Abstract
Type IB DNA topoisomerases cleave and rejoin one strand of the DNA duplex, allowing for the removal of supercoils generated during replication and transcription. In addition, electron microscopy of cellular and viral TopIB–DNA complexes has suggested that the enzyme promotes long-range DNA–DNA crossovers and synapses. Here, we have used the atomic force microscope to visualize and quantify the interaction between vaccinia topoisomerase IB (vTopIB) and DNA. vTopIB was found to form filaments on nicked-circular DNA by intramolecular synapsis of two segments of a single DNA molecule. Measuring the filament length as a function of protein concentration showed that synapsis is a highly cooperative process. At high protein:DNA ratios, synapses between distinct DNA molecules were observed, which led to the formation of large vTopIB-induced DNA clusters. These clusters were observed in the presence of Mg2+, Ca2+ or Mn2+, suggesting that the formation of intermolecular vTopIB-mediated DNA synapsis is favored by screening of the DNA charge.
INTRODUCTION
Type IB DNA topoisomerases (TopIB) remove the torsional stress that accumulates in double-stranded DNA molecules during replication and transcription. TopIB cleaves and rejoins one strand of the DNA duplex (1), allowing for the removal of positive or negative supercoils from supercoiled DNA. The topoisomerase from vaccinia virus (vTopIB) is the smallest topoisomerase known (314 amino acids), suggesting that vTopIB constitutes the minimal functional unit of a type IB enzyme (2). vTopIB binds non-specifically to duplex DNA (3), although cleavage preferentially occurs at the target sequence 5′-(T/C)CCTT↓, where ↓ denotes the cleavage site (4,5). The sequence-specific cleavage of vTopIB has allowed for accurate determinations of the rate constants for cleavage and religation, hence its use as a prototype. Sedimentation analysis, gel filtration, N-terminal sequencing of the native protein and conceptual translation of the open reading frame of the vTopIB gene indicate that vTopIB is a monomeric protein with a molecular mass of 36.7 kDa (6–8). vTopIB consists of two domains joined through a hinge and binds circumferentially to DNA (2,9,10). DNaseI and exonuclease III footprinting give an upper bound to the number of base pairs covered by vTopIB when bound to DNA (25 bp, ∼8.5 nm) (5,11).
The binding of eukaryotic Type IB topoisomerase to DNA duplexes has been the subject of two previous studies that relied on visualization using electron microscopy (12,13), both of which demonstrated binding of topoisomerases at intramolecular DNA crossovers. vTopIB was found to form intramolecular loop structures in which non-contiguous DNA segments were synapsed within filamentous protein stems (13). Shuman et al. (13) suggested that these loops arise through protein–protein-mediated DNA synapsis. The presence of filament-like structures at high topoisomerase:DNA ratios and individual intramolecular nodes at low topoisomerase:DNA ratios led to the suggestion that binding of vTopIB might be cooperative. However, the experimental strategy, which included glutaraldehyde fixation, prevented reliable quantification of the binding vTopIB to DNA.
Atomic force microscopy (AFM) (14) presents several advantages over electron microscopy techniques, most notably that no fixation procedures are needed to adsorb biological molecules on a surface. Furthermore, AFM allows rapid visualization of many individual molecules and the measurements of heights and volumes. Indeed, AFM has proven to be a very suitable technique to characterize and quantify protein–protein and DNA–protein interactions (15–18), and therefore we employed this technique to study the interaction between vTopIB and DNA. AFM revealed individual vTopIB binding events, intramolecular synapsis and filament-like structures on individual DNA molecules at vTopIB:DNA ratios from 10:1 to 60:1. Our data are supported by a model in which DNA-bound vTopIB protein forms a single intramolecular node in the DNA. From this single intramolecular node, a filament-like structure extends by the binding of new vTopIB protomers. We have used the theory published by McGhee and von Hippel (19,20) to quantify the generation of filaments by vTopIB on DNA. By measuring the length of the filaments and the fractional saturation of the DNA as a function of the concentration of vTopIB, the cooperativity parameter ω and the association constant K* to form a node were determined. At higher protein:DNA ratios, intermolecular synapses of DNA molecules were observed in the presence of divalent ions, such as Mg2+, Ca2+ and Mn2+.
MATERIALS AND METHODS
DNA constructs
In all experiments, the pGEM-3Z plasmid (2743 bp) was used in nicked-circular or in linear form. The nicked-circular DNA was generated by relaxing the plasmid using the enzyme N.BstNB I (New England Biolabs). This enzyme nicks the plasmid at five positions. To generate a linear fragment, the plasmid was digested with BamHI (New England Biolabs). The nicked or linearized DNA molecules were then purified using a DNA purification kit (MoBio Laboratories), checked by gel electrophoresis and inspected by AFM. This method resulted in nicked-circular and linear DNA molecules with mean contour lengths of 919 ± 69 nm and 928 ± 52 nm, respectively. These values agree with the expected B-form contour length of the plasmid. The cleavage consensus sequence of vaccinia topoisomerase [5′-(C/T)CCTT↓], occurred 16 times in the sequence of the DNA used.
