Abstract
The growth in popularity of RNA expression microarrays has been accompanied by concerns about the reliability of the data especially when comparing between different platforms. Here, we present an evaluation of the reproducibility of microarray results using two platforms, Affymetrix GeneChips and Illumina BeadArrays. The study design is based on a dilution series of two human tissues (blood and placenta), tested in duplicate on each platform. The results of a comparison between the platforms indicate very high agreement, particularly for genes which are predicted to be differentially expressed between the two tissues. Agreement was strongly correlated with the level of expression of a gene. Concordance was also improved when probes on the two platforms could be identified as being likely to target the same set of transcripts of a given gene. These results shed light on the causes or failures of agreement across microarray platforms. The set of probes we found to be most highly reproducible can be used by others to help increase confidence in analyses of other data sets using these platforms.
INTRODUCTION
The success of gene expression microarray technology has led to the production of multiple array platforms differing in the kind of probes used (short-oligonucleotide, long-oligonucleotide, cDNA, etc.), the hybridization paradigm (competitive versus non-competitive), the labeling method and the production method (in situ polymerization, spotting, microbeads, etc.). The diversity of microarray platforms has made it challenging to compare data sets generated in different laboratories, hindering multi-institutional collaborations and reducing the usefulness of existing experimental data. When comparing gene expression studies, we not only have to consider the interesting biological factors but a plethora of technical factors including diverse sample handling, target preparation and data processing methods, as well as microarray platform choice. In order for the benefits of comparisons between two laboratories to be realized, it is crucial to understand the benefits and limitations of each platform used as well as the cross-platform comparability.
At this writing, most published gene expression studies use Affymetrix GeneChips, spotted cDNA arrays or spotted long-oligonucleotide arrays. However, new approaches are still evolving, such as the recently introduced long-oligonucleotides bead-based array by Illumina, Inc. (1). Arrays produced by Affymetrix are fabricated by in situ synthesis of 25mer oligonucleotides (2) while the Illumina process involves using standard oligonucleotide synthesis methods as is used for spotted long-oligonucleotides arrays. However, on Illumina arrays the oligonucleotides are attached to microbeads which are then put onto microarrays using a random self-assembly mechanism (1). In addition to the difference in oligonucleotide physical attachment, the two platforms are also very different in probe selection and design procedure. Affymetrix uses multiple probes for each gene along with one-base mismatch probes intended as controls for non-specific hybridization. In contrast, the randomly generated Illumina arrays yield on the order of 30 copies of the same oligonucleotide on the array, which provide an internal technical replication that Affymetrix lacks. In addition, the Affymetrix arrays are constructed in a specific layout, with each probe synthesized at a predefined location (2), while individual Illumina arrays must undergo a ‘decoding’ step in which the locations of each probe on the array are determined using a molecular address (1). A final difference between the platforms is that in the current packaging, multiple Illumina arrays are placed on the same physical substrate, meaning that hybridization and other steps are performed in a parallel manner, while Affymetrix arrays are processed separately.
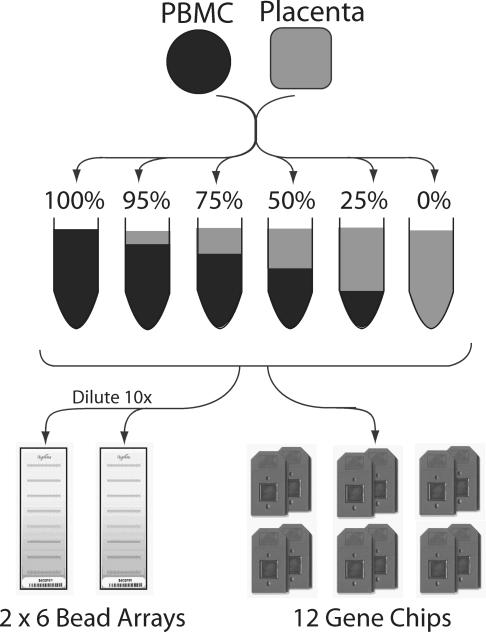
This paper details results from an experiment comparing Affymetrix HG-U133 Plus 2.0 microarrays with the Illumina HumanRef-8 BeadArrays. We used a dilution design, where two different RNA samples are mixed at known proportions, and the same RNA is analyzed in duplicate on each platform (see Figure 1). The advantage of this design is that it is very simple to prepare and the number of differentially expressed genes is large, allowing testing of many of the probes on each array. However, in contrast to spike-in studies, the identities of the genes expected to show differential expression are not definitely known ahead of time.
Figure 1.
Schematic representation of the experimental design. See Materials and Methods for details. The key features of the design are the use of a single pair of RNA samples for all analyses, mixed together in varying proportions and analyzed in technical replicates on each platform. Unlike the Affymetrix platform, each Illumina BeadArray slide contains multiple arrays, allowing us to analyze a complete dilution series on one slide. Note that the BeadArray slides actually contain eight arrays per slide, but we only used six for the data described here.
