Abstract
Through binding and fluorescence studies of oligonucleotides covalently attached to a pyrene group via one carbon linker at the sugar residue, we previously found that pyrene-modified RNA oligonucleotides do not emit well in the single-stranded form, yet the attached pyrene emits with a significantly high quantum yield upon binding to a complementary RNA strand. In sharp contrast, similarly modified pyrene–DNA probes exhibit very weak fluorescence both in the double-stranded and single-stranded forms. The pyrene-modified RNA oligonucleotides therefore provide a useful tool for monitoring RNA hybridization. The purpose of this paper is to present the structural basis for the different fluorescence properties of pyrene-modified RNA/RNA and pyrene-modified DNA/DNA duplexes. The results of absorption, fluorescence anisotropy and circular dichroism studies all consistently indicated that the pyrene attached to the RNA duplex is located outside of the duplex, whereas the pyrene incorporated into the DNA duplex intercalates into the double helix. 1H NMR measurements unambiguously confirmed that the pyrene attached to the DNA duplex indeed intercalates between the base pairs of the duplex. Molecular dynamics simulations support these differences in the local structural elements around the pyrene between the pyrene–RNA/RNA and the pyrene–DNA/DNA duplexes.
INTRODUCTION
Because pyrene can be chemically modified in various ways and because its fluorescence quantum efficiencies are high in both monomer and excimer emissions, a number of pyrene-modified oligonucleotides have been synthesized and exploited as fluorescent probes of DNA and RNA in hybridization assays (1–27). In addition, pyrene fluorescence is largely affected by environmental factors, such as solvent and nearby nucleotide bases, which has led to its development as a potential probe for nucleic acid structures (5–8). Although recent reports have shown that pyrene monomer emission is useful for fluorescent recognition of a given base at the target site in DNA (1), pyrene-oligonucleotide probes employing the monomer emission usually involve the disadvantage of fluorescence quenching. In oligonucleotides covalently attached via a flexible tether to pyrene at the internal phosphate (2), the nucleotide bases (3), and the terminus (4), fluorescence quenching of the pyrene strongly occurs both in the single-stranded and double-stranded forms via an electron migration between the excited pyrene and the nucleotide bases (28–30). To avoid these disadvantages, several attempts using the pyrene-excimer emission in place of the monomer emission have been made in the development of hybridization probes (9–22). Because pyrene-excimer emission is less sensitive to quenching by nucleobases than monomer emission, the pyrene-excimer-forming probes have been successfully used for DNA and RNA hybridization assays (13–22,27).
Our research efforts have been directed toward the design and synthesis of pyrene-modified oligonucleotides in which the sugar 2′-position has been used as the site of the covalent attachment (23–26). We have found that the incorporation of pyrene via one carbon linker into the sugar position of DNA resulted in probes displaying very weak monomer fluorescence because of severe quenching of the fluorescence. Thus, upon binding of the pyrene–DNAs to complementary DNA and to RNA sequences, the intensity of the pyrene fluorescence did not significantly change. The pyrene-modified RNAs also exhibited very weak monomer fluorescence, and the hybridization to DNA resulted in little fluorescence change. In sharp contrast to these observations, the fluorescence intensity of the pyrene-modified RNAs drastically increased upon hybridization to complementary RNA sequences. The resulting pyrene-modified RNA duplexes displayed fluorescence as intense as that of free pyrene in aqueous solution. The pyrene-modified RNA oligonucleotides therefore provide a useful tool for monitoring RNA hybridization (26).
In this paper, we discuss the local structure of the pyrene appended onto the sugar of the pyrene–RNA and the pyrene–DNA duplexes. All the absorption, fluorescence and circular dichroism (CD) spectral observations are consistent with attachment of the pyrene to the outside of the double helix in the RNA duplex, while the pyrene in the DNA duplex is placed inside the helix. Molecular dynamics (MD) simulation studies strongly support these differences of the local structure around the pyrene. 1H NMR measurements unambiguously confirmed that the pyrene attached to the DNA duplex intercalates between the base pairs of the duplex. The results of the present studies may contribute to further development of pyrene-labeled oligonucleotide probes for gene analysis (27).
MATERIALS AND METHODS
General methods
The synthesis of pyrene-modified oligonucleotides was carried out using 5′-O-dimethoxytrityl-2′-O-(1-pyrenylmethyl)uridine 3′-O-(2-cyanoethyl)-N,N′-diisopropylphosphoramidite as described previously (25,26). UV-visible spectra were recorded with a Hitachi U-3000 or Beckman DU-800 spectrophotometer equipped with a thermoelectrically controlled cell holder. Steady-state fluorescence and fluorescence anisotropy data were measured on a Hitachi F-2500 spectrofluorometer. Fluorescence quantum yields were estimated at 23°C on the basis of quinine sulfate in sulfuric acid (1.0 N) as a standard (26). Anisotropy was estimated by following equation, where I// and I⊥ are the fluorescence intensities of the parallel and perpendicular polarization components, respectively, and r is the fluorescence anisotropy (31): r = (I// − I⊥)/(I// + 2 I⊥). CD spectra were obtained on a JASCO CD J-805 spectrophotometer.
