Abstract
HeLa cytoplasmic extracts contain both 3′–5′ and 5′–3′ exonuclease activities that may play important roles in mRNA decay. Using an in vitro RNA deadenylation/decay assay, mRNA decay intermediates were trapped using phosphothioate-modified RNAs. These data indicate that 3′–5′ exonucleolytic decay is the major pathway of RNA degradation following deadenylation in HeLa cytoplasmic extracts. Immunodepletion using antibodies specific for the exosomal protein PM-Scl75 demonstrated that the human exosome complex is required for efficient 3′–5′ exonucleolytic decay. Furthermore, 3′–5′ exonucleolytic decay was stimulated dramatically by AU-rich instability elements (AREs), implicating a role for the exosome in the regulation of mRNA turnover. Finally, PM-Scl75 protein was found to interact specifically with AREs. These data suggest that the interaction between the exosome and AREs plays a key role in regulating the efficiency of ARE-containing mRNA turnover.
Keywords: AU-rich elements/exosome/mRNA stability/PM-Sc175
Introduction
mRNA turnover is a highly regulated process that plays an important role in regulating the levels of transcripts that encode a variety of proteins including cytokines, growth factors and proto-oncogenes (Mitchell and Tollervey, 2000a; Guhaniyogi and Brewer, 2001; Wilusz et al., 2001a). The decay of most mRNAs in mammalian cells is initiated by the shortening of the poly(A) tail (Wilson and Treisman, 1988; Shyu et al., 1991). Distal regions of the transcript can influence poly(A) shortening, as efficient deadenylation is mediated through an interaction between the deadenylase PARN and the 5′ mRNA cap (Dehlin et al., 2000; Gao et al., 2000; Martinez et al., 2001). Following deadenylation, the body of the transcript is decayed rapidly with no discernible intermediates. The pathway(s) involved in the subsequent turnover of deadenylated transcripts in mammalian cells, therefore, remains to be elucidated. Furthermore, an understanding of the factors involved in this step of mRNA decay is necessary to provide insights into regulatory targets and mechanisms.
There are two pathways that function to degrade mRNAs in the yeast Saccharomyces cerevisiae following poly(A) tail shortening by a complex involving Ccr4p and Caf1p/Pop2p (Daugeron et al., 2001; Tucker et al., 2001). The major pathway involves removal of the 5′ cap by Dcp1p followed by degradation of the mRNA body by the 5′–3′ exonuclease Xrn1p (Tucker and Parker, 2000). Mutations that block the decapping/5′–3′ pathway revealed an alternative decay pathway that involves a complex of 3′–5′ exonucleases called the exosome (van Hoof and Parker, 1999; Mitchell and Tollervey, 2000b). While a Dcp1p-like decapping activity (Gao et al., 2001) and homologs of the exosome complex (Brouwer et al., 2001b) have been identified in mammalian cells, their precise roles in mRNA turnover remain to be defined.
The exosome, a complex of 10 or more 3′–5′ exonucleases, is highly conserved over evolution (Mitchell et al., 1997; Chekanova et al., 2000; Brouwer et al., 2001a; Estevez et al., 2001) and is found in both the nucleus and cytoplasm. It plays an important role in the processing of rRNA (Mitchell et al., 1997; Briggs et al., 1998; Zanchin and Goldfarb, 1999; Allmang et al., 2000), as well as the 3′-end processing of numerous small nuclear and nucleolar RNAs (Allmang et al., 1999a; Kufel et al., 2000; van Hoof et al., 2000a). The exosome also plays a key role in the turnover of RNAs in both the nucleus (Bousquet-Antonelli et al., 2000) and the cytoplasm (Jacobs Anderson and Parker, 1998). Several components of the human exosome have been identified, including hRrp4p, hRrp40p, hRrp41p, hRrp46p, PM-Scl75 and PM-Scl100 (Mitchell et al., 1997; Allmang et al., 1999b; Brouwer et al., 2001a), the latter two being targets for autoantibody production in a subset of patients with myositis and scleroderma syndromes (Targoff, 2000). Recent data have also identified hRrp42p and hCsl4p as components of the human exosome (Raijmakers et al., 2002). A number of helicases and other factors involved in mRNA metabolism have been shown to interact with the yeast exosome functionally and/or physically (de la Cruz et al., 1998; Jacobs Anderson and Parker, 1998; Burkard and Butler, 2000; van Hoof et al., 2000b). This suggests that the exosome complex is likely to be a target for regulation and may also play an active role in the coordination of diverse processes in RNA metabolism. Finally, the mechanism by which the exosome distinguishes its substrates and specifically targets mRNAs for degradation remains to be elucidated.
AU-rich elements (AREs) present in the 3′-untranslated regions (3′-UTRs) of many mammalian mRNAs are responsible for targeting the transcripts for rapid decay (Chen and Shyu, 1995). Numerous proteins have been identified that specifically interact with AREs (Brewer, 1991; Levine et al., 1993; Ma et al., 1996; Lai and Blackshear, 2001), some of which appear to stabilize (Fan and Steitz, 1998; Peng et al., 1998; Ford et al., 1999) or destabilize (Lai et al., 1999; Loflin et al., 1999) mRNAs. How these ARE-binding proteins interface with the mRNA turnover machinery currently is unclear. Phosphokinase signaling pathways (Winzen et al., 1999; Ming et al., 2001) and ubiquitylation/proteasome activity (Laroia et al., 1999) also influence mRNA decay, providing numerous avenues for changes in the turnover rate of specific mRNAs in response to changes in the cellular environment. How AREs attract or repel the turnover machinery from selected mRNAs is a pivotal question in the post-transcriptional regulation of gene expression.
The general pathways and many of the regulatory aspects of mRNA deadenylation and decay can be reproduced using S100 cytoplasmic extracts from mammalian cells (Ford and Wilusz, 1999; Ford et al., 1999; Chen et al., 2000; Gao et al., 2000, 2001). In this study, we have characterized the major pathway of turnover of mRNAs following deadenylation in S100 extracts to be 3′–5′ decay mediated by exosomal proteins. This establishes a role for the exosome in mRNA turnover in mammalian systems. Interestingly, exosome-mediated 3′–5′ decay was found to be stimulated specifically by AREs. While addressing the question of how the exosome distinguishes an ARE-containing RNA substrate, we determined that the exosomal protein PM-Scl75 interacts with AREs with high specificity. These data suggest a mechanism by which the exosome selectively interacts with unstable RNAs and a model for the overall regulation of mRNA turnover by AREs.
