIV in L929 cells (lane 1), but the interaction is dramatically weaker in mtDNA-depleted L929 cells (lane 2). To make sure the differential interaction is caused by mitochondrial inhibition, we compared the co-immunoprecipitated PP2A in L929 cells treated with the different inhibitors. As shown in Figure 7A, antimycin A, FCCP and oligomycin are all able to impair the interaction between both enzymes. These results suggest that PP2A interaction with CaMKIV is weaker in mtDNA-depleted L929 cells and in cells submitted to the metabolic inhibitors. A very important question to answer is whether or not the weaker association of CaMKIV with PP2A observed in mtDNA-depleted cells is caused by CaMKIV’s increased activity as a result of the higher calcium concentration or if the impaired association with PP2A is reponsible for causing the increase in CaMKIV. To address this question, we performed co-immunoprecipitation experiments between CaMKIV and PP2A in L929 cells treated with ionomycin, a calcium ionophore, or in mtDNA-depleted cells treated with KN-93 to inhibit CaMKIV activity. The results presented in Figure 7B clearly show that ionomycin treatment inhibits the interaction while the association is increased in mtDNA-depleted L929 cells treated with KN-93. These results support the first hypothesis that the interaction can be modulated by the CaMKIV activity status.
Fig. 7. (A) Co-immunoprecipitation of PP2A catalytic C subunit with CaMKIV. CaMKIV was immunoprecipitated (IP) from L929 cells (lanes 1 and 3) treated or not with mitochondrial inhibitors (8 µM oligomycin, 1 µM antimycin A and 10 µM FCCP) (lanes 4–6) or from mtDNA-depleted L929 cells (lane 2). (B) Co-immunoprecipitation of PP2A catalytic C subunit with CaMKIV. CaMKIV was immuno precipitated from L929 cells (lane 3) treated or not with ionomycin at 10, 100 or 500 nM for 16 h (lanes 4–6) or from mtDNA-depleted L929 cells treated or not with 50 µM KN-93 for 16 h (lanes 1 and 2). Immune complexes were analyzed by immunoblotting with an anti-PP2A C subunit antibody (anti-PP2A) or an anti-CaMKIV antibody (anti-CaMKIV). Expression levels of the PP2A C subunit in the lysates are also shown.
Involvement of phosphorylated CREB in the cell proliferation defect
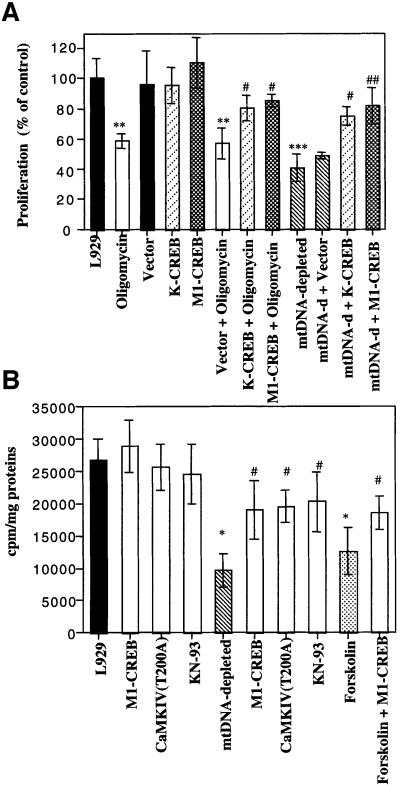
A slower proliferation of mtDNA-depleted cells than parental cells has been observed in several cell lines and is usually attributed to the lower ATP content in these cells (King and Attardi, 1996; Morais, 1996), but the molecular pathway is unknown. As stimulation of the cAMP pathway inhibits proliferation of some tumoral cell lines (Yokozaki et al., 1992), we hypothesized that phosphorylated CREB contributes to the lower proliferation of the mtDNA-depleted L929 or L929 cells treated with an inhibitor of mitochondrial activity. As shown in Figure 8A, the [3H]thymidine incorporation into mtDNA-depleted L929 cells and in oligomycin-treated L929 cells was strongly reduced as compared with L929. Therefore, we assessed the role of phosphorylated CREB on cell entry into S phase. L929 cells were transiently transfected with a control vector or with constructs encoding CREB dominant-negative mutants K-CREB and M1-CREB, 24 h before the addition of 8 µM oligomycin for 6 h. Oligomycin decreases [3H]thymidine incorporation by almost 40–50% in both untransfected control cells and transfected control cells. However, this effect is largely reduced when cells overexpressed either K-CREB or M1-CREB (Figure 8A). Similarly, mtDNA-depleted L929 cells incorporated only 50% of [3H]thymidine compared with parental cells, and the decrease can be significantly reduced by K-CREB or M1-CREB overexpression (Figure 8A). A reduced decrease in the [3H]thymidine incorporation was also observed when mtDNA-depleted L929 cells were transiently transfected with dominant-negative CaMKIV or treated with KN-93 (Figure 8B), two conditions that reduce the phosphorylation of CREB on Ser133 (Figure 5A). As a positive control, we showed that M1-CREB is also able to reduce the decrease of [3H]thymidine incorporation in forskolin-treated L929 cells (Figure 8B). In conclusion, mtDNA depletion or mitochondrial inhibitors such as oligomycin appear to inhibit cell cycle entry into S phase through the activation of CREB transcription factor.
