Abstract
Transforming growth factor-β (TGF-β)/activin-induced Smad2/Smad4 complexes are recruited to different promoter elements by transcription factors, such as Fast-1 or the Mix family proteins Mixer and Milk, through a direct interaction between Smad2 and a common Smad interaction motif (SIM) in the transcription factors. Here we identify residues in the SIM critical for Mixer–Smad2 interaction and confirm their functional importance by demonstrating that only Xenopus and zebrafish Mix family members containing a SIM with all the correct critical residues can bind Smad2 and mediate TGF-β-induced transcriptional activation in vivo. We identify significant sequence similarity between the SIM and the Smad-binding domain (SBD) of the membrane-associated protein SARA (Smad anchor for receptor activation). Molecular modelling, supported by mutational analyses of Smad2 and the SIM and the demonstration that the SARA SBD competes directly with the SIM for binding to Smad2, indicates that the SIM binds Smad2 in the same hydrophobic pocket as does the proline-rich rigid coil region of the SARA SBD. Thus, different Smad2 partners, whether cytoplasmic or nuclear, interact with the same binding pocket in Smad2 through a common proline-rich motif.
Keywords: Mixer/SARA/Smad2/Smad interaction motif/TGF-β signalling
Introduction
Signals from transforming growth factor-β (TGF-β) family members are transduced to the nucleus by members of the Smad family. Upon ligand binding, an active complex is formed comprising activated receptor-regulated Smads (R-Smads), such as Smad2 or Smad3 in the case of the ligands TGF-β and activin, and a co-Smad (Smad4; Massagué and Wotton, 2000). R-Smads are activated directly by phosphorylation at their extreme C-terminus by the type I receptor kinase (Massagué and Wotton, 2000). Smad2 and Smad3 are specifically recruited to the receptor complex by an FYVE domain protein called SARA (Smad anchor for receptor activation; Tsukazaki et al., 1998). After ligand stimulation, active Smad complexes rapidly accumulate in the nucleus, where they are directly involved in gene regulation.
Smads bind DNA very weakly with limited specificity and thus cooperate with other transcription factors, which target them to specific DNA-binding sites (Massagué and Wotton, 2000; ten Dijke et al., 2000; Shi, 2001). For example, in early Xenopus embryos, Smad complexes activated by activin-related ligands (consisting of XSmad2 and XSmad4β; Howell et al., 1999; Masuyama et al., 1999) are recruited to DNA by transcription factors, such as the forkhead/winged helix protein, XFast-1 (Chen et al., 1996, 1997), or a subset of paired-like homeodomain transcription factors of the Mix family, Mixer and Milk (Germain et al., 2000). These Smad-interacting transcription factors are key determinants of specificity since they have different DNA-binding domains and thus recruit a common activated Smad complex to different promoter elements to activate distinct sets of target genes. Consistent with this idea, Mix and Fast family members are expressed in different regions of the embryo: XFast-1 is highly expressed in prospective ectoderm and mesoderm, whereas Mixer and Milk are expressed in a ring in deeper layers of the mesoderm and endoderm (Hill, 2001).
Many different proteins at all levels of the TGF-β signalling pathways have been reported to interact with different combinations of Smads. For example, proteins such as SARA recruit Smad2 and Smad3 to the receptors for phosphorylation, transcription factors recruit different Smad complexes to DNA, co-activators and co-repressors are involved in modulating transcriptional responses, and E3 ubiquitin ligases, such as the Smurfs, bind Smads, which target them or associated proteins for degradation (ten Dijke et al., 2000). To understand these processes, it is important to define Smad interaction motifs in the partner proteins and to understand how these motifs interact specifically with different Smads.
We demonstrated previously that members of the Mix and Fast families that bind active Smad complexes do so through a common 25 amino acid Smad interaction motif (SIM), characterized by a conserved core (P-P-N-K-S/T-I/V), present in their C-terminal regions (Germain et al., 2000). The SIM binds to the C-terminal (MH2) domain of phosphorylated Smad2, which in turn binds Smad4. The SIM displays a high degree of binding specificity, interacting only with the MH2 domains of Smad2 and Smad3, and not with those of the BMP-activated R-Smads or Smad4 (Germain et al., 2000).
Here we set out to determine what constitutes a functional SIM, to understand the molecular basis for the interaction of the SIM with Smad2, and to determine whether it is unique to these Smad2/Smad3-interacting transcription factors or whether it may represent a generic Smad interaction motif. Using a combination of in vitro and in vivo binding and transcription assays, we have identified the conserved residues of the Mixer SIM critical for interaction with Smad2. The results demonstrate the importance of the very conserved P-P-N-K-S/T-I/V core and two C-terminal flanking residues, and indicate that the SIM is an extended motif. These preferences are observed in the other Xenopus Mix family members and in zebrafish Mixer [bonnie and clyde (bon); Alexander et al., 1999; Kikuchi et al., 2000], since only those family members that contain a SIM with all the correct critical residues bind Smad2 and mediate TGF-β-induced transcriptional activation in vivo.
Of the many reported Smad-interacting proteins, few bind the MH2 domain of Smad2 and show exactly the same specificity of Smad binding as the SIM. The cytoplasmic protein SARA is the best understood Smad partner with these characteristics. We find significant sequence similarity between the SIM and the proline-rich rigid coil region of the Smad-binding domain (SBD) of SARA. We demonstrate that the SIM binds to a region of the Smad2 MH2 domain, which also binds the rigid coil region of the SARA SBD (Wu et al., 2000), and that the SARA SBD competes with the SIM for binding to Smad2. We propose a molecular model in which a shallow hydrophobic groove on the surface of the MH2 domain of Smad2 is responsible for recruiting different Smad2 partners, such as SARA in the cytoplasm or SIM-containing transcription factors in the nucleus, via a defined proline-rich motif.
Results
Definition of the residues in the SIM critical for interaction with Smad2
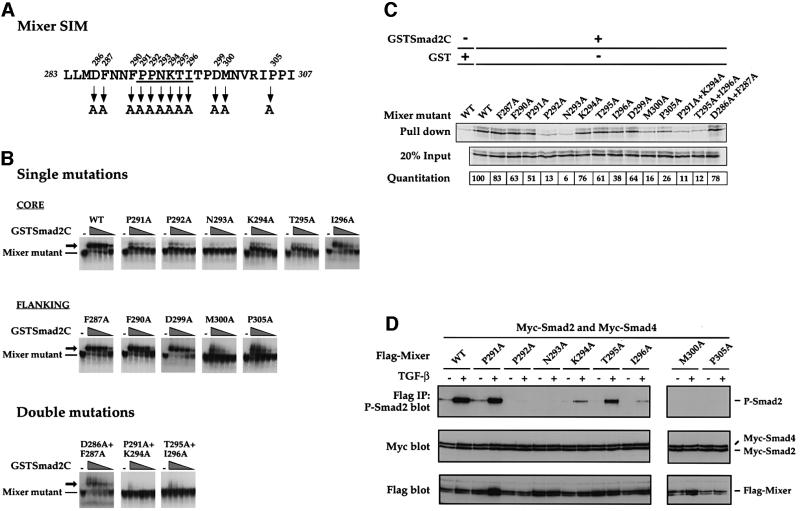
Alignment of the known functional SIMs in Fast and Mix family members reveals the characteristic conserved core P-P-N-K-S/T-I/V, flanked by other highly conserved residues (Germain et al., 2000). To understand how the SIM interacts with the Smad2 MH2 domain, we determined which of the conserved residues are absolutely required for interaction with Smad2. Selected amino acids of Mixer were mutated to alanine, either alone or in pairs (Figure 1A), and the resulting mutants were assayed for their ability to interact with Smad2.
