Abstract
Reverse gyrase is the only topoisomerase known to positively supercoil DNA. The protein appears to be unique to hyperthermophiles, where its activity is believed to protect the genome from denaturation. The 120 kDa enzyme is the only member of the type I topoisomerase family that requires ATP, which is bound and hydrolysed by a helicase-like domain. We have determined the crystal structure of reverse gyrase from Archaeoglobus fulgidus in the presence and absence of nucleotide cofactor. The structure provides the first view of an intact supercoiling enzyme, explains mechanistic differences from other type I topoisomerases and suggests a model for how the two domains of the protein cooperate to positively supercoil DNA. Coordinates have been deposited in the Protein Data Bank under accession codes 1GKU and 1GL9.
Keywords: DNA topology/gyrase/helicase/supercoiling/topoisomerase
Introduction
Topoisomerases catalyse the conversion between different superhelical states of DNA (Champoux, 2001) using a three-step mechanism of cleavage, strand passage and religation. In the first step, the enzyme uses a tyrosine residue as a nucleophile to attack the phosphodiester backbone, resulting in cleavage of the DNA. Type I topoisomerases cleave one strand of the duplex; type II enzymes cleave both strands. The enzyme, now covalently attached to the cut DNA, separates the free ends of the cleaved strand(s) and allows the other strand of the duplex (type I), or another region of duplex (type II), to pass through this gap. The protein then reseals the phosphodiester backbone of the cleaved DNA and releases the product. Whether the direction of strand passage leads to an increase or decrease in the number of helical turns in the DNA determines whether the superhelicity changes positively or negatively, respectively.
Practically all DNA transactions, including transcription, replication and recombination, require single-stranded DNA. Negatively supercoiled (underwound) DNA favours these processes because it suffers local strand separation more frequently than relaxed DNA. This may explain why mesophiles keep their genomes negatively supercoiled (Déclais et al., 2001). Such strand separation is dangerous for hyperthermophiles, which grow at temperatures >70°C. To protect their genomes from denaturation, these organisms rely on the positive supercoiling (overwinding) activity of reverse gyrase (Kikuchi and Asai, 1984; Forterre et al., 1985). This enzyme is unique to hyperthermophiles (Bouthier de La Tour et al., 1990, 1991) and is the only topoisomerase known that can positively supercoil DNA. The precise role of reverse gyrase in vivo remains unclear, but it is thought to be involved in renaturing melted DNA and perhaps also in removing metastable DNA structures that could impede replication and transcription (Déclais et al., 2001).
While all topoisomerases can remove (relax) supercoils in DNA, reverse gyrase is one of only two topoisomerases capable of creating supercoils. The other is the prokaryotic type II enzyme called gyrase, which can negatively supercoil DNA. Both enzymes require ATP to drive supercoiling, but gyrase uses a type II mechanism, whereas reverse gyrase works as a type I enzyme. It is the only known example of an ATP-dependent type I topoisomerase.
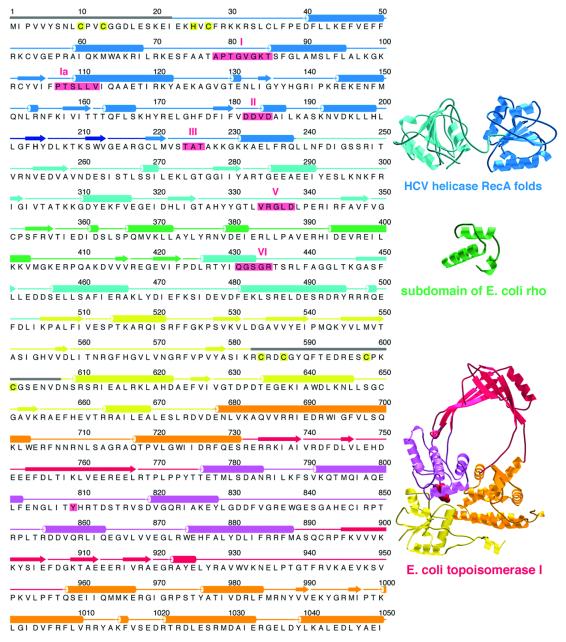
Reverse gyrase comprises two domains (Confalonieri et al., 1993): an N-terminal domain conserving sequence motifs from helicases of superfamilies I and II, and a C-terminal domain bearing 30% sequence identity to Escherichia coli topoisomerase I (Figure 1). On its own, the C-terminal domain functions like topoisomerase I, as an ATP-independent DNA relaxing enzyme (Déclais et al., 2000b). Yet full-length reverse gyrase differs from topoisomerase I in two fundamental ways. First, the latter does not require an external energy source, whereas reverse gyrase requires ATP both for DNA relaxation and positive supercoiling (Shibata et al., 1987; Krah et al., 1997). Secondly, topoisomerase I performs strand passage in either direction: it can relax both negatively and positively supercoiled DNA (Kirkegaard and Wang, 1985). Reverse gyrase, however, appears to perform strand passage exclusively in the direction of positive supercoiling. Thus, when presented with relaxed DNA in the presence of ATP, reverse gyrase gives only positively supercoiled product (Forterre et al., 1985).