Fig. 8. (A) Incorporation of [3H]thymidine into DNA was analyzed in L929 cells (treated or not with oligomycin) and compared with the incorporation for mtDNA-depleted L929 cells. In some conditions, these cells were transiently transfected before treatment with an empty vector or plasmids encoding dominant-negative CREB (K-CREB and M1-CREB). At 16 h post-transfection, oligomycin (8 µM) was added or not for 6 h and, the next day, cells were labeled with 2 µCi of [3H]thymidine for 4 h and processed accordingly. (B) Effect of M1-CREB and CaMKIV T200A overexpressions or KN-93 treatment on [3H]thymidine incorporation. L929 and mtDNA-depleted L929 cells were transiently transfected with an empty vector or plasmids encoding M1-CREB or CaMKIV T200A. The vector-transfected cells were treated or not with 50 µM KN-93 for 24 h before the [3H]thymidine incorporation assay. A control was perfomed with forskolin (10–5 M) for 24 h with or without M1-CREB overexpression. Values were determined and are expressed either as c.p.m./mg of proteins (B, n = 3) or as a percentage of [3H]thymidine incorporation for control L929 cells (A, n = 4). (*, **, *** or #, ##, ###, significantly different from L929 control cells or mtDNA-depleted L929 cells with P <0.05, P <0.01 or P <0.001).
Phosphorylated CREB modifies transactivation of p53 leading to p21 overexpression in mtDNA-depleted L929 cells
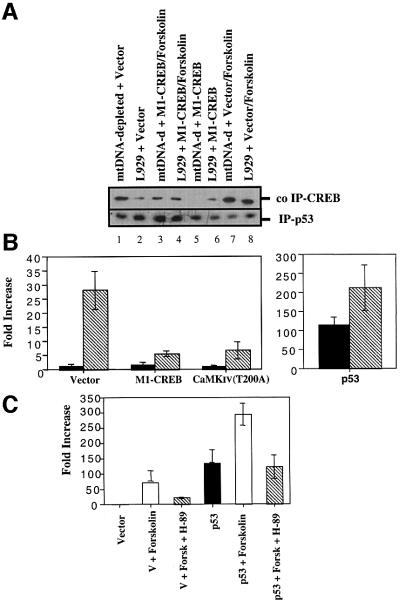
Finally, we delineated the mechanism by which phosphorylated CREB could control the cell cycle of mtDNA-depleted L929 cells. Inspired by the recent findings that phosphorylated CREB strongly and directly interacts with the N-terminus of the p53 tumor suppressor protein (Giebler et al., 2000), we tested the interaction between p53 and CREB as well as the transcriptional activity of p53 in mtDNA-depleted L929 cells. As shown in Figure 9A, CREB physically interacts with p53 in a phosphorylation-dependent manner. p53 tumor suppressor protein was immunoprecipitated from L929 and mtDNA-depleted L929 cells and the amount of co-immunoprecipitated CREB was analyzed by western blotting. The total CREB co-immunoprecipitated in mtDNA-depleted L929 cells is more abundant than in L929 control cells (Figure 9A, lanes 1 and 2). This protein–protein interaction is mediated by the phosphorylation status of CREB, as overexpression of M1-CREB, a non-phosphorylable CREB protein, completely abolished the interaction in mtDNA-depleted L929 cells (lane 5). In both cell lines, forskolin treatment to induce the phosphorylation of CREB results in an increase of CREB associated with p53 (lanes 7 and 8) while the overexpression of M1-CREB before forskolin stimulation decreases CREB protein binding to p53 (lanes 3 and 4). The physical interaction between phosphorylated CREB and p53 tumor suppressor protein observed in mtDNA-depleted L929 cells led us to address the question of whether or not the binding of phosphorylated CREB to p53 facilitates the p53-dependent transcription. We thus performed transient transfection assays with a p53-dependent luciferase reporter gene. L929 or mtDNA-depleted L929 cells were transiently transfected with a reporter plasmid carrying 13 copies of a p53-responsive promoter driving expression of the luciferase gene (pG13-Luc). As observed in Figure 9B, the luciferase activity is strongly increased in mtDNA-depleted L929 cells (28-fold increase) while co-transfection of either M1-CREB or CaMKIV (T200A) significantly inhibited p53-dependent luciferase activity. As expected, transfection of a p53 expression plasmid dramatically stimulated luciferase in L929 cells but, interestingly, the overexpression of p53 in mtDNA-depleted L929 cells resulted in an additional 2-fold stimulation (Figure 9B). As forskolin enhances CREB interaction with p53, we tested its effect on the expression of the p53 reporter gene in L929 cells. As observed in Figure 9C, forskolin also triggers p53-dependent luciferase expression (69-fold increase) and the stimulation can be partially inhibited by H-89, a PKA inhibitor. These effects are even stronger when L929 cells are transiently transfected with a plasmid encoding wild-type p53 before the stimulation with forskolin (Figure 9C).
