Abstract
STAT1 functions as both a constitutive transcriptional regulator and, in response to cytokine stimulation of cells, as an inducible tyrosine-phosphorylated transcription factor. Here, we identify and characterize a non-transferable nuclear targeting sequence in the STAT1 DNA-binding domain. This conserved signal is critical for the interferon-γ (IFN-γ)-induced nuclear import of phosphorylated STAT1 dimers and requires adjacent positively charged and hydrophobic residues for functioning. Additionally, the constitutive nucleocytoplasmic shuttling of STAT1 in the absence of IFN-γ stimulation is revealed. Nuclear import and export of unphosphorylated STAT1 are demonstrated to be sensitive towards wheat germ agglutinin and to occur independently of the import receptor p97. Loss-of-function mutations of the dimer-specific import signal block nuclear entry of tyrosine-phosphorylated STAT1, which in turn also prevents induction of cytokine-inducible target genes. Nevertheless, nuclear import of unphosphorylated STAT1 continues and the STAT1-dependent constitutive expression of caspases and the tumor necrosis factor-α-mediated induction of apoptosis proceed unaltered. Thus, tyrosine-phosphorylated and unphosphorylated STAT1 molecules shuttle via independent pathways to distinct sets of target genes.
Keywords: nuclear import/phosphorylation/shuttling/STAT/transcription
Introduction
Signal transducers and activators of transcription (STATs) constitute an ancient and evolutionarily conserved family of transcription factors comprising seven mammalian proteins with known homologs in all multicellular animal phyla investigated (Darnell, 1997a). The STATs are involved in the regulated expression of numerous genes underlying diverse cellular processes such as the immune response, antiviral protection, growth and apoptosis (Hoey, 1997; Stark et al., 1998; Leonard, 2001). Best characterized is their role in extracellular ligand-dependent gene induction, and it has become clear that cytokine-induced gene transcription depends in large part upon STAT signaling. The central event in cytokine-dependent transcriptional regulation is phosphorylation of STATs on a single tyrosine residue at their C-terminus (Darnell, 1997b). The reaction is catalyzed by cytokine receptor-associated tyrosine kinases of the Janus type (Jak) at the cell membrane and triggers the homo- and heterodimerization of STAT molecules via reciprocal phosphotyrosine–SH2 domain interactions (Darnell et al., 1994; Ihle, 1995). This transformation enables the STATs to bind to nonameric palindromes with relaxed sequence specificity, of the general form TTCNNNNAA, termed GAS (Decker et al., 1997), a process called ‘STAT activation’ since it rapidly induces transcription of genes harboring GAS sites in their promoters (Schindler and Darnell, 1995).
As has been reported by Stark and co-workers, these proteins can also exert constitutive functions in the nucleus, which do not require STAT activation (Ramana et al., 2000). Analysis of caspase gene expression in STAT1-deficient cells provided the first evidence for constitutive transcriptional activities of a STAT protein (Kumar et al., 1997). It was demonstrated that STAT1- deficient cells are insensitive to apoptosis-inducing signals due to very low or absent expression of caspases, but regained caspase expression with the reintroduction of STAT1. Remarkably, caspase expression was also rescued by STAT1 variants that cannot be phosphorylated and form homodimers. In the meantime, a large number of genes have been identified that are regulated by unphosphorylated STAT1 (Chatterjee-Kishore et al., 2000).