Fig. 1. Archaeoglobus fulgidus reverse gyrase comprises two domains with structural homology to helicases and topoisomerases. On the left is the amino acid sequence of reverse gyrase matched to its secondary structure as determined by crystallography (see Figure 2). To the right are protein folds homologous to reverse gyrase, as detected by DALI (Holm and Sander, 1993). The folds on the right have been coloured according to the corresponding regions in reverse gyrase. The reverse gyrase N-terminal domain contains the tandem RecA-like folds observed in the ATPase domain of helicases such as that of hepatitis C virus (Yao et al., 1997). Reverse gyrase has an insertion, termed subdomain H3, that is structurally homologous to part of the RNA-binding domain of E.coli rho protein (Allison et al., 1998). The helicase signature motifs conserved in reverse gyrase are boxed and numbered in red; apart from these motifs, reverse gyrase shows no significant sequence homology with helicases or with the rho fragment. The C-terminal domain shows both sequence and structural homology to E.coli topoisomerase I (Lima et al., 1994), here shown with its active-site Tyr in red. Highlighted in yellow in the reverse gyrase sequence are residues involved in a putative metal-binding site at the extreme N-terminus (Cys10, Cys13, His25, Cys27), and residues forming a Zn-finger motif in the topoisomerase domain (Cys584, Cys587, Cys598, Cys601). Regions disordered in the crystal structure are represented with grey bars for their secondary structure. The left part of the figure was produced with ALSCRIPT (Barton, 1993). The rest of the figures (except Figure 5) were generated with MOLSCRIPT (Priestle, 1991) or BOBSCRIPT (Esnouf, 1997), and rendered with POV-Ray (www.povray.org) or Raster3D (Merritt and Bacon, 1997).
Results and discussion
We cloned the reverse gyrase gene from the hyperthermophilic archaeon Archaeoglobus fulgidus and overexpressed it in E.coli. We determined the crystal structure of the protein, both on its own and complexed with a non-hydrolysable analogue of ATP [adenylylimidodiphosphate (ADPNP)]. Phases were determined using selenomethionine-substituted protein and multiwavelength anomalous dispersion (MAD) (Table I). Both models contain almost all of the 1054 residues of the protein. Because of poor electron density, some residues near the N-terminus were built as polyalanine and others were omitted from the models. The omitted region of residues 583–607 contains a Zn-finger motif (Jaxel et al., 1996) and may require DNA binding to become ordered. The region at the N-terminus modelled with polyalanine contains a potential metal binding site comprising three Cys (Cys10, Cys13, Cys27) and one His residue (His25), which may play a role in DNA binding. Other reverse gyrases have a Zn-finger motif in this region (Confalonieri et al., 1993).
Table I. Crystallographic data collection and refinement statistics.
| Data collection |
|
SeMeta |
|
Native |
ADPNP complex |
|---|---|---|---|---|---|
| Space group | P21 | P21 | P21 | ||
| Unit cell dimensions (a, b, c) (Å) | 63.9, 65.6, 133.4 | 65.2, 68.0, 129.7 | 132.4, 68.69, 134.0 | ||
| Unit cell angles (α, β, γ ) (°) |
|
90.0, 103.5, 90.0 |
|
90.0, 104.0, 90.0 |
90.0, 99.7, 90.0 |
| Peak | Inflection | Remote | |||
| Wavelength (Å) | 0.9795 | 0.9801 | 0.9686 | 0.9686 | 0.9686 |
| Resolution range (Å) | 34–2.8 | 34–2.8 | 34–2.8 | 41–2.7 | 34–3.2 |
| No. of unique reflections | 25 188 | 25 226 | 25 187 | 28 735 | 38 401 |
| I/σb | 9.5 (3.0) | 8.7 (2.8) | 9.3 (2.4) | 8.6 (2.6) | 5.2 (2.3) |
| Multiplicityb | 3.6 (3.6) | 3.6 (3.6) | 3.6 (3.6) | 3.2 (3.0) | 3.3 (2.9) |
| Percent completeness (anomalous) | 99.8 (90.4) | 99.8 (90.3) | 99.8 (90.5) | 99.3 (99.3)b | 96.7 (96.7)b |
| Rmerge (%)c | 5.7 (24.6) | 6.1 (26.5) | 6.5 (30.9) | 6.2 (28.8) | 10.2 (32.1) |
| Refinement | |||||
| resolution limit (Å) | 2.8 | 2.7 | 3.2 | ||
| R-factor (%)d | 23.1 | 22.6 | 25.6 | ||
| Rfreee | 30.8 | 29.5 | 33.2 | ||
| Ramachandran statistics | |||||
| most favoured (%) | – | 81.6 | 74.3 | ||
| additionally allowed (%) | – | 14.9 | 20.5 | ||
| generously allowed (%) | – | 2.9 | 3.6 | ||
| disallowed (%) | – | 0.5 | 1.6 | ||
| r.m.s.d. from ideality | |||||
| bond lengths (Å) | – | 0.007 | 0.008 | ||
| bond angles (°) | – | 1.3 | 1.3 | ||
| mean B-factor (Å2) | – | 55 | 76 |
aSeMet, selenomethionyl MAD dataset.