Fig. 9. (A) CREB directly interacts with p53 tumor suppressor protein in a phosphorylation-dependent manner. p53 protein was immunoprecipitated (IP) from L929 and mtDNA-depleted L929 cells transiently transfected or not with a plasmid encoding M1-CREB. When indicated, cells were treated with 10 µM forskolin for 24 h. Bound CREB protein was detected by western blot analysis. (B) p53-dependent transcription is increased in mtDNA-depleted L929 cells. The p53-responsive pG13-Luc reporter was co-transfected into L929 (black) or mtDNA-depleted L929 cells (hatched) with the β-galactosidase construct, together with 1 µg of an empty vector, of a plasmid encoding M1-CREB and CaMKIV T200A (left panel) or a p53 expression plasmid (pC53-SN3) (right panel). (C) p53-dependent transcription is stimulated by forskolin treatment in an H-89 inhibitable manner. The p53-responsive pG13-Luc reporter was co-transfected into L929 cells with the β-galactosidase construct, together with 1 µg of an empty vector or a plasmid encoding wild-type p53 (pC53-SN3). Cells were then stimulated with 10 µM forskolin with or without 10 µM H-89 for 16 h. Luciferase activity was measured 48 h post-transfection. Reported values are expressed in fold increase of L929 control cells transfected with an empty vector as means ± SD (n = 3).
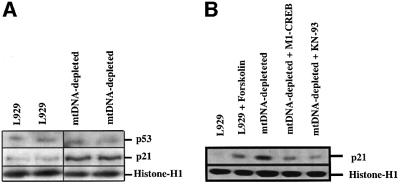
To address the biological relevance of the phosphorylated CREB pathway on p53-dependent transcription function, we analyzed the expression of an endogenous p53-responsive gene. We selected p21Waf1/Cip1, a cyclin-dependent kinase inhibitor gene, and examined its expression in L929 and mtDNA-depleted L929 cells. Western blot analysis revealed that the p21 protein level is increased in mtDNA-depleted L929 cells while little or no change in p53 tumor suppressor protein level was observed (Figure 10A). Furthermore, the level of p21 expression can be modulated by conditions that affect CREB phosphorylation status. Indeed, as observed in Figure 10B, forskolin stimulation increases p21 expression in L929 cells while inhibition of CaMKIV activity by KN-93 or overexpression of dominant-negative M1-CREB dramatically decreases p21 abundance in mtDNA-depleted cells. These results support a link between CREB activation and p21 expression.
Fig. 10. (A) p21 is overexpressed in mtDNA-depleted L929 cells. Western blotting analysis for p53 and p21 derived from L929 and mtDNA-depleted L929 cells. (B) p21 expression is stimulated in L929 cells treated with 10 µM forskolin for 16 h (lanes 1 and 2) while its expression is reduced in mtDNA-depleted cells transiently transfected with M1-CREB or incubated with 50 µM KN-93 for 16 h (lanes 3–5). Protein loading was controlled by the expression of histone H1.
Discussion
In this study, we report a new signaling pathway initiated by a dysfunction of the mitochondria and we analyzed its effect on transcription activation and cell proliferation. We used several models to study the cellular response to inhibition of mitochondrial activity: mtDNA-depleted L929 and ρ0 143B cell lines chronically treated with EtBr and a homoplasmic cybrid cell line relevant for MERRF disease which is mutated in a single nucleotide (A8344G) in the mtDNA-encoded tRNALys (King and Attardi, 1989). In all these cell lines, we found that CREB was phosphorylated on Ser133, localized in the nucleus and was able to trigger the transcription of a luciferase reporter gene driven by the authentic α-inhibin promoter containing several CRE consensus binding sites.
