Abstract
C1r is the modular serine protease (SP) that mediates autolytic activation of C1, the macromolecular complex that triggers the classical pathway of complement. The crystal structure of a mutated, proenzyme form of the catalytic domain of human C1r, comprising the first and second complement control protein modules (CCP1, CCP2) and the SP domain has been solved and refined to 2.9 Å resolution. The domain associates as a homodimer with an elongated head-to-tail structure featuring a central opening and involving interactions between the CCP1 module of one monomer and the SP domain of its counterpart. Consequently, the catalytic site of one monomer and the cleavage site of the other are located at opposite ends of the dimer. The structure reveals unusual features in the SP domain and provides strong support for the hypothesis that C1r activation in C1 is triggered by a mechanical stress caused by target recognition that disrupts the CCP1–SP interfaces and allows formation of transient states involving important conformational changes.
Keywords: activation/complement/innate immunity/modular structure/serine protease
Introduction
Complement is a major element of antimicrobial host defense, through its ability to recognize pathogens and limit infection in the early phase after exposure to microorganisms. It is also recognized that complement orientates and stimulates the subsequent adaptive immune response (Fearon and Locksley, 1996; Hoffmann et al., 1999). Paradoxically, due to either uncontrolled activity or adventitious recognition of antigens from self, complement activation may also be involved in various pathologies, including Alzheimer’s disease (Rogers et al., 1992) and prion diseases (Klein et al., 2001; Mabbott et al., 2001).
The classical pathway of complement is triggered by C1, a 790 kDa multimolecular complex comprising a recognition subunit C1q and two modular serine proteases (SPs), C1r and C1s, that are associated into a C1s–C1r–C1r–C1s tetramer in a Ca2+-dependent manner (Cooper, 1985; Arlaud et al., 1998). C1q is a hexamer of heterotrimers with N-terminal collagen-like fibers ending in C-terminal globular domains, and has the overall shape of a bunch of tulips (Kishore and Reid, 2000). Binding of the C1q subunit through its globular regions to a target pathogen triggers activation of the C1 complex, a two-step mechanism involving: (i) autolytic activation of C1r through cleavage of its Arg446–Ile447 bond; and then (ii) C1r-mediated activation of C1s through cleavage of its Arg426–Ile427 bond. Once activated, C1s will specifically cleave C4 and C2, the C1 substrates, and thereby initiate a series of proteolytic reactions resulting in diverse biological activities aimed at providing a first line of defense against infection.
C1r is a modular protease that comprises, starting from the N-terminal end, a CUB module (Bork and Beckmann, 1993), an epidermal growth factor (EGF)-like module, a second CUB module, two contiguous complement control protein (CCP) modules (Reid et al., 1986) and a chymotrypsin-like SP domain (Journet and Tosi, 1986; Leytus et al., 1986; Arlaud et al., 1987a). This type of modular architecture is shared by C1s, and by the mannan-binding lectin (MBL)-associated SPs (MASPs), a family of enzymes that, as established in the case of MASP-2, trigger complement activation through the MBL pathway (Sato et al., 1994; Thiel et al., 1997; Thielens et al., 2001). It is well established that C1r is composed of two functionally distinct regions (Arlaud et al., 1998): (i) the N-terminal CUB1–EGF interaction domain, which mediates the Ca2+-dependent association between C1r and C1s within the C1s–C1r–C1r–C1s tetramer (Thielens et al., 1999), and is also involved in the interaction between the tetramer and the collagen-like region of C1q (Arlaud et al., 1998); and (ii) the C-terminal CCP1–CCP2–SP catalytic domain, which associates as a non-covalent homodimer forming the core of the C1s–C1r–C1r–C1s tetramer and mediates the autolytic activation of C1r and the subsequent proteolytic cleavage of C1s (Villiers et al., 1985; Arlaud et al., 1986; Weiss et al., 1986; Lacroix et al., 1997).
Based on the above information, low-resolution models of the C1 complex have been proposed (Schumaker et al., 1986; Weiss et al., 1986; Arlaud et al., 1987b), in which the C1s–C1r–C1r–C1s tetramer folds into a compact ‘figure of eight’ shaped conformation that allows contact between the catalytic regions of C1r and C1s, and hence accounts for the two-step activation process of C1. Resolution of the crystal structure of the catalytic domain of C1s has provided a solid structural basis for the understanding of its substrate specificity and of its proteolytic function within the C1 complex (Gaboriaud et al., 2000). However, no equivalent information is currently available on either the molecular mechanisms that allow C1r to successively activate itself, and then to cleave C1s, or the nature of the conformational signal that triggers C1r activation when C1 binds to a target. Here, we describe the three-dimensional structure of a recombinant mutated proenzyme form of the CCP1–CCP2–SP segment of C1r and elaborate on the complex mechanisms involved in C1r activation.
Results and discussion
We have determined the three-dimensional structure of the catalytic domain of human C1r, comprising residues 280–688, which encompass the CCP1 and CCP2 modules and the C-terminal chymotrypsin-like SP domain. The segment was expressed in a baculovirus/insect cells system and stabilized in the proenzyme form by means of a mutation at the cleavage site (Arg446→Gln). The structure was solved by molecular replacement and refined at 2.9 Å resolution (see Materials and methods). The final Rwork and Rfree factors are 0.242 and 0.292, respectively, and the stereochemistry of the model is of good quality (Table I).
Table I. Refinement statistics for the mutated proenzyme structure.