Comparisons between long and short oligonucleotide arrays have been carried out in the past for other array types (3–7). To our knowledge, this study is the first to examine the comparison between in situ synthesized oligonucleotide arrays with bead-based oligonucleotide arrays. Our results show that these two completely different microarray technologies yield, on the whole, very comparable results. This is especially true once the factors of gene expression level and probe placement on the genome are considered.
MATERIALS AND METHODS
Samples, hybridization and quality control
Following informed consent (approved by Cincinnati Children's Hospital Medical Center Internal Review Board), ∼50 ml whole blood was collected from 30 adult, apparently healthy, volunteers using Acid Citrate Dextrose as an anti-coagulant. Samples were processed immediately following blood draw. Cells were isolated by Ficoll gradient centrifugation and total RNA was isolated using Trizol (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's protocol, aliquoted and stored at −80°C until use. Whole placenta was collected and immediately frozen in liquid nitrogen. RNA was extracted using TRI reagent, purified using RNeasy columns (Qiagen, Valencia, CA), aliquoted and stored at −80°C until use. RNA mixtures (100:0, 95:5, 75:25, 50:50, 25:75, 0:100; PBMC: placenta) were prepared in single aliquots. Portions of each aliquot were split in half for Affymetrix analysis as technical replicates. Another portion was diluted 1:10 to account for differences in RNA concentration used by each platform and then split into two samples for Illumina analysis as technical replicates. These four final samples were sent to the respective facilities for further processing as described below. Quality was assessed for each sample using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) at their respective core facilities according to internal quality control measures.
For Affymetrix microarray analysis, samples were run in the CCHMC Affymetrix core facility. Biotinylated cRNA was synthesized from total RNA (Enzo, Farmingdale, NY). Following processing according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA), labeled cRNA was quantitated by hybridization to Affymetrix U133plus2.0 GeneChips.
For Illumina microarray analysis, samples were prepared and analyzed in Illumina laboratories by Illumina personnel. Biotinylated cRNA was prepared using the Illumina RNA Amplification Kit (Ambion, Inc., Austin, TX) according to the manufacturer's directions starting with ∼100 ng total RNA. Samples were purified using the RNeasy kit (Qiagen, Valencia, CA). Hybridization to the Sentrix HumanRef-8 Expression BeadChip (Illumina, Inc., San Diego, CA), washing and scanning were performed according to the Illumina BeadStation 500× manual (revision C). Two BeadChips were used, each one containing eight arrays, so that each dilution series of six samples was run on an individual BeadChip.
Data extraction and normalization
Affymetrix data were extracted, normalized and summarized with the RMA method from Bioconductor's ‘affy’ package (8,9), using the default settings. The Illumina data were extracted using software provided by the manufacturer. Pilot studies indicated that background subtraction had a negative impact on the Illumina data quality, so we used data that had not been background subtracted. The Illumina data were then normalized using the ‘normalize.quantiles’ function from the ‘affy’ package. Other normalization methods yielded similar results (data not shown).
Probe annotation
Manufacturer's annotations for the Affymetrix platform were downloaded from the NetAffx web site (https://www.affymetrix.com/analysis/netaffx/) on March 15, 2005. Illumina also provided a table of annotations. We supplemented these annotations with our own sequence analysis based on comparing sequences with the Human genome sequence (assembly hg17, July 2004). For Affymetrix, we merged and joined the individual probe sequences to form a ‘pseudo-target’ sequence; we found that aligning these to the genome was much more effective and efficient than attempting to align individual probes or using the Affymetrix ‘target’ sequences (the merging procedure is depicted in a Supplementary Figure). For Illumina the input sequences were the 50 bp oligonucleotide sequences provided by the manufacturer. All sequences used are provided as Supplementary Data or are available from the manufacturers.
Each sequence was compared with the genome sequence using BLAT (10) with minimum score set to 20 and an initial minimum identity set to 0.5 (all other parameters were left to the default setting). Each hit was then given an overall score equal to (m − g)/s, where m is the number of matches, g is the number of gaps in the alignment, and s is the size of the query sequences. This score differs slightly from the default in the GoldenPath genome browser in that the gap penalty is lower, but we found it gave us higher sensitivity when aligning shorter sequences and permits good gapped alignments of the collapsed Affymetrix sequences. A threshold of 0.9 applied to this score yielded multiple BLAT hits for many of the probes. These hits were associated with genes as follows. The location of each hit was compared with the ‘refGene’ and ‘knownGene’ tables in the hg17 Golden Path database (11). A BLAT hit overlapping or falling within the annotated limits of a gene (on the correct strand) was retained as an initial hit. Each BLAT hit was further scored based on two criteria. First, the fraction of bases which overlapped with annotated exons or mRNAs (as represented in the hg17 database tables knownGene, refGene and all_mrna). Second, we computed the distance of the 3′ end of the BLAT hit from the 3′ end of the annotated transcript (using the center of the BLAT hit made no difference in the conclusions; see Supplementary Data).