We confirmed the direct role of a mitochondrial activity impairment in CREB activation by testing the effects of FCCP, oligomycin and antimycin A. All these metabolic mitochondrial inhibitors were able to trigger CREB-dependent luciferase activity. In the different models used to study mitochondrial dysfunction, CREB-sensitive luciferase expression is completely inhibited by K-CREB and M1-CREB, two CREB dominant-negative mutants (Bonni et al., 1999). Nuclear localization of phosphorylated CREB on Ser133 induced by metabolic inhibition was confirmed by confocal microscopy, while CREB expression was unchanged by the treatments, as analyzed by western blotting (data not shown).
Besides PKA, several kinases such as CaMKs, p70S6K, rsk2, MSK and MAPKAP-K2 are able to phosphorylate CREB on its transcriptional regulatory site, allowing interaction with the CREB-binding protein (CBP/p300) (De Cesare et al., 1999). Mitochondrial inhibition is not known to have any effect on the cAMP pathway and we did not observe any PKA activation in mtDNA-depleted L929 cells. Our results suggest an important role for a CaMK in CREB activation. Identification of the specifically activated CaMKIV by mitochondrial dysfunction and the fact that it is responsible for CREB phosphorylation was based on the results obtained from direct in vitro kinase assays and from the correlated inhibition of both CREB phosphorylation and luciferase activity by the overexpression of a dominant-negative mutant for CaMKIV.
Biswas et al. (1999) recently showed that elevated cytosolic calcium concentration observed after mitochondrial activity inhibition leads to increased transcription of the ryanodine receptor-1 Ca2+ release channel and COX Vb genes. We also found an increase in the [Ca2+]cyt in cells showing a mitochondrial activity impairment, and data obtained with mitochondrial aequorin overexpression suggest that the cytosolic increase induced by metabolic inhibitors is accompanied by a decrease in the mitochondrial calcium pool. Interestingly, hereditary defects in mitochondrial proteins such as pyruvate dehydrogenase (PDH) that decrease aerobic metabolism have been linked to reduced mitochondrial Ca2+ uptake and sequestration (Padua et al., 1998).
How do we explain the sustained CaMKIV activity observed in both mtDNA-depleted L929 cells and cells treated with metabolic inhibitors? The hypothesis of pre-existing complexes containing active protein kinases and phosphatases suggests an attractive mechanism by which the appropriate phosphorylation state of the kinase is regulated. Phosphorylation of CaMKIV is required to generate autonomous activity, and PP2A is a constitutively active phosphatase that turns down the activity of CaMKIV (Westphal et al., 1998). Immunoprecipitation of CaMKIV and analyses of the immune complexes by immunoblotting with an anti-PP2A antibody indicate that PP2A interaction with CaMKIV is very weak in both mtDNA-depleted L929 cells and cells treated with inhibitors. However, as the interaction can be inhibited by increasing the calcium concentration with ionomycin, it seems that the association is calcium dependent. Furthermore, KN-93 treatment that inhibits CaMKIV activity allows a stronger interaction in mtDNA-depleted cells. One can thus imagine that the physical interaction between PP2A and CaMKIV is modulated by calcium and the activity of CaMKIV.
CREB family members have been described as important transcription factors regulating cell proliferation and other nuclear responses (De Cesare et al., 1999). Stimulation of the cAMP/PKA signal transduction pathway inhibits proliferation of some tumoral cell lines (Yokozaki et al., 1992) and inhibits proliferation of activated hepatic stellate cells (Houglum et al., 1997). In culture, mtDNA-depleted cells or PDH-deficient fibroblasts replicated far more slowly than normal cells (Buchet and Godinot, 1998; Padua et al., 1998).
We first examined the role of CREB on cell proliferation by treating cells with forskolin, which inhibits L929 cell proliferation by 40–50%. We then confirmed that inhibition of mitochondrial respiratory function, induced by either oligomycin or mtDNA depletion, decreases cell [3H]thymidine incorporation and thus inhibits cell progression to G1 (Sweet and Singh, 1995). Here, we found that cells transfected with dominant-negative mutants K-CREB or M1-CREB are almost insensitive to cell proliferation inhibition induced by oligomycin. Further more, these mutants suppress the cell proliferation defect observed in mtDNA-depleted L929 cells. These results suggest that phosphorylated CREB contributes to the control of cell cycle progression. CREB activation could thus explain why doubling time is higher for mtDNA-depleted L929 cells, an observation commonly reported for other mtDNA-depleted cell lines (Buchet and Godinot, 1998).