NMR spectroscopy
The NMR sample was prepared in 10 mM phosphate buffer (pH 7.0) containing 100 mM NaCl and 0.2 mM EDTA. All the NMR spectra were recorded on a Bruker ARX-500 spectrometer (500.13 MHz for 1H). For the resonance assignment, 2D NMR spectra, TOCSY (mixing time 40 ms) and NOESY (mixing times 100, 200 and 350 ms) were measured at 25°C. All the proton resonances were assigned by the sequential procedure (32). 1H chemical shifts were referenced to sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4.
MD simulation
The modified nucleoside, 2′-O-(1-pyrenylmethyl)uridine, was constructed using a support program for AMBER on the basis of our originally developed molecular graphics program ‘Modrast-P’ (33,34). Formal charges for the modified nucleoside were calculated by MOPAC, and each atom in the pyrenyl residue was defined for molecular model building using AMBER (35). The starting structures for pyrene-modified RNA and DNA duplexes were constructed with the AMBER module and then optimized, thermalized (300 K), and equilibrated before MD simulation. MD simulations were performed using the ‘sander’ module of the AMBER 6.0 (36) software package with the ff94 force field (37) in which the systems were subjected to 1 ns (1 000 000 steps) of the simulations at constant temperature (300 K) and pressure (1 atm) with standard relaxation times. In these calculations, a suitable number of Na+ ions and ∼2200 of TIP3P water molecules were placed with the minimum distance from the duplex to the periodic boundary (10 Å).
RESULTS
Absorption spectral changes during the duplex melting
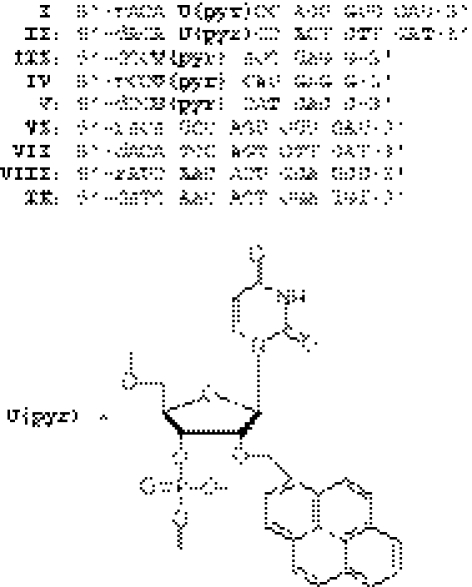
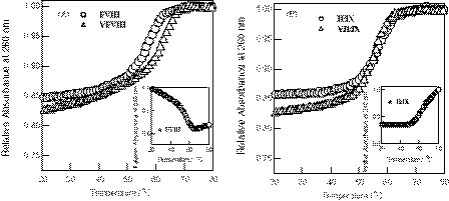
The sequences of the pyrene-modified oligonucleotides used in the present studies are listed in Scheme 1. The UV-melting curves monitored at 260 and 340 nm for pyrene-modified RNA/RNA, pyrene-modified DNA/DNA duplexes and their corresponding unmodified duplexes are shown in Figure 1. All the melting profiles for the pyrene-modified duplexes exhibit typical single-phase melting transitions that are similar to the unmodified duplexes. The Tm value estimated for pyrene-modified RNA duplex I/VIII was slightly lower than that for unmodified RNA duplex VI/VIII, while the pyrene–DNA/DNA duplex II/IX melted at a temperature a few degrees higher than the Tm of unmodified DNA duplex VII/IX.
Scheme 1.
The sequences of pyrene-modified oligonucleotides and unmodified oligomers used in this study.
Figure 1.
(A) UV-melting curves for the pyrene-modified RNA oligonucleotide duplex, I/VIII (circles) and unmodified RNA duplex, VI/VIII (triangles). (B) UV-melting curves for the pyrene-modified DNA oligonucleotide duplex, II/IX (circles) and unmodified DNA duplex, VII/IX (triangles). Measurements were carried out at 260 nm for a 1:1 mixture of oligonucleotides (33 µM) in a pH 7 buffer containing 10 mM sodium phosphate and 100 mM NaCl.
