Abstract
The regulatory mechanisms mediating basal and inducible platelet-derived growth factor (PDGF)-A expression have been the focus of intense recent investigation, but repression of PDGF-A expression is largely unexplored. Here we isolated a nuclear factor that interacts with the proximal region of the PDGF-A promoter using bulk binding assays and chromatography techniques. Peptide mass fingerprint and supershift analysis revealed this DNA-binding protein to be NF1/X. NF1/X repressed PDGF-A promoter-dependent transcription and endogenous mRNA expression, which was reversible by oligonucleotide decoys bearing an NF1/X-binding site. Mutation in the DNA-binding domain of NF1/X abolished its repression of PDGF-A promoter. NF1/X antagonized the activity of a known activator of the PDGF-A chain, Sp1, by inhibiting its occupancy of the proximal PDGF-A promoter. NF1/X physically and specifically interacts with Sp1 via its subtype-specific domain and blocks Sp1 induction of the promoter. NF1/X residues 311–416 mediated NF1/X suppression of basal PDGF-A transcription, whereas residues 243–416 were required for NF1/X repression of Sp1-inducible promoter activity. These findings demonstrate that repression of PDGF-A gene transcription is governed by interplay between NF1/X and Sp1.
Keywords: NF1/X/PDGF/Sp1/transcription
Introduction
NF1 represents a family of transcription factors that regulate the expression of a diverse range of cellular (Cereghini et al., 1987) and viral (Plumb et al., 1991; Mink et al., 1992; Kumar et al., 1993) genes. NF1 proteins were first cloned from human HeLa cells (Santoro et al., 1988) and subsequently isolated from hamster (Gil et al., 1988), rat (Paonessa et al., 1988), pig (Meisterernst et al., 1989) and chicken (Rupp et al., 1990). Sequence analysis led to the identification of NF1/A, NF1/B, NF1/C, NF1/L and NF1/X (Rupp et al., 1990; Kruse et al., 1991). This diversity is further increased by differential RNA splicing (Santoro et al., 1988) with >20 splicing isoforms of NF1 having been isolated (Osada et al., 1999). NF1 interacts with the 5′-TGG(N)6GCCAA-3′ consensus element (Nagata et al., 1983; Hay, 1985), which is often in close proximity to the binding sites of other transcription factors in promoter regulatory regions (Blomquist et al., 1996). Under the positive transcriptional control of NF1 are the mouse mammary tumour virus (MMTV) promoter (Nowock et al., 1985; Buetti and Kuhnel, 1986), the human papilloma virus type 16 enhancer (Apt et al., 1993), the liver-specific serum albumin enhancer (McPherson et al., 1993) and the CYP1A1 promoter (Wu and Whitlock, 1992). NF1 proteins can also repress the expression of certain genes. For example, NF1 inhibits human PITI/GHF1 expression by interacting with a distinct element in the distal regulatory region (Rajas et al., 1998). NF1/X represses α1B adrenergic receptor gene middle (P2) promoter activity in DDT1MF-2 cells (Gao and Kunos, 1998). NF1/L binds DNA and forms a multiprotein complex at a negative regulatory element in the peripherin gene (Adams et al., 1995). It can also down-regulate the rat poly(ADP-ribose) polymerase promoter in concert with Sp1 (Laniel et al., 2001). At present there are no reports of NF1 repression of a peptide growth factor gene.
The platelet-derived growth factor (PDGF) family consists of an A and a B chain (reviewed in Raines et al., 1990). Recently, the PDGF-C (Li et al., 2000) and -D chains (Bergsten et al., 2001; LaRochelle et al., 2001) were reported. Expression of the PDGF-A gene, which resides on human chromosome 7p21–p22 and spans ∼24 kb pairs of genomic DNA, is mediated by a single transcriptional start site located 36 bp downstream of the TATA box (Bonthron et al., 1988). The minimal promoter region, sufficient for optimal promoter activity in a number of cell lines (Kaetzel et al., 1993), consists of ∼100 bp. Previous studies by our group have demonstrated that PDGF-A gene expression in vascular smooth muscle and endothelial cells is under the transcriptional control of the zinc finger transcription factors Sp1, Sp3 and Egr-1, which interact with overlapping elements located at base pairs –71/–55 in the proximal PDGF-A promoter (Khachigian et al., 1995, 1997; Delbridge and Khachigian, 1997; Silverman et al., 1997). However, the PDGF-A promoter is also subject to negative regulation, although this is poorly understood at present. For example, the Wilms’ tumour suppressor gene product WT-1 represses activity of the PDGF-A promoter in murine fibroblasts and human kidney cells (Gashler et al., 1992; Wang et al., 1992) via the Sp1/Sp3/Egr-1-binding site. This element also mediates repression of PDGF-A expression by GC-factor 2 (GCF-2; Khachigian et al., 1999).
Investigations by our group and others have demonstrated the formation of a distinct nucleoprotein complex with prima-facie low relative molecular mass using the proximal region of the PDGF-A promoter (base pairs –76/–47) as an oligonucleotide probe in serial electrophoretic mobility shift assays (EMSA; Khachigian et al., 1996). Although competition studies indicate that the formation of this complex is sequence specific, the identity of the protein component of this complex has remained elusive to date. Here we have identified this to be the transcription factor NF1/X. We demonstrate that NF1/X interacts with a novel binding site in the proximal PDGF-A promoter, located at position –67/–60, and represses both basal and inducible PDGF-A transcription through a distinct repression domain located at its C-terminus (Roulet et al., 1995). These findings provide the first demonstration of transcriptional repression of PDGF by any member of the NF1 family.
Results
Identification of the protein component of complex A5
We demonstrated previously by EMSA, using a double-stranded 32P-labelled oligonucleotide bearing the –76/–47 element from the PDGF-A promoter ([32P]Oligo A), that this region supports the formation of several specific nucleoprotein complexes. The protein components of three slow mobility complexes, identified by supershift analysis, each transactivate the PDGF-A promoter and include Sp1 (Khachigian et al., 1995; Day et al., 1999), Egr-1 (Khachigian et al., 1995; Day et al., 1999) and Sp3 (Khachigian et al., 1995; Rafty and Khachigian, 1998; Day et al., 1999; Figure 1A). This analysis also revealed a distinct nucleoprotein complex (A5; Figure 1A) with faster mobility than the above, whose formation is sequence specific (Khachigian et al., 1995, 1997). The identity of the protein component of A5 has not yet been elucidated.
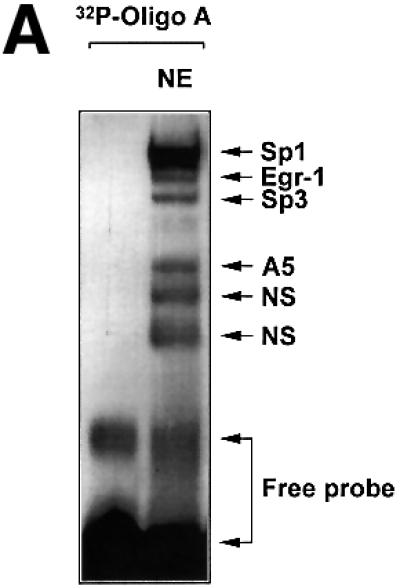
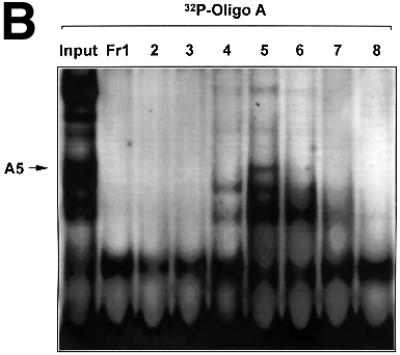
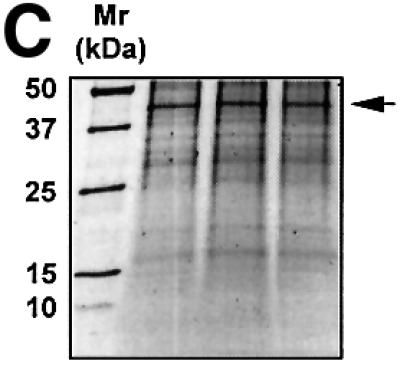
Fig. 1. The proximal PDGF-A promoter interacts with nuclear protein. (A) EMSA was performed using nuclear extracts from WKY12-22 cells and [32P]Oligo A (5′-GGGGGGGGCGGGGGCGGGGGCGGGGGAGG-3′, sense strand; Khachigian et al., 1995). The first lane contains probe alone. The identities of the protein components of the complexes have been delineated by supershift analysis in Day et al. (1999). NE, nuclear extract; NS, non-specific complexes based on oligonucleotide competition analysis. (B) EMSA was performed using [32P]Oligo A and fractions from size-exclusion chromatography. Unfractionated nuclear extract in EMSA is shown in the ‘Input’ lane. Fractions were collected in 1:1 mixture of buffers C and D prior to EMSA. Gel electrophoresis was performed as described in Materials and methods. The arrow indicates complex A5. (C) SYPRO-stained 1D SDS–PAGE of proteins eluted from EMSA gel fragments precipitated with methanol. The protein species indicated by the arrow (triplicate samples) were excised from each lane (three bands pooled) for MS/MS analysis [(see D)]. (D) MALDI-TOF analysis of complex A5. Peptides (in capitals) corresponding to Rattus norvegicus NF1/X.
