Abstract
We describe here the results of the first genome-wide survey of candidate exon repetition events in expressed sequences from human, mouse, rat, chicken, zebrafish and fly. Exon repetition is a rare event, reported in <10 genes, in which one or more exons is tandemly duplicated in mRNA but not in the gene. To identify candidates, we analysed database sequences for mRNA transcripts in which the order of the spliced exons does not follow the linear genomic order of the individual gene [events we term rearrangements or repetition in exon order (RREO)]. Using a computational approach, we have identified 245 genes in mammals that produce RREO events. RREO in mRNA occurs predominantly in the coding regions of genes. However, exon 1 is never involved. Analysis of the open reading frames suggests that this process may increase protein diversity and regulate protein expression via nonsense-mediated RNA decay. The sizes of the exons and introns involved around these events suggest a gene model structure that may facilitate non-linear splicing. These findings imply that RREO affects a significant subset of genes within a genome and suggests that non-linear information encoded within the genomes of complex organisms could contribute to phenotypic variation.
INTRODUCTION
The completion of the sequencing of the human genome (1,2) has raised more questions than it has answered, in regards to what it is that makes humans and other advanced organisms so complex. The lack of correlation between the number of genes and an organism's complexity raises the question of how complexity and diversity arise? Alternative splicing of mRNA molecules from expressed genes is now commonly thought to affect >70% of all human genes, suggesting that alternative splicing is one of the most significant processes in the functional complexity of the human genome (1,3–5). Alternative splicing contributes to functional complexity by increasing the protein diversity encoded from each gene and influencing protein expression regulation, via nonsense-mediated RNA decay, for example (6–8).
Most alternative splicing research to date has focussed on alternative cis-splicing, in which exons located within an individual pre-mRNA are differentially joined to generate mature mRNAs (9). This involves joining the exons to be included in the mature transcript in a linear contiguous manner, 5′–3′, in the order they are found in the genome. However, there is evidence that suggests there is an additional level of complexity in mammals. Instances of a phenomenon termed exon scrambling have been reported, whereby the order of the exons in the RNA does not reflect the linear order in the genome (10–13). However, it has been suggested that this results from splicing within the lariat produced when several exons are skipped, and it may not contribute to the pool of mRNA (13). Less easily explained is the discovery of tandemly repeated exons in mammalian mRNA in the absence of duplications in the genome (14–20), a phenomenon termed exon repetition. Exon repetition is allele-specific and operates strictly in cis, meaning that in heterozygotes only mRNA from the susceptible allele contains the repetition (20). Unlike exon scrambling, exon repetition occurs in mRNA and precise predictions about the arrangement of the exons can be made that allow it to be identified even where the cDNA sequences do not span the repeated sequences. For convenience, we refer to any departure of the order of exons in mRNA from the 5′–3′ order in the genome as rearrangements or repetition in exon order (RREO). This definition encompasses the phenomena of both exon scrambling and exon repetition. Currently, <10 genes have been discovered, mostly through serendipity, to exhibit RREO in mammals.
Expressed sequence tags (ESTs) provide an abundant source of information to study alternative splicing (4). ESTs are single pass reads obtained from either the 5′ or the 3′ end of a cDNA clone (21). As ESTs are derived from fully processed mRNA (after 5′ capping, splicing and polyadenylation), they provide snapshots of mRNA diversity. ESTs are therefore essential pieces of information that enable the investigation of the types of transcripts generated from a genome. Based on analyses of ESTs, many studies have shown that through the combination of bioinformatics methodology and genomic resources, much information about the extent of alternative cis-splicing can be obtained (22–26). However, it must be emphasized that the EST sequences catalogued in dbEST (27) are merely a sample of the transcriptome and have many limitations (21).
We describe here the first genome-wide survey of RREO events in publicly available expressed sequences from multiple species. The method we employed to detect these non-linear mRNA sequences is different from previous genome-wide surveys of linear-splicing, which have generally employed the strategy of aligning ESTs to the genome sequence (4). We have developed a computational approach based on the strategy taken previously by Hide et al. (16) in which they investigated linear exon skipping and single exon repetition events for human chromosome 22 (16). In our analyses, the complexity of each and every gene sequence was reduced to a set of possible non-linear exon splice junctions (100 bp in length) that were then used to search for ESTs or mRNA sequences spanning the non-linear exon–exon junctions (Figure 1). The exon sequences used in creating the non-linear exon splice junction probes were obtained from Ensembl, which currently provides the most definitive collection of gene structures for many species (28,29).
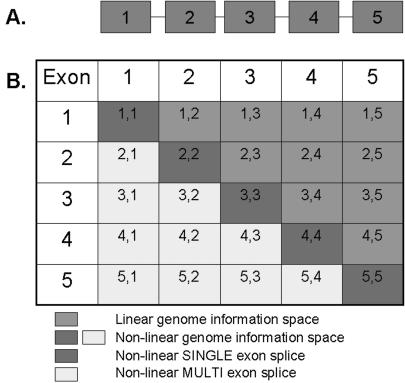
Figure 1.
An illustration of the linear and non-linear genome information spaces for a hypothetical gene that contains five exons. (A) A simple cartoon of a hypothetical gene containing five exons. (B) The total genome information space for a five-exon gene. Our approach to investigate the non-linear genome information space involved creating a set of possible non-linear exon–exon junction sequences of 100 bp (dark grey and light grey genome information spaces) for each gene. The first 50 bp are derived from the 5′ exon and the last 50 bp are derived from the 3′ exon in each possible non-linear exon–exon splice combination for all Ensembl exons from each gene.