| Resolution range (Å) | 12–2.9 |
| No. of unique reflections | 27 235 |
| Rwork (%) | 24.2 |
| Rfree (%) | 29.2 |
| No. of protein residues (A/B) | 369/367 |
| Average B-factor (Å2) | 57 |
| No. of carbohydrate residues | 8 |
| No. of water molecules | 18 |
| R.m.s.d. bond length (Å) | 0.015 |
| R.m.s.d. bond angle (°) | 1.9 |
A dimeric head-to-tail assembly
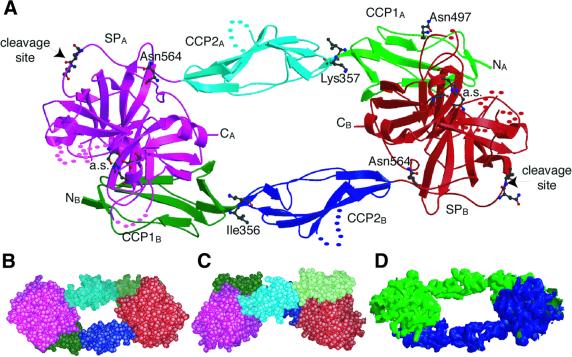
In agreement with previous observations (Villiers et al., 1985; Arlaud et al., 1986; Weiss et al., 1986; Lacroix et al., 1997), the catalytic domain of C1r associates as a homodimer (Figure 1). The two molecules interact in a head-to-tail fashion involving contacts between the CCP1 module of one monomer and the SP domain of the other, the resulting assembly displaying a pseudo 2-fold symmetry. The overall structure is 116 Å long, and 56 Å wide at its widest point. The most striking feature is a large opening, with estimated dimensions of 30 × 13 Å, found in the center of the dimer (Figure 1B). This opening could not be filled, even partially, by disordered oligosaccharide chains, because the two glycosylation sites of the SP domain lie elsewhere, one in an outer region of the structure (Asn497) and the other close to the interface with CCP1 (Asn564). Another remarkable feature of the assembly is the location of the SP catalytic sites at either end of the dimer, facing away from the central opening. The N-terminal end of each CCP1–CCP2–SP fragment, which normally connects to the CUB1–EGF–CUB2 interaction domain in intact C1r, also lies at each extremity of the structure. The mutated Gln446–Ile447 stretch, corresponding to the original Arg446–Ile447 cleavage site of each monomer, is located in a loop that protrudes from opposite corners of the dimer, ∼92 Å away from the catalytic Ser637 of the other monomer. Each monomer has a slightly twisted overall shape and, as a result, when seen edge-on, the dimer exhibits a butterfly-like shape (Figure 1C) that is reminiscent of electron microscopy pictures of the corresponding C1r fragment (Weiss et al., 1986).
Fig. 1. Homodimeric structure of the CCP1–CCP2–SP C1r catalytic domain. (A) Overall view of the structure of the zymogen. CCP1 modules are in green, CCP2 modules in blue and SP domains in magenta (molecule A) or red (molecule B). The residues at the catalytic sites (a.s.) and at the cleavage sites are shown, as are the residues Asn497 and Asn564, which bear oligosaccharide chains, and the residues Ile356 and Lys357 at the CCP1–CCP2 interface. NA, NB and CA, CB indicate the N- and C-terminal ends of molecules A and B. Dots represent disordered segments. (B and C) Space-filling representations of the bottom and side views of the structure. (D) Electron density map of the activated domain at 4 Å resolution. The map was contoured at the 1σ level and smoothed by solvent flattening (see Materials and methods).
Structure of the SP domain
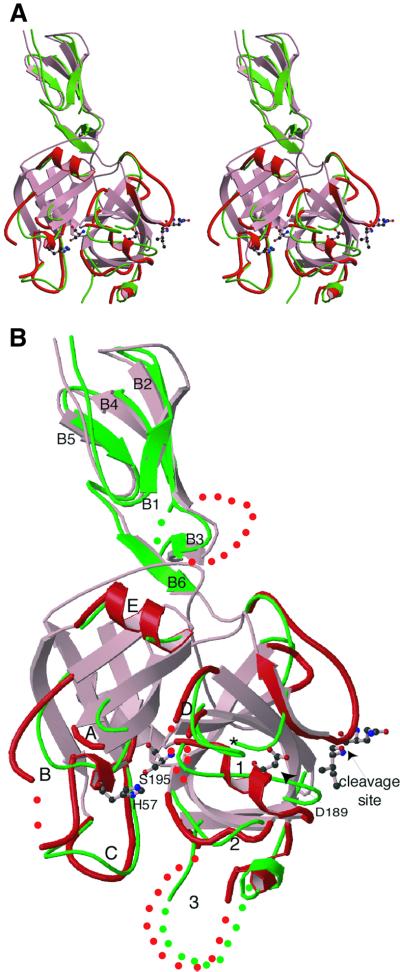
The core of the SP domain of C1r has the typical fold of chymotrypsin-like SPs, with two six-stranded β-barrels connected by three trans-segments and a C-terminal α-helix (Figure 2). Many of the surface loops of C1r differ in length and in conformation relative to the other SPs of known three-dimensional structure (see Table II; Figure 3A). The two major insertions occur in loops 3 and B, at opposite sides of the catalytic site entrance (Figure 2). A striking characteristic of the C1r SP domain is that loop E has an α-helical structure (Figure 2). This loop, usually involved in Ca2+ binding, has never been observed in such a conformation in any of the SPs of known structure. This helix largely contributes to the SP domain–CCP1 module interaction involved in the assembly of the dimer (see below).
Fig. 2. Three-dimensional structure of the CCP2–SP region of C1r and comparison with the homologous region of C1s: (A) stereoview; (B) detailed representation of the structures. The structures of C1r (red) and C1s (green) are superimposed. The loops in the SP domains are labeled according to Perona and Craik (1997), and the β-strands in the CCP module are numbered from B1 to B6. In the C1r structure, the His485(57) and Ser637(195) residues of the catalytic triad, the mutated Gln446(15)–Ile447(16) cleavage site, and Asp631(189) are shown with ball and sticks. The asterisk indicates the position of Ile423(16) in C1s after activation. Dots represent disordered segments. In the SP domain of C1s, only surface loops differing from the common core are shown for the sake of clarity.