Although little is known about the numerous mechanisms by which CREB affects the cell cycle, they could be mediated through molecular interactions between phosphorylated CREB and CBP, a key integrator of cellular functions (Kamei et al., 1996), or transcription factors such as p53 tumor suppressor protein (Giebler et al., 2000). Here, we demonstrate that, in mtDNA-depleted L929 cells, phosphorylated CREB physically interacts with p53 tumor suppressor protein and increases p53 transcriptional activity as assessed in a p53-dependent luciferase assay. p53 transcriptional activation in mtDNA-depleted cells can be suppressed by either M1-CREB or CaMKIV T200A overexpresssion. However, p53 protein expression is equal in both L929 and mtDNA-depleted cells. These results suggest that the increase of p53 transcriptional activity in mtDNA-depleted L929 cells is mediated through phosphorylated CREB, which recruits p53 protein and modifies its transcriptional activity. Among p53-inducible genes, the p21Waf1/cip1 gene encodes a protein that inhibits cyclin-dependent kinase activity and leads to G1 growth arrest (el-Deiry et al., 1993). Indeed, as evidence of p53 activation in mtDNA-depleted L929 cells, we found that p21Waf1/Cip1 protein is more abundant in this cell line. The mechanism by which phosphorylated CREB triggers the transactivation of p53 in mtDNA-depleted cells, while p53 protein abundance is equal, is unknown. However, CBP acetylates not only histones but also transcription factors such as p53, c-Myb, E2-F and GATA-1 and increases their DNA sequence-specific binding capacity (Sano and Ishii, 2001).
In conclusion, the results show that CaMKIV may be involved in the calcium-dependent transcriptional regulation of genes in mitochondria-deficient cells through phosphorylation of CREB. This transcription factor could mediate calcium-induced transcription of immediate-early genes or genes encoding mitochondrial proteins and containing CRE sequences (Soderling, 1999). Indeed, CREB has been suspected to play a role in the regulation of the expression of several mitochondrial proteins such as respiratory chain subunits (Scacco et al., 2000), cytochrome c (Gopalakrishnan and Scarpulla, 1994), manganese superoxide dismutase (Kim et al., 1999) and carnitine palmitoyltransferase (Brady et al., 1992). In this study, we demonstrated that phosphorylated CREB induced by a mitochondrial dysfunction is not only involved in transcriptional regulation of a target gene such as c-fos but can also be seen as a factor that causes a cell cycle defect in cells with impaired OXPHOS. CREB, a regulator of the cell cycle and cellular differentiation program, is thus a new factor involved in the ‘retrograde signaling’ and molecular communication between functionally impaired mitochondria and the nucleus in mammalian cells.
Materials and methods
Cell cultures
The 143B, ρ0 143B, wild-type MERRF cybrids, mutated (A8344G) MERRF cybrids, L929 and mtDNA-depleted L929 cell lines have been described previously (King and Attardi, 1989; Schulze-Osthoff et al., 1993). Human embryonic kidney 293 cells were from ATCC. The different cell lines were grown in high glucose (4.5 g/l) Dulbecco’s modified Eagle’s medium (DHG) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL) in a 5% CO2 incubator. mtDNA-depleted L929 and ρ0 143B cells were grown in the presence of 400 ng/ml EtBr (Sigma), 50 µg/ml uridine and 1 mM pyruvate.
Immunofluorescence confocal laser scanner microscopy
Cells were fixed and permeabilized with methanol/acetone (20/80%), rinsed three times with phosphate-buffered saline (PBS) and unspecific sites were blocked with 1% bovine serum albumin (BSA) in PBS. For phospho-CREB, cells were immunostained overnight at 4°C with a primary rabbit polyclonal antibody against phosphorylated CREB on Ser133 (NEBL) at 1:1000, while for COX I detection, cells were incubated for 1 h with a monoclonal antibody against mouse COX I protein (Molecular Probes) at a 1:20 dilution, washed three times with PBS, incubated with (1:1000) anti-rabbit or anti-mouse Alexa antibodies (Molecular Probes) for 1 h at 37°C and processed for confocal microscopy (Leica).
mtDNA analysis
Subcellular fractionation to obtain nuclear- and mitochondria-enriched fractions was performed and genomic DNA or mtDNA was prepared as previously described (Wiesner et al., 1991). A specific mitochondrial genome fragment of 823 bp was amplified by PCR using forward (5′-TTTGGGAACTACTAGAATTGATCA-3′) and reverse (5′-GCTGGTATTCTAATTAAACTACTT-3′) primers specific for the mouse mitochondrial D-loop. PCR was performed using 10 ng of mtDNA or genomic DNA for 30 cycles at the hybridization temperature of 50°C.