Our search for RREO patterns in publicly expressed sequences has yielded evidence for the occurrence of this phenomenon in ∼1% of mammalian genes, and also shows that this process is evolutionarily conserved for one gene between human and mouse, suggesting a biological function. We have conducted a manual assessment of 100 of the human RREO events, to assess the impact of the non-linear splice on the open reading frame (ORF) of the ESTs. A statistical analysis of the sizes of the exons and introns involved around these events has highlighted a significant characteristic associated with these genomic regions. These results imply that RREO is possible from a subset of genes within a genome and hints at the existence of non-linear information encoded within the genomes of complex organisms.
MATERIALS AND METHODS
Data sources
Our analysis is based on four major types of data: genome sequence assemblies, Ensembl exon sequence models, EST and mRNA sequences. Human genomic chromosome sequences and mRNA sequences were downloaded from the UCSC genome browser (UCSC hg17, May 2004, NCBI 35 assembly), (http://hgdownload.cse.ucsc.edu/goldenPath/hg17/bigZips/). Human genome exon sequence data were downloaded from Ensembl (v29.35b, NCBI 35 assembly), (http://www.ensembl.org/). Human EST sequences were downloaded from NCBI (May 2005), (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/). Mouse genomic chromosome sequences and mRNA sequences were downloaded from the UCSC genome browser (UCSC mm6, March 2005), (http://hgdownload.cse.ucsc.edu/goldenPath/mm6/bigZips/). Mouse genome exon sequence data were downloaded from Ensembl (v27.33c.1, NCBI m33 assembly). Mouse EST sequences were downloaded from NCBI (May 2005), (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/). Rat genomic chromosome sequences, EST and mRNA sequences were downloaded from the UCSC genome browser (UCSC rn3, June 2003) (http://hgdownload.cse.ucsc.edu/goldenPath/rnJun2003/bigZips/). Rat genome exon sequence data were downloaded from Ensembl (v27.3e.1, RGSC 3.1 assembly). Chicken genomic chromosome sequences, EST and mRNA sequences were downloaded from the UCSC genome browser (UCSC galGal2, February 2004), (http://hgdownload.cse.ucsc.edu/goldenPath/galGal2/bigZips/). Chicken genome exon sequence data were downloaded from Ensembl (v27.1d.1, WASHUC1 assembly). Zebrafish genomic chromosome sequences, EST and mRNA sequences were downloaded from the UCSC genome browser (UCSC danRer2, June 2004), (http://hgdownload.cse.ucsc.edu/goldenPath/danRer2/bigZips/). Zebrafish genome exon sequence data were downloaded from Ensembl (v27.4b.1, WTSI Zv4 assembly). Drosophila melanogaster genomic chromosome sequences, EST and mRNA sequences were downloaded from the UCSC genome browser (UCSC dm2, April 2004), (http://hgdownload.cse.ucsc.edu/goldenPath/dm2/bigZips/). Drosophila genome exon sequence data were downloaded from Ensembl (v27.3c.1, BDGP 3.1 assembly).
Detection of non-linear mRNA alternative splicing events in expressed sequences
A series of programs written in the Perl programming language (v5.8.5) were created to produce possible 100 bp non-linear exon–exon splice junction probe sequences for each gene (Figure 1). Separate programs were used to create the non-linear single exon splice sequences (dark grey space in Figure 1) and the non-linear multi-exon splice sequences (light grey space in Figure 1). For each species, all Ensembl exons were filtered so that only those genes with more than one exon and only exons >50 bp in length were used. Using this list of exon sequences, the non-linear single exon splice sequences were created for each gene by joining 50 bp from the 3′ terminus of each exon with 50 bp from the 5′ terminus of the same exon. The non-linear multi-exon splice sequences were created for each gene by using the following algorithm, ‘for each exon, starting with the most 3′ exon in the gene, take 50 bp from the 3′ terminus of the exon and join with the 50 bp from the 5′ terminus of each of the preceding exons in the gene’. The resulting list of 100 bp non-linear single and multi-exon splice sequences were submitted for similarity searching against all ESTs and mRNA sequences for the relevant species using megablast (30) (http://www.ncbi.nlm.nih.gov/blast/megablast.shtml). ESTs and mRNA sequences showing >95% similarity to the query sequence (E-value in megablast searches was 1 × 10−40) were extracted. All EST and mRNA sequences confirming non-linear exon splicing events were then mapped back to the relevant genome along with the original non-linear splice sequence by using the command line version of the BLAT program (31). We used BLAT version 32 for Linux (http://www.soe.ucsc.edu/~kent/exe/). Only those non-linear splicing events in which the EST or mRNA confirming the event was the best alignment with the highest BLAT score in the correct gene from which the original non-linear splice sequence was derived were kept. This ensured that we excluded ESTs or mRNAs from paralogous genes or which were derived from a different gene to the one under investigation. Also, we only kept those non-linear splicing events in which the original non-linear splice sequence produced only two alignments against the correct gene with a BLAT score of 45–55 for each half of the non-linear splice sequence. This ensured that we filtered out those non-linear splice sequences that were derived from genes, which contained exon duplications. All subsequent EST sequences confirming non-linear exon splicing events were then screened for vector sequence contamination by utilizing the VecScreen program at the National Centre for Biotechnological Information (NCBI, http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html). The resultant EST and mRNA sequences were catalogued as evidence confirming RREO for the relevant exons and genes. Evolutionarily conserved RREO events were detected by using gene orthologues information available from Ensembl and using programs written in Perl to find gene orthologue matches between species from our lists of genes found to be involved in RREO. Matches were inspected manually for exon orthologue verification, by ensuring that the exons involved were the best reciprocal hits using the BLASTview tool available at Ensembl (http://www.ensembl.org/Multi/blastview).