We prepared bulk nuclear extracts from the rat vascular smooth muscle cell line WKY12-22 (Rafty and Khachigian, 1998) and fractionated this mixture by size-exclusion chromatography. [32P]Oligo A-binding activity in small aliquots of the eluate, assessed by EMSA, revealed that fraction 5 contained complex A5, together with a number of other DNA-binding proteins (Figure 1B). All of fraction 5 was fractionated by multiple EMSA reactions with [32P]Oligo A. Complex A5 was carefully excised from this series of wet gels and analysed by MALDI-TOF mass spectroscopy (MS) to obtain a peptide mass fingerprint (PMF), which was searched against SWISS–PROT and TREMBL databases. The protein component of complex A5 had an Mr of ∼42 kDa (Figure 1C) whose sequence from peptide homology matched Rattus norvegicus NF1/X (Figure 1D).
NF1/X interacts with the proximal PDGF-A promoter
To provide independent evidence that NF1/X can bind to the proximal PDGF-A promoter, we generated NF1/X recombinant protein and performed EMSA with [32P]Oligo A. First, however, western blot analysis using polyclonal antibodies to NF1 was performed to demonstrate the capacity of the NF1/X-80L expression vector to generate immunoreactive protein (Figure 2A). EMSA then revealed the formation of a single discrete nucleoprotein complex (Figure 2B), which was apparent at concentrations as low as 50 ng (data not shown), but not detected when backbone vector (HisTag-80L) eluate was used (Figure 2B). The specificity of the complex was demonstrated by its abrogation in the presence of a 50-fold molar excess of unlabelled Oligo A (Figure 2C). In contrast, the same fold excess of an oligonucleotide targeting the Ets transcription factor E74 failed to affect the complex (Figure 2C). Antibody inhibition experiments were performed to confirm the identity of the protein component of this complex. NF1 antibodies eliminated this complex, whereas polyclonal Smad1 antibodies had no effect (Figure 2C). Finally, supershift analysis was performed to demonstrate that endogenous NF1 protein interacts with [32P]Oligo A. The intensity of complex A5 was strongly attenuated by pre-incubation of nuclear extract with NF1 antibodies, whereas Sp3 antibodies had no effect (Figure 2D). These latter findings show that both recombinant and nuclear NF1/X binds to the proximal region of the PDGF-A promoter in a specific manner, complementing our findings from MALDI-TOF analysis using nuclear extracts as the source of protein (Figure 1D).
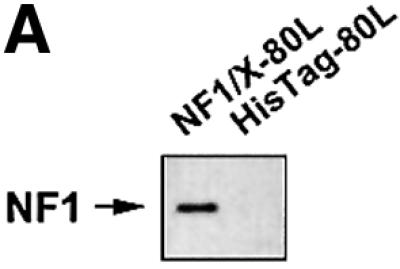
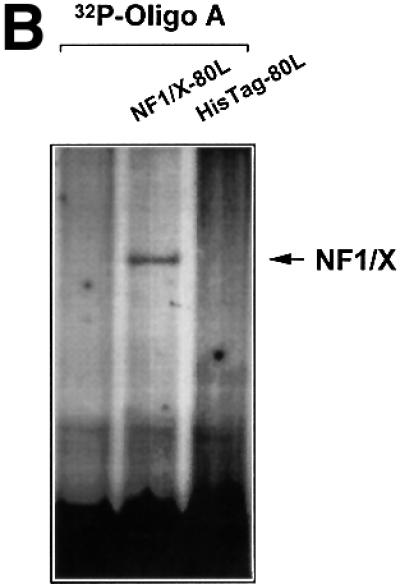
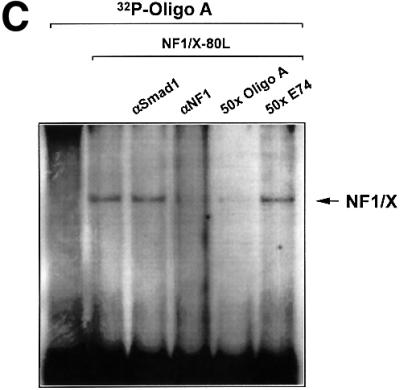
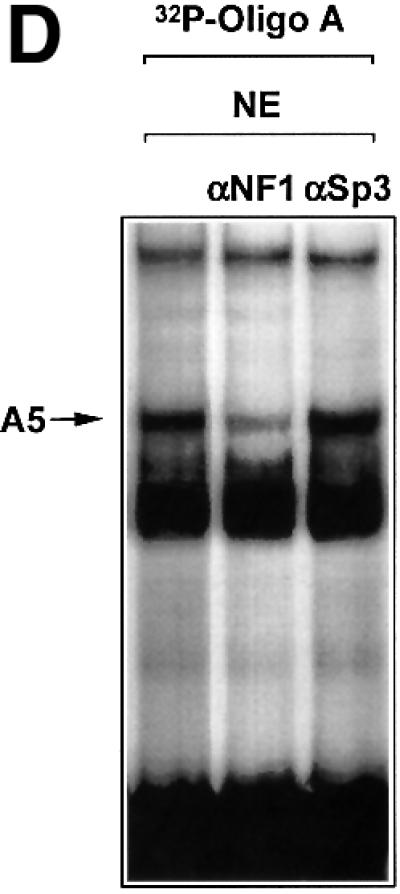
Fig. 2. NF1 interacts with the proximal PDGF-A promoter. (A) Recombinant NF1/X (1 µg) produced from construct NF1/X-80L and the control backbone HisTag vector (HisTag-80L) were analysed for NF1 immunoreactivity by western blot analysis using polyclonal antibodies to NF1. (B) Recombinant NF1/X protein (5 µg) and eluate from control backbone HisTag-80L vector were incubated with [32P]Oligo A prior to EMSA. The arrow represents Oligo A-bound recombinant NF1/X. (C) Competition and antibody elimination analysis. The indicated antibody (1 µg) was incubated with the binding mixture for 10 min before the addition of the [32P]Oligo A. For competition studies, a 50-fold molar excess of unlabelled oligonucleotide was incubated with the binding mixture for 10 min prior to the addition of the probe. The sequence of E74 is 5′-AGCTTCTCTAGCTGAATAACCGGAAGTAACTCATCGTCG-3′ (sense strand). (D) Interaction of endogenous NF1 protein with proximal PDGF-A promoter. Nuclear extracts from WKY12-22 cells were incubated with [32P]Oligo A with or without pre-incubation with 1 µg of polyclonal NF1 or Sp3 antibodies for 10 min prior to the addition of the 32P-labelled probe. Bound species were resolved by non-denaturing electrophoresis and visualized by autoradiography. The arrow denotes complex A5 containing NF1.
NF1/X represses PDGF-A promoter-dependent transcription
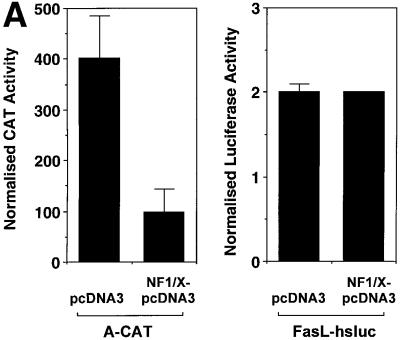
To determine whether NF1/X could influence expression driven by the PDGF-A promoter, we co-transfected WKY12-22 cells with a heterologous promoter– chloramphenicol acetyl-transferase reporter construct (A-CAT) in which the Oligo A sequence was cloned at the SmaI site of pCATprom (Khachigian et al., 1995) with an expression vector generating NF1/X or its control backbone, pcDNA3. NF1/X reduced CAT activity 4-fold compared with its backbone control (Figure 3A). To show specificity of PDGF-A promoter repression by NF1/X, we transfected the cells with a luciferase construct driven by 1.2 kb of the Fas ligand promoter (Kasibhatla et al., 1998). However, NF1/X had no effect on luciferase activity driven by this latter construct (Figure 3A).