Table II. Structural comparison of the C1r SP domain with homologous X-ray structures.
| (A) Overall structure | ||||||
|---|---|---|---|---|---|---|
| |
Chymotrypsinogen |
C1s |
Prethrombin-2 |
Plasminogen |
|
|
| Global r.m.s.d. (Å)/no. Ca | 1.38/190 | 1.06/190 | 1.26/203 | 1.37/185 | ||
| PDB code |
2cga |
1elv |
1mkw |
1ddj |
|
|
| (B) Surface segments of the C1r SP domain significantly different (D >1.5 Å) from the homologous segments of other SP domainsa | ||||||
| C1r numbering | Chymotrypsin numbering | Loop labelb | High B-factors | Disordered segments | Type of variation | More similar to: |
| 438–449 | 5–18 | act. pept.c | 443–445 | variable | ||
| 465–469 | 34–42 | A | deletion | C1s | ||
| 488–502 | 60–65 | B | 490–495 | 493–494 | insertion | prethrombin-2 |
| 510–517 | 75–82 | E | same length | |||
| 530–535 | 95–97 | C | 531–538 | insertion | ||
| 563–570 | 125–132 | variable | ||||
| 580, 586–587 | 142, 151–152 | D | 586–587 | 581–585 | deletion | |
| 599–616 | 164–175 | 3 | 604 | 605–617 | insertion | C1s |
| 626–635 | 185–192 | 1 | insertion | |||
| 644–650 | 202 | 647 | insertion | prethrombin-2 | ||
| 660–666 | 216–222 | 2 | 659–664 | deletion | C1s | |
aOther SP domains: C1s, chymotrypsin, trypsin, prethrombin-2, factor D, plasminogen.
bLoop labels as defined by Perona and Craik (1997), and used in Figure 2.
cActivation peptide.
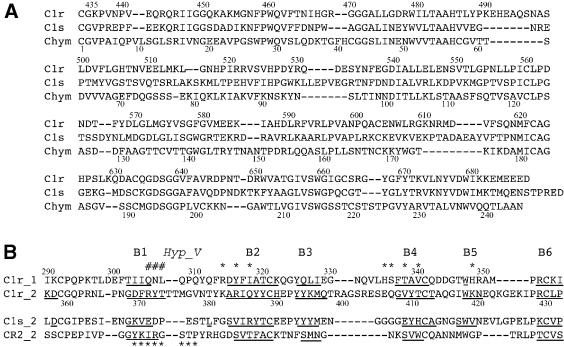
Fig. 3. Structural alignments of (A) the SP domains of C1r, C1s and chymotrypsinogen, and (B) the CCP modules of C1r, C1s and complement receptor 2. The residue numberings of C1r and chymotrypsinogen are shown in (A). In (B), the residues with a β-strand conformation are underlined, and the strand numbering is indicated above. The residues involved in the major CCP1–SP interaction in C1r, and in the interaction between the CCP2 module of CR2 and its C3d ligand, are indicated by asterisks. The residues involved in the additional interaction observed only at the CCP1A–SPB interface are indicated by a ‘#’. Hyp_V, hypervariable loop.
As expected because of the mutation at the cleavage site, the C1r structure exhibits key structural features typical of the zymogen conformation. In the crystal, the activation site 444(13)–448(17) segment (C1r numbering, followed by the chymotrypsinogen numbering in brackets) interacts mainly with exposed hydrophobic residues of neighboring molecules. These contacts are quite different in molecules A and B of the asymmetric unit and the segment exhibits two different conformations in these molecules. In the same way, Ile447(16) adopts two opposite orientations, with a distance of 7 Å between the corresponding Cα positions when the two molecules are superimposed (Figure 1). The SP domain of C1r has many flexible surface segments, which display high temperature factors after the crystallographic refinement. Some simply lack matching electron density and could not be traced (Table II). This has been observed in other zymogen structures, including chymotrypsinogen (Freer et al., 1970; Wang et al., 1985), trypsinogen (Bode et al., 1978), prethrombin-2 (Malkowski et al., 1997), profactor D (Jing et al., 1999) and plasminogen (Wang et al., 2000). When compared with the activated conformation of C1s (Figure 2), major structural differences are located in loops 1, 2 and D, which form the ‘activation domain’ of SPs and undergo conformational changes upon activation. Among the ordered residues in this domain, Asp636(194) occupies the same position as in chymotrypsinogen. Stabilization of the carboxyl group of Asp636(194) occurs through interaction with main-chain N atoms. Consequently, as in prethrombin-2 and tissue plasminogen activator (Vijayalakshmi et al., 1994), C1r does not show the ‘zymogen triad’ [Ser(32), His(40), Asp(194)] typical of the chymotrypsin family. A further characteristic of the C1r zymogen conformation is that residues 635(193)–637(195) exhibit a distorted oxyanion hole configuration that, when compared with other zymogens, is closest to that of chymotrypsinogen.
A specific feature of C1r is that Asp631(189), the well-established determinant of trypsin-like cleavage specificity, is exposed at the surface of the protein, at the position commonly occupied by residue Gly18 in active serine proteases (Figure 2). Thus, access of the cleaved N-terminal Ile447(16)–Ile448(17) dipeptide to its binding pocket would not be possible without displacing this residue. This orientation of Asp189 in C1r arises from its location in an insertion with an unusual local 310 helix conformation (Table II; Figure 2). Consequently, it differs very significantly from those in other zymogens, with distances between Cαs and Oδ1s of 3.6 and 7.3 Å, respectively, compared with trypsinogen, and of 2.5 and 5.3 Å, respectively, compared with prethrombin-2, which is the closest structure to C1r in this respect.
The SP domain bears two N-linked oligosaccharides, and the structurally better defined chain is that linked to Asn497 of SPB, where two N-acetyl-glucosamines, with a fucose attached to the proximal one, and a mannose were built into a well-defined electron density.
The CCP2 module and its interface with theSP domain
Both C1r CCP modules show a fold comparable to that described for other modules of this type, consisting of six β-strands enveloping a compact hydrophobic core. The N- and C-termini lie at opposite ends of the long axis of the modules, and the β-strands are approximately aligned with this axis (Figure 1). The C1r CCP2 module has a structure very close to that of its counterpart in C1s, with a root mean square deviation (r.m.s.d.), based on 69 Cα positions, of only 0.74 Å (Figure 2). This value is significantly lower than the r.m.s.d. of 1.06 Å, based on 42 residues, obtained when comparing C1r CCP2 with the contiguous CCP1 module. The CCP2 module of C1r exhibits the unusual large insertion between strands B5 and B6 previously observed in C1s (Gaboriaud et al., 2000) (Figures 2 and 3). The only important differences between the two CCP2 modules are two insertions: (i) residues 373–376 in the so-called ‘hypervariable loop’ (Wiles et al., 1997) connecting B1 to B2; and (ii) residues 398–407 in the loop between B3 and B4.