Analysis of the EST sequences involved in non-linear mRNA alternative splicing
Alignment of EST and mRNA sequences to genome sequences, in order to ascertain the exons covered in the expressed sequences was undertaken using the BLAT program available within the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgBlat). For ORF analysis, human EST sequences were submitted to the NCBI ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). We accepted the longest ORF in the correct direction for the EST and gene, with identity to the SWISS-PROT reference protein sequence for the gene. In the absence of any identity to the reference protein sequence, the longest ORF in the correct direction was taken. Alignments of ORF sequences with the SWISS-PROT reference sequence were undertaken using the program ClustalX (32).
Analysis of human genes, exons and introns involved in non-linear mRNA alternative splicing
Gene ontology annotation analysis of human genes was undertaken using the ‘GO::TermFinder’ program locally (33) and the web tool ‘Fatigo’ (http://www.fatigo.org/) (34). To estimate the relative expression of human exons involved in RREO, by their representation in dbEST, all human exon sequences (239 250) were downloaded from Ensembl. Exons <50 bp in length were filtered out, as these were not analysed for RREO. The resulting 225 252 exons (≥50 bp) were used in a megablast against all human ESTs, using an E-value of 1 × 10−40. Perl programs were used to parse the blast results, count the number of EST hits per exon and process the results. To undertake a statistical analysis of the lengths of the human exons and introns involved in RREO, a random set of 100 RREO events were generated from our list of all RREO human events. Also, a random set of 100 exons was selected from a list of all Ensembl human exons, which had been filtered to remove those genes for which we have evidence for involvement in RREO. All human exon and intron sizes were obtained from Ensembl. An independent samples t-test was used to calculate the P-values for the average sizes between these two samples with a 95% confidence interval. All statistical analyses were undertaken with the SPSS software package (v11.01 for Windows). Random sampling was undertaken using a Perl program with a random number generator.
RESULTS
We have undertaken a large-scale investigation of RREO events in all expressed sequences from GenBank for six species. As an example of the strategy of this work, ∼2.7 million 100 bp non-linear exon–exon splice junction probe sequences were generated using human exon sequences defined by the human Ensembl gene models. These 100 bp sequences were then used to search for exons rearranged out of linear order in ∼6.25 million human expressed sequences. From this search, a total of 263 human expressed sequences from 178 human genes were identified to confirm an RREO event. The results of the analyses of six species for RREO events in ESTs and mRNA sequences are summarized in Table 1. We have detected 245 genes in mammals and 264 in all six species that exhibit a signature for RREO in EST or mRNA sequences. The data in Table 1 shows the number of ESTs and mRNAs confirming the non-linear single exon splice events (dark grey non-linear genome information space in Figure 1) and the non-linear multi-exon splice events (light grey non-linear genome information space in Figure 1). An example of the frequency distribution of the number of ESTs per gene is shown in Figure 2, for the 170 human genes that exhibit RREO events in ESTs. Figure 2 shows that the majority (78.2%) of the 170 human genes are represented by a single EST, with 20.1% of the human genes represented by 2–4 ESTs and an additional 3 genes represented by 7, 9 and 19 ESTs. This pattern is typical of the other 5 species in Table 1.
Table 1.
A summary of the detection of RREO events in EST and mRNA sequences from six species
| Human | Mouse | Rat | Chicken | Zebrafish | Fruit fly | |
|---|---|---|---|---|---|---|
| Number of EST sequences in GenBank | 6 057 800 | 4 334 174 | 701 039 | 540 881 | 630 156 | 383 407 |
| Number of mRNA sequences in GenBank | 194 508 | 172 159 | 17 575 | 26 296 | 13 064 | 15 897 |
| No. of EST sequences confirming non-linear single exon splicing | 25 (Supplementary Table A)a | 28 (Supplementary Table E)a | 2 (Supplementary Table I)a | 0 | 3 (Supplementary Table N)a | 6 (Supplementary Table Q)a |
| No. of mRNA sequences confirming non-linear single exon splicing | 3 (Supplementary Table B)a | 9 (Supplementary Table F)a | 0 | 1 (Supplementary Table K)a | 1 (Supplementary Table O)a | 1 (Supplementary Table R)a |
| No. of EST sequences confirming non-linear multi-exon splicing | 221 (Supplementary Table C)a | 48 (Supplementary Table G)a | 15 (Supplementary Table J)a | 9 (Supplementary Table L)a | 23 (Supplementary Table P)a | 1 (Supplementary Table S)a |
| No. of mRNA sequences confirming non-linear multi-exon splicing | 14 (Supplementary Table D)a | 13 (Supplementary Table H)a | 0 | 2 (Supplementary Table M)a | 0 | 0 |
| Total no. of expressed sequences confirming non-linear splicing | 263 | 98 | 17 | 12 | 27 | 8 |
| Total no. of genes involved in non-linear splicing | 178 | 61 | 7 | 6 | 8 | 5 |
aEach supplementary table contains the EST or mRNA GenBank ID, Ensembl exon and gene identifiers as well as the sequence of the 100 bp non-linear splice sequence used to detect each event.
Figure 2.
Frequency distribution of the number of non-linear ESTs detected for each of the 170 human genes. The 170 human genes that exhibit non-linear splicing in EST sequences were assessed for the number of non-linear EST sequences within dbEST, which confirm each non-linear splice event.
Our analysis has identified two previously known genes that exhibit RREO in mRNA. The human FBXO7 gene (Ensembl ID ENSG00000100225) has been shown previously (16) to exhibit a single exon repetition of exon 2 in the EST AA569698, and our analysis also detects this same event. The Rat OCTC_RAT gene (Ensembl ID ENSRNOG00000006779), otherwise known as the COT gene, is one of the best-characterized examples of exon repetition. The Rat COT gene has been shown previously to exhibit single exon repetition of exon 2 (14,20), in liver and kidney tissues. We have discovered a single exon repetition event of exon 2 for the Rat COT gene in Brown Norway testis tissue (EST CK603740). The COT gene exon coverage of this EST is 1-2-2-3-4-5-6. We did not detect other previously known examples of RREO in ESTs or mRNAs because their signatures were not present in the sequences available in GenBank.