The final ‘best’ match for a probe was the transcript closest to the probe's 3′ end and with the largest non-intronic overlap. This means that probes falling entirely within introns were given similarity scores of zero, and when there were two alternative 3′ ends for a gene, the one with the 3′ end nearest to the probe was selected as the targeted gene. In cases where there were two or more equivalent ‘best’ hits to different sites in the genome (i.e. a tie), one was arbitrarily chosen (247 cases for Affymetrix, 231 cases for Illumina). These often represented alignments to sequences duplicated in the assembly (e.g. parts of chromosome 1 and chromosome 1_random; ∼10% of cases). Other ties often involved closely related genes, probably reflecting duplications (e.g. CGB and CGB5).
To analyze the relationship of cross-platform agreement with probe location, the distance between two probes was measured as the distance between the centers of their alignments on the genome.
Investigations where we varied these parameters or methodologies did not change our main conclusions, though the results for individual probes are naturally affected by the exact criteria used. The full sets of annotations we derived are available as Supplementary Data, along with additional details of the results of the BLAT analysis. A summary of the annotations used in this study is given in Table 1.
Table 1.
Summary statistics for array designs studied, comparing two annotation methods
| Array | Probes or probe sets | Probes for ‘known’ genesb | Known genes assayed | Unassigned probesb | Probes/gene (average) |
|---|---|---|---|---|---|
| Illumina (Mfg)a | 24 114 | 17 143 | 14 420 | 6971 | 1.19 |
| Illumina (BLAT) | 19 924 | 16 847 | 2978c | 1.18 | |
| Affymetrix (Mfg) | 54 675 | 34 089 | 16 610 | 20 586d | 2.05 |
| Affymetrix (BLAT) | 33 922 | 18 417 | 20 600c | 1.84 |
aMfg, Manufacturer's annotations; BLAT, our own annotations computed using BLAT alignments to the genomic sequence.
b‘Known genes’ are genes identified in the GoldenPath ‘refGene’ or ‘knownGene’ tables, including transcript information from the ‘all_mrna’ table to determine exon overlaps. We designate all other potential transcripts ‘unassigned probes’.
cIncludes probes not yielding BLAT results.
dIncludes 14 334 probes where no gene name is listed by the manufacturer.
Clustering
For clustering only, the data matrices for each platform were adjusted so the probe expression vectors had a mean of zero and variance one. A combined data matrix was constructed such that each probe for a gene on one platform was used to form new combined expression vectors with each probe for the same gene on the other platform. This means that if a gene appeared twice on each platform, a total of four new expression vectors were constructed. This final matrix had 36 024 rows (approximately a factor of two over what would have been obtained had we averaged the probes for each gene). The rows of this matrix were subjected to hierarchical clustering using XCluster (http://genetics.stanford.edu/~sherlock/cluster.html), with average linkage and Euclidean distance. The results were visualized with matrix2png (12).
Statistical analysis
Analyses were carried out in the R statistical language or using custom Java programs. The dilution profile was described as a simple factor in a linear model used to fit each gene. The correlation of each gene expression profile was used as a statistic for further analyses; P-values for the deviations from a correlation of zero were computed using standard normal assumptions. To establish statistical thresholds via false discovery rate (FDR) analysis (13), we used the ‘qvalue’ R package with default settings (14). To compare profiles across platforms, the Pearson correlation was used on non-log transformed data (the RMA data were transformed back from log2), though the Spearman rank correlation yielded very similar results (Supplementary Data). P-values for the test of the null hypothesis that a Pearson correlation was equal to zero were tested using the ‘cor.test’ function from R. As an alternative to presenting scatter plots of comparisons (which are available as Supplementary Data), we have presented results as stratified histograms. The thresholds for stratification were determined by inspection or from the statistical testing, and alternative reasonable thresholds do not change our findings. All R scripts and data files used in the analyses are available from the authors.
Tissue-specific genes
A search was performed of UniGene's EST database using the digital differential display tool to identify genes with differential expression (Fisher Exact Test, P < 0.05). A comparison was made between placenta genes (library IDs 13037, 13021, 10404, 10403, 10425, 10424 and 10405) and blood, lymphocytes and lymph nodes (library IDs 13050, 1317, 7038, 7037 and 10312).
RESULTS
Initial characterization
To analyze the ability of each platform to yield reproducible and accurate results, we used a dilution design, outlined in Figure 1 and detailed in Materials and Methods. An initial exploratory overview of the properties of the data is shown in Figure 2, which shows the results of hierarchical clustering of all genes that could be matched across platforms. It is apparent that there are many probes which show strong dilution effects, being overexpressed either in blood or in placenta, on both platforms. Furthermore, there are large numbers of probes which clearly agree across platforms. However, it is also possible to identify clusters of probes which seem to show dilution effects on one platform but not on the other (Figure 2, light bars). The rest of the results we describe first considers the dilution effect we observe within each platform and then the comparison across the platforms.