Figure 2 shows the temperature-dependent changes in the absorption spectra at the region between 300 and 360 nm. For the pyrene-modified RNA duplex I/VIII, the pyrene absorption band at 347 nm observed at the high temperature shifted to the shorter wavelength (342 nm) upon the duplex formation. The melting profile at 340 nm for the pyrene-modified RNA duplex I/VIII (Figure 1A, inset) exhibited a reverse sigmoid curve compared with that monitored at 260 nm. In contrast, the pyrene absorption band for the pyrene–DNA/DNA duplex II/IX showed the red shift from 347 to 352 nm with the duplex formation. The melting profile at 340 nm for DNA duplex II/IX (Figure 1B, inset) was thus similar to that obtained at 260 nm. These changes in the UV-spectra between the modified RNA and DNA strongly suggested that the structure or the environment around the pyrene in the modified RNA duplex I/VIII sharply differs from that of the DNA duplex II/IX. Absorption of polyaromatic intercalators involving pyrene shifts to a longer wavelength because of the π-stacking with nucleobases (38); thus, the observed spectral changes may be consistent with a pyrene intercalated between the bases or base pairs of the DNA duplex, while the pyrene in the RNA duplex is placed in a position relatively free from interaction with the bases.
Figure 2.
(A) Absorption spectra changes as a function of temperature for the pyrene-modified RNA oligonucleotide duplex, I/VIII. (B) Absorption spectra changes as a function of temperature for the pyrene-modified DNA oligonucleotide duplex, II/IX. Measurements were carried out for a 1:1 mixture of oligonucleotides (33 µM) in a pH 7 buffer containing 100 mM NaCl, 10 mM sodium phosphate and 1 mM EDTA·2Na.
Fluorescence anisotropy
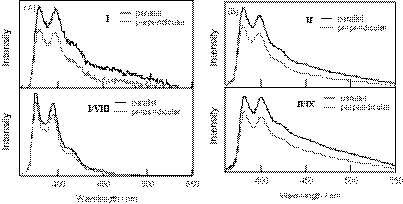
Figure 3 shows the steady-state fluorescence spectra of pyrene-modified RNA and DNA oligomers and their duplexes. The single-stranded pyrene–RNA I exhibited the typical pyrene monomer emission, and its fluorescence quantum yield was very low (quantum yield φ = 0.007) (26). The fluorescence of the pyrene–RNA/RNA duplex I/VIII was significantly enhanced with a φ value of 0.237 that is comparable with those for pyrene derivatives free in air-saturated aqueous solution, such as 1-pyrenebutyric acid (φ = 0.30) and 1-pyrenylmethanol (φ = 0.25). On the other hand, the pyrene-modified DNA II exhibited a pyrene monomer emission with a low quantum yield (φ = 0.006) both in the single-stranded and in the double-stranded forms.
Figure 3.
(A) Steady-state fluorescence spectra for pyrene-modified RNA oligonucleotide, I (thin line) and its duplex with RNA, I/VIII (bold line). (B) Steady-state fluorescence spectra for pyrene-modified DNA oligonucleotide, II (thin line) and its duplex with RNA, II/IX (bold line). The measurements were carried out at room temperature for a 1:1 mixture of oligonucleotides (6 µM) in the same buffer solutions used in Figure 2. The excitation wavelength was 350 nm.
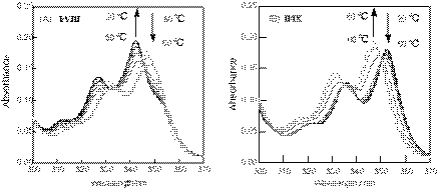
We then measured fluorescence anisotropy for the pyrene-modified RNA and DNA duplexes. Figure 4 represents the fluorescence anisotropy spectra for the single-stranded pyrene-modified oligomers and their duplexes. The anisotropy (r) value (3.85 × 10−2) at 380 nm for the pyrene-modified RNA duplex I/VIII was approximately one-third compared with that (12.01 × 10−2) of the single-stranded pyrene-modified RNA I. In contrast, the r-value (14.5 × 10−2) of the pyrene-modified DNA in the double-stranded form II/IX was slightly higher than that (12.16 × 10−2) of the single-stranded form II. These fluorescence anisotropy measurements indicate that the pyrene in the RNA duplex is relatively free from stacking within the bases compared with the single-stranded form. On the other hand, the pyrene is less mobile or more strongly stacked between the bases or the base pairs in the DNA duplex compared with the single-stranded form. Fluorescent anisotropy findings, therefore, are consistent with the observations from the UV-spectral analyses.
Figure 4.