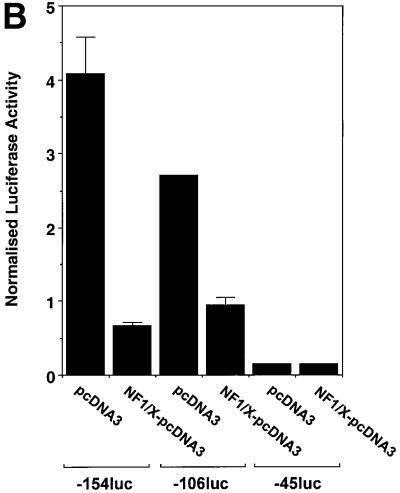
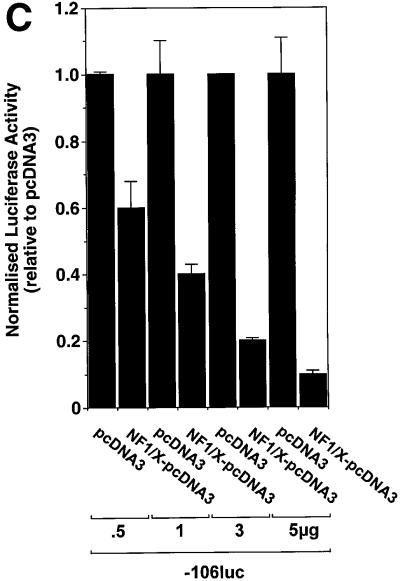
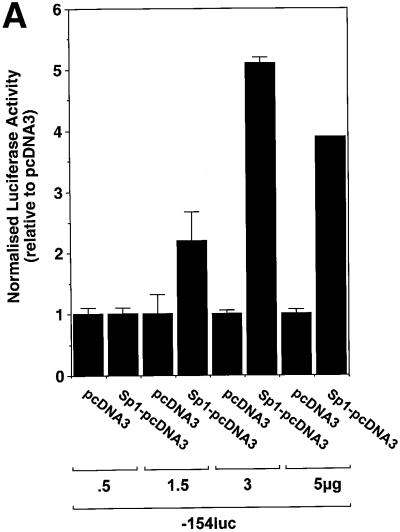
Fig. 3. Repression of PDGF-A promoter activity by NF1/X. (A) WKY12-22 cells were transiently co-transfected with 2 µg of A-CAT or 2 µg of a luciferase construct driven by 1.2 kb of the Fas ligand promoter, along with either 3 µg of NF1/X–pcDNA3 or pcDNA3. Twenty-four hours after transfection, the cells were lysed and assessed for CAT or luciferase activity. CAT activity was normalized to the concentration of protein in the lysate. (B) Serial 5′ deletion and transient transfection analysis using PDGF-A promoter constructs and NF1/X. COS-7 cells were transfected with the indicated PDGF-A promoter–luciferase constructs together with 3 µg of either NF1/X-pcDNA3 or pCDNA3. Twenty-four hours after transfection, the cells were lysed and assessed for luciferase activity. (C) Overexpression of NF1/X represses PDGF-A chain transcription in a dose-dependent manner. COS-7 cells were transiently transfected with 2 µg of –106luc and the indicated amounts of NF1/X or pcDNA3. Twenty-four hours post-transfection, the cells were lysed and assessed for luciferase activity. In experiments involving luciferase reporters, the cells were also transfected with 2 µg of pRL–TK to normalize for transfection efficiency. Firefly luciferase activity was normalized to Renilla. Identical results were obtained in WKY12-22 cells.
To define the region in the PDGF-A promoter mediating NF1/X suppression of PDGF-A transcription, transfection studies were performed using reporter constructs driven by various sized fragments of the PDGF-A promoter. Reporter activity in cells transfected with construct –154luc or –106luc, in which 154 and 106 bp, respectively, of the PDGF-A promoter sequence were cloned in front of firefly luciferase reporter cDNA was down-regulated upon overexpression of NF1/X (Figure 3B). NF1/X suppression of reporter activity was dose dependent (Figure 3C). In contrast, luciferase activity in cells transfected with –45luc, which has less basal activity than –154luc or –106luc, was not influenced by NF1/X (Figure 3B). These findings suggest that base pairs –106 and –45 in the PDGF-A promoter may mediate NF1/X repression.
NF1/X oligonucleotide decoys rescue the PDGF-A promoter and PDGF-A gene from repression
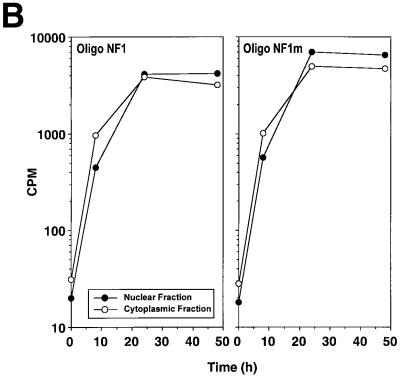
We next used double-stranded oligonucleotide decoys bearing the NF1/X consensus binding element to determine whether these could de-repress PDGF-A promoter activity. The decoy approach has previously been used to block the function of transcriptional activators, such as NF-κB (Morishita et al., 1997) and E2F (Morishita et al., 1995). Luciferase activity generated by construct –106luc increased in cells that were co-transfected with 1 µM Oligo NF1/X (Figure 4A). In contrast, PDGF-A promoter activity was only modestly affected by an identical concentration of Oligo NF1/Xm (Figure 4A), in which the NF1/X-binding site was mutated (G→T). We tracked the distribution of both 32P-labelled molecules over time to ensure intracellular localization. [32P]Oligo NF1/X and [32P]Oligo NF1/Xm first entered the cytoplasm, then the nucleus, with similar kinetics (Figure 4B). To demonstrate that NF1/X could repress the expression of endogenous PDGF-A, we performed northern blot analysis in cells transfected either with NF1/X–pcDNA3 or pcDNA3. Steady-state PDGF-A mRNA levels were abolished by NF1/X–pcDNA3; pcDNA3 had no effect (Figure 4C). Oligo NF1/X rescued PDGF-A expression back to basal levels, whereas Oligo NF1/Xm failed to have any effect (Figure 4C). These data demonstrate the capacity of the NF1/X decoy to rescue the PDGF-A promoter and endogenous PDGF-A gene from NF1/X repression.
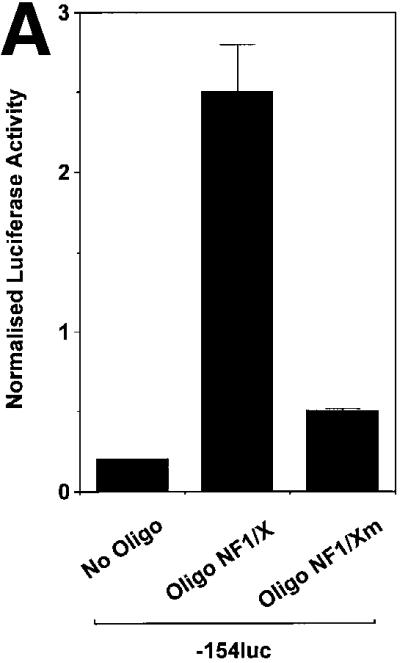
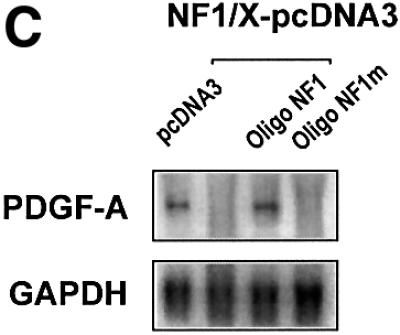
Fig. 4. NF1/X oligonucleotide decoys rescue the PDGF-A promoter from repression. (A) Cells were transfected with 8 µg of –106luc either alone or together with 1 µM Oligo NF1/X or Oligo NF1/Xm, as indicated in the figure. The cells were also transfected with 2 µg of pRL–TK to normalize for transfection efficiency. Firefly luciferase activity was normalized to Renilla. Luciferase activity in the cell lysates was determined after 24 h. (B) Oligo NF1/X and Oligo NF1/Xm localize in the nucleus upon transfection with similar kinetics. Nuclear and cytoplasmic extracts of cells that had previously been transfected with 32P-labelled Oligo NF1/X or Oligo NF1/Xm (500 fmol, 1 × 106 c.p.m.) were prepared 8, 24 and 48 h post-transfection. Radioactivity associated with each fraction was determined in a β-scintillation counter. (C) NF1/X represses endogenous PDGF-A expression and this can be reversed by Oligo NF1/X but not NF1/Xm. Northern blotting analysis was performed using total RNA of WKY12-22 cells 8 h after transfection with 20 µg of either NF1/X– pcDNA3 or pcDNA3 and 1 µM Oligo NF1/X or Oligo NF1/Xm. Hybridization was performed with 32P-labelled PDGF-A and GAPDH cDNA prior to washing and autoradiography. Sequence of Oligo NF1/X (5′-TATTTTGGATTGAAGCCAATATGATAATGA-3′) and Oligo NF1/Xm (5′-TATTTGTTATTGAAGCCAATATGATAATGA-3′).