While STAT1 resides mainly in the cytoplasm of unstimulated cells, interferon-γ (IFN-γ) stimulation with concurrent tyrosine phosphorylation results within minutes in a transient nuclear accumulation (Shuai et al., 1993b). It was shown recently that STAT1 and STAT3 are found generally at cell type-specific levels in the nuclei of unstimulated cells, and their nuclear presence was not dependent upon tyrosine phosphorylation (Chatterjee-Kishore et al., 2000; Meyer et al., 2002). However, the regulation of STAT target gene access is poorly understood, a situation caused partly by the absence of readily discernible nuclear export (NES) and nuclear localization signals (NLS) in the STATs. Despite this, it was shown that phosphorylated STAT1 associates with members of the importin superfamily of proteins to achieve accumulation in the nucleus (Sekimoto et al., 1997), a process that requires the integrity of the STAT DNA-binding domain (Herrington et al., 1999; Melen et al., 2001). Nuclear export of STAT1 seems to rely at least in part on the activity of the export receptor CRM1, as demonstrated by leptomycin B (LMB) sensitivity (Begitt et al., 2000; Mowen and David, 2000) and direct binding of recombinant STAT1 to CRM1 in vitro (McBride et al., 2000). Accordingly, leucine-rich sequence elements resembling canonical NESs have been mapped to different locations in the coiled-coil domain and the DNA-binding domain.
Here we show that the transcription factor STAT1 switches between two different nuclear import pathways in a tyrosine phosphorylation-dependent manner. Import as a tyrosine-phosphorylated molecule required a stretch of amino acids that constitute an unusual nuclear import signal. Loss-of-function mutations of this signal selectively abolished cytokine-induced gene activation, while constitutive transcriptional functions of STAT1 remained unaltered. Additionally, the constitutive and tyrosine phosphorylation-independent nucleocytoplasmic shuttling of STAT1 is uncovered, adding a new layer of complexity to our understanding of STAT regulation.
Results
A peptide from the STAT1 DNA-binding domain contains nuclear export activity
To identify peptide sequences in STAT1 that confer transport properties on a heterologous substrate, we expressed segments of ∼60 amino acids as GST–green fluorescent protein (GFP) fusion proteins and injected the affinity-purified products into the cytosol or nucleoplasm of HeLa-S3 cells. We had used this approach before to define an NES in the N-terminal region of STAT1 (Begitt et al., 2000). This procedure resulted in the identification of another nuclear export activity located in the STAT1 DNA-binding domain (see Supplementary data available at The EMBO Journal Online). Further analysis revealed a canonical leucine-rich NES between residues 400 and 410 (Figure 1). Expectedly, nuclear export of this fragment could be blocked with the export inhibitor leptomycin B (LMB) (Kudo et al., 1999). These findings confirm earlier results, which suggested a role in nuclear export of unphosphorylated STAT1 for this motif (McBride et al., 2000).
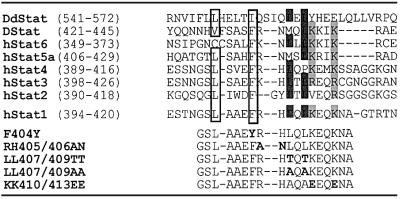
Fig. 1. Top: sequence alignment of a stretch from the DNA-binding domain of human STATs 1–6 (hStat), Drosophila STAT (Dstat) and Dictyostelium STAT (DdStat). This alignment reveals conservation of residues constituting the STAT1 dimer-specific NLS. Critical basic residues are highlighted in light gray and critical hydrophobic residues are marked with dark gray. Additionally, hydrophobic residues conferring export activity on the isolated STAT1 peptide are boxed. Bottom: overview over the mutations introduced into the STAT1 DNA-binding domain (bold letters).