Fig. 1. Identification of residues critical for SIM/Smad2 interaction. (A) The sequence of the Mixer SIM indicating the residues that have been mutated to alanine, either singly or in pairs. The conserved core of the SIM is underlined (Germain et al., 2000). (B) Analysis of the ability of in vitro translated Mixer mutants to interact with GSTSmad2C by bandshift assay using the MBS from the DE as probe. GSTSmad2C was titrated over the range 20, 10, 5, 2.5 and 1.25 ng. Mixer derivatives bound to DNA are indicated, as are ternary complexes containing GSTSmad2C (arrow). (C) Analysis of the ability of 35S-labelled in vitro translated Mixer mutants to interact with GSTSmad2C in a GST pull-down assay. Bound protein was visualized by SDS–PAGE and autoradiography. Lane 1 corresponds to the interaction between wild-type Mixer and GST to estimate non-specific binding. Twenty per cent input protein is shown. The amount of Mixer derivative bound is quantitated as a percentage of input and these values were normalized such that the binding of wild-type Mixer was 100. Three independent experiments gave similar results. (D) Analysis of the ability of Mixer mutants to interact with phosphorylated Smad2 in an IP western blot. Extracts were prepared from uninduced or TGF-β-induced NIH 3T3 cells that had been transfected with the different Flag-tagged Mixer mutants with Myc-tagged Smad2 and Smad4. Extracts were assayed by IP of complexes with anti-Flag antibody followed by western blotting with anti-phospho-Smad2 antibody (P-Smad2, top blot) or by western blotting the whole extract with anti-Myc antibody (middle blot) or with anti-Flag antibody (bottom blot).
First, in vitro synthesized Mixer mutants were assayed by bandshift for their ability to interact with a glutathione S-transferase (GST) fusion protein of the Smad2 MH2 domain (GSTSmad2C), using a probe corresponding to the Mixer-binding site (MBS) from the goosecoid DE (Figure 1B; Germain et al., 2000). The assay was made semi-quantitative by titrating GSTSmad2C over a range of concentrations. The single mutations that had the greatest effect on the ability of Mixer to interact with GSTSmad2C were P291A, P292A and N293A in the core, and M300A and P305A in the C-terminal flanking sequence. In all cases, either no supershift was seen with GSTSmad2C at any input concentration (N293A) or efficient supershifts were only detected at the higher concentrations of GSTSmad2C (P291A, P292A, M300A and P305A). The other single mutations either had no effect (F287A, F290A and D299A) or small effects (K294A, T295A and I296A). Mutants with double mutations in core residues (P291A+K294A and T295A+I296A) were almost completely defective for binding to GSTSmad2C, underlining the importance of the core residues for Smad2C binding (Figure 1B). A double mutation in the N-terminal flanking sequence (D286A+F287A) had no effect on GSTSmad2C binding, suggesting that these flanking residues do not play a role in the interaction with Smad2C. All Mixer derivatives bound DNA as well as wild-type Mixer, indicating that mutating the SIM had no impact on DNA binding.
The binding of the Mixer mutants to Smad2C was also assayed in the absence of DNA in a GST pull-down assay (Figure 1C). On the whole, the data agreed well with the bandshift assays. Mutation of N293 alone completely blocked Mixer interaction with GSTSmad2C. Other mutations that severely inhibited binding to GSTSmad2C were P292A, M300A, P305A and the combined mutations of P291A+K294A and T295A+I296A in agreement with the bandshift assays (Figure 1C). The P291A mutant was the only one that behaved significantly differently in the two assays; it interacted with GSTSmad2C more efficiently in the pull-down assay than would be expected from the bandshift assay (see below).
We then confirmed the relative importance of the core residues of the SIM and the C-terminal flanking residues, M300 and P305, for Smad2 interaction in vivo in an immunoprecipitation (IP) western assay, measuring the ability of the Mixer mutants to interact with phosphorylated Smad2 upon TGF-β induction in NIH 3T3 cells (Figure 1D). The results generally agreed with the in vitro binding analyses, and again indicated that P292 and N293 in the core and M300 and P305 in the C-terminal flanking region are absolutely required in vivo for Mixer to interact with phosphorylated Smad2. In addition, mutation of I296 also had a severe effect on the ability of Mixer to interact with phosphorylated Smad2.
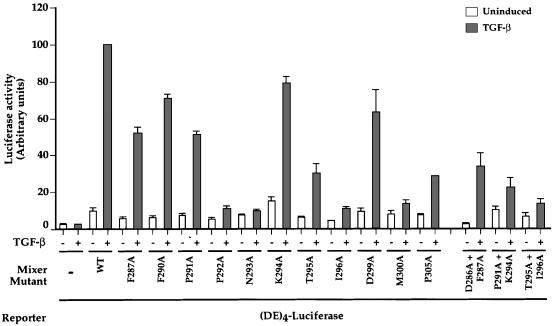
The Mixer mutants were then assayed for their ability to recruit active endogenous Smad2/Smad4 complexes to DNA by assessing their ability to mediate TGF-β-induced transcriptional activation via the goosecoid DE in NIH 3T3 cells (Figure 2). Wild-type Mixer mediated a 10-fold increase in transcriptional activation upon TGF-β stimulation due to recruitment of active endogenous Smad complexes (Figure 2; Germain et al., 2000). Mutants severely defective for interaction with Smad2 in some or all of the binding assays were either completely incapable or very poor at mediating TGF-β-induced transcriptional activation in vivo (Figure 2; P292A, N293A, I296A, M300A and P305A and the double mutants P291A+K294A and T295A+I296A). Control bandshifts and western blots of whole-cell extracts made from cells transfected with Flag-tagged wild-type or mutant Mixer derivatives demonstrated they were all efficiently expressed and bound DNA (Figure 1D and data not shown).
Fig. 2. The ability of Mixer derivatives to interact with Smad2C correlates with their ability to mediate TGF-β-induced transcription via the DE. NIH 3T3 cells were transfected with the (DE)4–luciferase reporter and plasmids expressing Flag-tagged wild-type Mixer or mutants. Cells were incubated for 8 h in the presence or absence of TGF-β. Luciferase was quantitated relative to β-Gal activity and the value for TGF-β-induced transcription using wild-type Mixer was set at 100. The data are the mean and standard deviation of three independent experiments.
We can now define residues in the SIM required for Smad2 binding. The conserved core residues P-P-N-K-T-I are important for interacting with Smad2 and recruiting activated Smad2/Smad4 complexes to DNA for transcriptional activation. Of these residues, P292 and N293 are the most critical for Smad2 interaction, since mutation of either one greatly reduced Smad2 binding. The importance of the other core residues only became apparent when double mutations were tested. The residues N-terminal to the core do not appear to play an important role in Smad2 interaction, but two flanking residues C-terminal to the core are critical for the function of the SIM; these are M300 and P305. This indicates that the SIM extends beyond the conserved core.
The Mixer SIM is necessary and sufficient for mediating TGF-β-inducible transcription
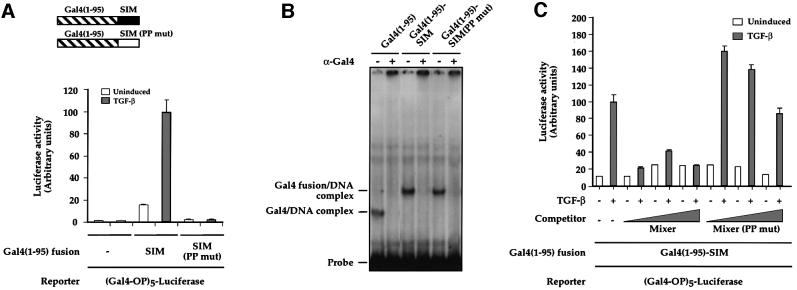
The Mixer SIM is required for mediating TGF-β-inducible transcription (Figure 2; Germain et al., 2000) but is it sufficient or are other sequences in Mixer also required? A fusion of the Mixer SIM (amino acids 283–307) with the Gal4 DNA-binding domain [Gal4(1–95)–SIM] was tested for its ability to confer TGF-β-inducible transcription in NIH 3T3 cells on (Gal4-OP)5–luciferase. The reporter alone was inactive and non-responsive to TGF-β (Figure 3A). Co-transfection of Gal4(1–95)–SIM conferred a low level of transcriptional activity on the reporter in the absence of TGF-β, which was increased ∼6-fold upon TGF-β stimulation (Figure 3A). A mutant SIM fusion protein, Gal4(1–95)–SIM(PP mut), which does not interact with Smad2, was completely inactive in the presence or absence of TGF-β (Figure 3A). Both Gal4 fusion proteins were equally well expressed and bound DNA efficiently (Figure 3B).