As shown in Figure 2, the orientation of the CCP2 module with respect to the SP domain in C1r is remarkably similar to the one previously observed in C1s (Gaboriaud et al., 2000), with an r.m.s.d. of 1.2 Å between the two CCP2–SP structures. Similarly, the interface shows interactions within a proline- and tyrosine-rich framework involving the same residues as in C1s, except for His390 and Lys395, which replace C1s Glu372 and Tyr377. The mean buried surface at the CCP2–SP interface in C1r is 700 Å2, close to the value of 756 Å2 determined for C1s. The remarkable similarity of the CCP2–SP assemblies in C1r and C1s provides strong support to the hypothesis that all members of the CCP–SP family exhibit a homologous rigid module–domain assembly (Gaboriaud et al., 1998).
The CCP1 module and its interface with CCP2
A particular feature of the CCP1 module is the different conformation of the insertion between B3 and B4 (residues 328–337), which is slightly shorter than its counterpart in CCP2, but is better defined, probably because it participates in the interaction with the SP domain (Figure 3). The 307–310 CCP1 segment in the hypervariable loop between B1 and B2 has a conformation close to the corresponding 18–21 segment of the N-terminal CCP module of β2-glycoprotein I (Bouma et al., 1999). The preceding residues 305–307 participate in the interaction with the SP domain only in molecule A (Figure 3).
The CCP1–CCP2 module pair shows an extended, approximately linear conformation, that is virtually identical in molecules A and B, with a distance of 57 Å between the Sγ positions of Cys292 and Cys430 in both cases, at opposite ends of the structure. Compared with known structures of CCP module pairs, the conformation in C1r is closer to that observed at the interface between domains II and III in β2-glycoprotein I (Bouma et al., 1999). The linker region between the last cysteine of CCP1 and the first cysteine of CCP2 contains four residues that form an extension of the β-strand B6 of CCP1 (Figure 1), as also observed at the interfaces between CCP modules II–III and III–IV of β2-glycoprotein I. The linker region stabilizes the CCP1–CCP2 interface by establishing several van der Waals contacts (Ile356) and hydrogen bonds (Lys357) with residues of both modules. The CCP1–CCP2 interface buries 469 Å2 of accessible surface (5% of the surface of each module), a value comparable to those determined for other CCP module pairs with an extended conformation (Bouma et al., 1999; Murthy et al., 2001).
Structure of the CCP1 module–SP domain interfaces
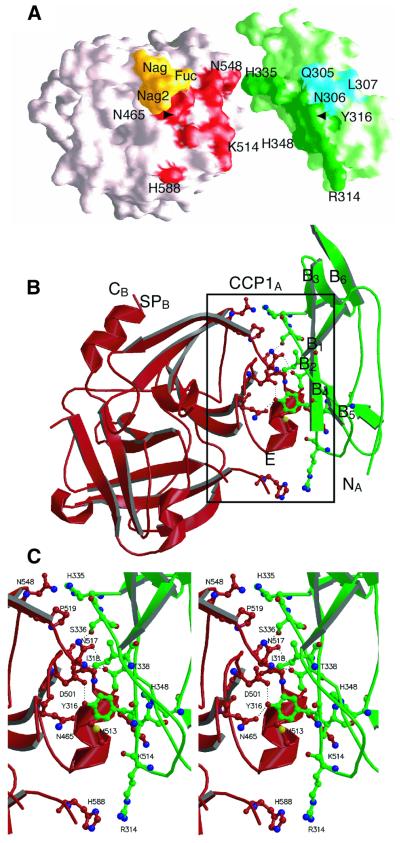
The CCP1–SP interfaces show extensive shape complementarity (Figure 4A), with total buried surfaces of 1457 Å2 between CCP1A and SPB, and 1175 Å2 between CCP1B and SPA. The latter value is similar to the 1154 Å2 (as measured by the same method) buried by the interaction between the CCP2 module of complement receptor 2 (CR2) and its C3d ligand (Szakonyi et al., 2001). Both CCP1–SP interfaces share a common, major interaction framework, consisting of five hydrogen bonds and numerous van der Waals contacts. Most of these interactions involve residues from strands B2 and B4 in CCP1, and residues within or after helix E in the SP domain (Figures 3 and 4C). The additional interactions specific to the more extensive CCP1A–SPB interface consist of three hydrogen bonds and several van der Waals contacts. These interactions are mediated by residues 305–307 in the hypervariable loop of CCP1A and by the distal N-acetyl glucosamine and the fucose of the oligosaccharide chain attached to Asn497, on the SPB domain (Figures 3 and 4A). The additional contacts thus involve a region of the CCP1A module homologous to that involved in the CR2–C3d interaction (Szakonyi et al., 2001), but the major CCP1–SP interaction common to both monomers is mediated by different areas of CCP1 (Figure 3). A further difference is that hydrogen bonds at the CCP1–SP interfaces in C1r involve both side-chain and main-chain atoms, whereas the CR2–C3d interaction shows extensive use of main-chain contacts.
Fig. 4. Structure at the CCP1A–SPB interface. (A) Surface representation illustrating shape complementarity between SPB (left) and CCP1A (right). CCP1A was shifted and rotated to the right for clarity. Areas involved in the major interaction common to both CCP1–SP interfaces are shown in red (SPB) and dark green (CCP1A). The areas involved in the additional interaction observed only at the CCP1A–SPB interface are colored yellow (SPB) and blue (CCP1A). (B) Overall structure of the assembly between SPB (red) and CCP1A (green). (C) Stereoview of the CCP1A–SPB interface. Only the major interaction is shown for clarity. The hydrogen bonding network at the interface is depicted by dotted lines. Strand B4 and the N-terminal part of strand B2 are not shown in ribbon representation for the sake of clarity.
Functional implications of the C1r catalytic domain structure
Several lines of evidence indicate that the crystal structure of the C1r catalytic domain determined here is physiologically relevant: (i) it is well established from various techniques, including electron microscopy (Villiers et al., 1985; Weiss et al., 1986) and sedimentation velocity analysis (Lacroix et al., 2001) that this domain associates as a homodimer; (ii) chemical cross-linking studies (Arlaud et al., 1986; Lacroix et al., 1997) are consistent with a head-to-tail configuration of the dimer; (iii) deletion of the CCP1 module prevents dimer formation (Lacroix et al., 2001); and (iv) the structure of the activated C1r catalytic domain solved at 4 Å resolution (Figure 1D) also exhibits a dimeric head-to-tail configuration stabilized through CCP1–SP interactions, very similar to that of the proenzyme species. This is despite the fact that the space group, and hence the packing, is different in the two structures. Therefore, it is most likely that the structure determined here corresponds to a resting, thermodynamically stable conformation of the C1r catalytic domain, and that activation does not influence the dimer configuration.