(A) Upper panel: parallel (bold line) and perpendicular (thin line) fluorescence anisotropy spectra of pyrene-modified RNA oligonucleotide, I; lower panel: parallel (bold line) and perpendicular (thin line) fluorescence anisotropy spectra of the pyrene-modified RNA oligonucleotide duplex, I/VIII. (B) Upper panel: parallel (bold line) and perpendicular (thin line) fluorescence anisotropy spectra of pyrene-modified DNA oligonucleotide, II; lower panel: parallel (bold line) and perpendicular (thin line) fluorescence anisotropy spectra of pyrene-modified DNA oligonucleotide duplex, II/IX. The measurements were carried out at room temperature with the same solutions used in Figure 2. The excitation wavelength was 350 nm.
Induced CD
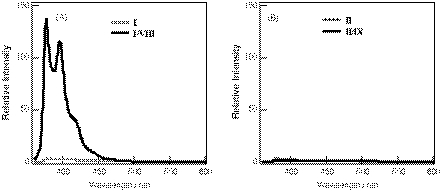
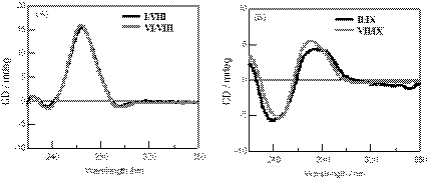
CD spectra for the pyrene-modified RNA and DNA duplexes and their corresponding unmodified duplexes are shown in Figure 5. The pyrene–RNA/RNA duplex I/VIII exhibits a CD profile that is very close to that of the unmodified RNA duplex VI/VIII of a typical A-form double helix. In this case, no induced CD signal resulting from the pyrenyl group at the region between 300 and 360 nm was observed. On the other hand, a negative-induced CD at the same region was observed for the pyrene–DNA/DNA duplex II/IX. The CD profile at ∼260 nm was slightly modified from that of the unmodified B-form DNA duplex VII/IX. Because pyrene is a planar achiral molecule that exhibits no intrinsic CD activity, the induced CD observed for the pyrene–DNA/DNA duplex strongly suggests the asymmetric interaction of the pyrene ring with the bases or base pairs of the duplex (39).
Figure 5.
(A) CD spectra for the pyrene-modified RNA oligonucleotide duplex, I/VIII (bold line) and unmodified RNA duplex, VI/VIII (thin line). (B) CD spectra for pyrene-modified DNA oligonucleotide duplex, II/IX (bold line) and unmodified DNA duplex, VII/IX (thin line). Measurements were carried out at room temperature for a 1:1 mixture of oligonucleotides (6 µM) in a pH 7 buffer containing 10 mM sodium phosphate and 100 mM NaCl.
NMR spectroscopy
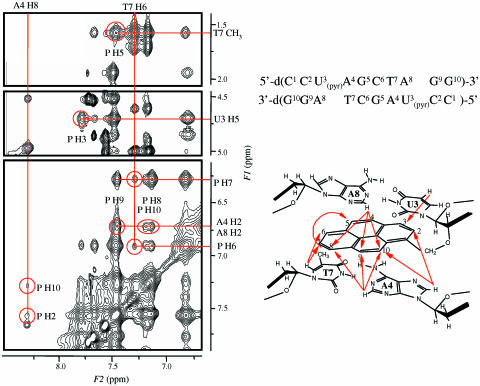
This interaction between the pyrene and bases or base pairs of the DNA duplex was investigated using 1H NMR measurements. The self-complementary pyrene-modified DNA duplex III was used for the measurements because the NMR signals are easily assignable owing to the symmetric nature of the DNA. The 2D NOESY spectrum of III and the assignment of the NOE cross-peaks are shown in Figure 6. The NOE cross-peaks were observed between the pyrene aromatic protons and the base protons of U3, A4, T7 and A8, which are summarized in the right lower panel of Figure 6. The NOE cross-peaks were observed between the proton pairs of H8 of A4 and the H2 and H10 aromatic protons of pyrene. Similarly, the cross-peaks were observed between the proton pairs of H5 of U3 and pyrene H3; H6 of T7 and pyrene H6 and H7; CH3 of T7 and pyrene H5; and H2 protons of A4 and A8 and pyrene H9 and H10. On the basis of these NMR data, the inference is that the intercalation of the pyrene ring occurs between the base pairs of U3-A8 and A4-T7. The pyrene protons of H3, H4, H5 and H6 are located in the major groove side of the DNA/DNA duplex, while those of H8, H9 and H10 are placed in the minor groove side.
Figure 6.
2D NOESY spectrum of [d(CCU(pyr)AGCUAGG)]2 III in D2O with the proton assignment (left). The numbering of the nucleotides in the DNA is shown in the upper right panel. The arrows indicate the observed NOE signals between the bases and pyrene protons (lower right panel).