The putative STAT1 export signal modulates nuclear import of activated STAT1
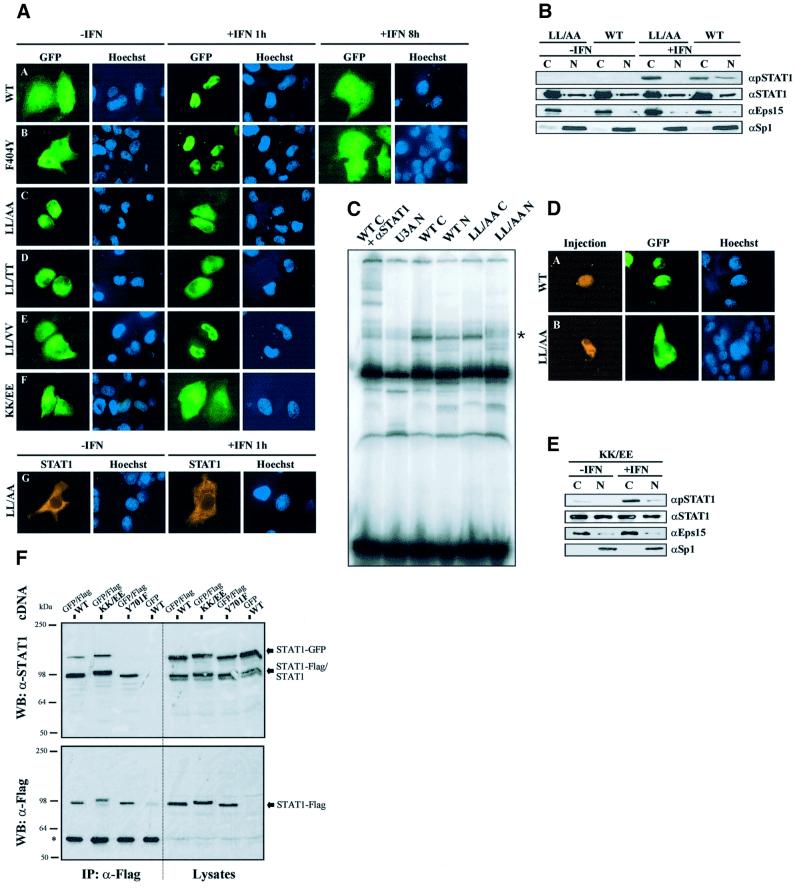
We then mutated in the full-length STAT1 those residues that were found to be critical for nuclear export of the peptides and examined the resulting subcellular STAT1 localization before and after treatment of cells with IFN-γ. Localization studies were performed with STAT1-negative U3A cells (Müller et al., 1993) complemented with wild-type or mutant STAT1. In addition, fusion proteins of STAT1 with GFP were studied, which are tyrosine phosphorylated and translocate into the nucleus similarly to untagged STAT1 (Begitt et al., 2000). As is shown in the fluorescence micrographs of Figure 2A, a complete cycle of IFN-γ-induced STAT1 nuclear accumulation with subsequent return to the initial distribution takes ∼8 h in HeLa-S3 cells. Surprisingly, none of the mutations that were shown to block export of the GST–STAT1367–427–GFP peptide (Phe404Tyr, Leu407Ala or Leu409Ala) had an effect on the nucleocytoplasmic translocation of full-length STAT1. Neither the resting distribution prior to cytokine stimulation nor the rate of nuclear clearance after cytokine-induced nuclear accumulation differed from wild-type STAT1 (shown for the mutant Phe404Tyr in Figure 2A, panels A and B).