RREO occurs in both the human and mouse CTBP2 genes (Ensembl ID ENSG00000175029 and Ensembl ID ENSMUSG00000030970, respectively). We have identified ESTs from both species that exhibit repetition of exon 3 (Ensembl human exon ENSE00001191630 and mouse exon ENSMUSE00000341464). Exon 3 from both species is 100% identical in its sequence composition. A human EST from a pooled pancreas and spleen tissue source (EST BI834017) exhibits the exon coverage pattern 1-2-3-3 when aligned to the genomic sequence of the human CTBP2 gene. Two mouse ESTs from a mammary tumour tissue sample (ESTs BI660369 and BI655703), both exhibit the exon coverage pattern 1-2-3-3-4 when aligned to the genomic sequence of the mouse CTBP2 gene. Interestingly, we also identified a mouse EST from a mammary tissue source (EST BY017851) that exhibits the exon coverage pattern 1-2-2. This EST was from a different clone library and separate laboratory to the previous two mouse ESTs.
A representative sample of 100 human RREO events in ESTs were examined manually, in order to elucidate the effect of RREO on the ORF of the EST sequences, when compared to the SWISS-PROT protein sequence, for the gene from which the EST was derived. The results of this analysis are documented in Supplementary Table T, as well as additional information on these 100 events. This more detailed analysis of non-linear expressed sequences enabled us to determine the exact exons involved. Due to the short nature of EST sequences, they are rather uninformative with regards to deciphering whether the non-linear splice event is due to the phenomenon of exon repetition or exon scrambling. In order for us to categorize the events in this manner with confidence, we would need the full transcript sequence. Nonetheless, even sequences that are not long enough to show the repetition of exons can provide evidence for exon repetition rather than scrambling. To illustrate this, consider a gene comprising 5 exons, in which exons 3 and 4 participate in exon repetition (i.e. mRNA is 1-2-3-4-3-4-5). The 4-3 junction shows RREO, being the site where the tandemly repeated blocks are juxtaposed. However, the exon arrangement to either side of this site is normal, with no other discontinuity from the linear order. In contrast, exon scrambling is thought to occur within circles or lariats containing only a subset of exons (12,13). Hence, the presence of terminal exons 1 and 5, or the 2-3 or 4-5 junctions, would argue strongly in favour of exon repetition.
Of the representative sample of 100 human RREO events in Supplementary Table T, 15 are examples of non-linear single exon splicing events (the non-linear exon splice being derived from the dark grey genome information space in Figure 1) and 85 are examples of non-linear multi-exon splicing events (light grey genome information space in Figure 1). Of these 100 events, 39 show a clear signature in the full EST sequence (as described above) of exon repetition. It was impossible to decipher whether the remaining 61 are a result of exon scrambling or exon repetition due to the short nature of the EST sequences, and we can only categorize these simply as non-linear mRNA transcripts. However, it is unlikely that these non-linear sequences are derived from the process of exon scrambling which produces circular mRNAs (13), because they would not contain polyA tails. Since most EST cDNA libraries are oligo(dT) primed and therefore select transcripts with a polyA tails (21), we would expect that non-linear transcripts produced as a result of exon scrambling to be very much a minor fraction of the non-linear transcript pool within dbEST.
Interestingly, four genes (SNX5, O60524, EYA1 and ARHGAP26) show evidence of two different regions of the gene being involved in non-linear splicing. For example, the gene SNX5 exhibits a non-linear splice between exons 12 and 3, and also between exons 4 and 3 in separate EST sequences.
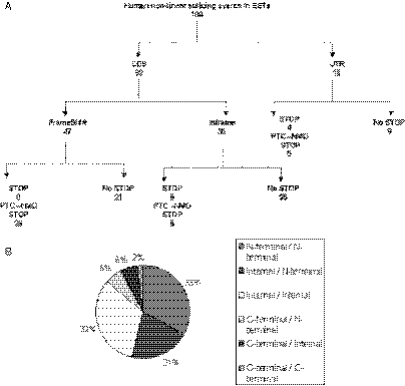
The results of the ORF analysis of 100 human RREO events in ESTs are summarized in Figure 3A. Of these 100 splicing events, 82 occur in the coding sequence region of the gene and 18 involve exons within the untranslated region (UTR) of the Ensembl gene models. Strikingly, we found no non-linear splicing events involving the first exon of a gene, suggesting that the processes that we are investigating are indeed alternative splicing mechanisms rather than an abnormality. We categorized EST sequences as candidates for nonsense-mediated RNA decay (NMD) if a termination codon was produced by RREO >50 nt upstream of the final exon (35). This produced 36 candidates among the 100 sequences. This is comparable with previous research, which suggests that approximately a third of alternative splicing events in humans might result in NMD candidate transcripts (6–8). Interestingly, 25/100 non-linear sequences exhibit a completely in-frame ORF when compared to the reference protein sequence, suggesting repetition of a portion of the original protein sequence. ORF analysis of the evolutionarily conserved RREO events from the CTBP2 gene were complicated by the fact that the exon 3 involved is not completely coding sequence. The first half of the CTBP2 exon 3 is 5′-UTR (according to current Ensembl gene model) and therefore comparison of the ORF with the reference protein sequence is uninformative after the non-linear splice site. We found the human CTBP2 non-linear EST to exhibit a frame shift (as expected) in the ORF with no premature stop codon, whereas the mouse CTBP2 non-linear EST sequences are frame shifted and are both candidate NMD transcripts with the PTC being derived from different regions of the repeated exon 3 sequence. As the EST sequencing error rate can be as high as 3% (36), it is possible that this is the cause of many PTCs in EST sequences and the false identification of NMD candidate sequences. Therefore, the functional analysis of the human and mouse EST transcripts derived from the CTBP2 gene is inconclusive. Figure 3B shows a summary of the non-linear splice locations within their respective protein sequences for the representative sample of 100 human RREO events in ESTs. The majority of the RREO events (54/100) involve the N-terminal region of the protein sequences encoded by these genes. A further 32/100 RREO events involve the internal regions of the proteins, with the minority (14/100) involving the C-terminal of the protein sequences. Therefore, the non-linear splicing phenomenon would seem to mostly affect the N-terminal regions of protein sequences.