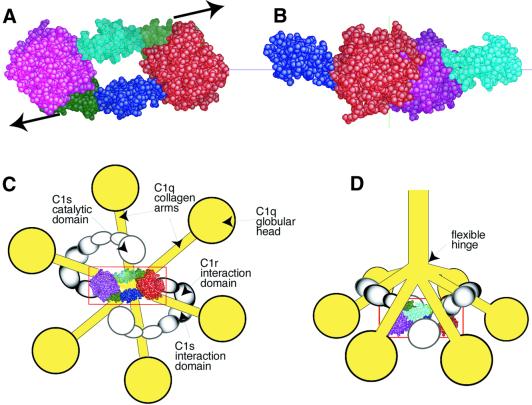
From a functional point of view, the most intriguing feature of this head-to-tail structure is that the catalytic site of one monomer and the activation site of the other lie at opposite ends of the dimer, ∼92 Å away from each other (Figure 1A). This configuration does not allow C1r self-activation, because this process requires cleavage of the susceptible Arg446–Ile447 bond of each monomer by the catalytic residue Ser637 of its counterpart (Lacroix et al., 2001). It follows from this requirement that C1r activation within the C1 complex must take place through transient conformational states, in which the SPA domain will cleave SPB, and vice versa (Figure 5). Both conformational states require a close interaction between the SPA and SPB domains, and this can only be achieved after disruption of the CCP1 module–SP domain interactions. We propose that this disruption is triggered by a mechanical stress that is transmitted from C1q to C1r when C1 binds to an activating surface. This hypothesis is based on the following: (i) each CCP1–CCP2–SP domain of C1r connects with the N-terminal CUB–EGF inter action domain, which itself is bound to the homo logous CUB–EGF domain of C1s, the resulting CUB–EGF heterodimer being attached to the C1q collagen ‘arms’ (Figure 5) (Strang et al., 1982; Busby and Ingham, 1990; Thielens et al., 1999); (ii) C1q possesses a semi-flexible hinge at the point where the six collagen arms join to the central portion of the molecule (Figure 5) (Schumaker et al., 1981; Poon et al., 1983); and (iii) the minimal complex required to achieve C1r activation is that formed in the presence of calcium ions from the association between intact C1q and C1r, and the C1s CUB–EGF fragment (Thielens et al., 1994; Tsai et al., 1997). Thus, binding of C1 to a target through multivalent binding of the C1q ‘heads’ to an irregular pattern of binding sites may be expected to increase the angle between the spreading arms of C1q and the central bundle, generating a tension that is transmitted to the catalytic domain of C1r and results in the dissociation of its dimeric structure. Whether the amplitude of the movement of the C1q arms is consistent with the significant distance (∼46 Å) to be covered by each SP domain to find its target site is difficult to determine, since we lack information about the region of the C1q arms where C1r attaches. On the other hand, it should be emphasized that the approaching of the two SP domains may be greatly facilitated by flexibility at inter-domain junctions. In particular, owing to the known ability of CCP module pairs to adopt different orientations (Wiles et al., 1997), the CCP1–CCP2 interface may adopt a different conformation after disruption of the stabilizing CCP1–SP interaction. At any rate, the large opening occurring in the central part of the dimer is a key structural feature, as it provides room for the conformational changes required for the activation process (Figure 5). It is known that C1 undergoes slow activation in vitro in the absence of activator, a process that is prevented when C1 inhibitor is present (Ziccardi, 1982). It may be hypothesized that this so-called ‘spontaneous activation’ arises from the intrinsic ability of the C1q arms to undergo slow motion about the semi-flexible hinge in the collagen-like region. When C1 binds to an activator, this ‘tick over’ mechanism would be greatly amplified, and also escape the control of C1 inhibitor, hence the much higher efficiency of the activator-mediated process. A physiologically relevant feature of the disruptive mechanism described above is that, due to the resting head-to-tail configuration of its catalytic domain, efficient C1r self-activation is subject to the recognition of a target by the C1 complex, which represents an efficient means of controlling the potentially harmful activities of complement.
Fig. 5. Functional implications of the dimeric structure of the C1r catalytic domain in the context of the C1 complex. (A) Resting head-to-tail configuration of the C1r catalytic domain. Arrows illustrate the triggering stress required to achieve the transient conformational state needed for activation of an SP domain by its counterpart (B). (C and D) The C1r catalytic domain in the context of the C1 complex: (C) bottom and (D) side views of a macroscopic model of C1 (modified from Arlaud et al., 1987b).
A further implication of the head-to-tail structure revealed in this study deals with the control of C1 proteolytic activity by C1 inhibitor. This member of the serpin (serine protease inhibitor) family reacts with the C1s and C1r active sites in the activated C1 complex, resulting in the dissociation of the C1s–C1r–C1r–C1s tetramer from C1q as C1 inhibitor–C1s–C1r–C1 inhibitor complexes (Sim et al., 1979; Ziccardi and Cooper, 1979). The first crystallographic structure of a serpin–protease assembly, the α1-antitrypsin–trypsin complex, has been solved recently, indicating that the interaction between the two molecules induces disorder in ∼37% of the trypsin structure (Huntington et al., 2000). A comparative analysis reveals that the segments of the C1r SP domain involved in the CCP1–SP interface correspond to areas of trypsin that become disordered upon reaction with α1-antitrypsin. Therefore, it may be hypothesized that the ability of C1 inhibitor to dissociate the C1s–C1r–C1r–C1s tetramer arises from the fact that it induces disorder in the region of the C1r SP domain that is involved in the CCP1–SP interaction, and thereby leads to irreversible disruption of the C1r–C1r interface. Thus, the rather peculiar dimeric structure of the C1r catalytic domain reported here provides a basis for the understanding of both the activation of the C1 complex and the control of its activity.