Fig. 3. The SIM is sufficient to confer TGF-β inducibility in vivo. (A) Schematics of the Gal4(1–95) fusion of the SIM (residues 283–307 of Mixer) or mutant SIM in which P291 and P292 are mutated to alanine. NIH 3T3 cells were transfected with (Gal4-OP)5–luciferase together with plasmids expressing Gal4(1–95), Gal4(1–95)–SIM or Gal4(1–95)–SIM(PP mut) as indicated; TGF-β inductions and luciferase assays were as above. The value for TGF-β-induced transcription using Gal4(1–95)–SIM was set at 100. (B) The Gal4(1–95) derivatives are equally expressed. Whole-cell extracts from NIH 3T3 cells transfected with the Gal4(1–95) derivatives as indicated were assayed by bandshift using a Gal4-binding site as a probe. The complexes were confirmed as Gal4(1–95) derivatives bound to DNA since they were all abolished by addition of the anti-Gal4 antibody, which recognizes the DNA-binding domain. (C) TGF-β-inducible transcription mediated by Gal4(1–95)–SIM results from recruitment of active Smad complexes. NIH 3T3 cells were transfected with (Gal4-OP)5–luciferase together with plasmids expressing Gal4(1–95)–SIM and plasmids expressing increasing amounts of Mixer, or Mixer(PP mut) which cannot bind Smad2. TGF-β inductions and luciferase assays were as above. The value for TGF-β-induced transcription using Gal4(1–95)–SIM was set at 100. In (A) and (C), the data are the mean and standard deviation of three independent experiments.
Overexpression of Mixer could compete with Gal4(1–95)–SIM for binding to endogenous Smads, confirming that the TGF-β-induced transcriptional activation was due to recruitment of active Smad complexes (Figure 3C). Competition with Mixer(PP mut) had no effect on the ability of the Gal4(1–95)–SIM fusion to confer TGF-β inducibility on the reporter.
Thus, the Mixer SIM is both necessary and sufficient to mediate TGF-β-induced transcription in vivo through its interaction with active Smad complexes.
Identification of Smad-interacting members of the Xenopus and zebrafish Mix families
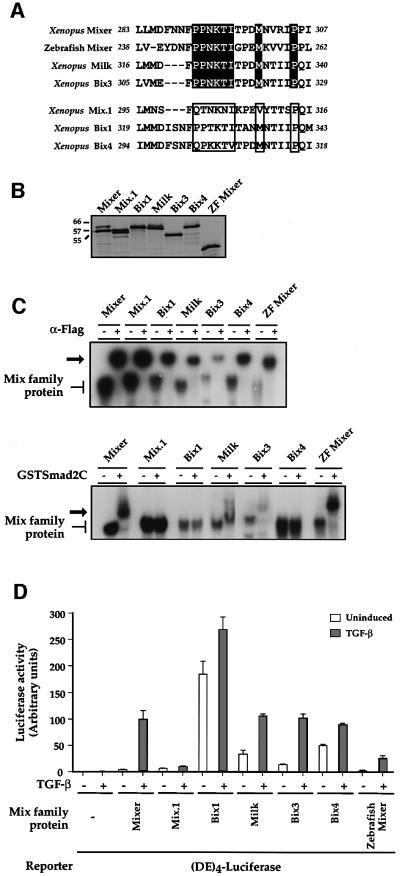
We have demonstrated the importance of the core residues of the SIM (in particular P292 and N293) and also two critical C-terminal flanking residues (M300 and P305) for interaction with Smad2. To determine whether these preferences are observed in other naturally occurring SIMs, we analysed other Mix family members to see which have functional SIMs. The SIM of Xenopus Mixer was aligned with the equivalent regions of the other Xenopus Mix family members, Mix.1, Bix1, Milk (Bix2), Bix3 and Bix4 (Rosa, 1989; Ecochard et al., 1998; Henry and Melton, 1998), and the zebrafish orthologue of Mixer, bon (Figure 4A; Alexander et al., 1999; Kikuchi et al., 2000). The Xenopus and zebrafish Mixers, Milk and Bix3 all have a recognizable SIM with the P-P-N-K-T-I core and the methionine and proline residues at positions equivalent to 300 and 305 in Xenopus Mixer, respectively. Based on our analysis of the SIM, we would predict that these would all interact with Smad2. Although Mix.1, Bix1 and Bix4 contain a subset of these residues, they do not contain all of them and we would predict that they would not interact with Smad2.
Fig. 4. Mix family members that contain a SIM interact with Smad2 and mediate TGF-β-induced transcriptional activation in vivo. (A) Alignment of the SIM regions of six Xenopus Mix family members and zebrafish Mixer. Above, family members that contain the P-P-N-K-T-I core and the C-terminal M and P known to be critical for Smad2 interaction (black shading). Below, family members that have no predicted SIM (open boxes). (B) Equal amounts of 35S-labelled in vitro translated Mix family members were analysed by SDS–PAGE and autoradiography. Molecular weights of marker proteins are indicated. (C) Mix family members that contain a SIM interact with GSTSmad2C in a bandshift assay. In vitro translated Flag-tagged Mix family members (as in B) were assayed by bandshift assay using the MBS probe in the presence or absence of antibody directed against the tag (upper panel) or with 20 ng of GSTSmad2C (bottom panel). Mix family members bound to DNA are indicated, as are antibody-supershifted complexes in the top panel and ternary complexes with GSTSmad2C in the bottom panel (arrow). (D) SIM-containing Mix family members mediate TGF-β-induced transcriptional activation. NIH 3T3 cells were transfected with (DE)4–luciferase together with plasmids expressing Mix family members. TGF-β inductions and luciferase assays were as above. The value for TGF-β-induced transcription using Mixer was set at 100. The data are the mean and standard deviation of three independent experiments.
The ability of the in vitro translated Mix family members (Figure 4B) to interact with GSTSmad2C was tested in a bandshift assay. All the proteins were supershifted by an antibody recognizing the N-terminal Flag tag on the proteins (Figure 4C, top panel), but only Xenopus and zebrafish Mixers, Milk and Bix3 interacted efficiently with GSTSmad2C and formed a ternary complex (bottom panel). This agrees with the presence of a predicted SIM containing all of the residues that we have shown to be critical. Moreover, the Mix family members that contain a SIM could mediate TGF-β-inducible transcription via the DE (6- to 10-fold for Xenopus Mixer, zebrafish Mixer and Bix3; ∼3-fold for Milk; Figure 4D). Mix.1, Bix1 and Bix4, which were not capable of interaction with Smad2, were unable to confer significant TGF-β-inducible transcription onto the DE (Figure 4D). The Mix family members were all synthesized efficiently in vivo (data not shown). The high basal level of transcription seen in Bix1, Milk, Bix4 and to a lesser extent Bix3, may be due to the presence of Q-rich transcriptional activation domains within the sequence of Bix1–4, which are absent from both Mixer and Mix.1 (Tada et al., 1998).