bValues in parentheses are for the highest resolution bin.
cRmerge = ΣΣi|Ih – Ihi|/ΣΣi Ih, where Ih is the mean intensity for reflection h.
dR-factor = Σ|Fo – Fc|/Σ|Fo|, where Fo and Fc are measured and calculated structure factors, respectively.
eRfree was calculated over 5% of reflections not used in the refinement.
Overall structure
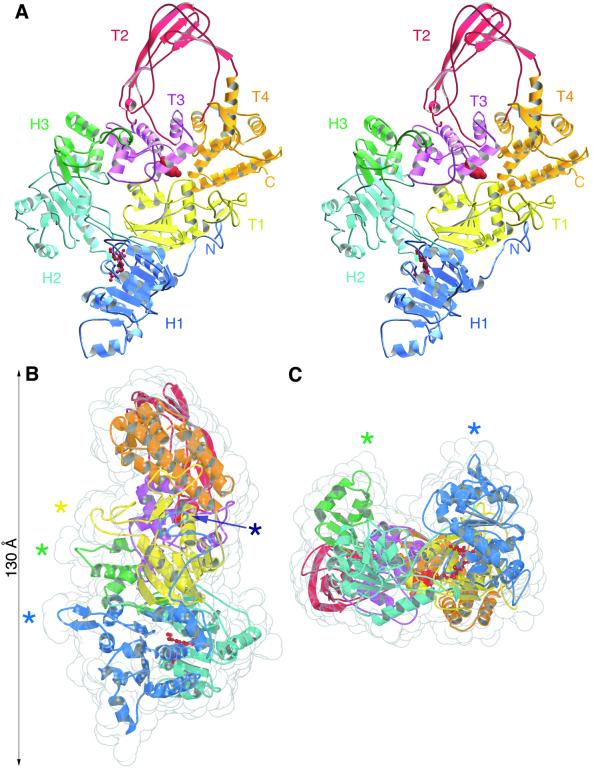
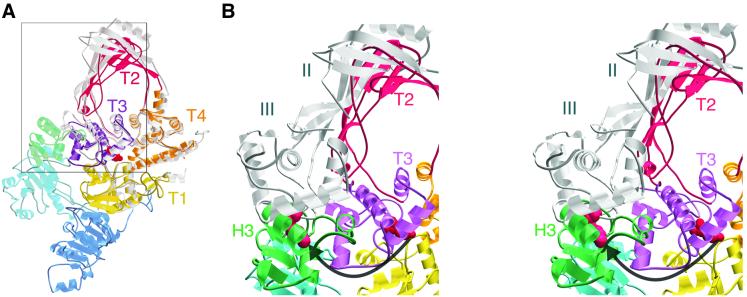
The crystal structure reveals reverse gyrase to have a thin, padlock-like shape (Figure 2). The N-terminal domain contains two folds (H1, H2) similar to the ATP-binding core of E.coli recombination protein RecA (Story et al., 1992). The C-terminal module comprises four subdomains (T1–T4) equivalent to domains I–IV of E.coli topoisomerase I (Lima et al., 1994) (Figure 3A). The C-terminal domain interacts with the N-terminal part through a latch-like insertion (H3) in subdomain H2 (Figure 3B). A search with DALI (Holm and Sander, 1993) revealed that H3 is structurally homologous to residues 1–46 of the E.coli rho transcription terminator. This region in rho is involved in RNA binding (Dombroski and Platt, 1988; Dolan et al., 1990) and may play an analogous role in reverse gyrase, as we postulate H3 to be involved in interacting with DNA (see below). The structures of the C-terminal domain and of a catalytic fragment of topoisomerase I (Lima et al., 1994) superimpose over 380 Cα atoms with a root mean square deviation (r.m.s.d.) of 1.4 Å (Figure 3A). The main difference between the two structures is the smaller size of the topoisomerase ‘hole’ in reverse gyrase (16 Å diameter) compared with that in topoisomerase I (25 Å), which might contribute to the former’s thermostability. The topoisomerase I fragment lacks the C-terminal tail of three contiguous Zn-finger motifs, which are thought to assist in DNA binding and at least one of which is required for relaxation activity (Lima et al., 1993). The C-terminal domain of reverse gyrase lacks such a tail but has a Zn-finger motif in subdomain T1 (Jaxel et al., 1996) (Figure 2). Additional DNA-binding sites may lie in the N-terminal domain, one at the extreme N-terminus (see above), and perhaps another in a β-hairpin (residues 201–217) that juts out from subdomain H1 (Figure 2).