Vaccinia topoisomerase IB
vTopIB was purified as described previously (7). vTopIB was aliquoted and stored at −80°C in 10 mM Phosphate Buffer (PB) at pH 7.4. For each experiment, a new aliquot was thawed and diluted in PB to the desired concentration.
AFM sample preparation
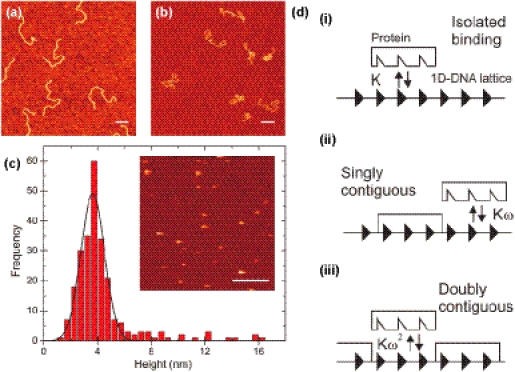
DNA molecules can be adsorbed on a mica surface using divalent ions (such as Mg2+, Mn2+ or Ca2+), or using polylysine (PL) (21,22). These two different adsorption methods are known to yield significantly different appearances of bare DNA on mica because in the first case DNA molecules equilibrate in a 2D configuration, while in the second case they are kinetically trapped onto the mica surface (23). Figure 1a shows DNA molecules adsorbed using 13 mM MgCl2 (similar concentrations were used in experiments involving MnCl2 or CaCl2, discussed below), and Figure 1b shows DNA molecules adsorbed on 0.01% polylysine-treated mica. In both cases, there was no vTopIB present in the solution and the length of the DNA used was identical. We observed individual DNA molecules homogeneously distributed over the mica surface in both cases. The polylysine-coated mica substrates were extensively rinsed with water and dried to remove any surface-mobile positive counterions (24,25). A control experiment was performed with vTopIB imaged in buffer in the absence of DNA (Figure 1c). The height distribution obtained from AFM (no. of events = 246) displayed a peak and was fitted to a Gaussian function, yielding an average height of 3.6 ± 0.9 nm, where we have quoted the standard deviation of the Gaussian function fit. From X-ray crystallography, the structure of the vTopIB catalytic domain shows it to be an oblong shaped globular protein with dimensions 5.6 × 2.8 × 2.6 nm (2). Our AFM height measurements agreed reasonably well with these dimensions given that they reflected both the average height of different orientations of the protein and the small deformation exerted by the tip in buffer (estimated at 15%, given that average heights of dsDNA molecules under the same conditions measured 1.7 ± 0.3 nm, compared with a nominal value of 2 nm). Modeling of the crystal structure of vTopIB onto B-form DNA indicated that vTopIB likely covers an 18 bp DNA segment (2). This value, n = 18 bp, will be used in our modeling below.
Figure 1.
Control experiments. DNA molecules are not observed to cluster in the absence of vTopIB when adsorbed on a mica surface using MgCl2 (a) or polylysine-coated mica (b). The appearance of the DNA molecules differs as a result of the adsorption processes involved. (c) Histogram of the height of vTopIB molecules measured in buffer in the absence of DNA. The average height of vTopIB in solution is 3.6 ± 0.9 nm, compatible with a monomeric state of the protein. An AFM image of vTopIB in buffer is shown as an inset in (c). Bar size is 200 nm. (d) Summary of cooperativity theory (19,20). Three different binding events can take place: isolated binding with an association constant K (d, i), singly contiguous binding with an association constant Kω (d, ii) and doubly contiguous binding with association constant Kω2 (d, iii). In our model an isolated binding event is attributed to the formation of an intramolecular node.
vTopIB–DNA binding reactions included 0.22 nM DNA (circular or linear) and 2.5–40 nM vTopIB in PB buffer, supplemented, when needed, with 13 mM MgCl2 to promote adsorption of the DNA molecules onto the mica. After mixing, the sample was incubated at 37°C for 15 min and deposited on a flat freshly cleaved mica surface, rinsed with water and dried under a gentle stream of nitrogen.
AFM imaging
Samples were imaged in air (except Figure 1c, which was taken in buffer solution) with a commercial AFM from Nanotec Electronica operating in dynamic mode, using soft cantilevers (0.39 N/m, Olympus OMCL-RC800PSA). Typical tapping amplitudes were 15–17 nm peak-to-peak at the resonance frequency of the cantilevers, ∼75 kHz. Image processing, including measurement of filament lengths, was carried out using WSxM freeware (www.nanotec.es). Standard image processing included plane subtraction and flattening.
Theoretical description of protein binding to DNA
We used the theory described by McGhee and von Hippel (19) to quantify the generation of filaments of vTopIB on DNA. We briefly summarize the theory's main elements here. The binding of a protein that covers n base pairs on a DNA is characterized by an association constant K (Figure 1d, i). The binding of a protein on one side of a previously bound protein (while on the other side there is no bound protein) has an association constant Kω. This is denoted singly contiguous binding (Figure 1d, ii). If a new protein binds between two previously bound proteins one speaks of a doubly contiguous binding event with an association constant Kω2 (Figure 1d, iii). ω is a measure for the degree of cooperativity of the binding process: ω = 1 is defined as non-cooperative binding whereas ω > 1 is defined as cooperative binding.