Swapping the SIM of Milk into Mix.1 is sufficient to convert Mix.1 into a Smad-interacting transcription factor
We next performed a gain-of-function experiment to determine whether it was possible to convert a non-Smad2-interacting Mix family member, Mix.1, into a Smad2-interacting protein. Instead of the PPNK core characteristic of all functional SIMs, the Mix.1 sequence is QTNK (Figure 4A). In initial experiments, we made four point mutations in Mix.1 to mutate Q300 and T301 to proline and N304 and K306 of Mix.1 to threonine, thus creating in Mix.1 the PPNKTIT of Mixer. This mutant did not interact with Smad2, confirming that additional residues in the SIM are required. Consistent with this, creating the PPNKTI motif in Bix1 also failed to generate a functional SIM (data not shown; see Discussion).
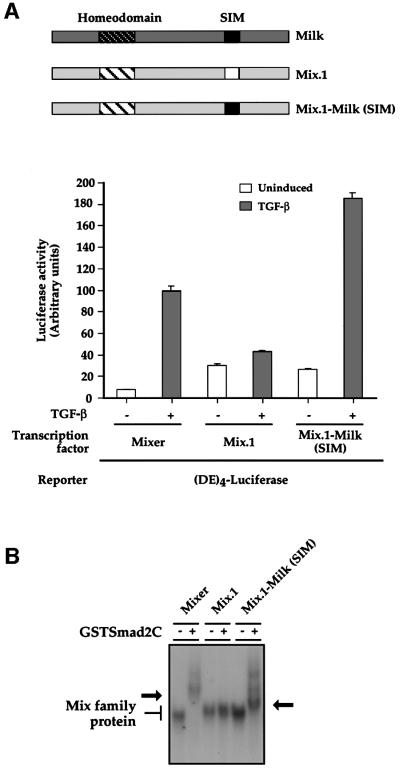
Mutations in Mix.1 outside the core must also be required to generate a functional SIM, and amino acids 299–314 of Mix.1 were therefore mutated to the corresponding residues of Milk, which contains a functional SIM, to generate Mix.1–Milk(SIM) (Figure 5A). This Mix.1 derivative conferred very strong TGF-β-inducible transcription on the DE (∼9-fold), comparable with that of Mixer. In contrast, Mix.1 was not TGF-β inducible in this assay. Mix.1–Milk(SIM) also interacted with GSTSmad2C in a bandshift assay (Figure 5B). Thus, swapping the functional Milk SIM into Mix.1 is sufficient to convert it into a Smad2-interacting transcription factor.
Fig. 5. Replacing the inactive SIM region of Mix.1 with the functional Milk SIM is sufficient to enable Mix.1 to mediate TGF-β-induced transcription activation and bind Smad2C. (A) NIH 3T3 cells were transfected with (DE)4–luciferase together with plasmids expressing Mixer, Mix.1 or the Mix.1–Milk(SIM) chimeric protein. TGF-β inductions and luciferase assays were as above. The value for TGF-β-induced transcription using Mixer was set at 100. The data are the mean and standard deviation of three independent experiments. Schematics of wild-type Mix.1, Milk and the Mix.1–Milk(SIM) chimera are shown. (B) Mixer, Mix.1 and Mix.1–Milk(SIM) were synthesized in vitro and analysed for their ability to interact with 20 ng of GSTSmad2C in a bandshift assay using the MBS as a probe. Both Mixer and Mix.1–Milk(SIM) interact with GSTSmad2C, as seen by the supershift (arrow).
Common residues in the Smad2 MH2 domain are required for interaction with the SIM and with SARA
Many proteins have been reported to interact with the different Smad family members, both in the nucleus and cytoplasm (ten Dijke et al., 2000), but other than the SIM-containing transcription factors, few interact with the Smad2 MH2 domain with the same specificity as the SIM. The best characterized is the membrane-bound FYVE domain protein SARA, which like the SIM-containing transcription factors, specifically recognizes Smad2 and Smad3, but not Smad1 or Smad4 (Tsukazaki et al., 1998). The structure of the SARA SBD with the Smad2 MH2 domain has been solved (Wu et al., 2000). The SARA SBD is in an extended conformation comprising a conformationally restrained proline-rich rigid coil, an α-helix and a β-strand. The rigid coil is responsible for the specificity of the interaction with Smad2 and makes contacts with Y366 in α-helix 2 of the Smad2 MH2 domain, with W368, T372 and C374 in the loop joining α-helix 2 to strand β-8, and with residues on strands β-8 and β-9 (Wu et al., 2000).
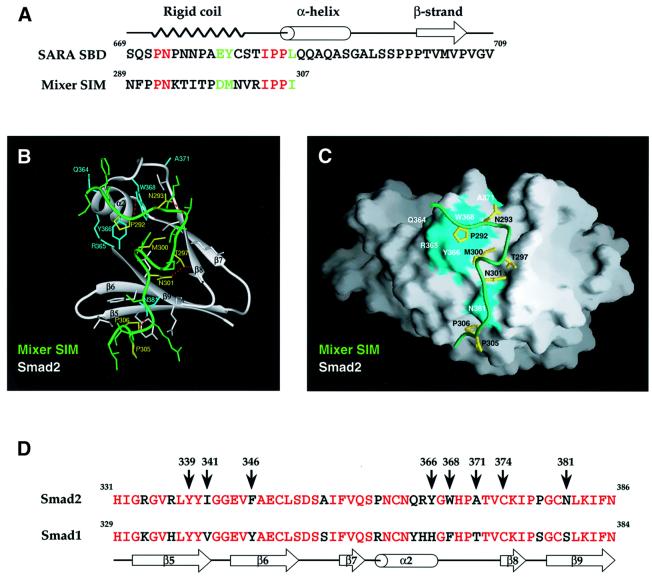
Previous work has demonstrated that residues in α-helix 2 of the Smad2 MH2 domain (including Y366) are also required to interact with Mixer, Milk and Fast-1 via the SIM (Chen et al., 1998; Germain et al., 2000), and dictate the specificity of the interaction such that the transcription factors bind Smad2 and not Smad1 (Chen et al., 1998). This suggests that the SARA SBD rigid coil and the SIM might bind Smad2 in a similar manner. Alignment of the two motifs revealed important similarities (Figure 6A). Strikingly, the residues in the Mixer SIM that we have demonstrated to be critical for Smad2 interaction (P292, N293, M300 and P305) are either identical in the two motifs or, in the case of M300, substituted by a residue with similar properties.
Fig. 6. The SIM binds to a region of the Smad2 MH2 domain that also binds the SARA SBD. (A) Alignment of the Mixer SIM with the rigid coil region of the SARA SBD (Wu et al., 2000). Identical residues are in red, conservative substitutions are in light green. (B) Model of the Mixer SIM bound to the Smad2 MH2 domain. Ribbon diagram of the region of Smad2 MH2 domain (grey) corresponding to α-helix 2 (α2) and β-strands (β5–β9) that interacts with the Mixer SIM (green). All side chains of the SIM peptide are shown. Only the side chains of Smad2 that make direct contact with the peptide are displayed. Residues of Smad2 coloured cyan have been mutated in this study (see text). Mixer SIM side chains are coloured green and yellow; yellow side chains are important for Smad2 interactions and for conserving the peptide conformation. Some key hydrogen bonds are indicated by dotted red lines (see Table I for a description of these interactions). (C) Molecular surface of Smad2 (grey) and a ribbon representation of the Mixer SIM (green) in the same orientation as in (B). The side chains of the Mixer SIM and Smad2, coloured yellow and cyan, respectively, are as in (B). The binding groove on Smad2 is generally shallow with larger indentations in the areas that contact M300 and P306 of the Mixer SIM. The figure was created using the program GRASP (Nicholls et al., 1991). (D) The region of Smad2 shown in the model is aligned with the analogous region of Smad1 (which does not interact with the SIM or with the SARA SBD). Red residues are identical between the two Smads and black residues are different. The assignment of structural motifs is based on Shi et al. (1997); α2 is α-helix 2 and β5–9 are β-strands. Eight residues are indicated, which are demonstrated from our mutation analysis and/or predicted from the model to be important for SIM–Smad2 interaction (B and C; Table I). Six of these are unique to Smad2.