Results
Both 3′–5′ and 5′–3′ exonuclease activities exist in HeLa cytoplasmic extracts
Following deadenylation in yeast, mRNAs can be degraded by either decapping/5′–3′ exonuclease or directly by a complex of 3′–5′ exonucleases (Wilusz et al., 2001a). Previous work has shown clearly that mammalian cells contain 3′–5′ exonucleases (Brewer, 1999; Ford et al., 1999) as well as a Dcp1p-like decapping enzyme (Gao et al., 2001). In order to test whether mammalian cells contain all of the enzymes necessary for the dual decay pathways of deadenylated mRNAs, we now examined HeLa cytoplasmic extracts for a 5′–3′ exonuclease activity.
In the absence of competitors that remove proteins from their 5′- or 3′-terminal structures, capped and polyadenylated RNA substrates are very stable in HeLa cytoplasmic extracts (Ford et al., 1999; Gao et al., 2001). In order to detect 5′–3′ exonuclease activities, we prepared a polyadenylated SV-A60 RNA substrate with pG at the 5′ end by transcribing the RNA in the presence of 5′-GMP. The pG RNA substrate is protected from 3′–5′ exonucleases due to the presence of poly(A)-binding proteins on its poly(A) tail (Bernstein et al., 1989; Ford et al., 1997, 1999). The lack of a 5′ cap or 5′-triphosphate, however, makes it an excellent substrate for 5′–3′ exonucleases. As seen in Figure 1, while capped and polyadenylated RNAs were very stable (lanes 7meGpppG), transcripts with a 5′-monophosphate end were degraded rapidly with no apparent intermediates. The activation of nuclease activity by exposing the 5′ end, as well as the lack of degradation intermediates, is very consistent with a processive 5′–3′ exonuclease being responsible for the observed decay. Furthermore, a decay intermediate consistent with 5′–3′ decay was identified in similar assays by incorporating phosphothioate modifications into RNA substrates as described below (data not shown). We conclude that both 3′–5′ and 5′–3′ exonucleases are present in HeLa cell cytoplasmic extracts. Since a Dcp1p-like decapping activity can also be demonstrated in these extracts under appropriate conditions (Gao et al., 2001), we suppose that either exonucleolytic pathway could be responsible for decay of the mRNA body following deadenylation in mammalian cells.
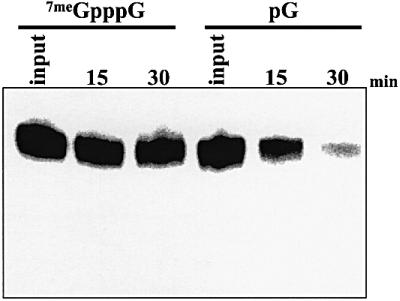
Fig. 1. 5′–3′ exonuclease activity can be identified in HeLa extracts. Polyadenylated SV-A60 RNA containing either a 5′ cap (7meGpppG lanes) or a 5′ monophosphate (pG lanes) were incubated in HeLa S100 cytoplasmic extract for the times indicated. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea.
3′–5′ exonucleolytic degradation is responsible for the decay of RNA substrates following deadenylation in HeLa cytoplasmic extracts
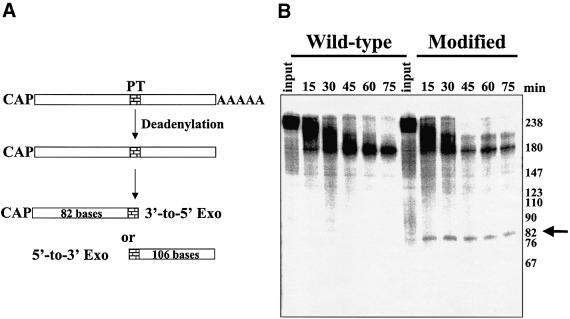
We previously have described a deadenylation-dependent in vitro RNA turnover assay using HeLa cytoplasmic extracts that faithfully reproduces aspects of regulated mRNA decay observed in vivo (Ford et al., 1999). While RNA turnover in the in vitro assay is initiated by deadenylation (Ford et al., 1999; Gao et al., 2000), the exonucleolytic pathway(s) involved in decay of the body of the transcript remains to be elucidated. The insertion of poly(G) sequences has been used successfully in yeast to trap mRNA degradation intermediates and identify turnover pathways (Muhlrad et al., 1994). Poly(G) tracts, however, failed to reveal mRNA turnover intermediates directly in mammalian systems either in vivo or in vitro (data not shown). As an alternative approach to identify mRNA turnover intermediates, we used phosphothioate derivatives to modify the backbone of RNA substrates. Synthetic RNA oligonucleotides that contained three consecutive phosphothioate derivatives at a selected site were prepared and reconfigured into a capped and polyadenylated RNA substrate by bridged ligation, using T4 DNA ligase, to a 5′-capped fragment and a 3′ RNA fragment that contained a 60-base poly(A) tail. Equimolar amounts of RNAs made in this fashion that either lacked (wild-type) or contained site-directed phosphothioates (modified lanes) were gel purified and incubated with HeLa extract in the in vitro mRNA turnover assay. As outlined in Figure 2A, following deadenylation, blockage of exonuclease activities at the phosphothioate modification in the RNA substrate would give an 82-base fragment for 3′–5′ exonucleases and a 106-base fragment for decapping/5′–3′ exonuclease decay. As seen in Figure 2B, while no consistent bands that accumulated with kinetics appropriate to be degradation intermediates could be detected with unmodified wild-type RNAs, phosphothioate-modified RNA substrates exclusively accumulated an 82-base fragment following deadenylation of the input transcript. The exclusive detection of an 82-base fragment is highly significant, as only 20% of the input radioactivity is present in this portion of the starting transcript, while 80% of the radioactivity is localized to the 106-base fragment that would have been produced as a result of 5′–3′ decay. All capped and polyadenylated phosphothioate-modified RNAs we have incubated in the in vitro RNA turnover assay have generated decay intermediates consistent with 3′–5′ exonucleolytic decay, regardless of the presence of an ARE (data not shown). We conclude that 3′–5′ exonucleolytic decay is the major, if not exclusive, pathway for turnover of mRNA following deadenylation in HeLa cytoplasmic extracts.