The parameters K and ω that characterize binding can be experimentally obtained from the AFM data by measuring the filament length distribution at a given DNA saturation θ, which is defined as the average fraction of DNA covered by the protein. The filament length c is defined as the number of proteins that are bound next to each other without a gap between them. θ can be experimentally determined from the images by calculating the fraction of the total contour length of all DNA molecules present in the sample that has proteins bound to it. At a given θ, the distribution of filament lengths depends on the values of n, K and ω (20). The normalized probability Pc of finding a filament of length c is
| 1 |
where P(b1|bn) is the probability of finding a protein bound right next to another one that is already bound to DNA (19). b1|bn denotes the protein b1 binding to the protein bn:
| 2 |
where
| 3 |
From experimental data, Pc can be calculated and plotted as a function of the filament length c. Using a maximum-likelihood (ML) data analysis (see below), a value for ω was obtained for different DNA saturations θ.
Once the cooperativity parameter ω is known, K can be deduced using Equation 4
| 4 |
where R and ν are defined in Equation 3, N is the total number of binding sites (bases of DNA) and L is the free protein concentration defined as L = LT − νN with LT being the total protein concentration (19). A fit of Equation 4 to the plot of θ versus the total concentration of vTopIB (LT) yields a value for the association constant K.
Data analysis
The length of the vTopIB filaments was measured by tracing the filaments with a segmented line. Tracing resulted in a height profile along the contour of the DNA molecule. The length of the filament was measured by attributing the beginning and the end of the filament to the positions where the height of the profile was reduced to half the maximum value. Next, the effect of tip convolution was taken into account by subtracting from the measured filament length the extra length induced by tip convolution, which was 13 nm. Tip convolution was estimated following ref. (26), taking into account the tip radius specified by the manufacturer, 10 nm, and the DNA radius, 1 nm.
The ML method was used to estimate the cooperativity parameter ω. The ML method is a powerful method to unambiguously estimate a parameter from a series of measurements (27). The ML method computes the distribution parameters that maximize the likelihood to observe a set of experimental outcomes given a known probability function. The likelihood function (LH) for the filament length distribution is defined as
| 5 |
where S is the number of data points in the data set and ci is the length of the ith filament. Equation 5 can be rewritten as
| 6 |
We then maximize this equation with respect to ω. Using the Maple software package together with Equations 2 and 3, an analytical solution for ω can be obtained:
| 7 |
Using Equation 7, ω can be calculated for different DNA saturations θ, which are experimentally obtained from the images. The uncertainty in ω corresponds to the places where ln LH is below its half maximum (28). Because LH is not symmetric around the peak value, the corresponding uncertainty is also asymmetric. However, due to the negligible difference between positive and negative errors found, we simply quote their mean.
RESULTS
vTopIB generates filament-like structures at low vTopIB:DNA ratios
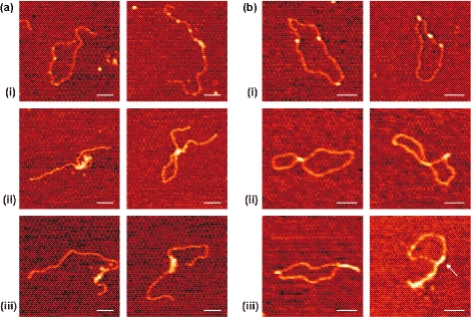
We characterized the binding of vaccinia topoisomerase IB to linear and circular DNA at vTopIB:DNA ratios from 10:1 to 60:1. These experiments are summarized in Figure 2a (linear DNA) and Figure 2b (circular DNA). We found three different types of binding events: (i) isolated binding, (ii) nodes and (iii) filament-like structures. We confirmed that binding of individual vTopIB proteins to DNA did not cause wrapping of the DNA around vTopIB as evinced by the absence of shortening of the DNA contour length upon vTopIB binding. The filament-like structures shown in Figure 2a (iii) and Figure 2b (iii) are qualitatively different from each other. In the first case, the filament-like structures involved a single DNA duplex while in the second case, with circular DNA, the filament-like structures involved two segments of the circular DNA, hence two DNA duplexes. This conclusion is further corroborated by measurements of the contour length of the uncovered and the protein-covered parts of the DNA. For instance, the full contour length of the DNA molecule in Figure 2b (iii) is only recovered if the length of the filament is counted twice, clearly showing that two duplexes participate in that filament. A small fraction of filaments (<10%) involving a single DNA duplex were also found when using circular DNA (see arrow in Figure 2). Although a tendency toward type (iii) events was observed with increasing vTopIB:DNA ratio, type (ii) or (iii) events were also found at low vTopIB:DNA ratios. This suggests cooperative behavior in the binding of vTopIB to DNA (13), which we quantify below.
Figure 2.
vTopIB–DNA complexes at low vTopIB:DNA values. The concentration of DNA is constant in all experiments and equal to 0.22 nM. (a) Gallery of AFM images of vTopIB bound to linear DNA. Three different types of complexes were found. (a, i) Individual vTopIB proteins bound to DNA ([vTopIB] = 4 nM), nodes (a, ii) and filament-like structures (a, iii) ([vTopIB] = 6.2 nM). (b) Gallery of AFM images of vTopIB bound to nicked-circular DNA. Similar types of complexes were found. (b, i) Individual vTopIB–DNA complexes ([vTopIB] = 4 nM), nodes (b, ii) and filament-like structures (b, iii) ([vTopIB] = 13.4 nM). Bar size is 100 nm.