NF1/X interacts with and inhibits Sp1 interaction with the PDGF-A promoter
The NF1/X-binding site in the PDGF-A promoter is located in a ‘hot spot’, which supports the interaction of several other nuclear factors (Figure 1A). The relatively loose NF1/X consensus binding element (Zorbas et al., 1992) appears in the Oligo A sequence. However, our attempts to selectively inhibit NF1/X binding to the PDGF-A promoter by mutational analysis, although subtle, also resulted in abrogation in the ability of other complexes to form. Since Sp1 regulates both basal and inducible PDGF-A expression (Khachigian et al., 1995) and produces a nucleoprotein complex using Oligo A with greatest intensity (Figure 1), we hypothesized that NF1/X repression of PDGF-A is mediated by interplay between NF1/X and Sp1. Gel-shift analysis revealed that recombinant Sp1 and NF1/X each bind [32P]Oligo A (Figure 5A). However, NF1/X blocked the formation of an Sp1 complex, whereas the same amount of bovine serum albumin (BSA) had no effect (Figure 5A).
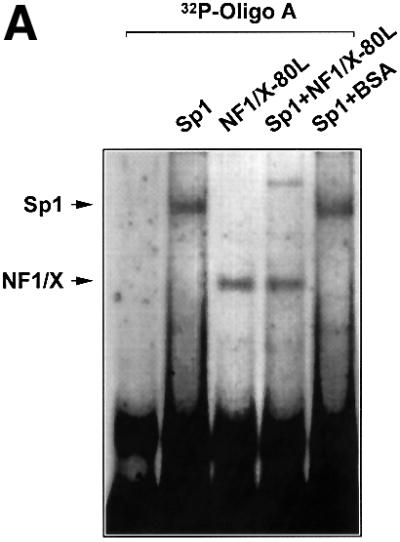
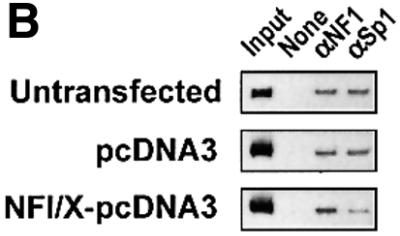
Fig. 5. NF1/X inhibits Sp1 occupancy of the proximal PDGF-A promoter. (A) EMSA was performed with [32P]Oligo A and 1 µg of recombinant Sp1 or NF1/X-80L, either alone or together. BSA was added in lieu of NF1/X where indicated. Sp1 and NF1/X complexes are indicated by arrows. (B) ChIP analysis in WKY12-22 cells transfected with pcDNA3, NF1/X–pcDNA3 or untransfected. Chromatin cross-linked protein–DNA complexes were immunoprecipitated using antibodies to NF1, Sp1 or no antibody and the PDGF-A promoter amplified by PCR.
To demonstrate interplay between these nuclear factors at the level of the endogenous gene, we next performed chromatin immunoprecipitation (ChIP) analysis using NF1 and Sp1 antibodies in combination with PCR in WKY12-22 cells transfected with pcDNA3, NF1/X– pcDNA3. Figure 5B demonstrates that NF1/X and Sp1 occupy the endogenous PDGF-A promoter in untransfected cells, consistent with EMSA (Figure 1), which was not altered by transfection with pcDNA3 (Figure 5B). Upon transfection with NF1/X–pcDNA3, interaction of Sp1 with the promoter was barely detectable (Figure 5B). These data show that NF1 and Sp1 co-occupy the PDGF-A promoter in vivo and, on NF1/X overexpression, inhibit Sp1 occupancy of the authentic PDGF-A promoter.
Since an interaction between NF1/X and Sp1 has not been reported previously, we performed precipitation experiments in which recombinant His-tagged NF1/X was incubated with nuclear extracts, precipitated with nickel–resin and any Sp1 bound to the His-tagged NF1/X detected by western blotting analysis. This produced a single band with an Mr of 100 kDa whose mobility under denaturing conditions was identical to that of immunoreactive Sp1 from unfractionated nuclear extracts (Figure 6). This demonstrates for the first time that NF1/X physically interacts with Sp1 and prevents its occupancy of the PDGF-A promoter.
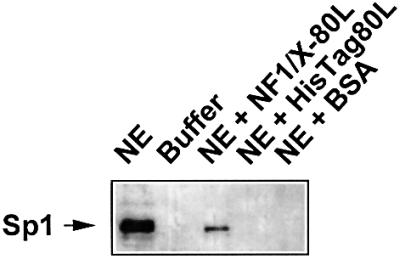
Fig. 6. NF1/X physically interacts with Sp1. Recombinant NF1/X-80L (or eluate from the HisTag backbone control or BSA) was incubated with COS-7 cell nuclear extract (NE) prior to precipitation with nickel–resin suspension and endogenous Sp1 detection by western blotting analysis and chemiluminescence. Lane 1 represents unfractionated NE containing Sp1 immunoreactivity; lane 2, NE buffer C/D; lane 3, Sp1 (out of NE) precipitated with NF1/X-80L; lane 4, no Sp1 detected using the HisTag-80L backbone; lane 5, no Sp1 detected when BSA used. The arrow indicates immunoreactive Sp1.
NF1/X antagonizes Sp1 activation of PDGF-A promoter
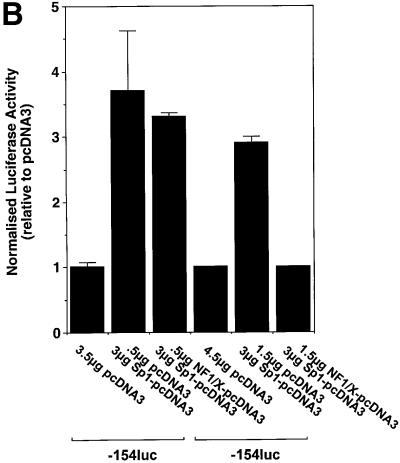
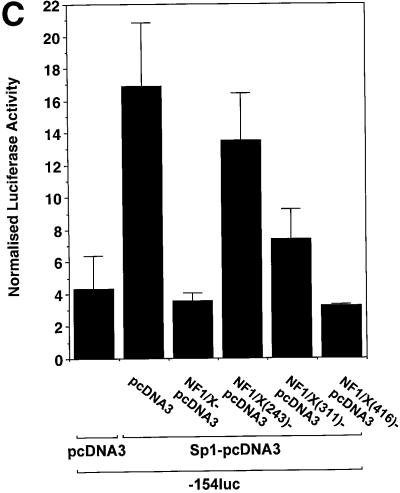
To demonstrate the capacity of NF1/X to repress inducible PDGF-A transcription, we overexpressed Sp1, which is a known positive regulator of PDGF-A transcription (Khachigian et al., 1995) to ascertain whether the inhibitory activity of NF1/X could override Sp1 transactivation. As expected, Sp1 induced PDGF-A promoter-dependent (–154luc) expression over a range of concentrations in a dose-dependent manner (Figure 7A). Co-expression of Sp1 (3 µg) with 0.5 µg of NFI/X–pcDNA3 produced modest inhibition (Figure 7B). Complete inhibition was observed with 1.5 µg of NFI/X–pcDNA3 (Figure 7B). These data demonstrate the capacity of NF1/X to antagonize the activity of a known transcriptional activator of PDGF-A transcription.
Fig. 7. NF1/X represses Sp1 induction of the PDGF-A promoter. (A) Sp1 transactivates the PDGF-A promoter. Cells were transiently transfected with 2 µg of –154luc and the indicated amounts Sp1–pcDNA3 or pcDNA3. (B) NF1/X inhibits Sp1 transactivation of the PDGF-A promoter. The cells were transiently transfected with 3 µg of Sp1–pcDNA3 and the indicated amount of NF1/X or pCDNA3. Twenty-four hours after transfection, the cells were lysed and luciferase activity determined. The cells were also transfected with 2 µg of pRL–TK to normalize for transfection efficiency. Firefly luciferase activity was normalized to Renilla.