Fig. 2. Phosphothioate-modified RNAs demonstrate that RNAs are degraded by a 3′–5′ exonuclease following deadenylation. A synthetic RNA containing three consecutive phosphothioate substitutions was prepared and ligated to RNA fragments containing a 5′ cap and a 3′ poly(A) tail as described in Materials and methods. Polyadenylated wild-type or phosphothioate-modified variants of GemARE-A60 RNA were incubated in the in vitro deadenylation/decay system using HeLa cytoplasmic extracts for the times indicated. As shown in (A), trapping of an 82-base intermediate would identify a block by the phosphothioate modification to 3′–5′ exonucleases, while trapping a 106-base fragment would be consistent with decay via a 5′–3′ exonucleolytic pathway. (B) Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The arrow on the right indicates the 82-base fragment that was trapped specifically by the phosphothioate modifications.
Exosomal proteins are required for 3′–5′ exonucleolytic decay in HeLa cytoplasmic extracts
The 3′–5′ exonucleolytic decay pathway in yeast is mediated by a large complex of exoribonucleases called the exosome (Mitchell et al., 1997; Jacobs Anderson and Parker, 1998). The exosome also plays a role in rRNA processing and maturation of small nuclear RNAs (Allmang et al., 1999a; van Hoof et al., 2000a). A similar complex of 11–16 proteins with significant homology to the yeast exonucleases has been observed in mammalian cells (Brouwer et al., 2001a,b). Several human exosomal homologs have been shown to complement yeast exosomal proteins and correct rRNA processing defects (Allmang et al., 1999b). Based on these striking similarities, we hypothesized that the human exosome is involved in 3′–5′ decay of mRNAs in cytoplasmic extracts.
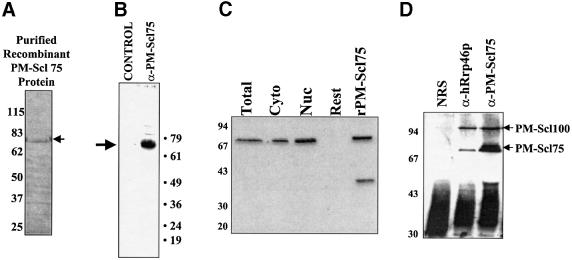
In order to test this hypothesis, we expressed the exosomal component PM-Scl75, a homolog of the yeast RNase PH family exonuclease Rrp45p, in bacteria and purified the recombinant His-tagged protein (Figure 3A). Polyclonal antibodies to PM-Scl75 protein were raised in mice. As seen in Figure 3B, this antibody detected endogenous PM-Scl75 protein in HeLa cytoplasmic extracts with high specificity. A variety of experiments were performed to confirm that PM-Scl75 was truly a component of the human exosome. First, in a manner identical to other antibodies to human exosomal components that have been analyzed (Alderuccio et al., 1991), PM-Scl75-specific antibodies efficiently stain the nucleoli of fixed Hep-2 cells (data not shown). Secondly, in order to confirm that PM-Scl75 exists in the cytoplasmic form of the exosome, western blots were performed on fractionated cell extracts using the modification of the Dignam procedure as described by Wahle and Keller (1994). As seen in Figure 3C, PM-Scl75 is present in both the nuclear and cytoplasmic fractions, but not in the nuclear fraction left behind following salt extraction (‘Rest’ lane) as has been seen for other exosomal proteins (Brouwer et al., 2001a). Finally, PM-Scl75 and PM-Scl100 specifically co-immunoprecipitated with hRrp46p, a known component of the exosome (Figure 3D). These data provide compelling evidence that PM-Scl75 is an exosomal protein and that the PM-Scl75 polyclonal antiserum we have developed can precipitate the exosomal complex.
Fig. 3. Preparation and characterization of recombinant PM-Scl75 protein and antibodies. (A) Purified recombinant His-tagged PM-Scl75 protein was prepared and analyzed by Coomassie Blue staining following electrophoresis on a 10% acrylamide gel containing SDS. The arrow indicates the position of the purified recombinant protein. (B) Polyclonal antibodies were raised against recombinant PM-Scl75 protein in mice and used in a western blot against HeLa cytoplasmic S100 extract to assess their specificity. The blot was developed using chemiluminescence. The arrow indicates the position of PM-Scl75 protein. (C) PM-Scl75 protein is present in both nuclear and cytoplasmic fractions. Fractionated HeLa cell extracts were probed for the presence of PM-Scl75 protein by western blotting using rabbit anti-PM-Scl75 antiserum. Cyto = cytoplasmic fraction; Nuc = nuclear fraction; Rest = membrane and nuclear remainder fraction; and rPMScl-75 = purified recombinant protein. (D) PM-Scl75 antiserum co-immunoprecipitates components of the exosome. Immunoprecipitations of S100 extract were performed using rabbit serum recognizing hRrp46p (lane anti-hRrp46p), PM-Scl75 (lane antiPM-Scl75) or normal rabbit serum (lane NRS). The precipitates were separated by SDS–PAGE and blotted and stained with a patient antiserum recognizing both PMScl-75 and PM-Scl100 proteins (indicated by the arrows on the right).