The heights of isolated DNA-bound proteins, node events and filament-like structures were measured in air. In every experiment, we measured the height of the DNA (hDNA) as a reference height. This is especially relevant for AFM measurements conducted in air, as height measurements are then sensitive to experimental imaging conditions (29). Values for hDNA measured in air ranged from 0.6 to 0.9 nm. Isolated DNA-binding events yielded a value for the height of a single DNA-bound protein of 2.1 ± 0.5 times hDNA (no. of events = 127); node events, a value of 2.4 ± 0.5 times hDNA (no. of events = 20); and filament-like structures, a value of 2.1 ± 0.6 times hDNA (no. of events = 66). The height of individual DNA-free vTopIB molecules deposited on the mica surface was 1.4 ± 0.3 times hDNA (no. of events = 63).The height distribution of isolated DNA-bound proteins consisted of a single peak in all tested experimental conditions (data not shown), consistent with the fact that vTopIB binds to DNA as a monomer (2).
Intermolecular synapsis of DNA molecules is found at high vTopIB:DNA ratios
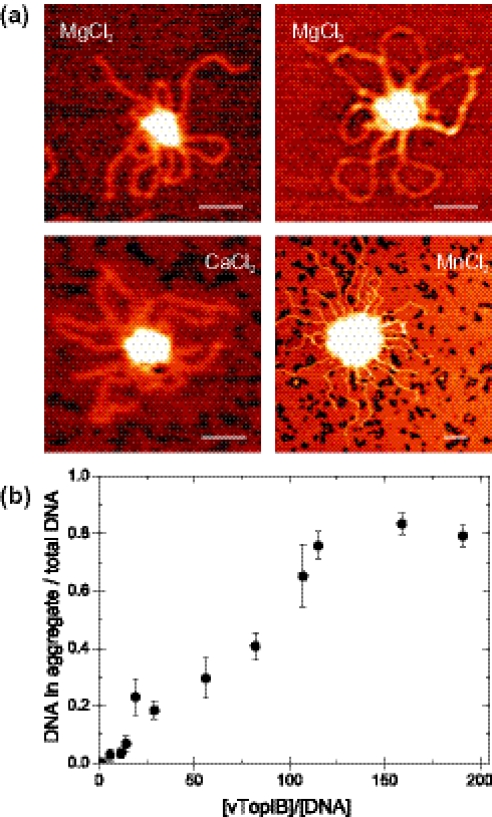
Focusing on the protein filament-like structures, we increased the concentration of vTopIB per DNA molecule in order to see if longer protein filaments would result. Instead, this resulted in large clusters containing many DNA molecules (Figure 3). Such intermolecular synapses were found on both linear and circular DNA in the presence of divalent ions, such as Mg2+, Mn2+ and Ca2+ (Figure 3a). Qualitatively, we did not note any cation-specific effect. Hence, we focused the remainder of our study on the biologically more relevant Mg2+. We quantified this phenomenon counting the number of DNA molecules involved in such clusters as a function of vTopIB concentration. Figure 3b plots the analysis of intermolecular synapsis using ∼1300 molecules, all adsorbed in the presence of MgCl2. The number of DNA molecules involved in a cluster increased monotonically with the vTopIB:DNA ratio (Figure 3b). This trend saturated at a protein:DNA ratio of 125:1. We expect the number of DNA molecules involved in a cluster to be underestimated at such high vTopIB:DNA ratios for two reasons. First, the identification of the individual DNA molecules becomes progressively more difficult. Second, since the number of DNA molecules was constant in all experiments the clusters were found increasingly far from each other. This results in uncertainties in determining the average number of DNA molecules per cluster because very large areas must be scanned with the AFM to yield enough statistics.
Figure 3.
Intermolecular synapsis of DNA at high vTopIB:DNA values. The concentration of DNA is constant in all experiments and equal to 0.22 nM. (a) AFM images of linear and circular DNA molecules incubated with [vTopIB] = 12.4 nM and [vTopIB] = 45 nM, respectively, showed clustering in the presence of Mg2+ cations (top). This effect was not cation-specific, as incubation of linear DNA and 12.4 nM [vTopIB] together with Ca2+ or Mn2+ likewise resulted in the formation of aggregates. Bar size is 100 nm. (b) Intermolecular synapsis of DNA was quantified by counting the number of DNA molecules involved in an cluster at different vTopIB:DNA ratios in the presence of MgCl2. The fraction of DNA molecules in a cluster increased linearly with increasing vTopIB concentration and saturated at very high vTopIB concentrations. This saturation was due to an underestimation of the number of DNA molecules involved (see text for details).