Materials and methods
Protein expression and purification
The wild-type and mutated recombinant fragments spanning the first and second CCP modules and the SP domain of C1r were expressed using a baculovirus/insect cells system and purified as described elsewhere (Lacroix et al., 2001). The proteins comprise the human C1r segment Gly280–Asp688 preceded by an Asp–His sequence added at the N-terminus due to introduction of a restriction site at the 5′ end of the cDNA, and contain either the wild-type sequence or a mutation (Arg446→Gln) at the activation site. Briefly, recombinant baculoviruses were generated using the Bac-to-Bac system (Life Technologies, Inc.) and used to infect High Five™ insect cells in serum-free medium. The recombinant proteins were isolated from the cell culture supernatant by anion-exchange chromatography on a Q-Sepharose-Fast Flow column followed by hydrophobic interaction chromatography on a TSK-Phenyl column.
Crystallization and data collection
Pooled fractions of the proenzyme CCP1–CCP2–SP C1r fragment were concentrated to 2–6 mg/ml in a solution containing 145 mM NaCl and 50 mM triethanolamine-hydrochloride buffer pH 7.4. Crystals were obtained at 20°C by the hanging drop vapor diffusion method by mixing equal volumes of the protein solution and of a reservoir solution composed of 1.5 M ammonium sulfate and 100 mM TAPS pH 8.5. A native data set was measured to a resolution of 2.9 Å on the ID14-EH3 synchrotron beamline at the ESRF, Grenoble, from a crystal of the proenzyme C1r fragment cooled to 100 K. Indexing using the MOSFLM software (Leslie, 1992) indicated that the crystal was orthorhombic, with a space group P212121. The data were scaled and reduced using the CCP4 suite (CCP4, 1994). Details are given in Table III. Crystals of the activated wild-type C1r fragment were obtained by the same method and used to collect two native data sets at the ESRF beamlines BM30 and BM14, at 6 and 4 Å resolution, respectively. The crystals belonged to the hexagonal space group P61 and data were reduced using the XDS package (Kabsch, 1993). Details are given in Table III.
Table III. Data collection and processing statistics for the mutated proenzyme and active forms.
| Proenzyme | Active | |
|---|---|---|
| Native 1 | Native 2 | |
| Unit cell parameters (Å) | a = 99.3 | a = 101.78 |
| b = 101.8 | b = 101.78 | |
| c = 122.4 | c = 461.57 | |
| Wavelength (Å) | 0.931 | 1.03 |
| Resolution (Å) | 2.9 | 4.0 |
| Redundancy | 5.1 (4.5)a | 4.0 (2.4)a |
| Completeness (%) | 99.8 (99.8)a | 97.8 (92.6)a |
| I/σ(I) | 11.0 (2.2)a | 8.57 (4.45)a |
| Rsym (%)b | 5.6 (34)a | 12.7 (19.5)a |
aNumbers in parentheses correspond to the highest resolution shell: 3.06–2.9 Å for native 1 and 4.2–4.0 Å for native 2.
bRsym = Σ|I – <I>|ΣI, where the summation is over all symmetry equivalent reflections.
Structure determination
Space group assignments were confirmed by molecular replacement searches using AMoRe (Navaza, 1994). The activated CCP2–SP fragment of C1s (Gaboriaud et al., 2000) was used as a search model to solve the structure of the mutated, proenzyme CCP1–CCP2–SP C1r fragment. This procedure resulted in a contrasted solution with a correlation value of 0.31 and an R-factor of 0.478 for the two molecules in the asymmetric unit, for data between 15 and 4 Å resolution. The refined proenzyme C1r CCP1–CCP2–SP monomeric structure was then used as a search model to solve the structure of the activated form. A unique, strongly contrasted solution was found, with a final correlation value of 0.47 and an R-factor of 0.454 for the two molecules in the asymmetric unit, for data between 15 and 4 Å resolution. The solvent content of this crystal form is slightly greater than 80%. A step of rigid body refinement using CNS (Brünger et al., 1998) was then applied, with final R-factor and Rfree values of 0.39 and 0.42, respectively. The phases were then improved using the procedure of solvent flipping combined with density truncation, as recommended in the CNS tutorial.
Structure refinement and analysis
The initial model of the proenzyme C1r fragment was corrected with the computer graphics program O (Jones et al., 1991) using (2Fo – Fc) electron density Fourier maps calculated from the molecular replacement solution. The C1r CCP1 module, absent from the C1s search model, could be built into well-defined matching electron density. Model building was alternated with crystallographic refinement using CNS (Brünger et al., 1998). All reflections from 12 to 2.9 Å resolution, except for the 5% reflections for the Rfree calculation (Brünger, 1992), were used during refinement.
The stereochemistry of the structure was assessed with PROCHECK (Laskowski et al., 1993). Details and statistics of the final refined model are presented in Table I. The atomic coordinates set of the proenzyme C1r fragment has been deposited in the Protein Data Bank (PDB), with accession code 1gpz.
The detailed analysis of the interfaces between the CCP modules and the SP domain was carried out using HBPLUS (McDonald and Thornton, 1994) and LIGPLOT (Wallace et al., 1995). The surface areas were calculated with NACCESS (Hubbard and Thornton, 1993). Figures were generated with several combined uses of MOLSCRIPT (Kraulis, 1991), BOBSCRIPT, GRASP (Nicholls et al., 1991) and Raster3D (Merritt and Bacon, 1997). The coordinates of known SP and CCP structures were taken from the PDB (Bernstein et al., 1977).
Quality of the model
The final refined model of the proenzyme C1r fragment comprises residues 290–685 in the two molecules of the asymmetric unit. Some flexible surface loops exhibit locally significantly different conformations in these molecules. Distances >2.5 Å are found for residues 296, 306, 422, 444–447, 565–568, 580 and 586. Besides these local differences, the two models are very similar, with an r.m.s. distance of 0.69 Å for 340 residue pairs. The following segments display highly flexible or disordered conformations and could not be fully modeled into matching electron density: 280–289, 399–405, 493–497, 581–585, 606–616 and 686–688 in molecule A; and 280–289, 399–407, 493–494, 581–585, 605–617 and 686–688 in molecule B. A zero occupancy was set to atoms lacking a clean matching electron density in the 3mFo – 2Fc map at a 1σ level in the following zones: 420–423, 442–446, 530–538 and 601–605 in molecule A; and 331–332, 421–422, 491–492, 532–536, 629–630 and 659–665 in molecule B. These ill-defined segments show the high intrinsic flexibility of some surface loops and are consistent with the high average B-factor value of the model (Table I). Of the 736 amino acid residues comprised in the C1r structure, >99% are in the favorable or additionally allowed regions in the Ramachandran plot. Only His588 is in the generously allowed region in molecule A. Six additional residues are found in this region in molecule B (Thr297, Leu307, Ser496, Phe537, Asp589, Lys629). Eight carbohydrate residues are defined in the structure, including two N-acetyl-glucosamines, a fucose and a mannose (Asn497 of SPB), two N-acetyl-glucosamines and a fucose (Asn564 of SPA), and one N-acetyl-glucosamine (Asn564 in SPB).