To determine whether the Mixer SIM could form a rigid coil similar to that in the N-terminal region of the SARA SBD and interact with Smad2 in an analogous way, we modelled the Mixer SIM onto the backbone of the SARA SBD. The resulting model indicated that this is feasible (Figure 6B and C; Table I). The Mixer SIM in this conformation forms a network of stabilizing intramolecular hydrogen bonds, with N293 being particularly important (Table I), as predicted by our mutational analysis. The interaction of the SIM with the Smad2 MH2 domain is mediated by a combination of hydrogen bonds and hydrophobic interactions (Table I). Key hydrogen bonds are between the carbonyl of P292 in the SIM with Nε1 of W368 in Smad2, the side chain of T297 in the SIM with the carbonyl of K375 in Smad2, and the carbonyl of I304 in the SIM with the side chain of N381 in Smad2 (Table I). Important hydrophobic interactions occur between P292 of the SIM and Y366 and W368 of Smad2, between M300 of the SIM and W368 and C374 of Smad2, P306 of the SIM and Y339 and F346 of Smad2, and between I307 of the SIM and I341 of Smad2 (Table I). Many of these interactions involve residues in the SIM that we have shown to be essential for Smad2 interaction (Figures 1 and 2); five out of these seven interacting residues are in Smad2 but not in Smad1 (Figure 6D).
Table I. Modelling the interactions of the Mixer SIM with Smad2 using the SARA SBD–Smad2 structure.
| SARA | Mixer | Comments |
|---|---|---|
| SBD | SIM | |
| S669 | N289 | no contact with Smad2 |
| Q670 | F290 | no contact with Smad2 |
| S671 | P291 | P of SIM makes the N-terminus of the peptide less flexible |
| P672 | *P292 | carbonyl of P H-bonds to Nε1 of W368 in Smad2. Ring of P makes hydrophobic contacts with Y366 and W368 of Smad2 |
| N673 | *N293 | N in SIM stabilizes peptide geometry by H-bonding with amide of T295 in SIM |
| P674 | K294 | charged side chain of K has potential to make salt bridge with E425 of Smad2 |
| N675 | T295 | no substantive contact with Smad2 |
| N676 | I296 | N in SARA forms intrapeptide H-bond with E679. I in SIM stabilizes peptide by virtue of β-branched side chain |
| P677 | T297 | T in SIM forms H-bonding network to N301 in SIM and to Smad2 at carbonyl of residue 375 |
| A678 | P298 | P makes peptide more rigid |
| E679 | D299 | E in SARA H-bonds to amide of residue N673 in SARA peptide. D in SIM is not optimal for H-bond geometry |
| Y680 | *M300 | Y in SARA involved in H-bonding network and hydrophobic contacts. M in SIM forms a number of hydrophobic contacts e.g. to W368 and C374 of Smad2 |
| C681 | N301 | N in SIM H-bonds to T297 of SIM which H-bonds to carbonyl of residue 375 of Smad2 |
| S682 | V302 | no contact with Smad2 |
| T683 | R303 | no contact with Smad2 |
| I684 | I304 | carbonyl of I forms stabilizing H-bond to N381 of Smad2 |
| P685 | *P305 | P maintains position of carbonyl of I304 to form H-bond |
| P686 | P306 | P forms hydrophobic interaction with Y339 and F346 of Smad2 |
| L687 | I307 | forms hydrophobic interaction with I341 of Smad2 |
See Figure 6A for alignment and B and C for model of the Mixer SIM with Smad2 based on the interactions of the SARA SBD rigid coil with Smad2 (Wu et al., 2000). Asterisks indicate residues of the SIM that we have shown in the mutational analysis to be particularly important for interaction with Smad2.
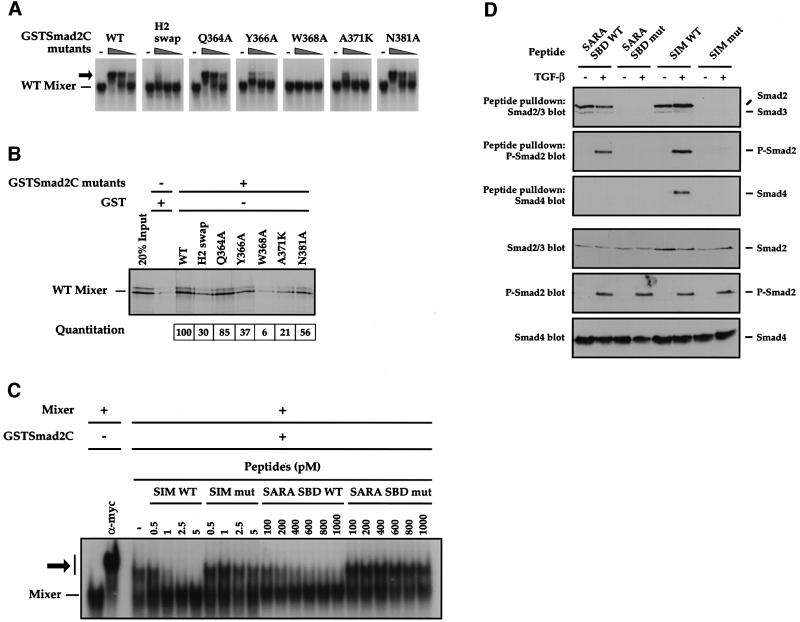
The feasibility of this model was tested by making point mutations in Smad2, focusing on surface residues to avoid indirect effects of distorting the Smad2 MH2 domain structure by mutating structurally important residues. Mutants were made in the context of GSTSmad2C and assayed for their ability to interact with Mixer in a bandshift assay (Figure 7A) and GST pull-down (Figure 7B). In the molecular model, W368 is a key residue in Smad2 for the interaction with the SIM, forming both a hydrogen bond to the carbonyl of P292 and hydrophobic interactions with the proline ring of P292 and with M300. As predicted, mutating W368 to alanine completely abolished the ability of GSTSmad2C to interact with Mixer in both assays (Figure 7A and B). The importance of the hydrogen bond was confirmed by the observation that mutating W368 to phenylalanine also abolished the interaction with Mixer (data not shown). Mutating Q364, R365 and Y366 to the residues that are present in the equivalent region of Smad1 (YHH; H2 swap) abolished the ability of GSTSmad2C to interact with Mixer (Germain et al., 2000). As predicted by the molecular model (Figure 6B and C; Table I), the key residue for this interaction is Y366, since mutating this residue to alanine substantially inhibited the ability of GSTSmad2C to interact with Mixer both on and off DNA, whereas mutating Q364 to alanine had little effect (Figure 7A and B). Mutating A371, a surface residue in the loop joining α-helix 2 to strand β-8, to lysine also has a severe effect on the ability of GSTSmad2C to interact with Mixer, probably by distorting this critical loop (Figure 7A and B). Finally, the model predicts that the side chain of N381 in Smad2 forms a hydrogen bond to the carbonyl of I304 in the SIM (Figure 6B and C; Table I). Mutating N381 to alanine, however, had little effect on the interaction with Mixer (Figure 7A and B), suggesting that other interactions may compensate for the loss of this hydrogen bond. As a control, we demonstrated that there were no gross structural changes in the GSTSmad2C mutants, since they could all interact efficiently with Smad4 (data not shown).