Figure 3.
An analysis of a representative sample of 100 human non-linear splicing events in EST sequences. (A) Open reading frame analysis of 100 human non-linear spliced EST sequences. CDS: non-linear splice site involves only exons within the coding sequence of the gene. UTR: non-linear splice site involves exons within the untranslated region of the gene. FrameShift: the non-linear splice introduces a frame shift in the open reading frame of the sequence when compared to the reference protein sequence for the gene. Inframe: the non-linear splice conserves the open reading frame of the sequence when compared to the reference protein sequence for the gene. STOP: the non-linear splice introduces a premature stop codon in the sequence. PTC-NMD STOP: the non-linear splice introduces a premature stop codon in the sequence that is >50 nt upstream of the final exon and is therefore a candidate sequence for nonsense-mediated RNA decay. (B) Summary of the non-linear splice locations in the proteins of 100 human events. The potential protein sequence regions affected by the 100 human non-linear human events in EST sequences. Each protein sequence was divided into thirds by the number of amino acids. The locations of the non-linear splice within the open reading frame of the protein were classified as N-terminal when occurring in the first third of the protein sequence, internal when occurring in the second third and C-terminal when occurring in the last third of the protein sequence.
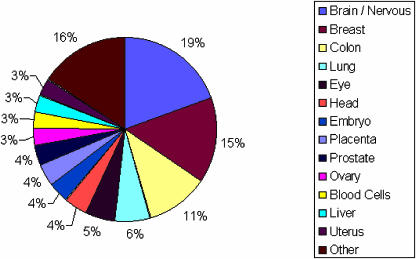
For the 170 human genes that exhibit RREO events in ESTs, Figure 4 illustrates the variety of tissue sources from which these ESTs were derived. The majority of tissues represented by these ESTs are brain/nervous tissue, breast, colon and lung tissues, with a diverse list of tissues representing the rest of the ESTs. This distribution reflects the frequency of tissues represented in dbEST and therefore suggests that there are no particular tissues that are associated with RREO. Therefore, RREO would appear to be a process that occurs throughout the human body. Also, we observed that 22.5% of the human non-linear spliced ESTs are derived from cancerous tissues. As it has been estimated that >50% of the ESTs in dbEST come from cancer cell lines or tumour tissues (37), this suggests that RREO is not an anomaly linked to cancerous tissues.
Figure 4.
A summary of the tissue sources from which the non-linear spliced ESTs for 170 human genes were derived. Tissue source information was obtained from the GenBank records of the EST sequences.
All the 178 human genes we found to be involved in RREO were analysed for functional annotation information. We used two different tools (see Materials and Methods) to assess the Gene Ontology (GO) terms associated with these 178 genes, in order to investigate whether there were any predominant gene functions associated with them. The only GO term that the ‘GO::TermFinder’ tool, found to be statistically significantly enriched in our gene list, was the GO cellular compartment term ‘intracellular’. Of the 178 human genes involved in RREO, we found 80 were annotated with GO terms at this level of the ‘cellular compartment’ GO model and 53/80 (66.2%) were annotated as being ‘intracellular’ (P = 0.00016), compared to the ‘genome frequency of use’ of this term. Use of the ‘Fatigo’ web tool found that the only other predominant GO term associated with this list of human genes was that of ‘binding’ from the ‘GO molecular function, level 2’ section of the ontology, wherein 106/178 genes were annotated with GO terms and 40.5% of these were annotated with the ‘binding’ term.
As many EST libraries are normalized, subtracted or enriched in certain clones, the frequency of representation of a gene in dbEST cannot be used to accurately predict its expression level (21,38). However, even with the process of library normalization, abundant transcripts are represented more frequently in dbEST than rare ones (21,38). Thus, the relative expression of human exons involved in RREO in mRNA can be roughly estimated by their representation in dbEST as compared to all other human exons. We undertook a large megablast of all human exons that were 50 bp or more in size (225 252/239 250 total exons) versus all human ESTs, as our RREO detection workflow only utilized exons that were 50 bp or more in length. Of the 225 252 exons that were used in the megablast, 56 809 were found to have no representation in dbEST. The resulting 168 442 exons were split into the three expression categories (Table 2), comprising approximately equal number of exons. The exons involved in RREO from all the 178 human genes that we found to exhibit RREO were then analysed. We found that the majority (17/24) of human exons involved in non-linear single exon splicing events were represented in the high category of Table 2. This suggests that these exons are highly represented by EST sequences in dbEST and are therefore likely to be highly expressed genes. The human exons involved in non-linear multi-exon splicing events were more evenly distributed amongst the three expression categories.
Table 2.