Acknowledgments
Acknowledgements
The authors are grateful to the local contacts at the ESRF: L.Dumont (ID14-EH3), J.L.Ferrer (BM30) and P.Carpentier (BM14). We thank J.-B.Reiser for his help with the use of the CNS software. This work was supported by the Commissariat à l’Energie Atomique and the Centre National de la Recherche Scientifique. M.B.-S. was supported by a grant from BASF (Ludwigshafen, Germany).
References
- Arlaud G.J., Gagnon,J., Villiers,C.L. and Colomb,M.G. (1986) Molecular characterization of the catalytic domains of human complement serine protease C1r. Biochemistry, 25, 5177–5182. [DOI] [PubMed] [Google Scholar]
- Arlaud G.J., Willis,A.C. and Gagnon,J. (1987a) Complete amino acid sequence of the A chain of human complement-classical-pathway enzyme C1r. Biochem. J., 241, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlaud G.J., Colomb,M.G. and Gagnon,J. (1987b) A functional model of the human C1 complex. Immunol. Today, 8, 106–111. [DOI] [PubMed] [Google Scholar]
- Arlaud G.J., Volanakis,J.E., Thielens,N.M., Narayana,S.V.L., Rossi,V. and Xu,Y. (1998) The atypical serine proteases of the complement system. Adv. Immunol., 69, 249–307. [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Bode W., Schwager,P. and Huber,R. (1978) The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. The refined crystal structures of the bovine trypsinogen–pancreatic trypsin inhibitor complex and of its ternary complex with Ile–Val at 1.9 Å resolution. J. Mol. Biol., 118, 99–112. [DOI] [PubMed] [Google Scholar]
- Bork P. and Beckmann,G. (1993) The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol., 231, 539–545. [DOI] [PubMed] [Google Scholar]
- Bouma B., de Groot,P.G., van den Elsen,J.M.H., Ravelli,R.B.G., Schouten,A., Simmelink,M.J.A., Derksen,R.H.W.M., Kroon,J. and Gros,P. (1999) Adhesion mechanism of human β2-glycoprotein I to phospholipids based on its crystal structure. EMBO J., 18, 5166–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Busby T.F. and Ingham,K.C. (1990) NH2-terminal calcium-binding domain of human complement C1s mediates the interaction of C1r with C1q. Biochemistry, 29, 4613–4618. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cooper N.R. (1985) The classical complement pathway: activation and regulation of the first complement component. Adv. Immunol., 37, 151–216. [DOI] [PubMed] [Google Scholar]
- Fearon D.T. and Locksley,R.M. (1996) The instructive role of innate immunity in the acquired immune response. Science, 272, 50–54. [DOI] [PubMed] [Google Scholar]
- Freer S.T., Kraut,J., Robertus,J.D., Wright,H.T. and Xuong,N.H. (1970) Chymotrypsinogen: 2.5 Å crystal structure, comparison with α-chymotrypsin and implications for zymogen activation. Biochemistry, 9, 1997–2009. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Rossi,V., Fontecilla-Camps,J.C. and Arlaud,G.J. (1998) Evolutionary conserved rigid module–domain interactions can be detected at the sequence level: the examples of complement and blood coagulation proteases. J. Mol. Biol., 282, 459–470. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Rossi,V., Bally,I., Arlaud,G.J. and Fontecilla-Camps,J.C. (2000) Crystal structure of the catalytic domain of human complement C1s: a serine protease with a handle. EMBO J., 19, 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J.A., Kafatos,F.C., Janeway,C.A. and Ezekowitz,R.A.B. (1999) Phylogenetic perspectives in innate immunity. Science, 284, 1313–1318. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J. and Thornton,J.M. (1993) ‘NACCESS’ computer program. Department of Biochemistry and Molecular Biology, University College London, London, UK.
- Huntington J.A., Read,R.J. and Carrell,R.W. (2000) Structure of a serpin–protease complex shows inhibition by deformation. Nature, 407, 923–926. [DOI] [PubMed] [Google Scholar]
- Jing H., Macon,K.J., Moore,D., DeLucas,L.J., Volanakis,J.E. and Narayana,S.V.L. (1999) Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. EMBO J., 18, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Journet A. and Tosi,M. (1986) Cloning and sequencing of full-length cDNA encoding the precursor of human complement component C1r. Biochem. J., 240, 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kishore U. and Reid,K.B.M. (2000) C1q: structure, function, and receptors. Immunopharmacology, 49, 159–170. [DOI] [PubMed] [Google Scholar]
- Klein M.A. et al. (2001) Complement facilitates early prion pathogenesis. Nature Med., 7, 488–492. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lacroix M., Rossi,V., Gaboriaud,C., Chevallier,S., Jaquinod,M., Thielens,N.M., Gagnon,J. and Arlaud,G.J. (1997) Structure and assembly of the catalytic region of human complement protease C1r: a three-dimensional model based on chemical cross-linking and homology modeling. Biochemistry, 36, 6270–6282. [DOI] [PubMed] [Google Scholar]
- Lacroix M., Ebel,C., Kardos,J., Dobo,J., Gal,P., Zavodszky,P., Arlaud,G.J. and Thielens,N.M. (2001) Assembly and enzymatic properties of the catalytic domain of human complement protease C1r. J. Biol. Chem., 276, 36233–36240. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1992) Molecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystallographic Computing 5: from Chemistry to Biology. Oxford University Press, Oxford, UK, pp. 50–61.