Figure 2.
Hierarchical clustering results of all 36 024 comparable pairs of probes. The dilution step is shown as a graph at the top of the figure (Blood/Placenta). Black bars at the side indicate large clusters of genes that appear to show clear dilution effects in both platforms. Gray bars indicate examples of clusters that appear to show dilution effects in one platform but not consistently in the other. Lighter colors indicate higher relative levels of expression on an arbitrary scale. Note that in this figure, if a gene occurs multiple times on one platform, it is shown in all possible valid comparisons with matching probes on the other platform.
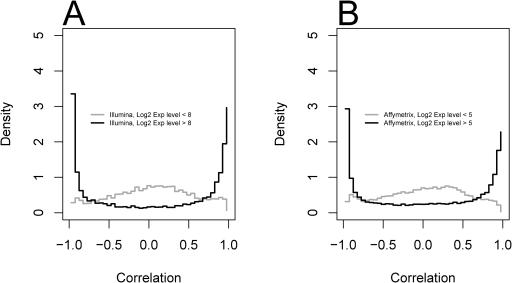
To quantify differential expression between placenta and blood, we measured the linear correlation of the designed dilution pattern to the expression pattern of each probe on each microarray (see Materials and Methods). We expected that large numbers of genes would show an effect of mixing of the RNA samples on their relative expression levels, while other genes expressed at equal levels (or not expressed) would not show such a pattern. Figure 3A and B shows the distributions of correlations for the two platforms. Both show pronounced peaks near correlations of −1 and 1, apparently reflecting probes whose targets are differentially expressed between the two samples. A ‘hump’ of points around a correlation of zero reflects probes which do not show a dilution effect. The Affymetrix and Illumina platforms yielded 35 and 33% of probes with very high dilution effects (absolute value correlations of greater than 0.8), respectively. In terms of tests of the null hypothesis that RNA concentration was not affected by dilution, 56% of Affymetrix and 50% of Illumina probes show significant effects [at an FDR < 0.05; the threshold correlations at this FDR are 0.53 (Affymetrix) and 0.55 (Illumina)].
Figure 3.
Distributions of correlations for the Illumina HumanRef-8 BeadArrays (A) and the Affymetrix HG-U133 Plus 2 arrays (B). Correlations near −1 and 1 reflect probes whose targets are differentially expressed between the two samples. Correlations near zero reflect probes which do not show a dilution effect. The dilution effect is more pronounced for probes targeted at ‘known’ genes. This effect is stronger for Illumina (C) than for Affymetrix (D), though the Illumina platform has fewer probes which cannot be assigned to known genes (Table 1). For complete data see the Supplementary Data.
We hypothesized that analyses focused on well-characterized genes would tend to yield better results. Other probes might target predicted genes that turn out to be false positives or pose extra challenges in probe design. Indeed, as shown in Figure 3C and D, probes targeted at known (annotated in RefSeq or in the ‘Known Gene’ table of the Golden Path database) genes have a stronger tendency to show a clear dilution effect. This is apparent by the lessening of the central hump in the known genes and its accentuation in the other probes. The effect appears to hold on both platforms, though the Affymetrix platform has many more probes which target other sites in the genome not containing annotated genes (‘unassigned’ probes, Table 1). Despite the overall difference from the ‘known genes’, many of these probe sets do yield strong correlations with the dilution profile, as evidenced by the smaller peaks near 1 and −1, suggesting that they have biologically meaningful targets (Figure 3C and D).
The genes that show no or weak dilution effects might sometimes correspond to genes that are not well measured. This is because noise will have a stronger influence on their measurement, making detecting a dilution effect difficult. Genes that are not expressed at all in either tissue studied will (by definition) be purely noise. Indeed, as shown in Figure 4, expression level is strongly associated with the measurement of a dilution effect on both platforms: many of the probes not showing dilution effects are expressed at lower levels, compared with the probes showing strong effects. As might be expected, there is an interaction of this effect with the known/unassigned gene distinction, and probes without known gene assignments tend to show lower expression levels (Supplementary Data).
Figure 4.
Distributions of correlations stratified by high and low expression levels (log2) for the Illumina HumanRef-8 BeadArrays (A) and the Affymetrix HG-U133 Plus 2.0 arrays (B). On both platforms, the probes not showing dilution effects tend to express at low levels, whereas highly expressed probes show strong dilution effects. For complete data see the Supplementary Data.