Fig. 2. Overall structure of reverse gyrase. (A) Stereo view of the molecule. The catalytic Tyr809 of the C-terminal domain is shown in red as a space-filled model, and helicase motif I (residues 78–85) in red ball-and-stick representation. The colouring of the subdomains of reverse gyrase is the same for all figures except Figures 4B and 5. (B) Side view of the molecule shown with a translucent space-filling envelope. Asterisks indicate four structural elements postulated to contact DNA: dark blue, a putative metal-binding site at the extreme N-terminus; light blue, a β-hairpin (residues 201–217); green, the ‘latch’ subdomain H3 (residues 352–427); yellow, a Zn-finger motif (residues 584–601). The conformation of the Zn-finger as shown is uncertain due to poor electron density, and has not been included in the refined model. Maximum dimensions of the molecule are 130 × 70 × 50 Å. (C) End-on view of the molecule, with the N-terminal domain towards the front.
Fig. 3. The C-terminal domain of reverse gyrase and its interaction with the N-terminal domain. (A) Superposition of the C-terminal domain with the 67-kDa catalytic fragment of E.coli topoisomerase I (Lima et al., 1994), shown in grey. The position of domains II and III of topoisomerase I correspond to the ‘closed’ form of the enzyme. A box encloses the region featured in (B). (B) Stereo view of reverse gyrase superimposed with domains II and III of topoisomerase I in the putative ‘open’ form (Feinberg et al., 1999). The catalytic Tyr in both enzymes is indicated in red space-filling representation. The arrow indicates the putative movement of reverse gyrase subdomains T2 and T3 during strand passage. This movement would be prevented by subdomain H3 in its current position.
The N-terminal domain
The pair of interacting RecA-like folds in the N-terminal domain has been observed in all helicase structures determined so far (Subramanya et al., 1996; Korolev et al., 1997; Yao et al., 1997; Kim et al., 1998; Singleton et al., 2000; Niedenzu et al., 2001). In helicases the dual RecA folds function as an ATP-dependent motor to move the enzyme along single-stranded DNA. Models describing this translocation have been developed for hepatitis C virus NS3 helicase (Kim et al., 1998) and for Bacillus stearothermophilus PcrA helicase (Velankar et al., 1999); mutagenesis studies have identified many of the residues involved (Lin and Kim, 1999; Dillingham et al., 2001). Structural superpositions indicate that reverse gyrase lacks these residues. The structure therefore explains why reverse gyrase does not translocate along DNA like a helicase (Déclais et al., 2000; A.C.Rodríguez, unpublished data).
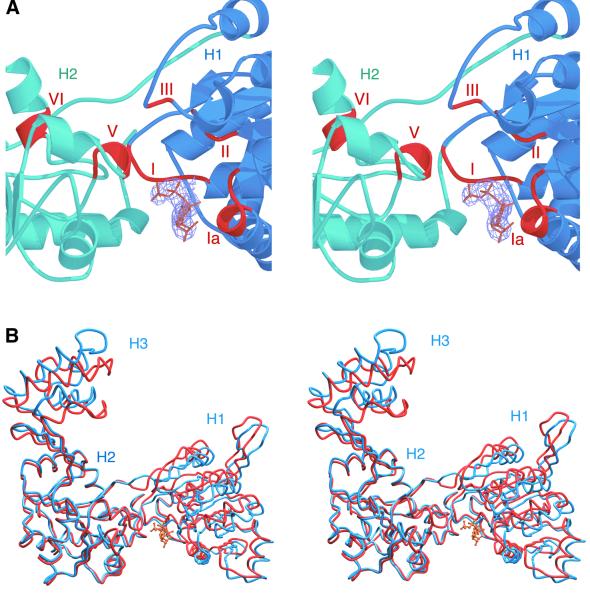
In general, the signature motifs of superfamily I and II helicases face into the cleft between the two RecA-like domains, and residues from both domains make contact with bound nucleotide (Korolev et al., 1997; Velankar et al., 1999; Singleton et al., 2000). Reverse gyrase conserves this spatial disposition of helicase motifs (Figure 4A). However, in both the apo and ADPNP structures, subdomains H1 and H2 are spread apart compared with their orientation in helicases. Conserved motifs on H2 are too far away from H1 to contact nucleotide. Consequently, the nucleotide in the co-crystal binds to only one side of the cleft (Figure 4A), overlapping almost exactly with where RecA binds ADP (Story and Steitz, 1992). The triphosphate moiety interacts with motif I (the P-loop) and the adenine ring interacts with Gln61. The interaction with Gln61, invariant among reverse gyrases, may explain the enzyme’s specificity for ATP/dATP (Shibata et al., 1987). The chief differences between the apo and ADPNP structures are a movement of subdomain H1 up and towards H2, and a shift in subdomain H3 (Figure 4B). Packing interactions are nearly identical in the apo and complex crystals, suggesting that at least the movement in H1 is induced by nucleotide binding. As a result of this movement, Asp182 and Asp183 of motif II are brought closer to the Mg2+ that interacts with the nucleotide. The shift in H3, however, is more difficult to explain, because it is not in direct contact with H1. It does indicate that H3 is capable of moving, and such flexibility appears necessary for catalysis (see below).