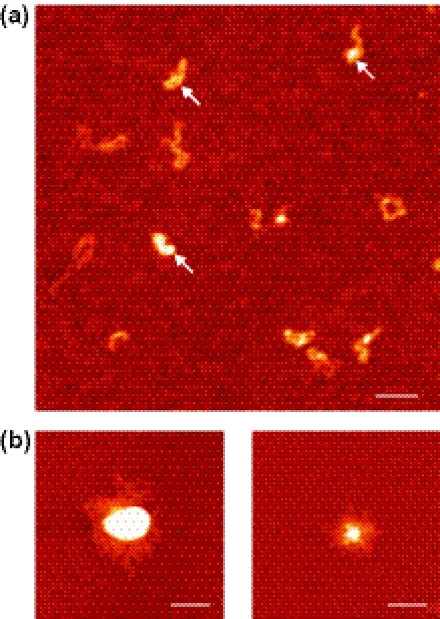
We also explored the effect of divalent ions in the generation of intermolecular synapses. Typically, vTopIB–DNA binding reactions included 13 mM MgCl2 in order to adhere the DNA to the mica (DNA does not adsorb on a bare mica surface). To isolate the effect of divalent ions, we instead treated the mica surface with polylysine, which is a polymer that positively charges the surface of the mica. Again we observed binding of vTopIB to DNA (Figure 4a, white arrows). Surprisingly, in the absence of divalent ions DNA molecules were always isolated from each other, even at high vTopIB concentrations (vTopIB:DNA = 123:1). To conclude that the observation of isolated DNA molecules was not caused by the chemistry of mica adsorption, we verified that adding 13 mM MgCl2 to the binding reaction and adsorbing the product on polylysine mica again resulted in the observation of DNA clusters (Figure 4b). Note that the difference in the physical appearance of the molecules and clusters shown in Figures 3 and 4 is due to the different adsorption processes on magnesium-coated and polylysine-coated mica (see Materials and Methods).
Figure 4.
Intermolecular synapsis of DNA is affected by the presence of MgCl2. (a) Linear DNA molecules were deposited on polylysine-coated mica and incubated with vTopIB in the absence of MgCl2 again resulting in the observation of vTopIB-bound DNA (white arrows). Under these conditions, however, the DNA molecules did not cluster. (b) Linear DNA molecules deposited on polylysine-coated mica showed intermolecular synapses when incubated with vTopIB and MgCl2. Bar size is 200 nm.
vTopIB generates filaments on DNA in a cooperative fashion
We used the AFM images obtained to quantify the binding of vTopIB to DNA. The quantification of the binding of vTopIB to DNA that follows was performed on circular DNA and restricted to low DNA saturation, θ < 0.25. We used circular DNA to favor protein–protein interactions and low θ to avoid intermolecular synapsis of DNA.
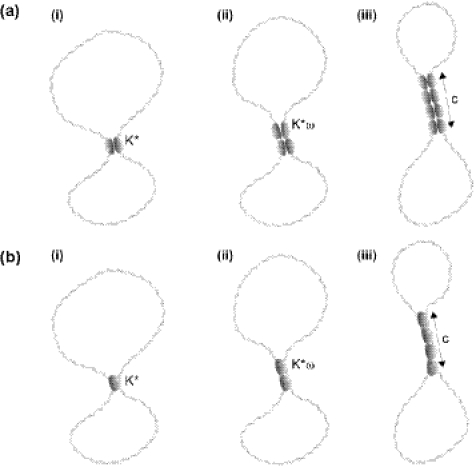
We now describe the extension of vTopIB filaments on DNA using the terminology of ref. (19), which is detailed in Materials and Methods. The formation of vTopIB filaments is likely to happen in the following steps (Figure 5). First, a single intramolecular node is formed (Figure 5a, i and Figure 5b, i). This intramolecular node may be composed of either a single vTopIB engaging two DNA duplexes or a dimer of vTopIB in which each monomer is bound to a single DNA duplex. We describe the association of a vTopIB monomer or dimer to the two DNA duplexes with the association constant K*. Second, the filament is extended by the addition of new vTopIB protomers (Figure 5, panel ii). The extension of the node by vTopIB has an association constant of K*ω, where ω is the cooperativity parameter for extending the filament. Values of ω larger than one indicate a cooperative process. Finally, a filament of length c is generated (Figure 5a, iii and Figure 5b, iii). Figure 5 shows a general cartoon depicting the formation and extension of a filament by either vTopIB monomers or dimers. The theory used here is applicable to both cases (see Discussion).
Figure 5.
Model describing the formation of filaments of vTopIB on DNA. The constituents of the filaments may be vTopIB dimers (a) or monomers (b). The formation of filaments requires several steps. (i) The formation of an intramolecular node with an association constant K*. (ii) The node is extended by binding of new vTopIB protomers with an association constant of K*ω. (iii) A filament of length c is generated.