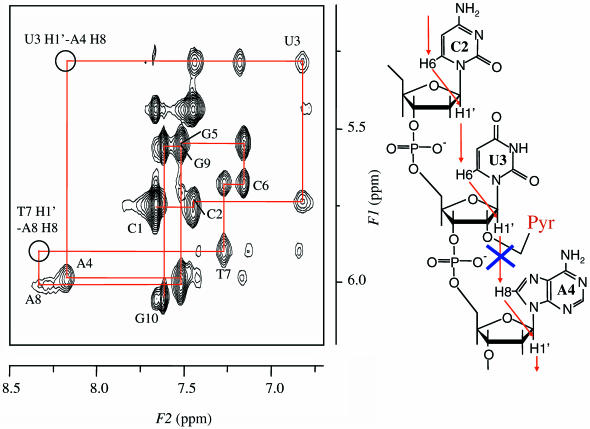
Figure 7 shows the NOE cross-peaks at the region between the sugar H1′ protons and base protons of III. The sequential NOE cross-peaks were not observed between H1′ of U3 and H8 of A4, and H1′ of T7 and H8 of A8; however, the cross-peaks between the sugar H1′ proton and the base proton at the 3′-side were observable for the other possible combinations of the bases and the sugars, such as the H1′ of C2 and H6 of U3. These observations provide the evidence that the base-pair pocket of U3-A8 and A4-T7 was locally enlarged along the axial of the DNA duplex because of the intercalation of the pyrene group, while retaining the global conformation of B-form double helix. The slight modification of the global conformation of the duplex is consistent with the CD spectral observation.
Figure 7.
H8/H6 and H1′ region of 2D NOESY spectrum of III in D2O with the proton assignment (left) and sequential NOEs in the segment of C2-U3-A4 (right).
MD simulations
We carried out MD simulations for the pyrene-modified RNA and DNA duplexes using an AMBER force field. The starting structures for pyrene-modified RNA IV and DNA V duplexes were constructed using the AMBER module on the basis of an A-form and B-form double helix, respectively, in which the pyrene group was placed at the inside of the model duplexes. The model duplex structures were optimized and equilibrated before MD simulation at 300 K. We found that after the energy minimization, in the pyrene–RNA duplex the pyrene ring was pushed to the outside of the duplex, while the pyrene was still found inside the helix in the pyrene–DNA duplex.
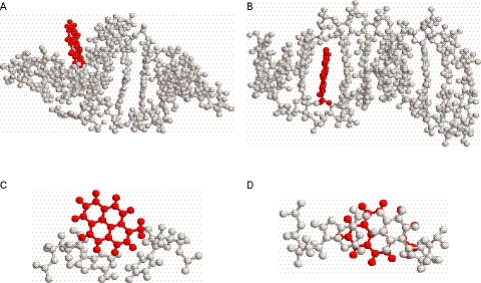
The MD simulations were performed using the AMBER 6.0 software package with an appropriate force field (36,37) in which the systems were subjected to 1 ns (1 000 000 steps) of the simulations at constant temperature (300 K) and pressure (1 atm) with standard relaxation times. Figure 8 shows the final structures of the pyrene–RNA/RNA IV and pyrene–DNA/DNA V duplexes obtained after the MD simulations. As shown in Figure 8A, the structure of the pyrene-modified RNA duplex retained its original A-form double helix in which the pyrene ring extends prominently from the helix. Figure 8B shows the top view of the pyrene plane together with the A-U pair of nucleosides, one of which was covalently attached to the pyrene. It can be seen that the pyrene aromatic ring is free from π-stacking interaction of the nucleobases.
Figure 8.
(A) The structures obtained from molecular dynamics simulation for the pyrene-modified RNA duplex, IV. (B) The top view of the pyrene aromatic plane together with A-U nucleoside pair of duplex IV. (C) The structures obtained from molecular dynamics simulation for pyrene-modified DNA duplex V. (D) The top view of the pyrene aromatic plane together with A-U nucleoside pair of duplex V.
On the other hand, in the pyrene–DNA/DNA structure (Figure 8C), the pyrene ring is intercalated into the designated base pairs, A-U and G-C pairs, in the double helix. The uracil and adenine bases were found to be well stacked with the pyrene, as shown in Figure 8D. The MD simulations show that, while the pyrene is indeed intercalated into the base pairs of the helix, the pyrene-modified DNA duplex was not significantly distorted and retained the global conformation of the B-form structure. This finding is consistent with the CD and NMR observations.
DISCUSSION
In this paper, we report the difference in structure between duplex DNA versus RNA possessing a single modified uridine with a 2′-(1-pyrenylmethyl) substituent. NMR spectra and MD simulations revealed that the pyrene-modified DNA duplex has an intercalated structure of pyrene. An intercalated structure is consistent with the spectral observations, such as the blue shift in pyrene absorption, small changes in the fluorescence anisotropy during the duplex melting and the induced CD in the duplex observed at the pyrene absorption band.