Fig. 4. Co-crystal structure of reverse gyrase with ADPNP. (A) Close-up stereo view of the ATP-binding site. The electron density for the nucleotide is shown, and the conserved helicase motifs are indicated in red. The C-terminal domain has been omitted. (B) Stereo view of the superimposed apo enzyme (blue) and ADPNP complex (red). Although the superposition was performed over the entire structures (2.1 Å r.m.s.d. over all Cα atoms), only the N-terminal domains are shown. No significant changes are observed in the C-terminal domains.
Nucleotide binding therefore seems to be insufficient for bringing the two RecA-like folds together. The ADPNP structure probably reflects a non-productive binding mode in the absence of DNA, as observed for PcrA helicase (Soultanas et al., 1999). Upon DNA binding, subdomain H2 is likely to move towards H1 to create a pocket for ATP hydrolysis. This would explain why reverse gyrase shows ATPase activity only in the presence of DNA (Shibata et al., 1987; A.C.Rodríguez, unpublished data).
Signalling between the two domains
The conformational changes in the N-terminal domain upon ATP and DNA binding must be transmitted to the C-terminal domain to induce positive supercoiling. The structure reveals a possible mechanism for this signalling. Subdomain H3 forms a latch that rests on top of residues 856–870 of subdomain T3 (Figure 3B). Crystallographic and biochemical studies with topoisomerase I suggest that subdomains II and III (T2 and T3 in reverse gyrase) rotate out and away from the protein to allow the intact strand to enter the central hole during strand passage (Feinberg et al., 1999; Li et al., 2001). In the reverse gyrase crystal structure, these subdomains cannot move because they are locked down by the latch. We predict that this latch pulls away from the C-terminal domain during catalysis. In this way, the latch functions as the regulatory module that prevents the C-terminal domain from working as an ATP-independent relaxing enzyme and instead constrains it to positively supercoil. The ADPNP complex shows that nucleotide binding is insufficient to open the latch. Nevertheless, reverse gyrase relaxes negatively supercoiled DNA in the presence of nucleotides that it can bind but not hydrolyse (Shibata et al., 1987). Latch opening is therefore likely to require binding of both nucleotide and DNA, and may occur as a result of the closing of the cleft between subdomains H1 and H2 to form the ATP hydrolysis pocket.
Possible mechanism for DNA unwinding
The structure of reverse gyrase is, to our knowledge, only the second view of a protein conserving helicase sequence motifs that does not display processive helicase activity. The first such view came with the structure of UvrB, a central player in nucleotide excision repair catalysed by the UvrABC system (Machius et al., 1999; Nakagawa et al., 1999; Theis et al., 1999). UvrB conserves the motif of dual RecA folds, together with the ATPase signature motifs, but it does not function as a helicase. When complexed with UvrA, UvrB shows a non-processive strand displacement activity that opens up the duplex around the site of DNA damage (Gordienko and Rupp, 1997; Zou and Houten, 1999).
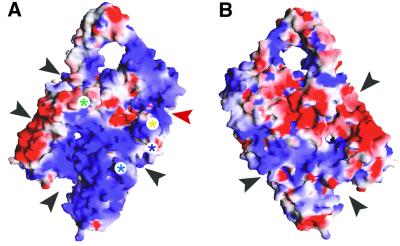
Like UvrB, reverse gyrase lacks the translocation activity of helicases, but it does cause local unwinding of the duplex (Déclais et al., 2000). Analysis of the electrostatic surface potential of reverse gyrase suggests the possibility of extensive interactions with DNA (Figure 5). A large region of positive potential extends across the N-terminal domain on the side of the protein containing the latch. In addition, a number of potential binding grooves run between the N- and C-terminal domains (Figure 5). A Zn-finger motif sits where one channel leaves the C-terminal domain. This groove grips single-stranded DNA, as shown in a recent co-crystal structure of E.coli topoisomerase III with oligonucleotide (Changela et al., 2001). An extensive binding surface may help reverse gyrase to unwind DNA. This unwinding may be catalysed by the RecA-like subdomains, as RecA can melt oligonucleotides shorter than ∼30 bases (Bianchi et al., 1985). It is tempting to speculate that the latch and possibly the β-hairpin (residues 201–217) wedge themselves between the strands of the duplex, as suggested for the β-hairpin of UvrB (Theis et al., 1999).
Fig. 5. The putative DNA-binding surface of reverse gyrase. The standard front view is shown in (A) and the rear view in (B). Electropositive surface potential appears blue and electronegative potential is red. Possible DNA-binding elements are marked with asterisks as in Figure 2B. Black arrows mark possible DNA-binding grooves. The red arrow marks the groove shown to bind single-stranded DNA in topoisomerase III (Changela et al., 2001). This figure was generated using GRASP (Nicholls et al., 1991).