Comparisons between the platforms
The key question we wanted to answer was whether these two platforms, measuring the same samples, yield the same results. We therefore identified probes which map to common genes between the platforms. Based on our own sequence analysis (Table 1 and Materials and Methods), we identified 28 383 Affymetrix probe sets and 17 711 Illumina probes that could be matched to a common gene (83 and 89% of probes mapped to genes), covering 14 929 known genes altogether. If there was more than one probe(set) for a given gene, when doing comparisons we considered all possible combinations (to avoid repetition, we will use the term ‘probe’ even when we mean Affymetrix ‘probe set’, unless stated explicitly). Thus, if both platforms had two probes for a single gene, there were four comparative values generated. Note that many probes are not specific for a single transcript of a gene. Probes that could not be matched across platforms represent probes which did not map to a ‘known’ gene, or to genes represented on only one platform.
A simple hypothesis is that the actual profile of gene expression should agree across platforms for all probes which can be matched to a common gene. As shown in Figure 5A, there is a remarkable level of agreement for many probes by this measure (Pearson correlation was used for this analysis; similar results are obtained with the rank correlation, see Supplementary Data). However, a large population of genes shows poor correlations (peak near zero in Figure 5A). At an FDR of 0.05, 37% of the cross-platform comparisons result in rejection of the null hypothesis of no correlation (the threshold correlation to achieve significance is ∼0.56).
Figure 5.
Cross-platform agreement for all ‘known’ genes (A), stratified by differential expression (B) and for placenta/blood specific genes (C). For complete data see the Supplementary Data.
A more refined analysis takes into account the fact that genes that are not differentially expressed between the two tissues would present noisy expression profiles that would not be predicted to be reproducible across platforms. Figure 5B suggests that a large fraction of the ‘failures’ of the platforms to agree can be accounted for by probes which show a weak or no dilution effect. Indeed, if we first filter the data to remove comparisons between probes, of which at least one do not show a significant dilution effect (FDR 0.05, removing 48% of the comparisons), rejection of 88% null hypotheses yields an FDR of <0.05 (i.e. 88% of the remaining comparisons are significant). Using a more stringent Bonferroni correction on this filtered data, 23.7% of the comparisons are considered significant at an alpha of 0.05, compared with 8.4% for the unfiltered data. This enrichment shows that when the dilution effect is considered, the agreement between the platforms rises substantially.
A difficulty with the analysis shown in Figure 5B and described above is that it relies on the arrays themselves to identify genes that might show a differential expression effect: an independent ‘gold standard’ would be desirable. While we are not aware of any large validated set of placenta- or blood-specific genes, from public databases we were able to obtain a set of 174 genes that are predicted to be placenta or blood specific (see Materials and Methods), and should therefore show a strong dilution effect. As shown in Figure 5C, these genes show excellent agreement across the platforms, with many fewer disagreements than the data considered at large (Figure 5A). Very similar results overall were obtained when using annotations provided by the manufacturers (Supplementary Data).
As mentioned, the level of expression would be an important factor in making a good comparison: if a gene is simply not present in the samples, the measurements will be just noise, and we do not expect noise to be similar across platforms (by definition). More generally, we expect higher expression levels to be associated with less noisy measurements, and therefore would yield better agreement across platforms. That this is indeed the case is shown in Figure 6A; the rank correlation of expression level to measure cross-platform agreement is 0.37–0.43 (depending on whether the Illumina or Affymetrix expression levels, or their means, are used for evaluation). Genes that are expressed at low levels are not as likely to be reproducible across platforms. This effect is maintained even after filtering out genes that show no or weak dilution effects as described above (comparisons were retained if at least one probe showed a significant dilution effect at an FDR of 0.05; Supplementary Data). This is because many of the weakly expressed probes do show (apparent) dilution effects.
Figure 6.
Cross-platform agreement measured by the rank correlation of expression levels as a function of expression level (log2) (A) and distance between probes in base pairs (B). For complete data see the Supplementary Data.
We further hypothesized that for two probes to agree across platforms, they should be measuring the same biological entity (transcript or set of transcripts). While our annotation system tries quite hard to identify which transcript or set of transcripts a probe is likely to hybridize to, thereby identifying cases where we believe the platforms are measuring the same RNA, the resolution of this approach is limited. Specifically, probes that are expected to hybridize to different parts of the same transcript might yield different signals. This could be due to differential detection of degraded messages, or limitations in annotations. As an example of the latter situation, if one probe targets a previously unannotated transcript while another does not, our system might measure them as assaying the same transcripts while there is in fact a difference.
To test this idea in a simple way, we plotted the relationship between cross-platform agreement and cross-platform probe spacing. As shown in Figure 6B, when Affymetrix and Illumina probes align to very close or overlapping locations in the genome, they have a tendency to agree more, whereas probes that hybridize to distinct locations, even along the same gene, tend to disagree more. Compared with the effect of expression level, the effect is small though still highly statistically significant, with a rank correlation of 0.18. However, this conclusion is complicated by the fact that expression level is also affected by distance from the 3′ end (rank correlation −0.15), so the measure of probe location difference is not independent of the level of expression. If we analyze only probes that have higher expression levels (e.g. Affymetrix expression level log2 > 7), the effect of location on agreement is enhanced slightly (rank correlation 0.25), indicating that the effect cannot be completely attributed to associated differences in expression level. As for the effect of expression level, the effect remains after removing probes which failed the dilution effect filter.