On the basis of the intercalated structure, the fluorescence properties of pyrene-modified DNA can be explained as follows. The incorporation of pyrene via one carbon linker into the sugar position of DNA produces the oligonucleotide probes that display very weak monomer fluorescence resulting from severe fluorescence quenching. The fluorescence quenching of pyrene has been generally observed for the single-stranded form of oligonucleotides covalently attached to the pyrene chromophore (1–4,29); the nucleobases can serve as efficient quenchers for pyrene fluorescence via electron transfer from the excited pyrene to nucleobases (28–30). The π-stacking interaction between the pyrene and nucleotide bases should facilitate the electron transfer and should occur even in single-stranded oligomers. In the present DNA duplex, pyrene intercalation is a favorable factor for electron transfer, resulting in a pyrene that displays very weak fluorescence. Similar to the DNA binding, the pyrene fluorescence does not always change much on binding of the pyrene-modified DNA to RNA sequences. The only exception has been observed in the pyrene-modified DNA probes containing dC at the 3′-site of the modification that showed a remarkable increase in pyrene monomer fluorescence upon binding to complementary RNA (25).
The pyrene-modified RNA duplex completely differs from the pyrene-modified DNA duplex in terms of local structure around the pyrene. The non-intercalated structure for the RNA duplex is strongly supported by the red shift in pyrene absorption and the decrease in fluorescence anisotropy during the duplex melting and the lack of induced CD at the pyrene absorption band. The MD simulations further support the structural features of the RNA duplex. The pyrene-modified RNA duplex retained its original A-form double helix in which the pyrene ring extends prominently from the helix. Importantly, the pyrene aromatic ring is totally free from the π-stacking interaction with the nucleobases. With these structural features, the electron transfer from the excited pyrene to the nucleobases may become unfavorable, and the pyrene is highly emissive in spite of the covalent attachment to the RNA duplex. The emission intensity of the pyrene in the RNA duplex is as strong as that of pyrene chromophores free in an aqueous solution, while single-stranded RNA with the pyrene exhibits very weak fluorescence (two orders of magnitude less than the duplex). In contrast to the RNA binding, the pyrene-modified RNA probes showed little change in fluorescence on binding to DNA sequences (26).
The above structural data do not appear to provide a reasonable explanation for why the pyrene is intercalated in the DNA duplex and not in the RNA duplex. However, the model building and energy minimization studies showed that the pyrene ring in the pyrene–RNA duplex was pushed to the outside from the duplex, although the initial structure had the pyrene at the inside of the helix. In contrast, in the pyrene–DNA duplex the pyrene was found to be still inside the helix even after the energy minimization. Thus, it may be highly likely that an intercalated structure for duplex RNA covalently attached to pyrene is not energetically favorable, probably because of the structural constraints resulting from the short-chain carbon linker. On the other hand, the structural constraint may be compensated by the energy gain from the pyrene intercalation in the duplex DNA. The UV-melting studies showing that pyrene-modified DNA is a little more stable than the unmodified duplex appear to support these ideas.
CONCLUSION
In summary, we conclude that an intercalated structure or π-stacking interaction in 2′-pyrene-modified oligonucleotides plays a key role in determining pyrene fluorescence efficiency. Duplex RNA has pyrene bound outside the helix, while duplex DNA has a pyrene-intercalated structure. The pyrene is therefore highly emissive when attached to duplex RNA but not to duplex DNA. Based on the outside binding structure and fluorescence properties of pyrene in the RNA duplex, dual-pyrene-labeled RNA probes have already been designed to emit excimer fluorescence and are used for RNA assays (27). In addition, the present findings can be applied to development of useful materials involving conjugation of aromatic hydrocarbons and RNA.