An estimation of the relative expression of human exons involved in non-linear splicing by their representation in dbEST
| Expression category | Non-linear single exon (n = 24) | Non-linear 5′ multi-exon (n = 152) | Non-linear 3′ multi-exon (n = 151) |
|---|---|---|---|
| High | 17 (70.8%) | 50 (32.9%) | 46 (30.5%) |
| Medium | 4 (16.7%) | 64 (42.1%) | 63 (41.7%) |
| Low | 3 (12.5%) | 38 (25%) | 42 (27.8%) |
The expression category HIGH includes exons (56 591 exons) which are represented by ≥34 ESTs in dbEST, with MEDIUM exons (53 837 exons) being represented by ≥9 and ≤33 ESTs and low exons (58 014 exons) being represented by ≥1 and ≤8 ESTs in dbEST. Non-linear single exons are those exons that are involved in non-linear single exon splicing (dark grey genome information space in Figure 1). Non-linear 5′ multi-exons are those exons that are the 5′ exon in a non-linear multi exon splicing event (light grey genome information space in Figure 1). Non-linear 3′ multi-exons are those exons that are the 3′ exon in a non-linear multi exon splicing event (light grey genome information space in Figure 1).
We investigated the sizes of exons and introns involved in human RREO events by comparing them with a random sample of exons that exhibit no evidence of RREO in human EST or mRNA sequences. The results of this analysis (Table 3) suggest that genes that contain a region with longer than average exons and introns are more likely to undertake RREO in mRNA. The intron that precedes the exon found in the mRNA at the 3′ side of the discontinuity (e.g. the intron preceding exon 2 in a non-linear splice involving exons 4-2) seems particularly significant, with an average length of >20 000 bases compared to an average of ∼3000 bases for those exons that have no evidence of being involved in this phenomenon. The exonic and intronic size distributions that were obtained from our small random sample of all human exons for which there is no evidence of RREO is comparable with those that have been reported previously for humans (1), where a mean intron size of 3365 and mean exon size of 145 was observed. We also observed the exon sizes from Table 3 for a multiple of three (their ability to preserve the ORF), of both the random exons that exhibit no evidence of RREO and the exons involved in RREO. We found that 48/100 of the 5′ exons involved in RREO are a multiple of 3. The 3′ exons involved in RREO show that 45/100 are a multiple of 3 in exon size. In contrast, 30/100 random exons with no evidence of RREO are a multiple of 3.
Table 3.
A statistical analyses of exon/intron sizes of a random sample of 100 human non-linear splicing events versus a random sample of 100 exons from all human exons, for which there is no evidence of non-linear splicing
| Exons with evidence for non-linear splicing | Random exons with no evidence of non-linear splicing | P-value | |
|---|---|---|---|
| A | 20 628 (± 3053) | 3159 (± 523) | 1.5 × 10−7 |
| B | 12 556 (± 2839) | 3159 (± 523) | 0.002 |
| C | 10 067 (± 1973) | 3191 (± 644) | 0.001 |
| D | 15 135 (± 2158) | 3191 (± 644) | 5.4 × 10−7 |
| E | 320 (± 58) | 153 (± 16) | 0.007 |
| F | 280 (± 49) | 153 (± 16) | 0.017 |
Numbers shown are the Mean length ± the standard error of the mean (n = 100). An independent samples t-test was used to calculate the P-values for the average sizes between these two samples with a 95% confidence interval. A: comparison of the 5′ intron of random exons, to the 5′ introns of the 3′ exon involved in a non-linear splicing event; B: comparison of the 5′ intron of random exons, to the 5′ introns of the 5′ exon involved in a non-linear splicing event; C: comparison of the 3′ intron of random exons, to the 3′ introns of the 3′ exon involved in a non-linear splicing event; D: comparison of the 3′ intron of random exons, to the 3′ introns of the 5′ exon involved in a non-linear splicing event; E: comparison of random exons, to the 5′ exon involved in a non-linear splicing event; F: comparison of random exons, to the 3′ exon involved in a non-linear splicing event.
DISCUSSION
Our computational approach has enabled us to identify precisely expressed sequences that contain a signature of RREO. However, this strategy is limited in its power to detect all instances of RREO for a number of reasons. Computational detection of splicing in non-linear genome information spaces is dependent on EST data sources as the largest pool of transcript data currently available. However, ESTs are a problematic source of information and they are merely a sample of the whole transcriptome. The detection of a particular splice variant in dbEST is possible only if its transcription level is sufficiently high in a tissue type (dbEST is highly biased for a limited number of tissue types) for which an EST library has been prepared. Also, as most ESTs are generated from the 5′ and the 3′ termini of the transcript, dbEST is highly biased towards under-representation of splice variants involving exons that are in the middle of long transcripts (4). Indeed, EST coverage in the 5′-UTR is often much lower than in the 3′-UTR (39). As we found most RREO events tend to occur in the 5′ half of the gene, this could contribute to why we have detected so few RREO events. In addition, ESTs contain a high sequence error rate of up to 3%, they are short, averaging ∼400 bp in length and they contain artefacts such as vector and bacterial sequence contamination (36). As our strategy depends on using stringent sequence matching parameters around the non-linear splice site, it obviously decreases the chances of detecting non-linear splice forms if there is a high degree of error in the sequences we are searching. The short length of most ESTs also limits our detection rate, as many ESTs are so short that they do not span an exon junction and are thus uninformative. We have not taken into account in this study the possibility that RREO may use alternative 5′ and 3′ splice sites to those currently defined by the Ensembl gene models. As this is a common mechanism in linear splicing, it is entirely likely that this also occurs in the non-linear genome information space. Future research could incorporate this into the computational analysis to enable a more thorough search of the non-linear genome information space. Our computational analysis pipeline filtered out those ESTs, which were derived from genes that have paralogues, or contain intragenic exon duplications, as these could not be discerned to be the result of linear or non-linear alternative splicing. All these factors taken together suggest that there may be many more genes than we have detected that undertake RREO.