Within-platform reproducibility
Some additional insight into the reproducibility problem comes from looking at reproducibility within each platform. On the Affymetrix platform especially, there are often multiple probes per gene (Table 1). It is expected that in many cases this redundancy is intended to address transcript diversity within a gene, but these data points provide a situation where we can assess impact on reproducibility of the same parameters discussed so far (expression level and probe placement) while disregarding platform differences. We hypothesized that reproducibility within each platform (for those genes with multiple probes predicted to target them) would show the same trends as reproducibility across platforms. We first found that within-platform ‘reproducibility’ was substantially lower on the Illumina array than for Affymetrix or between-platform reproducibility (9.5% of 4312 correlations over 0.8 on Illumina; 27.2% of 30 384 comparisons for Affymetrix, compared with 24% between platforms; see Supplementary Data for details). We interpret this as indicating that the Affymetrix array contains more probe sets that are ‘truly redundant’, at least as reflected in our tissue samples. We also confirmed that the expression level and probe location appear to play a similar role in reproducibility within platforms as they do between platforms, i.e. two probes targeting the same gene within a platform were more likely to yield concordant results if they exhibited stronger expression and were targeting nearby sites in the genome (see Supplementary Data for details).
‘Unexplained’ cases of disagreement
There are probes which, based on dilution effect, location and expression level criteria, would be predicted to yield reproducible results, but do not. This suggests that other factors are influencing the results, and further examination of these cases is warranted. To examine this in more detail, we sought to identify provisionally ‘unexplained’ cases of disagreement by filtering the full set of results, using partly arbitrary criteria. We removed genes for which the 3′ ends of the probes were located further apart than 100 bases (similar in size to the average human exon) between the two platforms. We then filtered out probes which were expressed at low levels on both platforms (medians below the 25th percentile). Finally, we removed pairs which showed good agreement across platforms (as these need no further explaining), setting a maximum correlation threshold of 0.5 (close to that which maintained an FDR of 0.05), and also required that at least one of the probes show a strong dilution effect (again using the threshold of 0.5, but as a lower limit). This leaves a set of 940 pairs of probes for further study, or ∼3% of all comparisons. We do not suggest this set represents all the disagreements, just a subset of harder-to-explain disagreements.
By far the most striking disagreements among the selected probes are those exhibiting strong negative correlations across platforms, i.e. both platforms indicate a strong dilution effect for the gene, but in the opposite direction. There are 41 pairs of probes in this set which show cross-platform rank correlations of <−0.5. These probes appear wholly unremarkable based on the parameters we have focused on (expression level and location), compared with the 899 other probe pairs (the complete list of the 940 probe pairs are provided as Supplementary Data). Most disagreements are more subtle. If one focuses on probe pairs that show cross-platform correlations of <0.2, the number of selected probes is reduced by about a factor of two (that is to say, quite a few of the ‘unexplained disagreements’ involve marginal cases with correlations between 0.2 and 0.5).
To attempt to further explain the 940 cases, we first hypothesized that despite having similar genomic locations of the centers of the targeted sequences, there might be larger differences in the sequences assayed on the two platforms. We considered this plausible because Affymetrix probe sets assay more sequence and often include probes spread fairly widely (a median of 481 genomic bases from the 5′ end of the 5′-most probe to the 3′ end of the 3′-most probe) compared with the Illumina platform, which use a single 50 bp probe that almost always maps to 50 contiguous bases in our analysis. This could lead to different populations of transcripts being assayed in some cases. However, there was no significant enrichment in alignments shorter or longer than the median in the set of 940 pairs (P > 0.05, Fisher's exact test), suggesting that there is no overall pattern of alignment statistics that can explain the relatively anomalous behavior of these 940 pairs.
Next, we counted the number of different transcripts predicted to be hybridized by each probe (assuming for the moment that all RNAs are equally likely to be detected, regardless of 3′ location of the probe). If these values are different on the two platforms, then agreement may be lower if the predicted transcripts are indeed present in the tissues we studied. However, there was no overall difference between Affymetrix and Illumina in the number of transcripts assayed among the set of 940 (P ∼ 0.3, paired t-test).
A potential remaining source of ‘disagreement’ could be differential cross-hybridization. Similar to transcript specificity, cross-hybridization is difficult to analyze computationally because it could involve platform-specific differences that might not be reflected in the probe sequences alone (e.g. synthesis efficiency on the Affymetrix platform, or the effect of the probe identification sequences and linker for Illumina probes), and other unknowns such as the impact of highly expressed but weakly cross-hybridizing transcripts. There are ∼250 probes on each platform that have very high potential for cross-hybridization based on our sequence analysis (see Materials and Methods). If anything these are slightly underrepresented among the 940 strongest disagreements (Fisher's exact test, P = 0.036, Illumina; 0.07, Affymetrix). We also note that on the Affymetrix platform, where the manufacturer ‘flags’ probe sets with the potential for various types of non-specificity, there is no difference in the proportion of flagged probe sets among the 940 compared with the rest of the probes (38 versus 37%).