Acknowledgments
The authors thank Fumika Tokuriki for preliminary experiments with UV and fluorescence spectroscopy studies. This research was in part supported by SORST and CREST of Japan Science and Technology Corporation. Funding to pay the Open Access publication charges for this article was provided by University of Hyogo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Okamoto A., Kanatani A., Saito I. Pyrene-labeled base-discriminating fluorescence DNA probes for homogeneous SNP typing. J. Am. Chem. Soc. 2004;126:4820–4827. doi: 10.1021/ja039625y. [DOI] [PubMed] [Google Scholar]
- 2.Yamana K., Letsinger R.L. Synthesis and properties of oligonucleotides bearing a pendant pyrene group. Nucleic Acids Res. Symp. Ser. 1985;16:169–172. [PubMed] [Google Scholar]
- 3.Telser J., Cruckshank K.A., Morrison L.E., Netzel T.L. Synthesis and characterization of DNA oligomers and duplexes containing covalently attached molecular labels: comparison of biotin, fluorescein, and pyrene labels by thermodynamic and optical spectroscopic measurements. J. Am. Chem. Soc. 1989;111:6966–6976. [Google Scholar]
- 4.Mann J.S., Shibata Y., Meehan T. Synthesis and properties of an oligodeoxynucleotide modified with a pyrene derivative at the 5′-phosphate. Bioconjug. Chem. 1992;3:554–558. doi: 10.1021/bc00018a015. [DOI] [PubMed] [Google Scholar]
- 5.Bevilacqua P.C., Kierzek R., Johnson K.A., Turner D.H. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 6.Kierzek R., Li Y., Turner D.H., Bevilacqua P.C. 5′-Amino pyrene provides a sensitive, nonperturbing fluorescent probe of RNA secondary and tertiary structure formation. J. Am. Chem. Soc. 1993;115:4985–4992. [Google Scholar]
- 7.Silverman S.K., Cech T.R. RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry. 1999;38:14224–14237. doi: 10.1021/bi991333f. [DOI] [PubMed] [Google Scholar]
- 8.Mahara A., Iwase R., Sakamoto T., Yamaoka T., Yamana K., Murakami A. Detection of acceptor sites for antisense oligonucleotides on native folded RNA by fluorescence spectroscopy. Bioorg. Med. Chem. 2003;11:2783–2790. doi: 10.1016/s0968-0896(03)00227-x. [DOI] [PubMed] [Google Scholar]
- 9.Tong G., Lawlor J.M., Tregear G.W., Haralambidis J. Oligonucleotide-polyamide hybrid molecules containing multiple pyrene residues exhibit significant excimer. J. Am. Chem. Soc. 1995;117:12151–12158. [Google Scholar]
- 10.Langenegger S.M., Haner R. Excimer formation by interstrand stacked pyrenes. Chem. Commun. 2004:2792–2793. doi: 10.1039/b412831a. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto A., Ochi Y., Saito I. Fluorometric sensing of the salt-induced B Z DNA transition by combination of two pyrene-labeled nucleobases. Chem. Commun. 2005:1128–1130. doi: 10.1039/b416965d. [DOI] [PubMed] [Google Scholar]
- 12.Kosuge M., Kubota M., Ono A. Multiple-pyrene residues arrayed along DNA backbone exhibit significant excimer fluorescence. Tetrahedron Lett. 2004;45:3945–3947. [Google Scholar]
- 13.Kitamura M., Nimura A., Yamana K., Shimidzu T. Oligonucleotides with bis-pyrene adduct in the backbone: synthesis and properties of intramolecular excimer forming probe. Nucleic Acids Res. Symp. Ser. 1991;25:67–71. [PubMed] [Google Scholar]
- 14.Yamana K., Takei M., Nakano H. Synthesis of oligonucleotide derivatives containing pyrene labeled glycerol linkers: enhanced excimer fluorescence on binding to a complementary DNA sequence. Tetrahedron Lett. 1997;38:6051–6055. [Google Scholar]
- 15.Lews F.D., Zhang Y., Letsinger R.L. Bispyrenyl excimer fluorescence: a sensitive oligonucleotide probe. J. Am. Chem. Soc. 1997;119:5451–5452. [Google Scholar]
- 16.Paris P.L., Langenhan J.M., Kool E.T. Probing DNA sequences in solution with a monomer-excimer fluorescence color change. Nucleic Acids Res. 1998;26:3789–3793. doi: 10.1093/nar/26.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuko M., Ohtani H., Ebata K., Shimadzu A. Optimization of excimer-forming two-probe nucleic acid hybridization method with pyrene as a fluorophore. Nucleic Acids Res. 1998;26:5409–5416. doi: 10.1093/nar/26.23.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostenko E., Dobrikov M., Pyshnyi D., Petyuk V., Komarova N., Vlassov V., Zenkova M. 5′-Bis-pyrenylated oligonucleotides displaying excimer fluorescence provide sensitive probes of RNA sequence and structure. Nucleic Acids Res. 2001;29:3611–3620. doi: 10.1093/nar/29.17.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamana K., Iwai T., Ohtani Y., Nakamura M., Nakano H. Bis-pyrene-labeled oligonucleotides: sequence specificity of excimer and monomer fluorescence changes upon hybridization with DNA. Bioconjug. Chem. 2002;13:1266–1273. doi: 10.1021/bc025530u. [DOI] [PubMed] [Google Scholar]