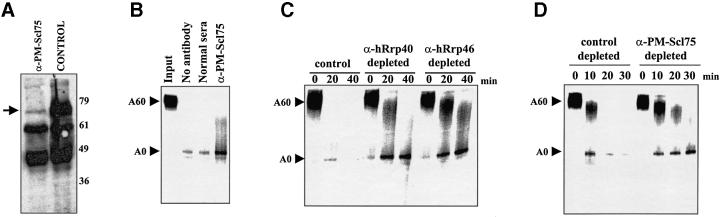
Immunodepletion experiments were performed using HeLa S100 cytoplasmic extract and either PM-Scl75-specific antibodies or normal mouse serum. As seen in Figure 4A, endogenous PM-Scl75 protein was depleted significantly by α-PM-Scl75 mouse serum, but not by normal serum (compare lanes α-PM-Scl75 and control). The bands of 61 and 49 kDa observed on this blot represent serum proteins present in the polyclonal antiserum used for the immunodepletions. We next tested these immunodepleted extracts for activity in in vitro mRNA turnover assays. As seen in Figure 4B, the polyadenylated GemARE-A60 transcript is deadenylated and degraded very efficiently in our standard assay (‘No antibody’ lane). Incubation of extracts with normal mouse sera had no effect on the rate of turnover in vitro (‘Normal sera’ lane). Extracts that had been immunodepleted with α-PM-Scl75 antibodies, however, showed a dramatic (∼6-fold) reduction in the efficiency of turnover in the in vitro assay (α-PM-Scl75 lane). The residual amount of PM-Scl75 (and associated exosomal components) that could not be removed by immunodepletion is probably responsible for the low levels of decay still observed in these treated extracts. Immunodepletion of extracts using antisera specific for hRrp40 and hRrp46, two other exosomal components, gave a similar reduction in the efficiency of RNA turnover (Figure 4C). Surprisingly, time course experiments revealed that the rate of deadenylation was also reduced reproducibly by immunodepletion using α-PM-Scl75, α-hRrp40 or α-hRrp46 antibodies (Figure 4C and D). A functional association therefore appears to exist between the exosome and the process of deadenylation. We conclude that components of the human exosome are likely to be responsible for the 3′–5′ exonucleolytic decay of deadenylated mRNAs in cytoplasmic extracts. The exosome appears to play precisely the same role in the mammalian cytoplasm as it does in the yeast S.cerevisiae.
Fig. 4. The exosome is required for 3′–5′ exonucleolytic decay of the body of RNA substrates in in vitro deadenylation/decay assays. HeLa S100 cytoplasmic extracts were immunodepleted using anti-PM-Scl75 mouse antiserum or pre-immune serum prior to their use in in vitro RNA deadenylation/decay assays. (A) Western blotting with PM-Scl75 antiserum demonstrates that most of the endogenous PMScl-75 protein (indicated by the arrow) was removed by immunodepletion with anti-PM-Scl75 antiserum (lane α-PM-Scl75) but not by normal mouse serum (control lane) in the extracts used for the assay in (B). (B) Capped and polyadenylated GemARE-A60 RNA was incubated in the in vitro deadenylation/decay assay for 30 min using either untreated S100 extract (lane no antibody), extract that was treated with normal serum (lane normal sera) or extract that had been immunodepleted with antibodies specific for PM-Scl75 protein (lane α-PM-Scl75). Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The positions of the polyadenylated input and deadenylated RNAs are indicated on the left. (C and D) Capped and polyadenylated GemARE-A60 RNA was incubated in the in vitro deadenylation/decay assay for the times indicated using either extract that was untreated (control lanes), extract that was treated with normal serum (control depleted lanes) or extract that had been immunodepleted with antibodies specific for PM-Scl75 protein (lanes α-PM-Scl75 depleted), hRrp40 protein (lanes α-hRrp40 depleted) or hRrp46 protein (lanes α-hRrp46 depleted). Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The positions of the polyadenylated input and deadenylated RNAs are indicated on the left.
Exosome activity is regulated by AU-rich elements in mammalian cells
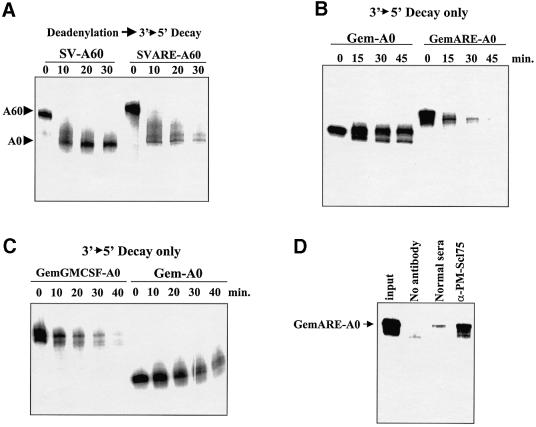
AREs stimulate the turnover of RNAs both in vivo and in our in vitro assay (Ford et al., 1999). We have shown previously that AREs stimulate the rate of deadenylation ∼2-fold in HeLa cytoplasmic extracts. AREs, however, stimulate the overall rate of turnover of RNAs in cytoplasmic extracts 4- to 8-fold (Ford et al., 1999; Figure 5A). Since we demonstrated above that 3′–5′ exonucleolytic decay is the major pathway of turnover of RNAs in vitro following deadenylation, we tested whether AREs could also stimulate the activity of the exosome on RNA substrates. Capped but non-polyadenylated RNAs were prepared that lacked (Gem-A0) or contained the tumor necrosis factor-α (TNF-α) AU-rich element (GemARE-A0) and incubated in HeLa S100 extracts. As seen in Figure 5B, the Gem-A0 RNA substrate that lacked an ARE showed only a small amount of 3′ trimming but was generally stable over the time course of incubation in the extract. The addition of an ARE to an RNA substrate (GemARE-A0), however, dramatically increased the rate of 3′–5′ exonucleolytic decay (∼10-fold). Similar results were obtained with the insertion of the granulocyte–macrophage colony-stimulating factor (GM-CSF) ARE (Figure 5C). Finally, immunodepletion of HeLa extracts with antibodies against the exosomal protein PM-Scl75 dramatically and specifically reduced the efficiency of 3′–5′ exonucleolytic decay in these assays (Figure 5D). We conclude that the presence of an ARE strongly stimulates 3′–5′ exonucleolytic decay mediated by the exosome in HeLa extracts.
Fig. 5. 3′–5′ Decay is regulated by AU-rich instability elements. (A) Polyadenylated SV-A60 RNA or a derivative that contains the 34 base ARE from TNF-α (SVARE-A60) was incubated with HeLa S100 cytoplasmic extracts for the times indicated. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The positions of the polyadenylated input RNAs and deadenylated intermediates are indicated on the left. Note that while the rate of deadenylation is only moderately affected, the presence of an ARE greatly stimulates the decay of the SVARE-A60 transcript. (B) Gem-A0 RNA [which lacks a poly(A) tail] or a derivative that contains the 34 base ARE from TNF-α (GemARE-A0) was incubated with HeLa S100 cytoplasmic extracts for the times indicated. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. (C) Gem-A0 RNA or a derivative that contains the 51 base ARE from GMCSF (GemGMCSF-A0) was incubated with HeLa S100 cytoplasmic extracts for the times indicated. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. (D) GemARE-A0 RNA was incubated in the in vitro decay assay for 30 min using either untreated S100 extract (lane no antibody), extract that was treated with normal serum (lane normal sera) or extract that had been immunodepleted with antibodies specific for PM-Scl75 protein (lane α-PM-Scl75). Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea.