Possible mechanism for positive supercoiling
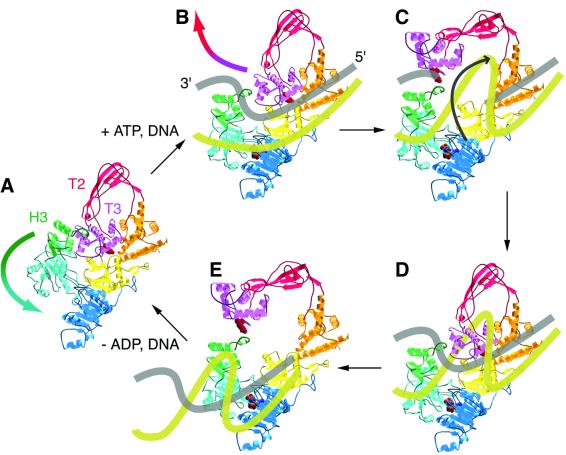
The structure leads us to propose the following mechanism for reverse gyrase (Figure 6). DNA binds to the protein and the latch (subdomain H3) intercalates between the strand to be cleaved and the strand to be passed, such that the scissile strand lies on the side facing subdomain T3. When both nucleotide and DNA are bound, subdomain H2 moves toward H1, pulling the latch away from subdomains T2 and T3. These swing out to allow the passed strand, probably guided by the latch, to enter the topoisomerase gate. Strand cleavage probably precedes any large conformational changes in the protein, consistent with the observation that cleavage occurs in the absence of ATP (Jaxel et al., 1989). The model stipulates that the latch and the Zn-finger motif in the topoisomerase domain are directly involved in keeping the cleaved and passed strands separated. The β-hairpin in subdomain H1 may also be involved in this process.
Fig. 6. Speculative model for positive supercoiling of DNA by reverse gyrase. The active-site Tyr809 is shown in space-filling representation, as is the bound ATP molecule. Binding of both DNA and ATP to the enzyme in (A) triggers closure of the cleft between subdomains H1 and H2, which pulls H3 away from the T2/T3 gate (B). Once DNA cleavage has occurred, the gate opens and the uncleaved strand passes into the central cavity in a right-handed direction (C). The topoisomerase gate closes and the break in the cleaved strand is resealed (D). Finally the gate opens a second time to release the product DNA (E), now containing one more right-handed turn than before. The conformational changes in H2 and H3 were derived from a superposition of reverse gyrase onto the structure of PcrA helicase complexed with DNA and ADPNP (Velankar et al., 1999), and the open form of the topoisomerase gate was derived from a superposition with the putative open form of topoisomerase I (Feinberg et al., 1999).
DNA unwinding probably underlies reverse gyrase’s specificity for supercoiling in the positive direction. An early mechanistic model for reverse gyrase accounted for this unidirectionality by proposing that the N-terminal domain acts like a helicase (Confalonieri et al., 1993). However, biochemical (Déclais et al., 2000) and now structural results indicate that this is unlikely. An alternative explanation is that reverse gyrase unwinds DNA in a controlled fashion to ensure that strand passage always occurs in the direction of positive supercoiling. A mechanism of controlled strand passage is also postulated for E.coli gyrase. This enzyme wraps ∼140 base pairs of DNA around itself, which constrains the protein to pass the duplex preferentially in the direction of negative supercoiling (Champoux, 2001). The small size of reverse gyrase (123 kDa) compared with gyrase (374 kDa) was thought to preclude a wrapping mechanism (Jaxel et al., 1989). However, the recent discovery that UvrB (75 kDa) wraps about seven helical turns of DNA around itself (Verhoeven et al., 2001) raises intriguing possibilities for the mechanism of reverse gyrase. The grooves and electropositive potential on both sides of the protein would be consistent with DNA wrapping (Figure 5).
The crystal structure of reverse gyrase provides the first high-resolution picture of an enzyme that positively supercoils DNA. The structure suggests that reverse gyrase supercoils through a mechanism of controlled strand passage, analogously to DNA gyrase. The lack of a crystal structure of the entire DNA gyrase tetramer has hindered detailed understanding of its mechanism. The structure of reverse gyrase is the first view of an intact supercoiling enzyme, and should provide a framework for further studies into how a protein can act as a DNA supercoiling machine.