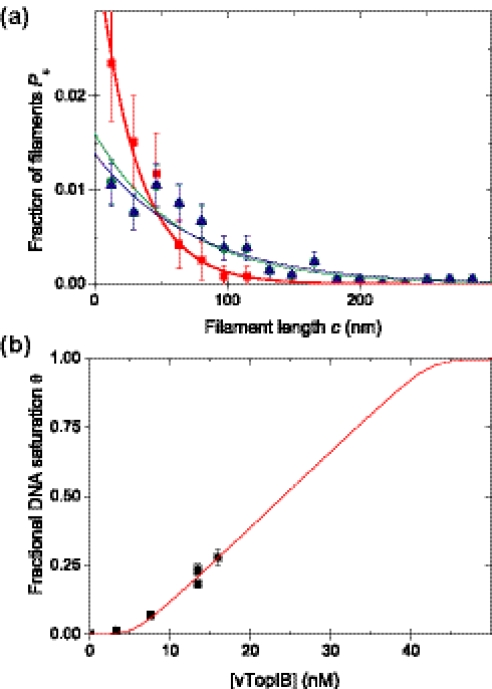
In order to determine the association constant K* and the cooperativity parameter ω of filament extension by vTopIB, we measured the length of filaments for several hundreds of DNA molecules at different vTopIB:DNA ratios. An average filament length was calculated and the DNA saturation θ was deduced. The distribution of filament lengths Pc is shown in Figure 6a for three different DNA saturations (θ = 0.07, 0.18 and 0.23). For each data set, the cooperativity parameter ω was estimated using the ML method described in Materials and Methods. The results are summarized in Table 1. Subsequently, Equation 1 was plotted in Figure 6a using the ω obtained for each data set. The error-weighted average cooperativity parameter was 7.7 ± 1.1 × 103. This value shows the generation of filaments on DNA by vTopIB is highly cooperative.
Figure 6.
Quantitative analysis of the binding of vTopIB to DNA. (a) Filament length distribution for three different data sets. For each set ω was estimated using the ML method resulting in ω = 5.6 ± 1.4 × 103 for DNA saturation θ = 0.07 (closed squares); ω = 9.1 ± 1.7 × 103 for DNA saturation θ = 0.18 (closed circles); and ω = 8.8 ± 1.8 × 103 for DNA saturation θ = 0.23 (closed triangles). The solid lines in (a) are plots of Equation 1 using the obtained ω. (b) Fractional DNA saturation as a function of total vTopIB concentration. The fit of Equation 4 to these data using n = 18, N = 2743 and ω = 7.7 ± 1.1 × 103 yielded a value for K* = 4.0 ± 0.4 × 104 M−1.
Table 1.
Cooperativity parameters for different DNA saturations considering n = 18 bp
| Data set | Number of filaments | Number of DNA | θ | ω |
|---|---|---|---|---|
| 1 | 70 | 107 | 0.07 | 5.6 ± 1.4 × 103 |
| 2 | 123 | 116 | 0.18 | 9.1 ± 1.7 × 103 |
| 3 | 95 | 98 | 0.24 | 8.8 ± 1.8 × 103 |
We also estimated the binding constant K* of the formation of a vTopIB–DNA node. Figure 6b shows DNA saturations (θ) as a function of total vTopIB concentrations. This data was fitted to Equation 4 using the least squares fitting method, yielding K* = 4.0 ± 0.6 × 104 M−1 given ω = 7.7 ± 1.1 × 103. The product K*ω was found to equal 3.1 ± 0.4 × 108 M−1.
DISCUSSION
We quantified the generation of filaments of vTopIB on DNA using a theory of DNA binding (19,20). vTopIB was found to form filaments on DNA in a highly cooperative fashion. Our analysis gave a value of 7.7 ± 1.1 × 103 for the cooperativity parameter ω, and 4.0 ± 0.6 × 104 M−1 for the association constant to generate a single intramolecular node K*. Kowalczykowski et al. (20) defined sigmoidal (cooperative) filament growth by the criterion ω > 10n. Our measurement of ω exceeds this threshold 40-fold. Our values are similar to those obtained for different cooperative binding proteins. For example, the gene 32 protein binds cooperatively to poly(rA) with parameters ω = 1.2 × 103, n = 7.5 and K = 9 × 103 M−1 (30); and The T4 UvsX protein forms filaments on single-stranded DNA with parameters ω = 1.0 × 102, n = 4.0 and K = 4.3 × 104 M−1 (31). The values for K* and ω quoted in this work may have a slightly different interpretation compared to other reported values. The reason for this is that in our work, the parameters K* and ω relate to the respective formation and extension of a node [consisting of two monomers of vTopIB (Figure 5a) or a single monomer (Figure 5b)], whereas the parameters obtained in the binding of gene 32 protein and the T4 UvsX-protein relate to single monomers binding to their substrate.
We now discuss whether vTopIB monomers or dimers are the minimal constituents of a single intramolecular synapsis. AFM-volume analysis has been proven to be useful in distinguishing between proteins adsorbed as monomers or as dimers (32). However, the volume analysis becomes less straightforward when proteins are small, as in the case of 36.7 kDa vTopIB, and becomes especially non-transparent when they are bound to DNA, because the proteins are of comparable height to the bare DNA. We found that, within the error, the heights of isolated binding events and filament-like structures were identical and equal to 2.1 ± 0.5 times hDNA. In the case of vTopIB binding to DNA, it was therefore impossible to unambiguously determine the number of vTopIB monomers that make up the intramolecular node, and hence we cannot rule out the possibility of a monomer of vTopIB being involved in the synapse of two DNA molecules.