Luciferase assay
293, 143B, ρ0 143B, wild-type MERRF cybrids, mutated (A8344G) MERRF cybrids, L929 and mtDNA-depleted L929 cells seeded in 12-well plates were transiently transfected by the calcium phosphate method or Superfect reagent (Qiagen) with a luciferase reporter construct (0.5 µg/well) and a β-galactosidase (0.25 µg/well) expression vector. In some experiments, cells were treated with 20 µM BAPTA-AM (Molecular Probes) for 6 h or co-transfected with 0.5 µg/well of an expression vector encoding K-CREB, M1-CREB, full-length CaMKIV or a dominant-negative form, CaMKIV T200A. For p53 activity, cells were transiently co-transfected with 0.5 µg/well of a p53-responsive (pG13-Luc) reporter with the β-galactosidase construct, together with 1 µg of an empty vector, or of a plasmid encoding M1-CREB, CaMKIV T200A or a p53 expression plasmid (pC53-SN3). In some experiments, cells were treated with 10 µM forskolin for 16 h in the presence or absence of 10 µM H-89. Cells were then harvested and luciferase activity was determined in cleared lysates using the commercial Reporter™ assay system (Promega) and normalized for β-galactosidase activity. Inhibitors were added for 16 h.
Cell proliferation index
L929 and mtDNA-depleted L929 cells were seeded in 12-well dishes at 50 000 cells/well. After a 24 h period of serum starvation, proliferation was triggered by the addition of fresh medium supplemented with 10% FBS, and the next day cells were incubated with 2 µCi of [3H]thymidine for 4 h. Cells were harvested and [3H]thymidine incorporation was determined as previously described (Buck et al., 1994) and expressed as c.p.m./mg of proteins. In some conditions, 24 h before the addition of 8 µM oligomycin for 6 h or 10 µM forskolin for 24 h, L929 and mtDNA-depleted L929 cells were transiently transfected using Superfect reagent with 0.5 µg/well of a control vector, with constructs encoding M1-CREB, K-CREB or CaMKIV T200A. KN-93 (50 µM) was added to empty vector-transfected L929 and mtDNA-depleted L929 cells for 24 h.
Western blotting analysis
Immunoblotting analyses were performed with 10 µg of protein of cleared lysates after separation on a 10 or 12% SDS–polyacrylamide gel. PVDF membranes were blocked overnight at 4°C in 5% non-fat dry milk or BSA-TBST. Primary antibodies were either anti-c-fos (Santa Cruz) at 1:1000, anti-CaMKIV or anti-CaMKII (Santa Cruz) at 1:100, anti-PP2A C subunit (Santa Cruz) at 1:1000, anti-phospho-CREB (Upstate Biotechnology), anti-CREB (NEBL) (1 µg/ml) or anti-p21 (Santa Cruz) at 1:2500, anti-histone-1 (Santa Cruz) at 1:1000 and anti-p53 (PharMingen) at 2 µg/ml. For loading controls, immunoblotting for α-tubulin or histone H1 was performed on the same membranes after stripping or not, followed by incubation with horseradish peroxidase-coupled secondary antibodies (Dako), and were detected by chemiluminescence (NEN).
Co-immunoprecipitation experiments
Endogenous CaMKII and IV were immunoprecipitated respectively with 10 µg of an anti-CaMKII or IV antibody (Santa Cruz) from clear lysates of mtDNA-depleted L929 cells or L929 cells treated or not for 6 h with 1 µM antimycin A, 8 µM oligomycin or 10 µM FCCP. Immune complexes were immobilized by adding 75 µl of a protein G + protein A–agarose mixture (Oncogene), washed three times and 40 µl of sample buffer was added. Western blot analyses for the PP2A C subunit in lysates, immunoprecipitated CaMKIV and co-immunoprecipitated PP2A C subunit were performed. p53 was immunoprecipitated from the lysates of L929 and mtDNA-depleted cells with 10 µg of an anti-p53 (Ab-1/epitope amino acids 371–380) before p53 and CREB were analyzed by western blotting.
In vitro kinase assays
CaMKII or IV activity was determined after immunoprecipitation by measuring its ability to phosphorylate a synthetic CREB phosphopeptide with the sequence KRREILSRRPS(p)YR (NEBL) according to the procedure described in the Supplementary data.