Subtype-specific domain of NF1/X mediates repression of basal and inducible PDGF-A transcription
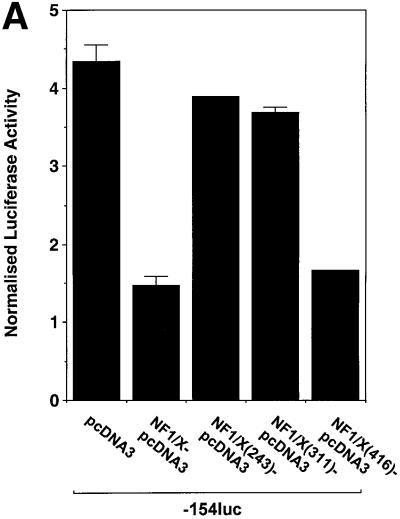
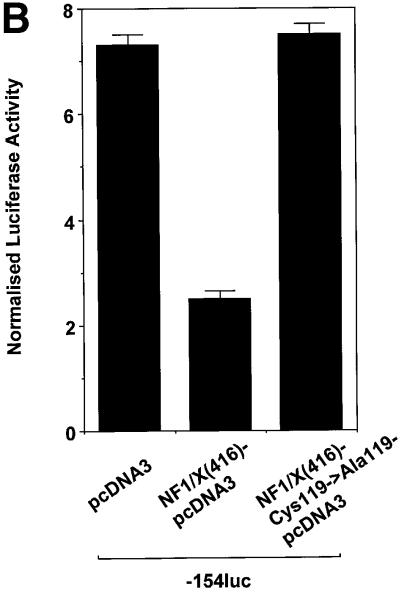
To gain insight into the region in NF1/X that mediates suppression of PDGF-A promoter-dependent expression, we performed transient transfection analysis using 3′ truncations of full-length NF1/X cDNA. As expected, full-length NF1/X inhibited basal PDGF-A promoter-dependent expression (Figure 8A). Overexpression of NF1/X(416), which comprises the first 416 of the 505 amino acids in NF1/X (Figure 8E), like NF1/X, completely inhibited luciferase expression (Figure 8A). In contrast, NF1/X(311) and NF1/X(243), which carry deletions of residues 312–505 and 244–505 (Figure 8E), respectively, failed to repress the PDGF-A promoter (Figure 8A). These findings show that residues 311–416, which span the C-terminal end of the NF1/X subtype-specific domain, are critical for NF1/X repression of basal PDGF-A transcription. A point mutation was introduced into the DNA-binding domain of NF1/X(416) that changed Cys at position 119 into Ala, a mutation that impairs its ability to bind DNA (Novak et al., 1992) and completely abrogated its capacity to repress the PDGF-A promoter (Figure 8B).
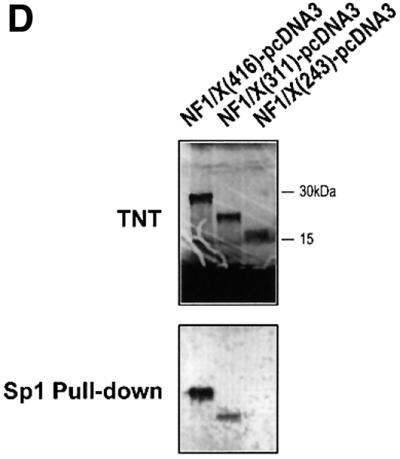
Fig. 8. Repression domain of NF1/X is mediated by the subtype-specific domain. (A) Residues 311–416 in NF1/X mediate inhibition of basal PDGF-A expression. Cells were transiently transfected with 2 µg of –154luc and 1.5 µg of NF1/X, NF1/X(243), NF1/X(311) or NF1/X( 416) in pcDNA3. (B) Mutation of Cys119 (to Ala) in NF1/X abrogrates its repression of the PDGF-A promoter. Transfections were performed with 2 µg of –154luc and 3 µg of NF1/X(416)–pcDNA3 or NF1/X(416)-Cys119 (to Ala)–pcDNA3. (C) Amino acids 243–416 in NF1/X mediate inhibition of Sp1-inducible PDGF-A expression. Transfections were performed with 2 µg of –154luc, 3 µg of Sp1–pcDNA3 and 1.5 µg of either NF1/X, NF1/X(243), NF1/X(311) or NF1/X(416) in pcDNA3. Twenty-four hours post-transfection, the cells were lysed and luciferase activity determined. The cells were also transfected with 2 µg of pRL–TK to normalize for transfection efficiency. Firefly luciferase activity was normalized to Renilla. (D) Immunoprecipitation experiments using in vitro transcribed/translated [35S]Met-labelled NF1/X(416), NF1/X(311) and NF1/X(243) in rabbit reticulocyte lysates and Sp1 antibodies. The top blot demonstrates the electrophoretic mobility of all three proteins on denaturing SDS–PAGE; the bottom blot is the product of immunoprecipitation analysis with Sp1 antibodies and electrophoretic resolution. (E) Schematic representation of NF1/X truncations used in this study. The various domains within NF1/X and their lengths (not to scale) are indicated in the schematic. The numbered residues indicate the last NF1/X amino acid in each construct.
Under conditions of Sp1 transactivation, NF1/X(416), like full-length NF1/X, blocked PDGF-A promoter activity, whereas NF1/X(243) had no effect (Figure 8C). Interestingly, unlike its inability to inhibit basal expression (Figure 8A), NF1/X(311) partially suppressed Sp1 induction of the PDGF-A promoter (Figure 8C). This indicates that different subdomains in NF1/X mediate PDGF-A repression depending on the activation state of the PDGF-A promoter. NF1/X amino acids 244–416 are required for NF1/X repression when the PDGF-A promoter is transactivated by Sp1, whereas a smaller region in NF1/X (amino acids 311–416) is required for complete inhibition of basal PDGF-A transcription. To com plement these studies, we performed immunoprecipitation experiments with Sp1 and in vitro transcribed/translated [35S]Met-labelled NF1/X(416), NF1/X(311) and NF1/X(243) in unfractionated reticulocyte lysates (Figure 8D, top blot). Sp1 antibodies pulled down NF1/X(416) (Figure 8D, bottom blot). However, NF1/X(311) was only partially precipitated using this strategy and NF1/X(243) was not detected (Figure 8D, bottom blot). These findings demonstrate that NF1/X repression of the PDGF-A promoter is dependent upon direct protein– protein interactions with Sp1.
Discussion
In this paper, we isolated a nuclear factor that interacts with the base pairs –70/–47 region of the PDGF-A promoter using bulk EMSAs and chromatography techniques. PMF and supershift analysis revealed this DNA-binding protein to be NF1/X. Both recombinant and endogenous NF1/X bound the PDGF-A promoter in a specific manner. NF1/X repressed PDGF-A promoter-dependent transcription and endogenous mRNA expression, which was rescued by double-stranded oligonucleotide decoys bearing an intact, but not mutant NF1/X-binding site. A point mutation that converts Cys119 to Ala in the DNA-binding domain of NF1/X (Novak et al., 1992) abolished its repression of PDGF-A promoter. EMSA and ChIP analysis revealed that NF1/X antagonized the activity of Sp1 by inhibiting its occupancy of the PDGF-A promoter. We also show here for the first time that NF1/X physically and specifically interacts with Sp1 through its subtype-specific domain and blocks Sp1 induction of PDGF-A promoter-dependent expression. Deletional analysis demonstrated that amino acids 311–416 in NF1/X subtype-specific domain mediated NF1/X suppression of basal PDGF-A transcription, whereas residues 243–416 were required for repression of Sp1-inducible promoter activity, findings supported by immunoprecipitation experiments with Sp1 antibodies and 35S-labelled NF1/X proteins bearing these C-terminal truncations. This is the first report of the transcriptional repression of a growth factor gene by any member of the NF1 family.
The NF1 family of nuclear regulatory factors can both activate (Shaul et al., 1986; Jones et al., 1987; Inoue et al., 1990) and repress (Rossi et al., 1988; Roy and Guerin, 1994; Adams et al., 1995; Szabo et al., 1995) gene transcription. NF1/X acts as a transcriptional silencer for genes such as those encoding retinol-binding protein (Colantuoni et al., 1987), von Willebrand factor (Jahroudi et al., 1996), peripherin (Adams et al., 1995) and mouse α2(I) collagen (Rossi et al., 1988). Conversely, NF1/X can activate the α-globin (Jones et al., 1987), human hepatitis B virus S (Shaul et al., 1986), p53 (Furlong et al., 1996) and myelin basic protein (Inoue et al., 1990) genes. Interestingly, NF1/X can repress the α1B adrenergic receptor middle (P2) promoter in DDY1MF-2 cells and activate the same promoter in Hep3B cells (Gao and Kunos, 1998). The specific sites in NF1/X protein mediating its ability to modulate transcription are poorly understood at the present time. However, it is clear that NF1/X contains both positive and negative regulatory domains (Osada et al., 1999). Examination of the influence of NF1 on α1B adrenergic receptor promoter activity revealed a positive domain that is located between amino acids 416 and 505, and a repression domain located between amino acids 243 and 416 (Gao and Kunos, 1998). Additional mechanisms mediating NF1 multiplicity of function may involve collaboration with other nuclear factors, local chromatin architecture and/or the activation state of intracellular signalling pathways. The present report extends these findings by demonstrating that NF1/X repression is mediated by residues 311–416 when the PDGF-A promoter is basally active, and residues 243–416 when the PDGF-A promoter is transactivated by Sp1.