The DNA aggregation found at high vTopIB:DNA ratios can be understood as an extension of the cooperative binding behavior presented above. Essentially, at similar protein:DNA ratios we found that intermolecular synapses are favored on linear DNA molecules while intramolecular synapses are favored in circular ones. This is unsurprising considering that the radius of gyration is larger for linear DNA than for circular DNA. Topological constrains of circular DNA could facilitate the propagation of a filament built up of vTopIB. Hence, interactions between two duplexes of the same molecule are more likely to occur in circular than in linear DNA. As expected, at very high protein–DNA ratios we also found clusters of circular DNA.
Interestingly, we found a correlation between DNA aggregation and the presence of the divalent ions Mg2+, Ca2+ or Mn2+. The fact that the aggregation was not an effect only caused by one specific ion supports the notion that DNA aggregation is a more general effect. In general, divalent ions reduce the Debye length, which is the characteristic length scale of the electrostatic interaction between two charged objects in an electrolyte solution (33). Since vTopIB does not bind divalent ions (34), their presence in the binding reaction has the effect of reducing the electrostatic repulsion between two DNA molecules and, as a consequence, in favoring interactions between two DNA-bound vTopIB molecules. Divalent ions consequently favor the formation of intermolecular synapsis.
Although our measurements provide clear evidence for cooperative extension of vTopIB filaments on DNA duplexes, the question remains what mechanism gives rise to the observed cooperativity. This question is especially intriguing given the fact that vTopIB is most likely a monomer in solution (6), in accordance with Figure 1. The clustering reported here is strictly DNA mediated. We speculate that a conformational change in the vTopIB due to DNA binding may be responsible for the cooperativity observed here. Such conformational changes of proteins upon DNA binding have been previously reported (35,36). Specifically, dimerization of the DNA-binding domains of the retinoid X receptor has been shown to be induced by DNA binding (37). Similarly, a conformational change in vTopIB upon DNA binding may trigger multimerization of vTopIB. Such a conformational change in vTopIB may seem reasonable given its clamp-like structure and the presence of a hinge domain (2,10)
Besides altering the number of supercoils in DNA molecules, Type IB topoisomerase have been shown to possess strand transferase activity that can result in DNA recombination (38–42). Furthermore, Type IB topoisomerase and site-specific tyrosine recombinases have been shown to share structural features in their catalytic domains and descend from a common ancestor (2). The vTopIB-mediated DNA synapsis shown and quantified here may be the first step in its strand transferase activity and we speculate that it may have been inherited from its ancestor. A filament structure built up of vTopIB dimers similar to those reported here could help to keep the two DNA duplexes to be joined in close proximity. In addition, our observation of vTopIB-mediated DNA condensation is consistent with the suggestion that this topoisomerase assists packaging the DNA into progeny virus particles (13), where it plays a role in the transcription of viral early genes (43).
In summary, we have presented the first quantitative AFM study of vTopIB binding to DNA. AFM images showed individual vTopIB proteins binding to DNA, single intramolecular synapsis and filament-like structures. vTopIB generated filament-like structures on individual DNA molecules at protein:DNA ratios between 10:1 and 60:1. We calculated the association constant of creating an intramolecular node by vTopIB and the cooperativity parameter from the filament-lengths distribution at different DNA saturations. From these values, we conclude that the formation of filaments on DNA by vTopIB is a highly cooperative process. In addition, intermolecular synapses of DNA were observed at high vTopIB:DNA ratios. Interestingly, this interaction was promoted by divalent ions.
Acknowledgments
F.M.-H. is supported by a postdoctoral fellowship from La Fundación Ramón Areces. The authors acknowledge Dick Korbee for technical support in the mechanical and acoustical isolation of the AFM. C.D. and N.H.D. acknowledge financial support from FOM (Dutch Foundation for Fundamental Research on Matter) and NWO (Netherlands Organization for Scientific Research). S.S. acknowledges support from NIH grant GM46330. Funding to pay the Open Access publication charges for this article was provided by FOM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Cheng C., Kussie P., Pavletich N., Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 3.Sekiguchi J., Shuman S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 1994;22:5360–5365. doi: 10.1093/nar/22.24.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- 5.Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J. Biol. Chem. 1991;266:11372–11379. [PubMed] [Google Scholar]
- 6.Shaffer R., Traktman P. Vaccinia virus encapsidates a novel topoisomerase with the properties of a eucaryotic type I enzyme. J. Biol. Chem. 1987;262:9309–9315. [PubMed] [Google Scholar]
- 7.Shuman S., Golder M., Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J. Biol. Chem. 1988;263:16401–16407. [PubMed] [Google Scholar]
- 8.Shuman S., Moss B. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc. Natl Acad. Sci. USA. 1987;84:7478–7482. doi: 10.1073/pnas.84.21.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma A., Hanai R., Mondragon A. Crystal structure of the amino-terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution. Structure. 1994;2:767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- 10.Sekiguchi J., Shuman S. Vaccinia topoisomerase binds circumferentially to DNA. J. Biol. Chem. 1994;269:31731–31734. [PubMed] [Google Scholar]