Another problem with ESTs that is particularly relevant to this study is that of chimeric artefact sequences, which are the result of concatenation of two or more expressed sequences from different regions of the genome. It can be an artefact of cDNA cloning, sequencing (40) or abnormal reverse-transcription (41). However, because the portions of a chimeric EST sequence are joined at random, they are generally from different chromosomes or distant regions of the same chromosome. Therefore, artefacts such as chimeric sequences are unlikely to join by chance at exact exon–exon splice sites. Our strategy was based on the use of computationally generated non-linear exon–exon splice junction sequence probes from established Ensembl gene models for searching dbEST. This provides us with confidence that we are not selecting for artefacts such as chimeric sequences. Moreover, our approach ensures that we only detect those ESTs, whose sequence spans a canonical non-linear exon–exon boundary in a ‘sense/sense’ manner. Chimeric transcripts generated as a result of artefacts would be expected to randomly generate ‘sense/antisense’ sequences in one half of the cases.
Our estimate of the relative expression of exons involved in RREO (by their representation in dbEST) suggests that a substantial portion of these exons are highly expressed, indicating that non-linear splice forms may be a minor component of the transcriptome compared to linear alternative splice forms, as most of the genes we found to exhibit RREO were represented by 1–2 ESTs. A number of previous in silico approaches that have searched ESTs for linear splicing patterns have required that the discovery of alternative splice forms are observed in multiple ESTs (often from different libraries), so as to increase confidence that they are not low frequency error products (4). However, it is also recognized that this stringency is unsuitable for the detection of minor splice forms (4). Our research suggests that non-linear splice forms are indeed a minor component of the transcriptome compared to linear splice forms. Recent research has suggested that approximately one-third of functional linear splice forms are represented by only one EST in dbEST (42). Indeed, recent gene expression experiments have revealed that half of all transcripts within a cell are present in fewer than 10 copies (43). Therefore, the RREO events we have detected from a single EST cannot be dismissed easily as error products or splicing noise. Besides, our analysis has found genes that provide evidence for non-linear splicing from multiple ESTs and from different cDNA libraries. However, it is possible that some of the RREO events may represent species-specific ‘splicing noise’, but the existence of such noise might be evidence of the flexibility of the splicing mechanism that could enable the evolution of new functional forms from non-linear genome information spaces.
The significance of RREO in mRNA has yet to be fully established. However, several lines of evidence suggest biological function. We found evidence of an evolutionarily conserved exon repetition event, involving exon 3 from the CTBP2 gene in both mouse and human. The conservation of such an event in both human and mouse species, which diverged from their common ancestor 75–110 million years ago, suggests some functional importance of this non-linear splice form to both species.
There are several routes by which RREO might contribute to a selectable phenotype. A quarter of the ORFs that we analysed were preserved after a non-linear exon splice, such that the effect of RREO would be to introduce a duplicated section into the protein. An example of this has been noted already by Claudevilla et al. (14), who have directly shown that that the COT gene exon repetition of exons 2 and 3 resulted in the predicted larger protein. In other cases, RREO introduced frame shifts and premature truncation of the protein sequences, which would increase protein diversity. In over a third of the RREO events, the resulting sequences would be expected to be potential targets for NMD, which is recognized as a means of regulating gene expression at a post-transcriptional point (6–8). Interestingly, whereas the Sprague–Dawley rats studied by Claudevilla et al. (14) repeat COT exons 2 and 3, resulting in the synthesis of a longer polypeptide, the WKY strain produces repetition of exon 2, which creates an upstream out-of-frame initiation codon (20) that reduces expression substantially in reporter assays (R. Rigatti, N. J. Samani and I. C. Eperon, unpublished data) and might make the mRNA susceptible to NMD. The same repeat was found in the database for Brown Norway rat testis RNA. Hence, the different preferences for patterns of exon repetition among different alleles of one gene (20) can result in the extreme alternatives of longer polypeptides or the attenuation of expression. Since the majority of RREO events involve the protein-coding region, as others have found for linear alternative splicing events (26), it seems likely that RREO is subject to selection, with effects on biological function or expression that contribute to phenotypic variation.
It is currently unclear how the mechanism of RREO in mRNA occurs. In our analysis, we have detected expressed sequences, which exhibit a non-linear exon order according to the current Ensembl gene models. Some of the ESTs we have detected are clearly the result of exon repetition. It has been suggested that exon repetition is the result of trans-splicing (3,14). However, research on the two best-characterized examples of exon repetition suggests that this phenomenon is restricted to specific alleles of the affected genes and is determined in cis rather than as a result of interallelic trans-splicing (20). Therefore, if exon repetition is the result of trans-splicing it must be from transcripts from the same allele. Our analysis of the size distributions of exons and introns involved in non-linear splicing suggests that genes, with longer than average exons and introns are more likely to be involved in RREO. Recent research on the human genome has found that <10% of all introns are >11 000 bp in length and <15–20% of all exons are >200 bp in length (44). As we found most exons and introns involved in RREO are significantly longer than this size range, it suggests that only a minor fraction of all genes could undertake RREO, which contributes to explaining why non-linear splice forms are a minor component of the transcriptome. This characteristic of the genes involved in RREO suggests an intron/exon arrangement which may facilitate the mechanism of RREO in mRNA. Our observations, in particular that the length of the intron preceding the exons involved in the exon repetition is longer than average, is consistent with work on the mechanisms of exon repetition (J.-H. Jia, N. J. Samani and I. C. Eperon, unpublished data) and with some recent experiments on trans-splicing (45).
Future analysis could search for genes with this particular characteristic and enable a more directed investigation for genes susceptible to RREO. We intend to create an integrated computational pipeline and database to automatically detect and store further RREO events, as more expressed sequences from many species become publicly available. This will provide a rich repository of information on the genes involved in RREO and enable further research into the sequences around these genomic regions to provide insights into the mechanisms of RREO.