We performed a similar analysis for the within-platform analyses. Within-platform reproducibility showed many fewer hard-to-explain failures of reproducibility. For the Illumina platform, application of the same filter that yielded 940 pairs of probes (2.6% of the total) for the cross-platform comparison yielded 10 pairs of probes (0.2%), while for Affymetrix it yielded 103 pairs (0.3%). Interestingly, 3 of these Illumina probes and 37 of the Affymetrix probes appear in the list of 940 probes involved in poor ‘unexplained’ cross-platform comparisons. This enrichment is highly significant (P = 0.0039 and P < 10−15 for Illumina and Affymetrix, respectively). Our interpretation of this finding is that these probes are somehow inherently ‘poorly behaved’ and we predict that they will not yield biologically useful results.
DISCUSSION
Our main conclusion from this study is that the Affymetrix and Illumina platforms yield highly comparable data, especially for genes predicted to be differentially expressed. Beyond this conclusion, two more specific findings we wish to highlight in the discussion are that expression level plays a major role in determining reproducibility across platforms, and that the precise location of the probe on the genome affects the measurements to a substantial degree, such that two probes which do not map to the same location cannot be assumed to be measuring the same thing. When these two factors are taken into account, the agreement of the results across platforms is very high, though still not perfect.
Contrary to our general findings, a number of groups have found that concordance of results across expression analysis platforms is low (4,5,15–18). The reason for the discordance between such findings and ours, as well as that of a number of other groups (19–26) is not always clear, especially as in some cases the data are not publicly available. At least one group (24) has reported higher reproducibility than in a previous analysis of the same data (15), suggesting that data treatment and choice of comparison metric plays a role. One group reported high reproducibility of Affymetrix and long oligonucleotide arrays (which share similarities to the BeadArrays in the type of sequence used), but not of cDNA arrays (21), suggesting that there could be real differences in the reproducibility of different platforms, and that arrays based on long clones may have particular problems with specificity (16,17).
Our study reinforces the idea that a failure to consider annotations and expression levels sufficiently carefully can help explain some of the observed differences. For example, we note that Tan et al. (who compared three platforms) (4) relied on GenBank or UniGene identifiers to match genes across platforms. We believe this approach may be unsuitable for high-sensitivity comparisons across platforms, because of the coarseness of resolution of UniGene or GenBank IDs compared with the actual probes used on the arrays. The citation of the particular accession number only indicates the source of the probe sequence and does not imply that that GenBank sequence is specific for a particular gene. In extreme cases, the GenBank accession number referenced by the manufacturer includes multiple genes. Thus, even when two manufacturers cite the same GenBank accession number, there is no guarantee that the same transcripts are being assayed. For this reason, we have discarded identifier cross-references as a primary means of matching probes across platforms. Tan et al. (4) also do not document any consideration of the impact of expression level on agreement. In contrast, Park et al. (5) reported both an expression level and a probe specificity effect. It will be of interest to re-examine other cases of poor agreement across platforms in light of such considerations.
In our study, we identified thousands of probes on both platforms which show extremely good confirmation of results. In contrast to studies where a few results are checked by quantitative PCR, we have built-in cross-validation of a huge fraction of the results of the experiment. This set of cross-validated probes, though identified using conservative criteria on only one set of samples, could be considered as a starting point for identifying probes that perform well on these platforms. It is likely that many other probes on both platforms also perform well, but could not be evaluated due to insufficient signals in the tissues we studied. The complete list of probes on both platforms, with their agreement statistics across platforms, is included as Supplementary Data.
Our recommendation for groups which plan to compare or combine data across platforms (whether array-based or using another technology), or even across laboratories using the same platform, is to take the following issues into consideration. First, not surprisingly, genes which give weak signals are hard to confirm. Therefore the failure of one platform to confirm a result on a rare transcript should be interpreted cautiously. Second, careful bioinformatics analysis of each platform is necessary to maximize the precision of the comparison. At the first level of analysis, the manufacturer's annotations should be evaluated for comparability or replaced with a customized analysis that uses a common approach, to avoid conflicts due to differences in annotation methodology. At a second level of analysis, one can consider a finer level of stratification of probes based on their relative locations.
Some questions still remain. There are a still fairly numerous probes which, based on dilution effect, location and expression level criteria, would be predicted to yield reproducible results, but do not. As mentioned, we could not fully address the possibility that cross-hybridization may play an important role (16). Resolving this will likely require additional data. We also cannot eliminate the possibility that refinements of transcript assignment would resolve some cases of ‘disagreement’. Our assignment of probes to genes was based on limited databases of mRNAs and known genes, and transcripts not represented in these databases would not be reflected in our analysis. This could lead to incorrectly predicting the same hybridization pattern for two probes located at nearby locations in the genome. A likely explanation for some of the effects we see have to do with differences in the technologies, such as differences in RNA labeling protocols, or the linker and ‘bar code’ sequences on the Illumina arrays compared with the direct attachment of the Affymetrix sequences to the substrate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online and at http://microarray.cu-genome.org/platformCompare.