- 11.Cheng C., Shuman S. Site-specific DNA transesterification by vaccinia topoisomerase: role of specific phosphates and nucleosides. Biochemistry. 1999;38:16599–16612. doi: 10.1021/bi992001d. [DOI] [PubMed] [Google Scholar]
- 12.Zechiedrich E.L., Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuman S., Bear D.G., Sekiguchi J. Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J. 1997;16:6584–6589. doi: 10.1093/emboj/16.21.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binnig G., Quate C.F., Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 15.Viani M.B., Pietrasanta L.I., Thompson J.B., Chand A., Gebeshuber I.C., Kindt J.H., Richter M., Hansma H.G., Hansma P.K. Probing protein–protein interactions in real time. Nature Struct. Biol. 2000;7:644–647. doi: 10.1038/77936. [DOI] [PubMed] [Google Scholar]
- 16.Hansma H.G. Surface biology of DNA by atomic force microscopy. Annu. Rev. Phys. Chem. 2001;52:71–92. doi: 10.1146/annurev.physchem.52.1.71. [DOI] [PubMed] [Google Scholar]
- 17.van Noort S.J., van der Werf K.O., Eker A.P., Wyman C., de Grooth B.G., van Hulst N.F., Greve J. Direct visualization of dynamic protein–DNA interactions with a dedicated atomic force microscope. Biophys J. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno-Herrero F., Herrero P., Colchero J., Baro A.M., Moreno F. Imaging and mapping protein-binding sites on DNA regulatory regions with atomic force microscopy. Biochem. Biophys. Res. Commun. 2001;280:151–157. doi: 10.1006/bbrc.2000.4074. [DOI] [PubMed] [Google Scholar]
- 19.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA–protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 20.Kowalczykowski S.C., Paul L.S., Lonberg N., Newport J.W., McSwiggen J.A., von Hippel P.H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 21.Bussiek M., Mucke N., Langowski J. Polylysine-coated mica can be used to observe systematic changes in the supercoiled DNA conformation by scanning force microscopy in solution. Nucleic Acids Res. 2003;31:e137. doi: 10.1093/nar/gng137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams R.C. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc. Natl Acad. Sci. USA. 1977;74:2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 24.Fang Y., Hoh J.H. Cationic silanes stabilize intermediates in DNA condensation. FEBS Lett. 1999;459:173–176. doi: 10.1016/s0014-5793(99)01237-5. [DOI] [PubMed] [Google Scholar]
- 25.Crampton N., Bonass W.A., Kirkham J., Thomson N.H. Formation of aminosilane-functionalized mica for atomic force microscopy imaging of DNA. Langmuir. 2005;21:7884–7891. doi: 10.1021/la050972q. [DOI] [PubMed] [Google Scholar]
- 26.Bustamante C., Vesenka J., Tang C.L., Rees W., Guthold M., Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992;31:22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- 27.Chatfield C. Statistics for Technology. 3rd edn. Chapman & Hall; 1996. [Google Scholar]
- 28.Brock I. Statistical Methods of Data Analysis. 2001. Chapter 6 Retrieved June, 2005, from the University Bonn website: http://www-zeus.physik.uni-bonn.de/~brock/stat_ws0001/ [Google Scholar]
- 29.Jiao Y., Schaffer T.E. Accurate height and volume measurements on soft samples with the atomic force microscope. Langmuir. 2004;20:10038–10045. doi: 10.1021/la048650u. [DOI] [PubMed] [Google Scholar]
- 30.Lonberg N., Kowalczykowski S.C., Paul L.S., von Hippel P.H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III) J. Mol. Biol. 1981;145:123–138. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 31.Ando R.A., Morrical S.W. Single-stranded DNA binding properties of the UvsX recombinase of bacteriophage T4: binding parameters and effects of nucleotides. J. Mol. Biol. 1998;283:785–796. doi: 10.1006/jmbi.1998.2124. [DOI] [PubMed] [Google Scholar]
- 32.Ratcliff G.C., Erie D.A. A novel single-molecule study to determine protein–protein association constants. J. Am. Chem. Soc. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- 33.Israelachvili J. Intermolecular and Surface Forces. London: Academic Press; 1991. [Google Scholar]
- 34.Stivers J.T., Shuman S., Mildvan A.S. Vaccinia DNA topoisomerase I: single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry. 1994;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 35.Boskovic J., Rivera-Calzada A., Maman J.D., Chacon P., Willison K.R., Pearl L.H., Llorca O. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J. 2003;22:5875–5882. doi: 10.1093/emboj/cdg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Herrero F., de Jager M., Dekker N.H., Kanaar R., Wyman C., Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon DNA binding. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 37.Holmbeck S.M., Dyson H.J., Wright P.E. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J. Mol. Biol. 1998;284:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- 38.Shuman S. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:3489–3493. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuman S. Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific. Proc. Natl Acad. Sci. USA. 1991;88:10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J. Biol. Chem. 1992;267:8620–8627. [PubMed] [Google Scholar]
- 41.Shuman S. Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J. Biol. Chem. 1992;267:16755–16758. [PubMed] [Google Scholar]
- 42.Christiansen K., Westergaard O. Characterization of intra- and intermolecular DNA ligation mediated by eukaryotic topoisomerase I. Role of bipartite DNA interaction in the ligation process. J. Biol. Chem. 1994;269:721–729. [PubMed] [Google Scholar]
- 43.Da Fonseca F., Moss B. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proc. Natl Acad. Sci. USA. 2003;100:11291–11296. doi: 10.1073/pnas.1534874100. [DOI] [PMC free article] [PubMed] [Google Scholar]