Materials and methods
Expression and purification
Reverse gyrase was cloned by PCR using genomic DNA from A.fulgidus (DSMZ) and overexpressed in pRET3a (Tan et al., 2000) in E.coli C41(DE3) (Miroux and Walker, 1996). The native Met1 at the N-terminus was replaced with a FLAG tag (MDYDDDDK), and the native C-terminus was extended with a His6 tag. Overexpressing cells were boiled for 5 min, and the supernatant was purified on Ni2+-nitrilotriacetic acid resin (Qiagen), followed by Sephacryl S300 gel filtration (Pharmacia). Protein was stored in 20 mM Tris–HCl pH 8.0, 200 mM NaCl, 10 mM MgCl2, 1 mM EDTA and 0.02% NaN3. Reverse gyrase substituted with selenomethionine was prepared as described elsewhere (van Duyne et al., 1993) and purified as for the native protein, except that overexpressing cells were not boiled but sonicated and all buffers contained reducing agents. Sequencing of the expression construct indicated two PCR-induced mutations (Pro719→Leu and Leu1046→Met), but the recombinant protein behaves similar to other reverse gyrases in positive supercoiling and ATP hydrolysis assays (Forterre et al., 1985; Shibata et al., 1987; Déclais et al., 2000b).
Crystallization
Sitting drops were prepared by 1:1 mixing of 10 mg/ml reverse gyrase in storage buffer (see above) with 15% polyethylene glycol (PEG) 1000, 15% ethylene glycol and 100 mM cacodylate pH 6, and incubated at 18°C. Dithiothreitol (DTT) was included for selenomethionyl crystallization and 2 mM ADPNP was included for co-crystallization. Reverse gyrase crystallizes in two different forms, regardless of nucleotide content. One crystal form has a monomer in the asymmetric unit, and this form was used for solving the native and selenomethionyl structures. The other crystal form, used to solve the ADPNP structure, contains a dimer in the asymmetric unit. Native crystals were cryoprotected with 25% PEG 1000 and 25% ethylene glycol; 5 mM DTT was also included for selenomethionyl crystals.
Structure determination
Two single-wavelength datasets and one three-wavelength MAD dataset were collected at –180°C on beamline ID14–4 at the ESRF (Grenoble) from a native crystal, an ADPNP co-crystal and a selenomethionyl crystal. Data were integrated with MOSFLM (Leslie, 1991) and scaled using SCALA (CCP4, 1994). Selenium sites were identified and refined using SOLVE (Terwilliger and Berendzen, 1999), and density modification of experimental maps was performed with RESOLVE (Terwilliger, 1999). We solved and partially refined the structure of the selenomethionyl protein, then used molecular replacement in CNS (Brünger et al., 1998) to solve the native and complex structures. Model building was carried out using O (Jones and Kjeldgaard, 1997) and MAIN (Turk, 1992), and refinement was performed with CNS.
Coordinates
Atomic coordinates and structure factors for the native apo structure and ADPNP complex have been deposited in the Protein Data Bank under accession codes 1GKU and 1GL9, respectively.
Acknowledgments
Acknowledgements
We are indebted to Andrew Travers for many helpful discussions. We also thank him, together with Andrew Leslie, Daniela Rhodes and Andrew Carter, for critical reading of the manuscript. We thank Gordon Leonard (ESRF) for his assistance with data collection. This work was supported by a Medical Research Council Career Development Award to D.S. and by a Howard Hughes Predoctoral Fellowship to A.C.R.
References
- Allison T.J., Wood,T.C., Briercheck,D.M., Rastinejad,F., Richardson, J.P. and Rule,G.S. (1998) Crystal structure of the RNA-binding domain from transcription termination factor rho. Nature Struct. Biol., 5, 352–356. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Bianchi M., Riboli,B. and Magni,G. (1985) E.coli recA protein possesses a strand separating activity on short duplex DNAs. EMBO J., 4, 3025–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de C., Portemer,C., Nadal,M., Stetter,K.O., Forterre,P. and Duguet,M. (1990) Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J. Bacteriol., 172, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de C., Portemer,C., Huber,R., Forterre,P. and Duguet,M. (1991) Reverse gyrase in thermophilic eubacteria. J. Bacteriol., 173, 3921–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Champoux J.J. (2001) DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem., 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Changela A., DiGate,R.J. and Mondragón,A. (2001) Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature, 411, 1077–1081. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Confalonieri F., Elie,C., Nadal,M., Bouthier de La Tour,C., Forterre,P. and Duguet,M. (1993) Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl Acad. Sci. USA, 90, 4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déclais A.-C., Marsault,J., Confalonieri,F., Bouthier de La Tour,C. and Duguet,M. (2000) Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem., 275, 19498–19504. [DOI] [PubMed] [Google Scholar]
- Déclais A.-C., Bouthier de La Tour,C. and Duguet,M. (2001) Reverse gyrases from Bacteria and Archaea. Methods Enzymol., 334, 146–162. [DOI] [PubMed] [Google Scholar]
- Dillingham M.S., Soultanas,P., Wiley,P., Webb,M.R. and Wigley,D.B. (2001) Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl Acad. Sci. USA, 98, 8381–8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J.W., Marshall,N.F. and Richardson,J.P. (1990) Transcription termination factor rho has three distinct structural domains. J. Biol. Chem., 265, 5747–5754. [PubMed] [Google Scholar]