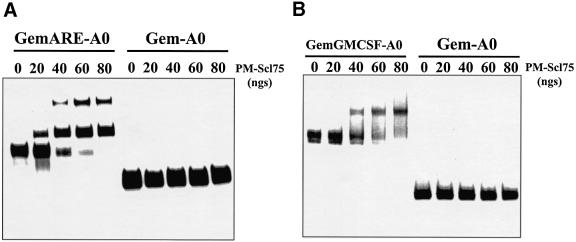
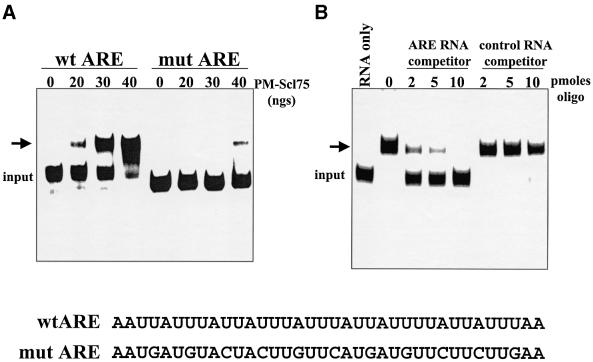
AREs probably stimulate 3′–5′ exonucleolytic decay by promoting loading of the exosome onto RNA substrates. Loading of the exosome could be promoted by ARE-binding proteins, or perhaps exosomal protein(s) may be capable of directly interacting with AREs. In order to address the latter hypothesis, we added increasing amounts of purified PM-Scl75 protein to RNAs that either contained or lacked AREs, and assessed protein–RNA interactions by gel-shift analysis. As seen in Figure 6, PM-Scl75 interacted specifically with RNAs that contained either the TNF-α or GM-CSF ARE. The slowest migrating band representing PM-Scl75-RNA complexes was not observed reproducibly in all experiments and may be due to secondary structure in the RNA substrate or multimers of protein. Since the addition of spermidine in subsequent experiments focused the gel shift into a single band (i.e. Figures 6B and 7), the extra band was probably due to RNA secondary structure. Matched RNAs that lacked an ARE failed to interact with PM-Scl75 protein. The interaction between PM-Scl75 and the ARE was highly specific, as mutations in the ARE significantly reduced binding (Figure 7A), and synthetic RNAs that contained the TNF-α ARE selectively competed for PM-Scl75 binding (Figure 7B). Recombinant PM-Scl75 protein interacted with ARE-containing RNA substrates with a Kd of ∼2 × 10–8 M. In addition, poly(A) also failed to compete for PM-Scl75 protein interactions (data not shown). We conclude that the exosomal protein PM-Scl75 is an ARE-binding protein, suggesting a mechanism for direct loading of the exosome onto unstable ARE-containing mRNAs.
Fig. 6. PM-Scl75 protein interacts with RNAs that contain AU-rich instability elements. (A) Gem-A0 RNA or a derivative that contains the 34-base AU-rich element from TNF-α (GemARE-A0) was incubated with the indicated amounts of purified recombinant PM-Scl75 protein. Heparin-resistant protein–RNA complexes were resolved on a 5% native acrylamide gel. (B) Gem-A0 RNA or a derivative that contains the 51-base ARE from GMCSF (GemGMCSF-A0) was incubated with the indicated amounts of purified recombinant PM-Scl75 protein. Heparin- and spermidine-resistant protein–RNA complexes were resolved on a 5% native acrylamide gel.
Fig. 7. The exosomal protein PM-Scl75 specifically binds to AU-rich instability elements. (A) RNAs that contain either the wild-type TNF-α ARE (lanes wt ARE) or a mutated variant (lanes mut ARE) were incubated with the indicated amounts of purified recombinant PM-Scl75 protein. Heparin-resistant protein–RNA complexes were resolved on a 5% native acrylamide gel. The sequence of the wild-type and mutated versions of the TNF-α ARE are shown at the bottom of the figure. (B) GemARE-A0 RNA was incubated with 50 ng of purified PM-Scl75 protein in the presence of the indicated amounts of a synthetic competitor RNA that contained either the TNF-α ARE (lanes ARE RNA competitor) or an unrelated sequence (lanes control RNA competitor). Heparin-resistant protein–RNA complexes were resolved on a 5% native acrylamide gel.
Discussion
With the discovery of a 5′–3′ exonuclease activity in Figure 1, we have now shown that HeLa cytoplasmic S100 extracts contain all of the primary enzymatic activities that have been shown to play a role in mRNA turnover in yeast (Caponigro and Parker, 1996). Xrn1p homologs previously have been shown to exist in Drosophila and mouse and can complement yeast lacking the 5′–3′ exonuclease (Shobuike et al., 1997; Till et al., 1998). Our in vitro approach therefore provides a versatile means of focusing on and dissecting aspects of any specific decay pathway through the introduction of inhibitors (i.e. cap analog to block deadenylation; Gao et al., 2000) or by immunodepletion of components of competing pathways.