Statistical analysis
All experiments were performed at least twice and one set of representative results is shown. Results are expressed as means ± SD. Statistical significance was determined by Scheffé’s contrasts after analysis of variance (ANOVA1).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The professors and doctors who we thank for material supply and plasmids used in this study are mentioned in the Supplementary data. T.A. is a Research Associate of FNRS (Fonds National de la Recherche Scientifique, Brussels, Belgium). This work was partly supported by FRFC and SSTC. This text presents results of the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming. The scientific responsibility is assumed by its authors.
References
- Biswas G., Adebanjo,O.A., Freedman,B.D., Anandatheerthavarada,H.K., Vijayasarathy,C., Zaidi,M., Kotlikoff,M. and Avadhani,N.G. (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J., 18, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A., Brunet,A., West,A.E., Datta,S.R., Takasu,M.A. and Greenberg,M.E. (1999) Cell survival promoted by the Ras–MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science, 286, 1358–1362. [DOI] [PubMed] [Google Scholar]
- Brady P.S., Park,E.A., Liu,J.S., Hanson,R.W. and Brady,L.J. (1992) Isolation and characterization of the promoter for the gene coding for the 68 kDa carnitine palmitoyltransferase from the rat. Biochem. J., 286, 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchet K. and Godinot,C. (1998) Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted ρ° cells. J. Biol. Chem., 273, 22983–22989. [DOI] [PubMed] [Google Scholar]
- Buck M., Turler,H. and Chojkier,M. (1994) LAP (NF-IL-6), a tissue-specific transcriptional activator, is an inhibitor of hepatoma cell proliferation. EMBO J., 13, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila T., Anderson,K.A., Ho,N. and Means,A.R. (1996) A unique phosphorylation-dependent mechanism for the activation of Ca2+/calmodulin-dependent protein kinase type IV/GR. J. Biol. Chem., 271, 21542–21548. [DOI] [PubMed] [Google Scholar]
- De Cesare D., Fimia,G.M. and Sassone-Corsi,P. (1999) Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem. Sci., 24, 281–285. [DOI] [PubMed] [Google Scholar]
- el-Deiry W.S. et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- Epstein C.B., Waddle,J.A., Hale,W.,IV, Dave,V., Thornton,J., Macatee,T.L., Garner,H.R. and Butow,R.A. (2001) Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell, 12, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebler H.A., Lemasson,I. and Nyborg,J.K. (2000) p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol. Cell. Biol., 20, 4849–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan L. and Scarpulla,R.C. (1994) Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. Role of cyclic AMP in cytochrome c and COXIV gene expression. J. Biol. Chem., 269, 105–113. [PubMed] [Google Scholar]
- Hall J., Thomas,K.L. and Everitt,B.J. (2001) Fear memory retrieval induces CREB phosphorylation and Fos expression within the amygdala. Eur. J. Neurosci., 13, 1453–1458. [DOI] [PubMed] [Google Scholar]
- Heddi A., Lestienne,P., Wallace,D.C. and Stepien,G. (1993) Mito chondrial DNA expression in mitochondrial myopathies and coordinated expression of nuclear genes involved in ATP production. J. Biol. Chem., 268, 12156–12163. [PubMed] [Google Scholar]
- Houglum K., Lee,K.S. and Chojkier,M. (1997) Proliferation of hepatic stellate cells is inhibited by phosphorylation of CREB on serine 133. J. Clin. Invest., 99, 1322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y. et al. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- Kim H.P., Roe,J.H., Chock,P.B. and Yim,M.B. (1999) Transcriptional activation of the human manganese superoxide dismutase gene mediated by tetradecanoylphorbol acetate. J. Biol. Chem., 274, 37455–37460. [DOI] [PubMed] [Google Scholar]
- King M.P. and Attardi,G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- King M.P. and Attardi,G. (1996) Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol., 264, 304–313. [DOI] [PubMed] [Google Scholar]
- Komeili A., Wedaman,K.P., O’Shea,E.K. and Powers,T. (2000) Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol., 151, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Neufer,P.D. and Williams,R.S. (1995) Nuclear responses to depletion of mitochondrial DNA in human cells. Am. J. Physiol., 269, C1265–C1270. [DOI] [PubMed] [Google Scholar]