- Leytus S.P., Kurachi,K., Sakariassen,K.S. and Davie,E.W. (1986) Nucleotide sequence of the cDNA coding for human complement C1r. Biochemistry, 25, 4855–4863. [DOI] [PubMed] [Google Scholar]
- Mabbott N.A., Bruce,M.E., Botto,M., Walport,M.J. and Pepys,M.B. (2001) Temporary depletion of complement component C3 or genetic deficiency of C1q significantly delays onset of scrapie. Nature Med., 7, 485–487. [DOI] [PubMed] [Google Scholar]
- Malkowski M.G., Martin,P.D., Guzik,J.C. and Edwards,B.F. (1997) The co-crystal structure of unliganded bovine α-thrombin and prethrombin-2: movement of the Tyr-Pro-Pro-Trp segment and active site residues upon ligand binding. Protein Sci., 6, 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I.K. and Thornton,J.M. (1994), Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Murthy K.H.M., Smith,S.A., Ganesh,V.K., Judge,K.W., Mullin,N., Barlow,P.N., Ogata,C.M. and Kotwal,G.J. (2001) Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell, 104, 301–311. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Perona J.J. and Craik,C.S. (1997) Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem., 272, 29987–29990. [DOI] [PubMed] [Google Scholar]
- Poon P.H., Schumaker,V.N., Phillips,M.L. and Strang,C.J. (1983) Conformation and restricted segmental flexibility of C1, the first component of human complement. J. Mol. Biol., 168, 563–577. [DOI] [PubMed] [Google Scholar]
- Reid K.B.M., Bentley,D.R., Campbell,R.D., Chung,L.P., Sim,R.B., Kristensen,T. and Tack,B.F. (1986) Complement system proteins which interact with C3b and C4b. A superfamily of structurally related proteins. Immunol. Today, 7, 230–234. [DOI] [PubMed] [Google Scholar]
- Rogers J. et al. (1992) Complement activation by β-amyloid in Alzheimer disease. Proc. Natl Acad. Sci. USA, 89, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Endo,Y., Matsushita,M. and Fujita,T. (1994) Molecular characterization of a novel serine protease involved in activation of the complement system by mannose-binding protein. Int. Immunol., 6, 665–669. [DOI] [PubMed] [Google Scholar]
- Schumaker V.N., Poon,P.H., Seegan,G.W. and Smith,C.A. (1981) Semi-flexible joint in the C1q subunit of the first component of human complement. J. Mol. Biol., 148, 191–197. [DOI] [PubMed] [Google Scholar]
- Schumaker V.N., Hanson,D.C., Kilchherr,E., Phillips,M.L. and Poon,P.H. (1986) A molecular mechanism for the activation of the first component of complement by immune complexes. Mol. Immunol., 23, 557–565. [DOI] [PubMed] [Google Scholar]
- Sim R.B., Arlaud,G.J. and Colomb,M.G. (1979) C1 inhibitor-dependent dissociation of human complement component C1 bound to immune complexes. Biochem. J., 179, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang C.J., Siegel,R.C., Phillips,M.L., Poon,P.H. and Schumaker,V.N. (1982) Ultrastructure of the first component of human complement: electron microscopy of the crosslinked complex. Proc. Natl Acad. Sci. USA, 79, 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi G., Guthridge,J.M., Li,D., Young,K., Holers,M. and Chen,X.S. (2001) Structure of complement receptor 2 in complex with its C3d ligand. Science, 292, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Thiel S. et al. (1997) A second serine protease associated with mannan-binding lectin that activates complement. Nature, 386, 506–510. [DOI] [PubMed] [Google Scholar]
- Thielens N.M., Illy,C., Bally,I.M. and Arlaud,G.J. (1994) Activation of human complement serine protease C1r is down-regulated by a Ca2+-dependent intramolecular control that is released in the C1 complex through a signal transmitted by C1q. Biochem. J., 301, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielens N.M., Enrié,K., Lacroix,M., Jaquinod,M., Hernandez,J.-F., Esser,A.F. and Arlaud,G.J. (1999) The N-terminal CUB-EGF module pair of human complement protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J. Biol. Chem., 274, 9149–9159. [DOI] [PubMed] [Google Scholar]
- Thielens N.M., Cseh,S., Thiel,S., Vorup-Jensen,T., Rossi,V., Jensenius,J.C. and Arlaud,G.J. (2001) Interaction properties of human mannan-binding lectin (MBL)-associated serine proteases-1 and -2, MBL-associated protein 19, and MBL. J. Immunol., 166, 5068–5077. [DOI] [PubMed] [Google Scholar]
- Tsai S.W., Poon,P.H. and Schumaker,V.N. (1997) Expression and characterization of a 159 amino acid, N-terminal fragment of human complement component C1s. Mol. Immunol., 34, 1273–1280. [DOI] [PubMed] [Google Scholar]
- Vijayalakshmi J., Padmanabhan,K.P., Mann,K.G. and Tulinsky,A. (1994) The isomorphous structures of prethrombin-2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci., 3, 2254–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiers C.L., Arlaud,G.J. and Colomb,M.G. (1985) Domain structure and associated functions of subcomponents C1r and C1s of the first component of human complement. Proc. Natl Acad. Sci. USA, 82, 4477–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Wang D., Bode,W. and Huber,R. (1985) Bovine chymotrypsinogen A: X-ray crystal structure analysis and refinement of a new crystal form at 1.8 Å resolution. J. Mol. Biol., 185, 595–624. [DOI] [PubMed] [Google Scholar]
- Wang X., Terzyan,S., Tang,J., Loy,J.A., Lin,X. and Zhang,X.C. (2000) Human plasminogen catalytic domain undergoes an unusual conformational change upon activation. J. Mol. Biol., 295, 903–914. [DOI] [PubMed] [Google Scholar]
- Weiss V., Fauser,C. and Engel,J. (1986) Functional model of subcomponent C1 of human complement. J. Mol. Biol., 189, 573–581. [DOI] [PubMed] [Google Scholar]
- Wiles A.P., Shaw,G., Bright,J., Perczel,A., Campbell,I.D. and Barlow,P.N. (1997) NMR studies of a viral protein that mimics the regulators of complement activation. J. Mol. Biol., 272, 253–265. [DOI] [PubMed] [Google Scholar]
- Ziccardi R.J. (1982) Spontaneous activation of the first component of human complement (C1) by an intramolecular autocatalytic mechanism. J. Immunol., 128, 2500–2504. [PubMed] [Google Scholar]
- Ziccardi R.J. and Cooper,N.R. (1979) Active disassembly of the first complement component, C1, by C1 inactivator. J. Immunol., 123, 788–792. [PubMed] [Google Scholar]