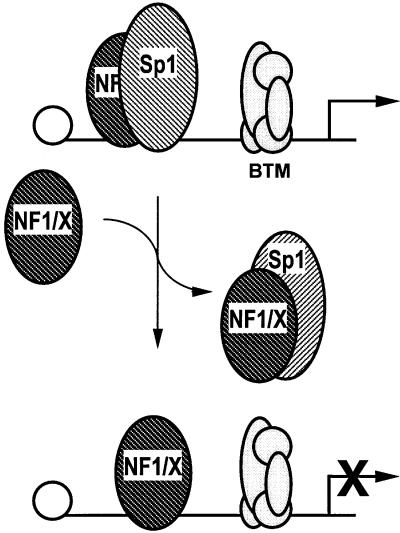
Transcriptional repressors regulate gene expression by at least two distinct mechanisms. Passive repression involves nuclear proteins competing with positive transcriptional regulators for common DNA-binding sites. Passive repressors are also capable of interacting with transcriptional activators and, as a result, destabilize bound factors or prevent binding of such factors. An example of a likely passive repressor of PDGF-A promoter is GCF-2, because it competes with at least three transcriptional activators (Sp1, Sp3 and Egr-1) for common binding sites in the PDGF-A promoter (Khachigian et al., 1999). Active repressors, on the other hand, possess intrinsic repressor activity and the capacity to down-regulate transcription through modular domains. An active repressor of the PDGF-A promoter is WT-1, which binds the G+C-rich region in the proximal promoter and inhibits PDGF-A promoter-dependent gene expression (Wang et al., 1992; Gashler et al., 1993). Here we have identified a potent repressor of basal and inducible PDGF-A transcription. The extent to which any given gene is transcribed is dependent upon the net influence of both activators and repressors on that gene. It is not surprising that complex A5 (NF1/X) is a minor band compared with the sum of the intensities of the (positive regulatory) Sp1/Sp3/Egr-1 complexes, given the high level expression of PDGF-A in vascular smooth muscle cells (Majesky et al., 1988). Forcing NF1/X expression would alter this activator–repressor balance in favour of the latter, as shown by ChIP analysis; indeed, northern blotting revealed that endogenous PDGF-A expression is shut down as a result of altering this balance. Whether NF1/X functions as an active or passive repressor in the context of PDGF-A is not entirely clear. However, since the subtype-specific domain in NF1/X located at residues 243–416 is capable of negatively regulating transcription in the context of GAL4 (Roulet et al., 1995; Gao and Kunos, 1998) and this study demonstrates that the same region mediates NF1/X repression of (basal and Sp1-inducible) PDGF-A expression, NF1/X may inhibit PDGF-A expression by active means. Alternatively, or in addition since NF1 and Sp1 can independently bind the same region in the proximal PDGF-A promoter, NF1/X could, as a passive repressor, displace Sp1 from its recognition element. The fact that NF1/X physically interacts with Sp1 and prevents Sp1 occupancy of the PDGF-A promoter lends support to both possibilities (Figure 9). Previous studies by our group demonstrated that Egr-1 can displace Sp1 and Sp3 from the PDGF-A promoter and activate gene expression (Khachigian et al., 1995; Silverman et al., 1997). NF1/X in the present study antagonizes occupancy of the same region in the promoter by Sp1, leading to suppression of gene expression. Thus, two functionally different displacement mechanisms are involved in the transcriptional regulation of the PDGF-A gene.
Fig. 9. Model of NF1/X repression of PDGF-A promoter activity. NF1/X physically interacts with Sp1 (which regulates basal and inducible PDGF-A transcription) and prevents Sp1 occupancy of the PDGF-A promoter, shutting down gene expression as a consequence. The influence of NF1/X on other promoter-bound nuclear factors is presently unclear. BTM, basal transcriptional machinery.
Thus, building on a large body of literature that has focused largely on the mechanisms governing inducible PDGF-A promoter-dependent expression, the present study, which now defines NF1/X as a novel DNA-binding repressor of the PDGF-A promoter by antagonizing Sp1, indicates that PDGF-A expression is tightly controlled by positive and negative regulatory factors, and that the stoichiometric relationship between these proteins will probably influence the expression pattern of this gene.
Materials and methods
Cell culture
WKY12-22 rat pup aortic smooth muscle cells (Rafty and Khachigian, 1998) were cultured in Waymouth’s MB752/1 medium (Life Technologies Inc.) supplemented with 10% fetal calf serum (FCS), 30 µg/ml l-glutamine, 10 U/ml penicillin and 10 µg/ml streptomycin at 37°C and 5% CO2. COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Inc.) supplemented with 10% FCS, 30 µg/ml l-glutamine, 10 U/ml penicillin and 10 µg/ml streptomycin at 37°C and 5% CO2.
Transient transfection analysis
WKY12-22 cells and COS-7 cells were seeded in 6-well tissue culture plates for 24 h prior to transfection. When ∼70% confluent, the cells were transfected with 2 µg of the indicated promoter–reporter plasmids. Transfections were performed using FuGENE6 (Roche Molecular Biochemicals). A precipitate was formed using 3 µl of FuGENE6/µg of transfected DNA and the transfection mix was made up to 1 ml with serum-free medium. After incubation at 22°C for 10 min, the DNA/FuGENE6 mixture was added to cells containing 2 ml of complete medium. Two days post-transfection, the cell lysates were prepared for assessment of either CAT or luciferase activity.
Plasmid constructs
Mutant forms of the plasmid, –154luc (Day et al., 1999), –154lucAm1 and –154lucAm2, were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer’s instructions by cloning OligoAm1 and OligoAm2 (sense orientation only) into the –76/–47 region of the PDGF-A promoter (relative to the TATA box). Mutant NF1/X(416)-Cys119→Ala119 was constructed using the QuikChange site-directed mutagenesis kit (Stratagene), in accordance with the manufacturer’s instructions, using 5′-CGGATTGACTCCCTGCGCCAG-3′ as the forward primer.
Nuclear extract preparation and EMSA
Nuclear extracts were prepared and EMSA performed as described previously (Day et al., 1999).
Complex A5 isolation
Size exclusion of nuclear proteins was performed with a Bio-Gel P60 gel (Bio-Rad) in accordance with the manufacturer’s instructions. The eluate used was buffers C and D (see preparation of nuclear extracts) in a ratio of 1:1. Proteins were eluted from gel pieces in 100 mM sodium acetate, 0.1% SDS and 10 mM dithiothreitol (DTT) at 37°C overnight. Proteins were then precipitated at –80°C overnight and spun at 12 000 g for 1 h to pellet. The pellet was resuspended in 50 mM DTT, 2% SDS, 20% glycerol, 375 mM Tris and boiled for 5 min. The sample was run over three lanes of a 10–20% T-mini-gel with one lane of protein standards. The gel was stained with SYPRO Ruby and visualized using a Bio-Rad Molecular Imager FX. The two major bands were excised from each lane for MS/MS analysis.
Q-TOF MS/MS
Samples were subjected to a 16 h tryptic digest at 37°C. The resulting peptides were purified using a ZipTip to concentrate and desalt the sample. The samples were then analysed by ESI-TOF using a MicromassQ-TOF MS equipped with a nanospray source, using either flow injection coupled to a Waters CapLC or manually acquired using borosilicate capillaries for nanospray acquistion. After peptides were selected, the MS was switched to MS/MS mode and data collected over the m/z range 50–2000 Da with variable collision energy settings.
MALDI-TOF PMF
The sample was analysed by MALDI-TOF MS to obtain a PMF. The peptide masses were searched against SWISS-PROT and TREMBL.
Recombinant NF1/X preparation
NF1/X recombinant protein was made by subcloning the EcoRI-digested pNF1/X cDNA fragment into the cis-repressed pQE vector set (Qiagen). Recombinant protein was purified by Ni-NTA–agarose in accordance with the manufacturer’s instructions (Qiagen).