Unlike yeast which predominantly use the decapping/5′–3′ pathway for mRNA decay (Caponigro and Parker, 1996), the 3′–5′ pathway is the major mechanism for the turnover of RNAs following deadenylation in HeLa cytoplasmic extracts. Several models could explain the differences observed between the two experimental systems. First, the accessory role of the 5′ cap in mRNA turnover could be different in the two organisms. In mammalian cells, the 5′ cap specifically interacts with the deadenylase PARN (Dehlin et al., 2000; Gao et al., 2000; Martinez et al., 2001). Furthermore, PARN–cap interactions prohibit the mammalian Dcp1p-like decapping enzyme from gaining access to the cap (Gao et al., 2001). Since the 5′ cap in cis or trans does not appear to influence the rate of yeast deadenylation in vitro (Wilusz et al., 2001b), the yeast deadenylase does not appear to interact stably with the cap nor interfere with the decapping/5′–3′ exonuclease pathway. Secondly, it is possible that independent deadenylases are responsible for targeting mRNAs for decay by the 5′–3′ or 3′–5′ pathways. Perhaps the Ccr4p homolog that exists in mammalian cells (Albert et al., 2000) specifically targets mRNAs to the decapping/5′–3′ exonuclease pathway while PARN selectively targets transcripts to the exosome. Finally, the predominance of the 3′–5′ decay pathway we observe in extracts may not reflect the relative usage of the two pathways in vivo. This question cannot be addressed adequately until a method is found to trap true mRNA turnover intermediates reliably in vivo. However, the ability to analyze the 3′–5′ decay pathway in mammalian cell extracts has generated numerous insights regarding the enzymes involved and their regulation.
The co-immunoprecipitation and localization data shown in Figure 3, along with the observation that our anti-PM-Scl75 antiserum stains the nucleoli of methanol/acetone-fixed Hep-2 cells (Alderuccio et al., 1991; data not shown), suggest that PM-Scl75 is a bona fide component of the human exosome. While PM-Scl75 protein is a homolog of the Escherichia coli RNase PH exonuclease, it does not contain any discernible homology to known RNA-binding domains that would explain its sequence-specific binding with AREs. PM-Scl75 and a second protein, PM-Scl100 (the human Rrp6p homolog), represent major autoantigens in polymyositis–scleroderma overlap syndrome (Targoff, 2000; Brouwer et al., 2001c). While PM-Scl100 is localized predominantly to the nucleus (Allmang et al., 1999b), PM-Scl75 is found in both compartments, consistent with a role for the protein in cytoplasmic mRNA turnover. Furthermore, yeast Rrp6p/PM-Scl100 has been shown to interact with poly(A) polymerase and Npl3p, a poly(A)+ mRNA-binding protein that may help target it to aberrantly processed RNAs in the nucleus (Burkard and Butler, 2000). While PM-Scl75 protein can target itself directly to specific RNA substrates that contain AREs, perhaps its activity is regulated by additional protein–protein interactions.
AREs have now been shown to regulate mRNA turnover at several levels. First, AREs stimulate deadenylation of RNAs in vivo (Wilson and Treismann, 1988) and in vitro (Voeltz and Steitz, 1998; Ford et al., 1999). While the mechanism behind this stimulation is unclear, it may reflect an interaction between the exosome and PARN that assists in loading the deadenylase onto ARE-containing RNA substrates. Consistent with this hypothesis, it is interesting to note that immunodepletion of exosomal components with PM-Scl75 antiserum consistently resulted in less efficient deadenylation of RNA substrates as well (Figure 4B). Secondly, AREs have been shown to influence the rate of decapping in vitro (Gao et al., 2001). This may reflect a separate, exosome/PARN-independent pathway of mRNA turnover. Alternatively, increased exosome activity on RNA substrates could stimulate decapping indirectly by the mammalian Dcp1p-like decapping enzyme. The mammalian decapping enzyme effectively processes short RNA substrates (Gao et al., 2001) that would not be decapped by the yeast Dcp1p enzyme (LaGrandeur and Parker, 1998). Thirdly, AREs can stimulate mRNA degradation of mRNAs with histone-like 3′ ends (Lai and Blackshear, 2001) or small nuclear RNAs (Fan et al., 1997) in the absence of deadenylation. This observation is very consistent with the loading of an active exosome complex near the 3′ end of these transcripts. Finally, ARE-binding proteins probably regulate the assembly of the exosome onto ARE-containing mRNAs. For example, the abundant nuclear ARE-binding proteins that have been identified may function to inhibit or regulate binding of the nuclear exosome to ARE-containing pre-mRNAs while they are being processed in the nucleus. The nucleo-cytoplasmic transport of these proteins, along with various post-translational modifications, would allow for numerous points for mRNA-specific regulation in response to changes in the cellular environment.
Our findings can be summarized in the model shown in Figure 8. First, the exosome complex efficiently loads onto short-lived mRNAs by interacting with AREs located in the 3′-UTR through the exosomal protein PM-Scl75. The relative efficiency of exosome loading may in fact determine the overall half-life of the mRNA. The loading of the exosome can be regulated by ARE-binding proteins, which may sterically block the exosome from interacting with the ARE, or, in other cases, perhaps assist in its loading via protein–protein interactions. The loaded exosome then may attract the PARN deadenylase to the RNA, helping to initiate efficient shortening of the poly(A) tail. Alternatively, the exosome itself may contribute to the deadenylation of the RNA substrate. After deadenylation, the mRNA is handed off efficiently from PARN to the exosome which is now in an optimal position to begin degrading the newly liberated 3′ end of the dying transcript. Whether or not allosteric changes occur in the exosome to activate its enzymatic activity on deadenylated RNA substrates remains to be determined.
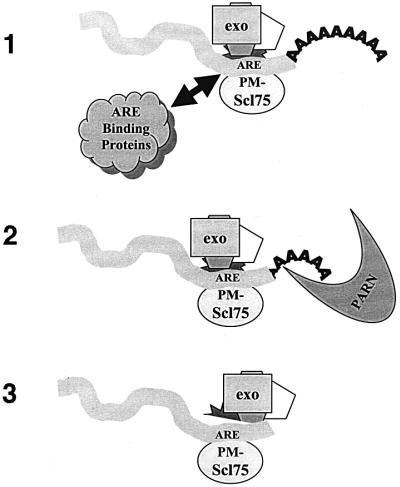
Fig. 8. A model for the regulation of mammalian mRNA turnover by the exosome. (1) Loading of the exosome complex onto mRNAs is promoted by AREs in the 3′-UTR and may be influenced by known ARE-binding proteins such as HuR, AUF-1/hnRNP D, etc. (2) Once loaded, the exosome may help promote mRNA deadenylation. (3) Following deadenylation, the transcript is handed off very efficiently to the exosome that was loaded onto the 3′-UTR, and is degraded rapidly.