This research suggests that there are genome information spaces, which need to be investigated if we are to fully understand how life orchestrates itself in health and disease. We suggest that genomes may contain information encoded in a non-linear manner. For us to understand this information and the role it plays in biology, we need to advance from analysing ESTs and do as researchers in the field of linear mRNA alternative splicing have done, by moving to more high throughput methods such as microarrays designed to detect splice forms. The use of such methods would enable the experimental verification of novel non-linear splice forms and provide data for their functional characterization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The work was supported by the Wellcome Trust Functional Genomics Initiative in Cardiovascular Genetics and a MRC Cooperative Grant on variability, instability and pathology of the human genome. N.J.S. holds a BHF Chair of Cardiology. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. [Google Scholar]
- 5.Johnson J., Castle J., Garrett-Engle P., Kan Z., Loerch P., Armour C., Santos R., Schadt E., Stoughton R., Shoemaker D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 6.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillman R.T., Green R.E., Brenner S.E. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lareau L.F., Green R.E., Bhatnager R.S., Brenner S.E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Black D. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 10.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 11.Cocquerelle C., Daubersies P., Majerus M., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surono A.Y., Takeshima T., Wibawa M., Ikezawa I., Nonaka I., Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum. Mol. Genet. 1999;8:493–500. doi: 10.1093/hmg/8.3.493. [DOI] [PubMed] [Google Scholar]
- 13.Zaphiropoulos P. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 1997;17:2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claudevilla C., Serra D., Miliar A., Codony C., Asins G., Bach M., Hegardt F.G. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl Acad. Sci. USA. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frantz S.A., Thiara A.S., Lodwick D., Ng L.L., Eperon I.C., Samani N.J. Exon repetition in mRNA. Proc. Natl Acad. Sci. USA. 1999;96:5400–5405. doi: 10.1073/pnas.96.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hide W.A., Babenko V.N., van Heusden P.A., Seoighe C., Kelso J.F. The contribution of exon-skipping events on chromosome 22 to protein coding diversity. Genome Res. 2001;11:1848–1843. doi: 10.1101/gr.188001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flouriot G., Brandt H., Seraphin B., Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J. Biol. Chem. 2002;277:26244–26251. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- 18.Takahara T., Kanazu S.I., Yanagisawa S., Akanuma H. Heterogenous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J. Biol. Chem. 2000;275:38067–38072. doi: 10.1074/jbc.M002010200. [DOI] [PubMed] [Google Scholar]
- 19.Akopian A.N., Okuse K., Souslova V., England S., Ogata N., Wood J.N. Trans-splicing of a voltage gated sodium channel is regulated by nerve growth factor. FEBS Lett. 1999;445:177–182. doi: 10.1016/s0014-5793(99)00126-x. [DOI] [PubMed] [Google Scholar]
- 20.Rigatti R., Jia J.H., Samani N.J., Eperon I.C. Exon repetition: a major pathway for processing mRNA of some genes is allele-specific. Nucleic Acids Res. 2004;32:441–446. doi: 10.1093/nar/gkh197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfsberg T.G., Landsman D. Expressed Sequence Tags (ESTs) In: Baxevanis A.D., Oulette B.F., editors. Bioinformatics—A Practical Guide to the Analysis of Genes and Proteins. New York: Wiley-Liss, Inc.; 2001. [Google Scholar]
- 22.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croft L., Schandorff S., Clark F., Burrage K., Arctander N., Mattick J.S. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet. 2000;24:340–341. doi: 10.1038/74153. [DOI] [PubMed] [Google Scholar]
- 24.Brett D., Hanke J., Lehmann G., Haase S., Delbruck S., Krueger S., Reich J., Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 25.Kan Z., Rouchka E.C., Gish W.R., States D.J. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modrek B., Resch A., Grasso C., Lee C. Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boguski M.S., Lowe T.M., Tolstoshev C.M. dbEST—database for ‘expressed sequence tags’. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 28.Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curwen V., Eyras E., Andrews D.T., Clarke L., Mongin E., Searle S., Clamp M. The Ensembl Automatic Gene Annotation System. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 31.Kent W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;4:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle E.I., Weng S., Gollub J., Heng J., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder—open source software for accessing Gene Ontolgy information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2005;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Shahrour F., Díaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 35.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 36.Wolfsberg T.G., Landsman D. A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res. 1997;25:1626–1632. doi: 10.1093/nar/25.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baranova A.V., Ivanov D.V., Krukovskaya L.L., Yankovsky N.K., Kozlov A.P. In silico screening for tumour-specific expressed sequences in the human genome. FEBS Lett. 2001;508:143–148. doi: 10.1016/s0014-5793(01)03028-9. [DOI] [PubMed] [Google Scholar]
- 38.Bains W. Virtually sequenced: the next genomic generation. Nat. Biotechnol. 1996;14:711–713. doi: 10.1038/nbt0696-711. [DOI] [PubMed] [Google Scholar]
- 39.Gupta S., Zink D., Korn B., Vingron M., Haas S.A. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics. 2004;20:2579–2585. doi: 10.1093/bioinformatics/bth288. [DOI] [PubMed] [Google Scholar]
- 40.Sorek R., Safer H.M. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 2003;31:1067–1074. doi: 10.1093/nar/gkg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brakenhoff R.H., Schoenmakers J.G., Lubsen N.H. Chimeric cDNA clones: a novel PCR artefact. Nucleic Acids Res. 1991;19:1949. doi: 10.1093/nar/19.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorek R., Shamir R., Ast G. How prevalent is functional alternative splicing in the human genome. Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Jongeneel C.V., Iseli C., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O'Hare M.J., Neville A.M., Simpson A.J.G., Strausberg R.L. Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc. Natl Acad. Sci. USA. 2003;100:4702–4705. doi: 10.1073/pnas.0831040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakharker M.K., Chow V.T.K., Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol. 2004;4:387–393. [PubMed] [Google Scholar]
- 45.Takahara T., Tasic B., Maniatis T., Akanuma H., Yanagisawa S. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol. Cell. 2005;18:245–251. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.