ChIP analysis
Six hours after transfection of WKY12-22 cells with NF1/X–pcDNA3 or pcDNA3 with FuGENE6, the cells were washed with phosphate-buffered saline (PBS) pH 7.4, incubated with 1% formaldehyde for 10 min and quenched with glycine (final concentration 0.1 M). The cells were washed with PBS, trypsinized, then resuspended and incubated in buffer A (100 mM Tris–HCl pH 9.4, 10 mM DTT) for 15 min at 30°C. The cells were precipitated by spinning at 14 000 g for 1 min, washed sequentially in cold PBS, buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES pH 6.5), buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES pH 6.5), lysed in lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 µg/ml leupeptin, 2 µg/ml aprotinin] and sonicated. After spinning, the supernatant was mixed with dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl pH 8.1). Protein G–Sepharose (previously incubated with salmon sperm DNA and pre-immune serum) was added to the supernatant and gently mixed at 4°C for 2 h. The suspension was evenly divided and 5 µg of rabbit polyclonal antibodies were added to Sp1 (Santa Cruz Biotechnology) and NF1 (Santa Cruz Biotechnology) or no antibody was added, and incubated overnight at 4°C. The suspensions were washed sequentially in TSEI (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), TSEII (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8.1), 1× TE and elution was performed in elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C for 4 h. The suspension was treated with proteinase K at 37°C for 4 h, prior to phenol–chloroform extraction and ethanol precipitation in the presence of tRNA. The PDGF-A promoter was amplified by PCR at 30 cycles, producing a 446 bp product.
Immunoprecipitation of recombinant NF1/X-bound nuclear Sp1
A nuclear extract (10 µg) was incubated with either 5 µg of recombinant His-tagged NF1/X-80L, 5 µg of HisTag-80L or 5 µg of BSA in 100 µl of buffer C/D for 1 h at 4°C. A nickel resin suspension (10 µl; Qiagen; previously spun and resuspended in buffer C/D) was added to the samples and incubated for 2 h at 4°C. The samples were spun/washed three times with buffer C/D. After the final wash, the resin was completely dried and resuspended in 20 µl of SDS sample buffer. The samples were boiled for 3 min prior to running on 8% PAGE and immunoreactive Sp1 was detected by western blot analysis.
Oligonucleotide decoy analysis
For reporter gene analysis, WKY12-22 cells were transfected with 8 µg of construct –106luc, either alone or in combination with 1 µM Oligo NF1/X or Oligo NF1/Xm using FuGENE6. The cells were also transfected with 2 µg of pRL-TK to normalize for transfection efficiency. Firefly luciferase activity was normalized to Renilla. Twenty-four hours post-transfection, cell lysates were assessed for luciferase activity. For northern blot analysis, the cells were transfected with 20 µg of pcDNA3 or NF1/X–pcDNA3 and either 1 µM Oligo NF1/X or NF1/Xm, using FuGENE6, and total RNA was prepared 24 h subsequently. To ensure that both oligonucleotides entered the nuclear compartment, radioactivity in the nuclear and cytoplasmic fractions was assessed 8, 24 and 48 h after transfection with 500 fmol (1 × 106 c.p.m.) of [32P]Oligo NF1/X or [32P]Oligo NF1/Xm.
Northern blot analysis
Northern blotting was performed with total RNA as described previously (Day et al., 1999).
Immunoprecipitation of in vitro transcribed/translated NF1/X truncated proteins with Sp1 antibodies
Various [35S]Met-labelled C-terminal truncations of NF1/X protein were generated using a commercial coupled transcription/translation system (Promega). Lysates were incubated with 1 µg of rabbit polyclonal Sp1 antibody (Santa Cruz Biotechnology) in 1:1 mix of buffers C (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA) and D (20 mM HEPES pH 7.9, 10 mM KCl, 0.2 mM EDTA, 20% glycerol). Samples were incubated for 1 h on a rotating wheel at 4°C. Sepharose G (previously washed with buffer C/D) was added to the samples and incubated for 3 h while rotating at 4°C. Samples were spun and the resin was washed in buffer C/D. On the final spin, the resin was resuspended in SDS sample buffer containing DTT. The samples were boiled for 5 min prior to loading on a polyacrylamide gel. Gels were fixed and dried before detection of 35S-labelled species by phosphoimaging.
Acknowledgments
Acknowledgements
The authors thank Dr Bin Gao (Virginia Commonwealth University) for the generous gift of the NF1/X expression vectors. This work was supported by grants from the NHMRC and NSW Department of Health. L.M.K. is a Principal Research Fellow of the NHMRC.
References
- Adams A.D., Choate,D.M. and Thompson,M. (1995) NF1-L is the DNA-binding component of the protein complex at the peripherin negative regulatory element. J. Biol. Chem., 270, 6975–6983. [DOI] [PubMed] [Google Scholar]
- Apt D., Chong,T., Liu,Y. and Bernard,H.-U. (1993) Nuclear factor 1 and epithelial cell-specific transcription of human papillomavirus type 16. J. Virol., 67, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten E., Uutela,M., Li,X., Pietras,K., Ostman,A., Heldin,C.-H., Alitalo,K. and Eriksson,U. (2001) PDGF-D is a specific, protease-activated ligand for the PDGF β-receptor. Nature Cell Biol., 3, 512–516. [DOI] [PubMed] [Google Scholar]
- Blomquist P., Li,Q. and Wrange,O. (1996) The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem., 271, 153–159. [DOI] [PubMed] [Google Scholar]
- Bonthron D.T., Morton,C.C., Orkin,S.H. and Collins,T. (1988) Platelet-derived growth factor A chain: gene structure, chromosomal location and basis for alternative mRNA splicing. Proc. Natl Acad. Sci. USA, 85, 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E. and Kuhnel,B. (1986) Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor promoter identified by linker scanning mutagenesis. J. Mol. Biol., 190, 379–389. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Raymondjean,M., Carranca,A.G., Herbomel,P. and Yaniv,M. (1987) Factors involved in control of tissue-specific expression of albumin gene. Cell, 50, 627–638. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Pirrozi,A., Blance,C. and Cortese,R. (1987) Negative control of liver-specific gene expression: cloned human retinol-binding protein gene is repressed in HeLa cells. EMBO J., 6, 631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F., Rafty,L.A., Chesterman,C.N. and Khachigian,L.M. (1999) Angiotensin II (ATII)-inducible platelet-derived growth factor A-chain gene expression is p42/44 extracellular signal-regulated kinase-1/2 and Egr-1-dependent and mediated via the ATII type 1 but not type 2 receptor. J. Biol. Chem., 274, 23726–23733. [DOI] [PubMed] [Google Scholar]
- Delbridge G.J. and Khachigian,L.M. (1997) FGF-1-induced PDGF A-chain gene expression in vascular endothelial cells involves transcriptional activation by Egr-1. Circ. Res., 81, 282–288. [DOI] [PubMed] [Google Scholar]
- Furlong E.E.M., Rein,T. and Martin,T. (1996) YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element and YY1 and E1A cooperate to amplify p53 promoter activity. Mol. Cell. Biol., 16, 5933–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B. and Kunos,G. (1998) Cell type-specific transcriptional activation and suppression of the α1B adrenergic receptor gene middle promoter by nuclear factor 1. J. Biol. Chem., 273, 31784–31787. [DOI] [PubMed] [Google Scholar]
- Gashler A.L., Bonthron,D.T., Madden,S.L., Rauscher,F.J., Collins,T. and Sukhatme,V.P. (1992) Human platelet-derived growth factor A chain is transcriptionally repressed by the Wilms tumor suppressor WT1. Proc. Natl Acad. Sci. USA, 89, 10984–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gashler A.L., Swaminathan,S. and Sukhatme,V.P. (1993) A novel repression module, an extensive activation domain and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol. Cell. Biol., 13, 4556–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G., Smith,J.R., Goldstein,J.L., Slaughter,C.A., Orth,K., Brown,M.S. and Osborne,T.F. (1988) Multiple genes encode nuclear factor 1-like proteins that bind to the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl Acad. Sci. USA, 85, 8963–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T. (1985) The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J., 4, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Tamura,T., Furuich,T. and Mikoshiba,K. (1990) Isolation of complementary DNAs encoding a cerebellum-enriched nuclear factor I family that activates transcription from the mouse myelin basic protein promoter. J. Biol. Chem., 265, 19065–19070. [PubMed] [Google Scholar]