Materials and methods
Plasmids and RNAs
SV-A0 RNA is a non-polyadenylated transcript derived from a BamHI–BclI fragment representing the 3′ portion of SV40 late mRNAs, as described previously (Ford et al., 1999). A derivative which contains a 60 base poly(A) tail at its 3′ end, SV-A60, was prepared by a ligation-PCR approach (Ford et al., 1999). SVARE-A60 RNA, which ends with a 60 base poly(A) tail and contains the 34-base ARE from TNF-α inserted into the BamHI–BclI fragment of the 3′ portion of SV40 late mRNAs, was prepared as described previously (Ford et al., 1999). Gem-A0 RNA was prepared by transcription of pGem4 templates that were cleaved by HindIII. GemARE-A0, a variant that contains the 34 base ARE from TNF-α inserted between the PstI and HindIII sites, was described previously (Ford et al., 1999). A version of this RNA that contains a 60-base poly(A) tail at its 3′ end was transcribed from templates generated by a ligation-PCR approach (Ford et al., 1997). Gem-mtARE-A60, which contains a mutated version of the TNF-α ARE in which approximately every third base of the element is altered, has been described previously (Gao et al., 2000). pGemGMCSF, which contains the 51-base ARE from the 3′-UTR of the GM-CSF mRNA, was prepared by inserting the GM-CSF ARE-specific PCR product generated from pGMCSF (Shaw and Kamen, 1986) with the primers 5′-GCGACTGCAGTAATATTATA and 5′-CCCAAGCTTTTAAATAAATAA into the PstI and HindIII sites of pGem4. Transcription of HindIII-linearized templates gave GemGMCSF-A0 RNA.
All RNAs were prepared by in vitro transcription using SP6 RNA polymerase and purified on denaturing acrylamide gels prior to use as described previously (Wilusz and Shenk, 1988). Transcripts containing a 5′-GMP were prepared by including 5 mM 5′-GMP in the transcription reaction.
Synthetic RNAs used in competition analyses were prepared by the Molecular Core Facility of New Jersey Medical School. The specific ARE competitor RNA contained the sequence 5′-AUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUUA, while the non-specific competitor RNA was 5′-GGAUUAACUAAUUGAUACCGCGUAUACACGCGG.
Preparation of RNAs containing phosphothioate substitutions
To generate phosphothioate-modified RNA substrates for turnover assays, a three-fragment bridged ligation approach was used (Moore and Query, 2000). The capped and radiolabeled 5′ fragment was transcribed from EcoRV-linearized pBluescript II KS using T3 polymerase in the presence of cap analog and [32P]UTP. The middle fragment in the ligation was a synthetic RNA oligonucleotide 5′-GCAAGTAGAACCTCAGTG[GCT]PTTATGGCATG in which the bracketed nucleotides were either wild-type or modified by phosphothioate incorporation during synthesis. The 3′ fragment in the ligation contained a 5′-monophosphate and a 60-base poly(A) tail and was prepared by transcribing GemARE-A60 PCR products in the presence of 500 µM GMP. The bridge ligation reaction contained 50 mM Tris pH 7.5, 10 mM MgCl2, 1 mM KCl, 10 mM dithiothreitol (DTT), 25 mg/ml bovine serum albumin (BSA) and 2 pmol of each RNA segment. The reaction mixture was heated to 90°C for 2 min and allowed to cool to room temperature. RNasin, 1 mM ATP and T4 DNA ligase were then added and the reaction mixture was incubated at 30°C for 2–4 h. Ligated RNA products were purified on acrylamide/urea gels prior to use.
Preparation of PM-Scl75 protein and antibodies
The KpnI–PstI fragment of a plasmid containing the open reading frame of PM-Scl75 protein (Alderuccio et al., 1991; obtained from Dr Edward Chan) was inserted into pTrcHisA at the same sites. His-tagged fusion protein was purified from BL21LysS transformed cell lysates using His-bind resin (Novagen). A 50 µg aliquot of purified protein was mixed with Freund’s adjuvant and injected intraperitoneally into female ICR mice four times at 2 week intervals. Mice were terminally bled 4 days after the last injection and serum was collected by centrifugation. Rabbit polyclonal antibodies from recombinant PM-Scl75 protein were commercially prepared at Pocono Rabbit Farm. Western blots were performed using PVDF membranes and antisera at a 1:1000 or 1:5000 dilution. In order to determine the subcellular localization of PM-Scl75, cellular fractionation of HeLa cells was performed as described (Brouwer et al., 2001a). Co-immunoprecipitation experiments of exosome components from HeLa extracts were performed as described previously (Brouwer et al., 2001a).
In vitro mRNA deadenylation/decay assays
Polyadenylated RNAs that were internally labeled with [32P]UTP were incubated in HeLa S100 cytoplasmic extracts at 30°C as described previously (Ford et al., 1999). Reaction products were analyzed on 5% acrylamide gels containing 7 M urea and visualized by phosphoimaging and/or autoradiography. Immunodepletion of extracts with PM-Scl75-specific or control antibodies was performed using protein A–Sepharose as described previously (Gao et al., 2000). The extent of immunodepletion was assessed by western blotting with anti-PM-Scl75 antiserum at a 1:1000 dilution as described previously (Gao et al., 2000).
Exonuclease assays
3′–5′ exonucleases were assayed using non-polyadenylated transcripts that contained 5′ 7meGpppG caps (to inhibit 5′–3′ exonuclease access). 5′–3′ exonucleases were assayed using transcripts that contained a 5′-monophosphate (prepared as described above) along with a 3′ 60-base poly(A) tail (which, in conjunction with PABP found in cytoplasmic extracts, inhibits 3′–5′ exonucleases). Reaction products were assayed on 5% acrylamide gels containing 7 M urea and quantitated by phosphoimaging.
Protein–RNA interactions
Interactions of purified proteins with RNA were assayed by electrophoresis on 5% native acrylamide gels using heparin, as described previously (Bagga et al., 1998). EDTA was added to all assays to inhibit exonuclease activity.
Acknowledgments
Acknowledgements
We wish to thank Dr Ed Chan (Scripps) for the kind gift of pPM-Scl75, Bob Donnelly and the NJMS Molecular Resource Facility for DNA and RNA oligonucleotides, Carol Wilusz for critical comments, and members of the Wilusz laboratory for helpful discussions. This work was supported by grants from the National Institutes of Health (grant CA80062 to J.W.), Scleroderma Foundation (J.W., C.S.L.), American Cancer Society (C.S.L.) and the Netherlands Organization for Scientific Research NOW-CW (G.P.).