Fig. 7. Substantiating the SIM–Smad2 model. (A and B) W368 in Smad2 is essential for binding the Mixer SIM. In vitro translated wild-type Mixer was analysed for its ability to interact with GSTSmad2C and point mutant derivatives in a bandshift assay using the MBS as a probe (A) or in a GST pull-down (B). In (A), GSTSmad2C derivatives were titrated over the range 12, 6 and 3 ng and the Mixer–DNA complex is indicated, as is the ternary complex with GSTSmad2C (arrow). In (B), bound 35S-labelled Mixer was visualized by SDS–PAGE and autoradiography. Lane 1 shows 20% input protein and lane 2 corresponds to the interaction between wild-type Mixer and GST to estimate non-specific binding. The amount of Mixer bound by each GSTSmad2C mutant was quantitated and normalized such that the binding of Mixer to WT GSTSmad2C was 100. (C) Peptides corresponding to the Mixer SIM or to the SARA SBD peptide disrupt the Mixer–GSTSmad2C interaction. In vitro translated Myc-tagged Mixer was incubated with MBS probe alone or with anti-Myc antibody or 3 ng of GSTSmad2C, as indicated. Wild-type (WT) or mutant (mut) SIM or SARA SBD peptides were used at the concentrations shown. Mixer complexed with the probe is indicated; ternary complex with GSTSmad2C or anti-Myc antibody (arrow). (D) Peptides corresponding to the Mixer SIM or to the SARA SBD peptide bind both Smad2 and phosphorylated Smad2, but only the SIM can bind a heteromeric complex of Smad2 and Smad4. The peptides used in (C) were immobilized and incubated with whole-cell extracts from uninduced or TGF-β-induced NIH 3T3 cells. Associated proteins were separated by SDS–PAGE and western blotted for Smad2/Smad3, phospho-Smad2 (P-Smad2) and Smad4 (upper panels). Whole-cell extracts were also blotted with the same antibodies as a control (lower panels).
The Mixer SIM and SARA SBD compete for binding to Smad2
We tested directly whether the Mixer SIM interacts with Smad2 in the same region as does the rigid coil of the SARA SBD, as predicted, using a peptide competition assay. A peptide corresponding to the SARA SBD disrupted the interaction between Mixer and GSTSmad2C in a bandshift assay, as seen by the disappearance of the supershifted ternary complex of Mixer–GSTSmad2C–DNA and reappearance of the Mixer–DNA complex with increasing amounts of SARA SBD (Figure 7C). In contrast, a mutated SARA SBD peptide containing mutations in four residues known to be critical for interaction with Smad2 (Wu et al., 2000) had no effect on the Mixer–GSTSmad2C–DNA complex (Figure 7C). As a positive control, the Mixer SIM peptide could efficiently disrupt the Mixer–GSTSmad2C interaction and a mutant Mixer SIM peptide could not (Figure 7C).
Thus, the SARA SBD peptide competes directly with the Mixer SIM for interaction with the Smad2 MH2 domain, confirming that they interact with a common binding pocket in Smad2.
The Mixer SIM and SARA SBD bind phosphorylated Smad2, but only the SIM interacts with active Smad2/Smad4 complexes
In the cell, SARA and the SIM-containing transcription factors interact with Smad2 sequentially. SARA binds unphosphorylated Smad2 at the membrane to recruit it to the receptor complex, and the SIM-containing transcription factors interact with phosphorylated Smad2 complexed with Smad4 in the nucleus. To understand the molecular details of these interactions, the wild-type and mutant peptides used in Figure 7C were immobilized on beads and incubated with whole-cell extracts from uninduced and TGF-β-induced NIH 3T3 cells to determine their ability to interact with endogenous unphosphorylated Smad2, phosphorylated Smad2 and, through this interaction, with Smad4 (Figure 7D). Both the SIM and SARA SBD interact with unphosphorylated Smad2 in extracts from unstimulated cells (top panel). Both also interact with phosphorylated Smad2, although this interaction was stronger for the SIM (second panel; Xu et al., 2000). However, only in the case of the SIM can we detect an interaction with Smad4 (third panel).
Thus, the SIM binds activated Smad2/Smad4 complexes, consistent with its role in transcription factors in recruiting these complexes to DNA to mediate transcriptional activation. However, the SARA SBD cannot bind activated Smad2/Smad4, although it does bind both unphosphorylated and phosphorylated Smad2, suggesting that a region of the SARA SBD (not present in the SIM) competes with Smad4 for interaction with Smad2. We confirmed this with the demonstration that overexpression of Smad4 further reduced the amount of phosphorylated Smad2 that can be pulled down by the SARA SBD peptide, but had no effect on the amount bound by the SIM peptide (data not shown; see Discussion).
Discussion
Functional definition of the SIM
We originally identified the SIM as a common Smad-interacting motif found in a subset of Mix family members, Mixer and Milk and members of an unrelated family of transcription factors, the Fasts (Germain et al., 2000). We have now dissected the SIM in the context of Mixer and used this information to functionally characterize other Mix family members. We have demonstrated that the SIM is actually a specialized case of a more generic proline-rich Smad interaction motif that also exists in the membrane-bound protein SARA, and we propose that the SIM and the rigid coil of the SARA SBD interact with a common binding pocket in the Smad2 MH2 domain.
Our data underline the importance of the conserved P-P-N-K-S/T-I/V core of the SIM for Smad2 interaction, the two most critical residues being P292 and N293. It is easy to rationalize this with our molecular model, as P292 is involved in hydrophobic interactions with Smad2, as well as forming a hydrogen bond from its carbonyl oxygen to W368 in Smad2. N293 is critical because it is involved in a hydrogen bond that holds the SIM in the correct conformation to bind Smad2. Mutating other amino acids in the core singly had a limited effect on Smad2 binding, but mutating them in pairs revealed their importance. The SIM does not extend N-terminally beyond the core, since mutations in N-terminal residues had little or no effect on Smad2 interaction. However, our data demonstrate that the SIM extends C-terminally beyond the core, as residues M300 and P305 are also critical. The molecular model provides an explanation for this (Table I) and also suggests that P306 and I307 are important for Smad2 interaction.
The molecular model, as well as the sequences of Mix family members that do not interact with Smad2, also suggests the importance of T297, P298 and N301 in the Mixer SIM–Smad2 interaction. The threonine and asparagine are involved in a hydrogen bonding network that also involves the carbonyl of residue 375 of Smad2, and the proline will facilitate formation of these hydrogen bonds (Figure 6B; Table I). The importance of this proline is highlighted by the inability to bind Smad2 of a Bix1 mutant in which the PPNKTI core has been created, but which still contains an alanine instead of the proline. Similarly, a Mix.1 mutant in which the core PPNKTI and methionine equivalent to M300 in the Mixer SIM have been resubstituted still fails to bind Smad2, due to the presence of a lysine and a tyrosine in place of T297 and N301, respectively.
The Mix family of transcription factors
We have demonstrated that only Xenopus Mix family members [i.e. Mixer, Milk (Bix2) and Bix3] that contain a recognizable SIM bind Smad2 and mediate TGF-β-induced transcriptional activation in vivo. Bix1 and Bix4 are distinct in that they are constitutively active and their activity is not significantly TGF-β inducible. Mix.1 has a low basal activity and is also not inducible. The different subgroups of Mix family members are likely to have distinct roles in vivo. All the family members are expressed at approximately the same time, in broadly similar regions of the Xenopus embryo (Rosa, 1989; Ecochard et al., 1998; Henry and Melton, 1998; Tada et al., 1998; Germain et al., 2000) and their synthesis is induced by Nodal family members (Xanthos et al., 2001). To understand their functions in vivo, it will be important to discover exactly where the proteins are expressed relative to each other and to identify their optimal binding sites and their target genes.
Zebrafish bon, like Xenopus Mixer, has very low inherent transcriptional activity and requires ligand-induced Smad2/Smad4 complexes to mediate transcriptional activation. Interestingly, bon shares only two regions of sequence identity with Mixer, the paired-like homeodomain and the SIM, suggesting that these are the only important functional domains. The requirement of bon to recruit activated Smad2/Smad4 complexes for transcriptional activation fits well with recent functional data from zebrafish, which indicated that bon requires active Nodal signalling for its function (Kikuchi et al., 2000). Since Nodal is the only known inducer of Smad2/Smad4 complexes in zebrafish embryos (Schier and Shen, 2000), this provides good evidence for the requirement of activated Smad complexes recruited via the SIM for Mixer/bon transcriptional activity in vivo.