- Dombroski A.J. and Platt,T. (1988) Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc. Natl Acad. Sci. USA, 85, 2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Feinberg H., Lima,C.D. and Mondragón,A. (1999) Conformational changes in E.coli DNA topoisomerase I. Nature Struct. Biol., 6, 918–922. [DOI] [PubMed] [Google Scholar]
- Forterre P., Mirambeau,G., Jaxel,C., Nadal,M. and Duguet,M. (1985) High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. EMBO J., 4, 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordienko I. and Rupp,W.D. (1997) The limited strand-separating activity of the UvrAB protein complex and its role in the recognition of DNA damage. EMBO J., 16, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jaxel C., Nadal,M., Mirambeau,G., Forterre,P., Takahashi,M. and Duguet,M. (1989) Reverse gyrase binding to DNA alters the double helix structure and produces single-strand cleavage in the absence of ATP. EMBO J., 8, 3135–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaxel C., Bouthier de La Tour,C., Duguet,M. and Nadal,M. (1996) Reverse gyrase gene from Sulfolobus shibatae B12: gene structure, transcription unit and comparative sequence analysis of the two domains. Nucleic Acids Res., 24, 4668–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. and Kjeldgaard,M. (1997) Electron-density map interpretation. Methods Enzymol., 277B, 173–207. [DOI] [PubMed] [Google Scholar]
- Kikuchi A. and Asai,K. (1984) Reverse gyrase: a topoisomerase which introduces positive superhelical turns into DNA. Nature, 309, 677–681. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K. and Wang,J.C. (1985) Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol., 185, 625–637. [DOI] [PubMed] [Google Scholar]
- Korolev S., Hsieh,J., Gauss,G.H., Lohman,T.M. and Waksman,G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- Krah R., O’Dea,M.H. and Gellert,M. (1997) Reverse gyrase from Methanopyrus kandleri. J. Biol. Chem., 272, 13986–13990. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1991) Recent changes to the MOSFLM package for processing film and image plate data. CCP4 and ESF-EACMB Newsletters on Protein Crystallography. SERC Laboratory, Daresbury, UK.
- Li Z., Mondragón,A. and DiGate,R.J. (2001) The mechanism of type IA topoisomerase-mediated DNA topological transformations. Mol. Cell, 7, 301–307. [DOI] [PubMed] [Google Scholar]
- Lima C.D., Wang,J.C. and Mondragón,A. (1993) Crystallization of a 67 kDa fragment of Escherichia coli DNA topoisomerase I. J. Mol. Biol., 232, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Lima C.D., Wang,J.C. and Mondragón,A. (1994) Three-dimensional structure of the 67K N-terminal fragment of E.coli DNA topoisomerase I. Nature, 367, 138–146. [DOI] [PubMed] [Google Scholar]
- Lin C. and Kim,J.L. (1999) Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol., 73, 8798–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machius M., Henry,L., Palnitkar,M. and Deisenhofer,J. (1999) Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA, 96, 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: Photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker,J.E. (1996) Overproduction of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Sugahara,M., Masui,R., Kato,R., Fukuyama,K. and Kuramitsu,S. (1999) Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem., 126, 986–990. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Niedenzu T., Roleke,D., Bains,G., Scherzinger,E. and Saenger,W. (2001) Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol., 306, 479–487. [DOI] [PubMed] [Google Scholar]
- Priestle J. (1991) A program to produce both detailed and schematic drawings for protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Shibata T., Nakasu,S., Yasui,K. and Kikuchi,A. (1987) Intrinsic DNA-dependent ATPase activity of reverse gyrase. J. Biol. Chem., 262, 10419–10421. [PubMed] [Google Scholar]
- Singleton M., Sawaya,M., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Soultanas P., Dillingham,M.S., Velankar,S.S. and Wigley,D.B. (1999) DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol., 290, 137–148. [DOI] [PubMed] [Google Scholar]
- Story R.M. and Steitz,T.A. (1992) Structure of the recA protein–ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E.coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- Tan S., Hunziker,Y., Pellegrini,L. and Richmond,T.J. (2000) Crystallization of the yeast MATα2/MCM1/DNA ternary complex: general methods and principles for protein/DNA cocrystallization. J. Mol. Biol., 297, 947–959. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (1999) Reciprocal-space solvent flattening. Acta Crystallogr. D, 55, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis K., Chen,P.J., Skorvaga,M., Houten,B.V. and Kisker,C. (1999) Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J., 18, 6899–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekülgraphik und Elektronendichte-Manipulation und seine Anwendung auf verschiediene Protein-Strukturauflärungen. PhD thesis, Technische Universität München, Germany.
- van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Verhoeven E.E., Wyman,C., Moolenaar,G.F., Hoeijmakers,J.H. and Goosen,N. (2001) Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J., 20, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N., Hesson,T., Cable,M., Hong,Z., Kwong,A., Le,H. and Weber,P. (1997) Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol., 4, 463–467. [DOI] [PubMed] [Google Scholar]
- Zou Y. and Houten,B.V. (1999) Strand opening by the UvrA2B complex allows dynamic recognition of DNA damage. EMBO J., 18, 4889–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]