Acknowledgments
Thanks to Illumina for providing and running the BeadArrays as part of a Customer Service Evaluation. We thank Kiran Keshav for assistance in preparing the manuscript. This work was supported in part by the Children's Hospital Research Foundation of Cincinnati, the Schmidlapp Foundation, and National Institutes of Health Grants GM076880, AR47363, AR47784 and AR50688. Funding to pay the Open Access publication charges for this article was provided by NIH grant AR048929.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gunderson K.L., Kruglyak S., Graige M.S., Garcia F., Kermani B.G., Zhao C., Che D., Dickinson T., Wickham E., Bierle J., et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 3.Barczak A., Rodriguez M.W., Hanspers K., Koth L.L., Tai Y.C., Bolstad B.M., Speed T.P., Erle D.J. Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res. 2003;13:1775–1785. doi: 10.1101/gr.1048803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan P.K., Downey T.J., Spitznagel E.L., Jr, Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park P.J., Cao Y.A., Lee S.Y., Kim J.W., Chang M.S., Hart R., Choi S. Current issues for DNA microarrays: platform comparison, double linear amplification, and universal RNA reference. J. Biotechnol. 2004;112:225–245. doi: 10.1016/j.jbiotec.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Shippy R., Sendera T.J., Lockner R., Palaniappan C., Kaysser-Kranich T., Watts G., Alsobrook J. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics. 2004;5:61. doi: 10.1186/1471-2164-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Spletter M.L., Johnson J.A. Dissecting tBHQ induced ARE-driven gene expression through long and short oligonucleotide arrays. Physiol Genomics. 2005;21:43–58. doi: 10.1152/physiolgenomics.00214.2004. [DOI] [PubMed] [Google Scholar]
- 8.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 9.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent W.J. BLAT: the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlidis P., Noble W.S. Matrix2png: a utility for creating matrix visualizations. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 13.Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 14.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo W.P., Jenssen T.K., Butte A.J., Ohno-Machado L., Kohane I.S. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Pankratz M., Johnson J.A. Differential gene expression patterns revealed by oligonucleotide versus long cDNA arrays. Toxicol Sci. 2002;69:383–390. doi: 10.1093/toxsci/69.2.383. [DOI] [PubMed] [Google Scholar]
- 17.Kothapalli R., Yoder S.J., Mane S., Loughran T.P., Jr Microarray results: how accurate are they? BMC Bioinformatics. 2002;3:22. doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurata L.W., Bukhman Y.V., Charles V., Capriglione F., Bullard J., Lemire A.L., Mohammed A., Pham Q., Laeng P., Brockman J.A., et al. Comparison of microarray-based mRNA profiling technologies for identification of psychiatric disease and drug signatures. J. Neurosci. Methods. 2004;138:173–188. doi: 10.1016/j.jneumeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Petersen D., Chandramouli G.V., Geoghegan J., Hilburn J., Paarlberg J., Kim C.H., Munroe D., Gangi L., Han J., Puri R., et al. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6:63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvinen A.K., Hautaniemi S., Edgren H., Auvinen P., Saarela J., Kallioniemi O.P., Monni O. Are data from different gene expression microarray platforms comparable? Genomics. 2004;83:1164–1168. doi: 10.1016/j.ygeno.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Woo Y., Affourtit J., Daigle S., Viale A., Johnson K., Naggert J., Churchill G. A comparison of cDNA, oligonucleotide, and Affymetrix GeneChip gene expression microarray platforms. J. Biomol. Tech. 2004;15:276–284. [PMC free article] [PubMed] [Google Scholar]
- 22.Weis B.K. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 23.Huminiecki L., Lloyd A.T., Wolfe K.H. Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap and TissueInfo databases. BMC Genomics. 2003;4:31. doi: 10.1186/1471-2164-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.K., Bussey K.J., Gwadry F.G., Reinhold W., Riddick G., Pelletier S.L., Nishizuka S., Szakacs G., Annereau J.P., Shankavaram U., et al. Comparing cDNA and oligonucleotide array data: concordance of gene expression across platforms for the NCI-60 cancer cells. Genome Biol. 2003;4:R82. doi: 10.1186/gb-2003-4-12-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin J.E., Frank B.C., Gavras H., Sultana R., Quackenbush J. Independence and reproducibility across microarray platforms. Nature Methods. 2005;2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry R.A., Warren D., Spencer F., Kim I.F., Biswal S., Frank B.C., Gabrielson E., Garcia J.G., Geoghegan J., Germino G., et al. Multiple-laboratory comparison of microarray platforms. Nature Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]