Interaction of the SIM with Smad2: comparison with the binding of the SARA SBD
Significant sequence similarity between the rigid coil of the SARA SBD (Wu et al., 2000) and the SIM prompted us to model the SIM interaction with Smad2 using the SARA SBD–Smad2 structure. Extensive mutational analysis of both Smad2 and the SIM, as well as the peptide competition assays, strongly support the model. Both the SIM and the rigid coil region of the SARA SBD are conformationally constrained due to their high proline content and several key intramolecular hydrogen bonds. The rigid coil contacts Smad2 via hydrogen bonds and hydrophobic interactions. Our model explains the specificity of the SIM binding to Smad2 and not to Smad1 or Smad4 (Germain et al., 2000). Five of the seven residues in Smad2 that the model predicts directly interact with the SIM (I341, F346, Y366, W368 and N381) are unique to Smad2 and Smad3 and are not found in Smad1 or Smad4 (Figure 6D). The same five residues are responsible for the specificity of interaction of the SARA SBD with Smad2 (Wu et al., 2000), indicating that in the molecular model, although many of the side chains of the SIM are different to those in the SARA SBD, those important for interactions with the most critical residues in Smad2 are all conserved.
The sequence similarity between the SIM and the rigid coil of the SARA SBD led us to the view that they represented a common Smad interaction motif. However, the SIM binds Smad2 with higher affinity than does the rigid coil of the SARA SBD. The SARA SBD rigid coil together with the adjacent α-helix is not sufficient to bind Smad2 (Wu et al., 2000), whilst the rigid coil of the SIM is. Sequence comparison and molecular modelling suggest that it may be the absence of the hydrogen bonding network in the SARA SBD, involving residues T297 and N301 of the SIM and the carbonyl of residue 375 of Smad2, which might account for these differences (Figure 6B; Table I). In addition, C681 in SARA (equivalent to residue N301 in the SIM) appears to be suboptimal for Smad2 binding, since it is a polar residue in a hydrophobic pocket that can form no favourable interactions with any neighbouring residues (Wu et al., 2000).
The molecular model and the supporting data strongly suggest that the same hydrophobic pocket of Smad2 that binds the rigid coil region of the SARA SBD also binds the SIM. This pocket is well away from the putative interface between different Smads in the heterotrimer (Shi et al., 1997; Chacko et al., 2001), consistent with the ability of the SIM to bind activated Smad2/Smad4 complexes and its role in recruiting them to DNA. However, the SARA SBD is more extensive than the SIM, including an α-helix and β-strand, and our data now shed light on the role of this region of the SARA SBD. Our observation that, unlike the SIM, the SARA SBD binds phospho-Smad2, but not when complexed with Smad4, suggests that the residues in the region of the SARA SBD that are not similar to the SIM must be responsible for preventing interaction with Smad2/Smad4 complexes. The most likely region responsible is the β-strand, as it makes contacts with α-helix 5 and the adjacent strand β1′ of Smad2 (Wu et al., 2000) and residues in α-helix 5 are thought to be involved in contacts between Smad molecules in heterotrimers (Shi et al., 1997; Chacko et al., 2001). The role of SARA is to recruit unphosphorylated Smad2 (and Smad3) to the receptors for phosphorylation, which subsequently induces its dissociation from SARA (Tsukazaki et al., 1998; Xu et al., 2000). Our data suggest that the binding of Smad4 to activated Smad2 might play a role in Smad dissociation from SARA.
In conclusion, we propose that the rigid coil of the SARA SBD and the SIM can both be considered as related proline-rich Smad2-binding motifs that bind to a common shallow hydrophobic groove on Smad2. The proteins containing this motif have very different functions and exist in different compartments of the cell, but their common function is to interact specifically with Smad2 and we propose that they do so through a shared binding pocket.
Materials and methods
Plasmids
The following plasmids have been described: Mixer, Mixer(PP mut), Milk, Mix.1 and XFast-1 in pEF-Flag expression vectors (Germain et al., 2000), pGal4(1–95) (Sadowski and Ptashne, 1989), GSTSmad2C (Germain et al., 2000), EFLacZ (Bardwell and Treisman, 1994) and (DE)4–luciferase (Pierreux et al., 2000). Mixer, Milk and Mix.1 were subcloned into FTX9, a derivative of FTX5 (Howell and Hill, 1997) with the Flag tag replacing the Myc tag. The coding sequences of Bix1, Bix3, Bix4 (Tada et al., 1998) and bon (Alexander et al., 1999) were subcloned into pFTX9 and pEF expression vectors (Hill et al., 1995). pGal4(1–95)–SIM corresponds to the fusion of amino acids 283–307 of Mixer with Gal4 DNA-binding domain residues 1–95. pGal4 (1–95)–SIM(PP mut) is a mutant version of pGal4(1–95)–SIM with prolines 291 and 292 mutated to alanine. p(Gal4–OP)5–luciferase comprises five Gal4-binding sites fused to a luciferase reporter gene derived from pGL3-Enhancer (Promega). Point mutations in GSTSmad2C, full-length Xenopus Mixer and Mix.1 were made using PCR and are described in the text.
GST fusion protein purification, GST pull-downs and in vitro transcription and translation
Expression of GSTSmad2C and mutant GSTSmad2C fusion proteins and in vitro coupled transcription and translation in reticulocyte lysate were performed using standard methods. GST pull-down assays were as described (Germain et al., 2000), except those shown in Figure 7 where 100 ng of GST fusion protein were used and washes contained 250 mM NaCl.
Transfections, transcription assays and TGF-β induction
Maintenance of NIH 3T3 cells, transfection and transcription assays were as described (Pierreux et al., 2000). TGF-β1 (PeproTech) was used at 2 ng/ml. For transcription assays, cells were induced with TGF-β for 8 h; for all other assays, inductions were for 1 h.
Peptides and peptide pull-down assays
The wild-type SIM peptide and SIM mutant peptide, which contain an N-terminal biotin group, were as described (Germain et al., 2000). The SARA SBD peptide had the composition SQSPNPNNPAEYCSTIPPLQQAQASGALSSPPPTVMVPVGV and the mutant was SQSPNPNNPAEAESTIPELQQAQASGALSSPPPTAMVPVGV (Wu et al., 2000).
The SARA SBD peptides were biotinylated at the N-terminal amino group using EZ-Link™ NHS-LC-LC-Biotin (Pierce). Free biotin was removed using a D-Salt™ column (Pierce) and addition of a single biotin was confirmed by MALDI-TOF mass spectroscopy. Each of the peptides were attached to NeutrAvidin beads (Pierce) at saturation. Whole-cell extracts were made from uninduced or TGF-β-induced NIH 3T3 cells using buffer Y (Vastrik et al., 1999). The cell extract (400 µl) was incubated with 10 µl of peptide-conjugated beads for 1.5 h at 4°C, then washed four times with buffer Y. The beads were resuspended in SDS sample buffer and associated proteins analysed by western blotting for phospho-Smad2, Smad2 and Smad4.
Bandshift assays and IP western blots
Bandshift probes corresponding to the Mixer-binding site (MBS) of the DE- or the Gal4-binding site were generated by PCR (Germain et al., 2000) using the following oligonucleotides: 5′-CACCGTTAATCTG-3′ (MBS top) and 5′-CTAGCCATTAATCAGATTAACGGTG-3′ (MBS bottom) and 5′-GAATTCGAGCTCGTACCCGGGTCGGAGTACTGTCC-3′ (Gal4 top) and 5′-AAGCTTGCATGCCTGCAGTCGGAGGACAGTACTCCGACCCGGG-3′ (Gal4 bottom). Bandshift assays were as described (Germain et al., 2000). For in vitro translated Mix family members and mutants, bandshift conditions were as described (Wilson et al., 1993). IP westerns were performed as described (Pierreux et al., 2000). The antibodies used in bandshifts and westerns were anti-Flag, M2 (Sigma); anti-Smad2/3 (Transduction Laboratories); anti-Smad4, B8 and anti-Gal4 (Santa Cruz); anti-Myc, 9E10 and anti-phospho-Smad2 (a gift from Peter ten Dijke).