References
- Albert T.K., Lemaire,M., van Berkum,N.L., Gentz,R., Collart,M.A. and Timmers,H.T. (2000) Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res., 28, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderuccio F., Chan,E.K. and Tan,E.M. (1991) Molecular characterization of an autoantigen of PM-Scl in the polymyositis/scleroderma overlap syndrome: a unique and complete human cDNA encoding an apparent 75-kD acidic protein of the nucleolar complex. J. Exp. Med., 173, 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999a) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 19, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999b) The yeast exosome and human PM-Scl are related complexes of 3′–5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga P.S., Arhin,G.K. and Wilusz,J. (1998) DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res., 26, 5343–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P., Peltz,S.W. and Ross,J. (1989) The poly(A)–poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol., 9, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Brewer G. (1991) An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol., 11, 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G. (1999) Evidence for a 3′–5′ decay pathway for c-myc mRNA in mammalian cells. J. Biol. Chem., 274, 16174–16179. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard,K.T. and Butler,J.S. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brouwer R., Allmang,C., Raijmakers,R., van Aarssen,Y., Egberts,W.V., Petfalski,E., van Venrooij,W.J., Tollervey,D. and Pruijn,G.J. (2001a) Three novel components of the human exosome. J. Biol. Chem., 276, 6177–6184. [DOI] [PubMed] [Google Scholar]
- Brouwer R., Pruijn,G.J. and van Venrooij,W.J. (2001b) The human exosome: an autoantigenic complex of exoribonucleases in myositis and scleroderma. Arthritis Res., 3, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer R. et al. (2001c) Autoantibody profiles in the sera of European patients with myositis. Ann. Rheum. Dis., 60, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard K.T and Butler,J.S. (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol., 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova J.A., Shaw,R.J., Wills,M.A. and Belostotsky,D.A. (2000) Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem., 275, 33158–33166. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. and Shyu,A.B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi,R., Andersen,J.S., Gaietta,G., Jurchott,K., Royer,H.D., Mann,M. and Karin,M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev., 14, 1236–1248. [PMC free article] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion,F. and Seraphin,B. (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res., 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E., Wormington,M., Korner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la J., Kressler,D. Tollervey,D. and Linder,P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J., 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A.M., Kempf,T. and Clayton,C. (2001) The exosome of Trypanosoma brucei. EMBO J., 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C. and Steitz,J.A. (1998) Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C., Myer,V.E. and Steitz,J.A. (1997) U-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev., 11, 2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P. and Wilusz,J. (1999) An in vitro system using HeLa cytoplasmic extracts that reproduces regulated mRNA stability. Methods, 17, 21–27. [DOI] [PubMed] [Google Scholar]
- Ford L.P., Bagga,P.S. and Wilusz,J. (1997) The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol., 17, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Fritz,D.T., Ford,L.P. and Wilusz,J. (2000) Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell, 5, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Wilusz,C.J., Peltz,S.W. and Wilusz,J. (2001) A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J., 20, 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi J. and Brewer,G. (2001) Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23. [DOI] [PubMed] [Google Scholar]
- Jacobs Anderson J.S. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D.L. and Tollervey,D. (2000) Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol., 20, 5415–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Grandeur T.E. and Parker,R. (1998) Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme.EMBO J., 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S. and Blackshear,P.J. (2001) Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem., 276, 23144–23154. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G., Cuesta,R., Brewer,G. and Schneider,R.J. (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- Levine T.D., Gao,F., King,P.H., Andrews,L.G. and Keene,J.D. (1993) Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol., 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin P., Chen,C.Y. and Shyu,A.B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.J., Cheng,S., Campbell,C., Wright,A. and Furneaux,H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem., 271, 8144–8151. [DOI] [PubMed] [Google Scholar]
- Martinez J., Ren,Y.G., Nilsson,P., Ehrenberg,M. and Virtanen,A. (2001) The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem., 276, 27923–27929. [DOI] [PubMed] [Google Scholar]
- Ming X.F., Stoecklin,G., Lu,M., Looser,R. and Moroni,C. (2001) Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol., 21, 5778–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000a) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000b) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Moore M. and Query,C.C. (2000) Joining of RNAs by splinted ligation. Methods Enzymol., 317, 109–123. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Peng S.S., Chen,C.Y., Xu,N. and Shyu,A.B. (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R., Noordman,Y.E., van Venrooij,W.J. and Pruijn,G.J.M. (2002) Protein–protein interactions of hCs14p with other human exosome subunits. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- Shobuike T., Sugano,S. Yamashita,T. and Ikeda,H. (1997) Cloning and characterization of mouse Dhm2 cDNA, a functional homolog of budding yeast SEP1. Gene, 191, 161–166. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Targoff I.N. (2000) Update on myositis-specific and myositis-associated autoantibodies. Curr. Opin. Rheumatol., 12, 475–481. [DOI] [PubMed] [Google Scholar]
- Till D.D. et al. (1998) Identification and developmental expression of a 5′–3′ exoribonuclease from Drosophila melanogaster. Mech. Dev., 79, 51–55. [DOI] [PubMed] [Google Scholar]
- Tucker M. and Parker,R. (2000) Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem., 69, 571–595. [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- van Hoof A. and Parker R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz,P. and Parker,R. (2000a) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Staples,R.R., Baker,R.E. and Parker,R. (2000b) Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol., 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. and Steitz,J.A. (1998) AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol. Cell. Biol., 18, 7537–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. and Keller,W. (1994) 3′ End processing of mRNA. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing, A Practical Approach. Vol. II. Oxford University Press, Oxford, UK, pp. 1–34.
- Wilson T. and Treisman,R. (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature, 336, 396–399. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Wormington,M. and Peltz,S.W. (2001a) The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell. Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Gao,M., Jones,C.L., Wilusz,J. and Peltz,S.W. (2001b) Poly(A) binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA, 7, 1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. and Shenk,T. (1988) A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell, 52, 221–228. [DOI] [PubMed] [Google Scholar]
- Winzen R. et al. (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J., 18, 4969–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchin N.I. and Goldfarb,D.S. (1999) The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res., 27, 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]