- Liao X. and Butow,R.A. (1993) RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell, 72, 61–71. [DOI] [PubMed] [Google Scholar]
- Lombes A. et al. (1989) Myoclonic epilepsy and ragged-red fibers with cytochrome oxidase deficiency: neuropathology, biochemistry and molecular genetics. Ann. Neurol., 26, 20–33. [DOI] [PubMed] [Google Scholar]
- Mayfield S.P. (1990) Chloroplast gene regulation: interaction of the nuclear and chloroplast genomes in the expression of photosynthetic proteins. Curr. Opin. Cell Biol., 2, 509–513. [DOI] [PubMed] [Google Scholar]
- Moraes C.T., Shanske,S., Tritschler,H.J., Aprille,J.R., Andreetta,F., Bonilla,E., Schon,E.A. and DiMauro,S. (1991) mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet., 48, 492–501. [PMC free article] [PubMed] [Google Scholar]
- Morais R. (1996) Isolation of avian mitochondrial DNA-less cells. Methods Enzymol., 264, 296–304. [DOI] [PubMed] [Google Scholar]
- Padua R.A., Baron,K.T., Thyagarajan,B., Campbell,C. and Thayer,S.A. (1998) Reduced Ca2+ uptake by mitochondria in pyruvate dehydrogenase-deficient human diploid fibroblasts. Am. J. Physiol., 274, C615–C622. [DOI] [PubMed] [Google Scholar]
- Parikh V.S., Morgan,M.M., Scott,R., Clements,L.S. and Butow,R.A. (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science, 235, 576–580. [DOI] [PubMed] [Google Scholar]
- Park I.K. and Soderling,T.R. (1995) Activation of Ca2+/calmodulin-dependent protein kinase (CaM-kinase) IV by CaM-kinase kinase in Jurkat T lymphocytes. J. Biol. Chem., 270, 30464–30469. [DOI] [PubMed] [Google Scholar]
- Pei L., Dodson,R., Schoderbek,W.E., Maurer,R.A. and Mayo,K.E. (1991) Regulation of the α inhibin gene by cyclic adenosine 3′,5′-monophosphate after transfection into rat granulosa cells. Mol. Endocrinol., 5, 521–534. [DOI] [PubMed] [Google Scholar]
- Sano Y. and Ishii,S. (2001) Increased affinity of c-Myb for CBP after CBP-induced acetylation. J. Biol. Chem., 276, 3674–3682. [DOI] [PubMed] [Google Scholar]
- Scacco S., Vergari,R., Scarpulla,R.C., Technikova-Dobrova,Z., Sardanelli,A., Lambo,R., Lorusso,V. and Papa,S. (2000) cAMP-dependent phosphorylation of the nuclear encoded 18-kDa (IP) subunit of respiratory complex I and activation of the complex in serum-starved mouse fibroblast cultures. J. Biol. Chem., 275, 17578–17582. [DOI] [PubMed] [Google Scholar]
- Schulze-Osthoff K., Beyaert,R., Vandevoorde,V., Haegeman,G. and Fiers,W. (1993) Depletion of the mitochondrial electron transport abrogates the cytotoxic and gene-inductive effects of TNF. EMBO J., 12, 3095–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T., Thornton,J. and Butow,R.A. (2000) Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell, 11, 2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling T.R. (1999) The Ca–calmodulin-dependent protein kinase cascade. Trends Biochem. Sci., 24, 232–236. [DOI] [PubMed] [Google Scholar]
- Sweet S. and Singh,G. (1995) Accumulation of human promyelocytic leukemic (HL-60) cells at two energetic cell cycle checkpoints. Cancer Res., 55, 5164–5167. [PubMed] [Google Scholar]
- Vaillant F. and Nagley,P. (1995) Human cell mutants with very low mitochondrial DNA copy number (ρ d). Hum. Mol. Genet., 4, 903–914. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wang H. and Morais,R. (1997) Up-regulation of nuclear genes in response to inhibition of mitochondrial DNA expression in chicken cells. Biochim. Biophys. Acta, 1352, 325–334. [DOI] [PubMed] [Google Scholar]
- Westphal R.S., Anderson,K.A., Means,A.R. and Wadzinski,B.E. (1998) A signaling complex of Ca2+–calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science, 280, 1258–1261. [DOI] [PubMed] [Google Scholar]
- Wiesner R.J., Swift,H. and Zak,R. (1991) Purification of mitochondrial DNA from total cellular DNA of small tissue samples. Gene, 98, 277–281. [DOI] [PubMed] [Google Scholar]
- Yokozaki H., Tortora,G., Pepe,S., Maronde,E., Genieser,H.G., Jastorff,B. and Cho-Chung,Y.S. (1992) Unhydrolyzable analogues of adenosine 3′:5′-monophosphate demonstrating growth inhibition and differentiation in human cancer cells. Cancer Res., 52, 2504–2508. [PubMed] [Google Scholar]