Molecular modelling of the Smad–Mixer interface
The Mixer SIM sequence was aligned with the rigid coil region of the SARA SBD and the alignment used to replace the side chains of SARA with those of the Mixer SIM in the Smad2–SARA SBD complex (Wu et al., 2000), using the modelling program 3D-JIGSAW (Bates and Sternberg, 1999). Side chain conformations were allowed to vary on both the SIM and Smad2, keeping the protein backbones of both fixed. To remove the small number of steric clashes, 100 steps of steepest descents energy minimization (all atoms unrestrained) were run using the program CHARMM (Brooks et al., 1983). The overall quality of side chain packing and stereochemistry of the final model were checked using QUANTA (Molecular Simulations software, version 3.3.); no bad clashes or poor side chain packing at the Smad2–SIM interface were found. The side chain conformations of Smad2 at the Smad2–SIM interface were mainly conserved between the SARA SBD and the SIM; the few exceptions being side chains at the edge of the interface, e.g. K375 and C380 in Smad2. All key side chain conformers on Smad2 at the interface, such as W368 and N381, were conserved, indicating that the backbone of the SIM only need undergo minor adjustments relative to the SARA SBD to maintain a similar binding energy.
Acknowledgments
Acknowledgements
We thank Didier Stainier for zebrafish Mixer, Jim Smith for Bix1, Bix3 and Bix4, Peter ten Dijke for anti-phospho-Smad2 antibody, Nicola O’Reilly for peptide synthesis and purification, and Nick Totty and Sarah Hanrahan for MALDI-TOF mass spectroscopy. We are very grateful to Mike Howell for useful discussions and insights, and to Mike Howell, Neil McDonald, Sara Nakielny, Malcolm Parker and Mark Uden for constructive comments on the manuscript. The work was funded by the Imperial Cancer Research Fund.
References
- Alexander J., Rothenberg,M., Henry,G.L. and Stainier,D.Y. (1999) Casanova plays an early and essential role in endoderm formation in zebrafish. Dev. Biol., 215, 343–357. [DOI] [PubMed] [Google Scholar]
- Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Bates P.A. and Sternberg,M.J. (1999) Model building by comparison at CASP3: using expert knowledge and computer automation. Proteins, 37, 47–54. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri,R.E., Olafson,B.D., States,D.J., Swaminathan,S. and Karplus,M. (1983) CHARMM: a program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem., 4, 187–217. [Google Scholar]
- Chacko B.M., Qin,B., Correia,J.J., Lam,S.S., de Caestecker,M.P. and Lin,K. (2001) The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nature Struct. Biol., 8, 248–253. [DOI] [PubMed] [Google Scholar]
- Chen X., Rubock,M.J. and Whitman,M. (1996) A transcriptional partner for MAD proteins in TGF-β signalling. Nature, 383, 691–696. [DOI] [PubMed] [Google Scholar]
- Chen X., Weisberg,E., Fridmacher,V., Watanabe,M., Naco,G. and Whitman,M. (1997) Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature, 389, 85–89. [DOI] [PubMed] [Google Scholar]
- Chen Y.G., Hata,A., Lo,R.S., Wotton,D., Shi,Y., Pavletich,N. and Massagué,J. (1998) Determinants of specificity in TGF-β signal transduction. Genes Dev., 12, 2144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecochard V., Cayrol,C., Rey,S., Foulquier,F., Caillol,D., Lemaire,P. and Duprat,A.M. (1998) A novel Xenopus Mix-like gene Milk involved in the control of the endomesodermal fates. Development, 125, 2577–2585. [DOI] [PubMed] [Google Scholar]
- Germain S., Howell,M., Esslemont,G.M. and Hill,C.S. (2000) Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev., 14, 435–451. [PMC free article] [PubMed] [Google Scholar]
- Henry G.L. and Melton,D.A. (1998) Mixer, a homeobox gene required for endoderm development. Science, 281, 91–96. [DOI] [PubMed] [Google Scholar]
- Hill C.S. (2001) TGF-β signalling in early Xenopus development. Curr. Opin. Genet. Dev., 11, 534–541. [DOI] [PubMed] [Google Scholar]
- Hill C.S., Wynne,J. and Treisman,R. (1995) The Rho family GTPases RhoA, Rac1 and CDC42Hs regulate transcriptional activation by SRF. Cell, 81, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Howell M. and Hill,C.S. (1997) XSmad2 directly activates the activin-inducible, dorsal mesoderm gene XFKH1 in Xenopus embryos. EMBO J., 16, 7411–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M., Itoh,F., Pierreux,C.E., Valgeirsdottir,S., Itoh,S., ten Dijke,P. and Hill,C.S. (1999) Xenopus Smad4β is the co-Smad component of developmentally-regulated transcription factor complexes responsible for induction of early mesodermal genes. Dev. Biol., 214, 354–369. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Trinh,L.A., Reiter,J.F., Alexander,J., Yelon,D. and Stainier,D.Y. (2000) The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev., 14, 1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Massagué J. and Wotton,D. (2000) Transcriptional control by the TGF-β/Smad signaling system. EMBO J., 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama N., Hanafusa,H., Kusakabe,M., Shibuya,H. and Nishida,E. (1999) Identification of two Smad4 proteins in Xenopus. Their common and distinct properties. J. Biol. Chem., 274, 12163–12170. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Pierreux C.E., Nicolás,F.J. and Hill,C.S. (2000) Transforming growth factor-β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol., 20, 9041–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F.M. (1989) Mix.1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell, 57, 965–974. [DOI] [PubMed] [Google Scholar]
- Sadowski I. and Ptashne,M. (1989) A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res., 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A.F. and Shen,M.M. (2000) Nodal signalling in vertebrate development. Nature, 403, 385–389. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2001) Structural insights on Smad function in TGFβ signaling. BioEssays, 23, 223–232. [DOI] [PubMed] [Google Scholar]
- Shi Y., Hata,A., Lo,R.S., Massagué,J. and Pavletich,N.P. (1997) A structural basis for mutational inactivation by the tumour suppressor Smad4. Nature, 388, 87–93. [DOI] [PubMed] [Google Scholar]
- Tada M., Casey,E.S., Fairclough,L. and Smith,J.C. (1998) Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development, 125, 3997–4006. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Miyazono,K. and Heldin,C.H. (2000) Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci., 25, 64–70. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T., Chiang,T.A., Davison,A.F., Attisano,L. and Wrana,J.L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGF-β receptor. Cell, 95, 779–791. [DOI] [PubMed] [Google Scholar]
- Vastrik I., Eickholt,B.J., Walsh,F.S., Ridley,A. and Doherty,P. (1999) Sema3A-induced growth-cone collapse is mediated by Rac1 amino acids 17–32. Curr. Biol., 9, 991–998. [DOI] [PubMed] [Google Scholar]
- Wilson D., Sheng,G., Lecuit,T., Dostatni,N. and Desplan,C. (1993) Cooperative dimerization of paired class homeo domains on DNA. Genes Dev., 7, 2120–2134. [DOI] [PubMed] [Google Scholar]
- Wu G., Chen,Y.G., Ozdamar,B., Gyuricza,C.A., Chong,P.A., Wrana,J.L., Massagué,J. and Shi,Y. (2000) Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science, 287, 92–97. [DOI] [PubMed] [Google Scholar]
- Xanthos J.B., Kofron,M., Wylie,C. and Heasman,J. (2001) Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development, 128, 167–180. [DOI] [PubMed] [Google Scholar]
- Xu L., Chen,Y.G. and Massagué,J. (2000) The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nature Cell Biol., 2, 559–562. [DOI] [PubMed] [Google Scholar]