- Jahroudi N., Ardekani,A.M. and Greenberger,J.S. (1996) An NF1-like protein functions as a repressor of the von Willebrand factor promoter. J. Biol. Chem., 271, 21413–21421. [DOI] [PubMed] [Google Scholar]
- Jones K.A., Kadonaga,J.T., Rosenfeld,P.J., Kelly,T.J. and Tjian,R. (1987) A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell, 48, 79–89. [DOI] [PubMed] [Google Scholar]
- Kaetzel D.M., Coyne,D.W. and Fenstermaker,R.A. (1993) Transcriptional control of the platelet-derived growth factor subunit genes. Biofactors, 4, 71–81. [PubMed] [Google Scholar]
- Kasibhatla S., Brunner,T., Genestier,L., Echeverri,F., Mahboubi,A. and Green,D.R. (1998) DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activaton of NF-κB and AP-1. Mol. Cell, 1, 543–551. [DOI] [PubMed] [Google Scholar]
- Khachigian L.M., Williams,A.J. and Collins,T. (1995) Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J. Biol. Chem., 270, 27679–27686. [DOI] [PubMed] [Google Scholar]
- Khachigian L.M., Lindner,V., Williams,A.J. and Collins,T. (1996) Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science, 271, 1427–1431. [DOI] [PubMed] [Google Scholar]
- Khachigian L.M., Anderson,K.A., Halnon,N.J., Resnick,N., Gimbrone, M.A.,Jr and Collins,T. (1997) Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress response element in the PDGF A-chain promoter. Arterioscler. Thromb. Vasc. Biol., 17, 2280–2286. [DOI] [PubMed] [Google Scholar]
- Khachigian L.M., Santiago,F.S., Rafty,L.A., Chan,L.-W., Delbridge, G.J., Bobik,A., Collins,T. and Johnson,A.C. (1999) GC factor 2 represses platelet-derived growth factor A-chain gene transcription and is itself induced by arterial injury. Circ. Res., 84, 1258–1267. [DOI] [PubMed] [Google Scholar]
- Kruse U., Qian,F. and Sippel,A.E. (1991) Identification of a fourth nuclear factor 1 gene in chicken by cDNA cloning: NF1-X. Nucleic Acids Res., 19, 6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K.U., Pater,A. and Pater,M.M. (1993) Human JC virus perfect palindromic nuclear factor 1-binding sequences important for glial cell-specific expression in differentiating embryonal carcinoma cells. J. Virol., 67, 572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laniel M.A., Poirier,G.G. and Guerin,S.L. (2001) Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity. J. Biol. Chem., 276, 20766–20773. [DOI] [PubMed] [Google Scholar]
- LaRochelle W.J. et al. (2001) PDGF-D, a new protease-activated growth factor. Nature Cell Biol., 3, 517–521. [DOI] [PubMed] [Google Scholar]
- Li X. et al. (2000) PDGF-C is a new protease-activated ligand for the PDGF α-receptor. Nature Cell Biol., 2, 302–309. [DOI] [PubMed] [Google Scholar]
- Majesky M.W., Benditt,E.P. and Schwartz,S.M. (1988) Expression and developmental control of platelet-derived growth factor A-chain and B-chain/Sis genes in rat aortic smooth muscle cells. Proc. Natl Acad. Sci. USA, 85, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson C.E., Shim,E.Y., Friedman,D.S. and Zaret,K.S. (1993) An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell, 75, 387–398. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Rogge,L., Foeckler,R., Karaghiosoff,M. and Winnacker,E.L. (1989) Structural and functional organization of a porcine gene coding for nuclear factor I. Biochemistry, 28, 8191–8200. [DOI] [PubMed] [Google Scholar]
- Mink S., Hartig,E., Jennewein,P., Doppler,W. and Cato,A.C. (1992) A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NF1 and a novel transcription factor, mammary cell-activating factor. Mol. Cell. Biol., 12, 4906–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita R., Gibbons,G.H., Horiuchi,M., Ellison,K.E., Nakama,M., Zhang,L., Kaneda,Y., Ogihara,T. and Dzau,V.J. (1995) A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl Acad. Sci. USA, 92, 5855–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita R. et al. (1997) In vivo transfection of cis element ‘decoy’ against nuclear factor-κB binding site prevents myocardial infarction. Nature Med., 3, 894–899. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer,R.A. and Hurwitz,J. (1983) Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl Acad. Sci. USA, 80, 6177–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A., Goyal,N. and Gronostajski,R.M. (1992) Four conserved cysteine residues are required for the DNA binding activity of nuclear factor I. J. Biol. Chem., 267, 12986–12990. [PubMed] [Google Scholar]
- Nowock J., Borgmeyer,U., Puschel,A.W., Rupp,R.A. and Sippel,A.E. (1985) The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication and the BK virus enhancer. Nucleic Acids Res., 13, 2045–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S., Matsubara,T., Daimon,S., Terazu,Y., Xu,M., Nishihara,T. and Imagawa,M. (1999) Expression, DNA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem. J., 342, 189–198. [PMC free article] [PubMed] [Google Scholar]
- Paonessa G., Gounari,F., Frank,R. and Cortese,R. (1988) Purification of a NF1-like DNA binding protein from rat liver and cloning of the corresponding cDNA. EMBO J., 7, 3115–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M., Fulton,R., Breimer,L., Stewart,M., Willison,K. and Neil,J.C. (1991) Nuclear factor 1 activates the feline leukemia virus long terminal repeat but is posttranscriptionally down regulated in leukemia cell lines. J. Virol., 65, 1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafty L.A. and Khachigian,L.M. (1998) Zinc finger transcription factors mediate high constitutive platelet-derived growth factor-B expression in smooth muscle cells derived from aortae of newborn rats. J. Biol. Chem., 273, 5758–5764. [DOI] [PubMed] [Google Scholar]
- Raines E.W., Bowen-Pope,D.F. and Ross,R. (1990) Handbook of Experimental Pharmacology: Peptide Growth Factors and their Receptors. Springer-Verlag, Berlin, Germany, pp. 173–262.
- Rajas F., Delhase,M., de La Hoya,M., Verdood,P., Castrillo,J.L. and Hooghe-Peters,E.L. (1998) Nuclear factor 1 regulates the distal silencer of the human PIT1/GHF1 gene. Biochem. J., 333, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P., Karsenty,G., Roberts,A.B., Roche,N.S., Sporn,M.B. and de Crombrugghe,B. (1988) A nuclear factor 1 binding site mediates the transcriptional activation of a type 1 collagen promoter by transforming growth factor-β. Cell, 52, 405–414. [DOI] [PubMed] [Google Scholar]
- Roulet E., Armentero,M.-T., Krey,G., Corthesy,B., Dreyer,C., Mermod,N. and Wahli,W. (1995) Regulation of the DNA-binding and transcriptional activities of Xenopus laevis NF1-X by a novel C-terminal domain. Mol. Cell. Biol., 15, 5552–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R.J. and Guerin,S.L. (1994) The 30-kDa rat liver transcription factor nuclear factor 1 binds the rat growth-hormone proximal silencer. Eur. J. Biochem., 219, 799–806. [DOI] [PubMed] [Google Scholar]
- Rupp R.A., Kruse,U., Multhaup,G., Gobel,U., Beyreuther,K. and Sippel,A.E. (1990) Chicken NF1/TGGCA proteins are encoded by at least three independent genes: NF1-A, NF1-B and NF1-C with homologues in mammalian genomes. Nucleic Acids Res., 18, 2607–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod,N., Andrews,P.C. and Tjian,R. (1988) A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature, 334, 218–224. [DOI] [PubMed] [Google Scholar]
- Shaul Y., Ben-Levy,R. and De-Medina,T. (1986) High affinity binding site for nuclear factor I next to the hepatitis B virus S gene promoter. EMBO J., 5, 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E.S., Khachigian,L.M., Lindner,V., Williams,A.J. and Collins,T. (1997) Inducible PDGF A-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am. J. Physiol., 273, H1415–H1426. [DOI] [PubMed] [Google Scholar]
- Szabo P., Moitra,J., Rencendorj,A., Rakhely,G., Rauch,T. and Kiss,I. (1995) Identification of a nuclear factor-1 family protein-binding site in the silencer region of the cartilage matrix protein gene. J. Biol. Chem., 270, 10212–10221. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Madden,S.L., Deuel,T.F. and Rauscher,F.J. (1992) The Wilms’ tumor gene product, WT-1, represses transcription of the platelet-derived growth factor A-chain gene. J. Biol. Chem., 267, 21999–22002. [PubMed] [Google Scholar]
- Wu L. and Whitlock,J.P. (1992) Mechanism of dioxin action: Ah receptor-mediated increase in promoter accessibility in vivo. Proc. Natl Acad. Sci. USA, 89, 4811–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas H., Rein,T., Krause,A., Hoffmann,K. and Winnacker,E.L. (1992) Nuclear factor 1 (NF1) binds to an NF1-type site but not to the CCAAT site in the human α-globin promoter. J. Biol. Chem., 267, 8478–8484. [PubMed] [Google Scholar]