- 20.Yamana K., Fukunaga Y., Ohtani Y., Sato S., Nakamura M., Kim W.J., Akaike T., Maruyama A. DNA mismatch detection using a pyrene-excimer-forming probe. Chem. Commun. 2005:2509–2511. doi: 10.1039/b502033f. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto K., Shimizu H., Inouye M. Unambiguous detection of target DNAs by excimer-monomer switching molecular beacons. J. Org. Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto A., Ichiba T., Saito I. Pyrene-labeled oligonucleotide probe for detecting base insertion by excimer fluorescence emission. J. Am. Chem. Soc. 2004;126:8364–8365. doi: 10.1021/ja049061d. [DOI] [PubMed] [Google Scholar]
- 23.Yamana K., Ohashi Y., Nunota K., Kitamura M., Nakano H., Sangen O., Shimidzu T. Synthesis of oligonucleotide derivatives with pyrene group at sugar fragment. Tetrahedron Lett. 1991;32:6347–6350. [Google Scholar]
- 24.Yamana K., Gokota T., Ozaki H., Nakano H., Sangen O., Shimidzu T. Enhanced fluorescence in the binding of oligonucleotides with a pyrene group in the sugar fragment to complementary polynucleotides. Nucleosides Nucleotides. 1992;11:383–390. [Google Scholar]
- 25.Yamana K., Iwase R., Furutani S., Tsuchida H., Zako H., Yamaoka T., Murakami A. 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent prove of RNA. Nucleic Acids Res. 1999;27:2387–2392. doi: 10.1093/nar/27.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamana K., Zako H., Asazuma K., Iwase R., Nakano H., Murakami A. Fluorescent detection of specific RNA sequence using 2′-pyrene-modified oligonucleotides. Angew. Chem. Int. Ed. 2001;40:1104–1106. doi: 10.1002/1521-3773(20010316)40:6<1104::aid-anie11040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Mahara A., Iwase R., Sakamoto T., Yamana K., Yamaoka T., Murakami A. Bispyrene-conjugated 2′-O-methyloligonucleotide as a highly specific RNA-recognition probe. Angew. Chem. Int. Ed. 2002;41:3648–3649. doi: 10.1002/1521-3773(20021004)41:19<3648::AID-ANIE3648>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Shafirovich V.Y., Courtney S.H., Ya N., Geacintov N.E. Proton-coupled photoinduced electron transfer, deuterium isotope effects, and fluorescence quenching in noncovalent benzo[a]pyrenetetraol-nucleoside complexes in aqueous solutions. J. Am. Chem. Soc. 1995;117:4920–4929. [Google Scholar]
- 29.Manoharan M., Tivel K.L., Zhao M., Nafisi K., Netzel T.L. Base-sequence dependence of emission lifetimes for DNA oligomers and duplexes covalently labeled with pyrene: relative electron-transfer quenching efficiencies of A, G, C, and T nucleosides toward pyrene*. J. Phys. Chem. 1995;99:17461–17472. [Google Scholar]
- 30.Netzel T.L., Zhao M., Nafisi K., Headrick J., Sigman M.S., Eaton B.E. Photophysics of 2′-deoxyuridine (dU) nucleosides covalently substituted with either 1-pyrenyl or 1-pyrenonyl: Observation of pyrene-to-nucleoside charge-transfer emission in 5-(1-pyrenyl)-dU. J. Am. Chem. Soc. 1995;117:9119–9128. [Google Scholar]
- 31.Elbaum D., Nair S.K., Patchan M.W., Thompson R.B., Christianson D.W. Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor. J. Am. Chem. Soc. 1996;118:8381–8387. [Google Scholar]
- 32.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley InterScience; 1986. [Google Scholar]
- 33.Uno T., Hayashi H., Yamana K., Nakano H. Development of support program for AMBER. J. Chem. Softw. 1999;5:39–48. [Google Scholar]
- 34.Sasa K., Uno T., Hayashi H., Yamana K., Nakano H. Development of an assisting system to add new residues to the standard AMBER residue database. J. Comput. Chem. Jpn. 2003;2:135–142. [Google Scholar]
- 35.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 36.Case D.A., Pearlman D.A., Caldwell J.W., Cheatham T.E., III, Ross W.S., Simmerling C.L., Darden T.L., Marz K.M., Stanton R.V., Cheng A.L., et al. AMBER6. San Francisco, CA: University of California; 2000. [Google Scholar]
- 37.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Cho N., Asher S.A. UV resonance Raman studies of DNA-pyrene interactions: optical decoupling Raman spectroscopy selectively examines external site bound pyrene. J. Am. Chem. Soc. 1993;115:6349–6356. [Google Scholar]
- 39.Berova N., Nakanishi K., Woody R.W. Principles and Applications, 2nd edn. New York: Wiley-VCH; 2000. Circular dichroism. [Google Scholar]