Fig. 2. Mutations in the STAT1 DNA-binding domain prevent nuclear entry of tyrosine-phosphorylated STAT1. (A) Effects of IFN-γ on STAT1 distribution in HeLa-S3 cells transiently expressing full-length STAT1–GFP fusion proteins (A–F), and U3A cells expressing the untagged STAT1-LL407,409AA mutant (G). Shown are fluorescence micrographs of resting cells (–IFN) and cells after 1 (+IFN 1 h) or 8 h (+IFN 8 h) stimulation with IFN-γ. After the indicated incubation times, the cells were fixed, stained for DNA (Hoechst) and processed for detection of GFP fluorescence (A–F) or STAT1 by immunocytochemistry (G). The following STAT1 constructs are included: wild-type STAT1 (WT), and mutants F404Y (F404Y), LL407,409AA (LL/AA), LL407,409TT (LL/TT), LL407,409VV (LL/VV) and KK410,413EE (KK/EE). (B) Western blot of cytoplasmic (C) and nuclear (N) extracts from untreated (–IFN) or IFN-γ-treated (30 min) U3A cells transiently expressing the mutant STAT1-LL407,409AA or wild-type STAT1. Identical extracts were probed in parallel experiments with pY701-STAT1, STAT1, Eps15 or Sp1 antibodies (top to bottom). (C) Gel shift assay with extracts as described in (B) using a radiolabeled GAS oligonucleotide. The band position of the STAT1–DNA complex is indicated by an asterisk on the right. In the left lane, STAT1 antibody C24 was included in the DNA-binding reaction, resulting in shifting of the STAT1-specific band. (D) STAT1-LL407,409AA does not enter the nucleus in response to IFN-γ. GFP fusion proteins of STAT1 wild type or the LL407,409AA mutant were transiently expressed in HeLa-S3 cells. After nuclear microinjection of STAT1 antibody S1α/β, the cells were incubated for 30 min in the presence of IFN-γ before fixation. The injection site was marked with TRITC–BSA. (E) Western blot of cytoplasmic (C) and nuclear (N) extracts from untreated (–IFN) or IFN-γ-treated (30 min) U3A cells transiently expressing the mutant STAT1-KK410,413EE. Identical extracts were probed in parallel experiments as described in (B). (F) IFN-γ promotes dimerization of STAT1-KK410,413EE. HeLa-S3 cells were co-transfected with the indicated cDNAs for GFP- and Flag-tagged STAT1, and 24 h later the cells were treated as described with IFN-γ and vanadate for 60 min. Shown are western blots of whole-cell lysates before immunoprecipitation (Lysates), and the immunoprecipitates with Flag antibodies (IP: α-Flag). The blots were developed with STAT1 antibodies (upper panel) or Flag antibodies (lower panel). The positions of Flag-tagged (STAT1-Flag), GFP-tagged (STAT1-GFP) and endogenous wild-type STAT1 (STAT1) are indicated on the right. The positions and molecular weight of pre-stained standards are indicated. (*) denotes the position of antibody-decorated Ig heavy chains.
We additionally introduced double mutations in positions 407 and 409 of full-length STAT1. While exchange of the native leucines with valines or isoleucines (Figure 2A, panel E and data not shown) was without effect, replacement in these positions with alanines or threonines (termed LL407,409AA or LL407,409TT) significantly affected the subcellular distribution of STAT1. Astonishingly, nuclear accumulation after destruction of the NES-like signal was not observed; rather, the nuclear presence of the mutant forms of STAT1 was slightly diminished in unstimulated cells (Figure 2A, panels C and D). Even more strikingly, we were unable to induce nuclear accumulation in response to IFN-γ, as the mutant proteins appeared to remain in the cytoplasm. The lack of nuclear accumulation was not linked to the presence of a GFP tag, since untagged STAT1-LL407,409AA was equally incompetent for nuclear accumulation (Figure 2A, panel G).
Because tyrosine phosphorylation and DNA binding have been associated with STAT nuclear accumulation, the double mutant STAT1-LL407,409AA expressed in U3A cells was tested for these properties. We probed cytoplasmic and nuclear extracts of wild type and the import mutant for STAT1 immunoreactivity before and after stimulation with IFN-γ. Wild-type STAT1 as well as STAT1-LL407,408AA were found in both cytoplasmic and nuclear extracts irrespective of IFN-γ stimulation when we used a STAT1-specific antibody for detection. Probing of these extracts with a phosphotyrosine-specific STAT1 antibody revealed the absence of tyrosine phosphorylation prior to cytokine stimulation. Importantly, we were unable to detect significant quantities of the tyrosine-phosphorylated STAT1 mutant in the nuclear extracts of IFN-γ-stimulated cells (Figure 2B). Nevertheless, the mutant was tyrosine phosphorylated overall to a similar degree as wild-type STAT1, as is revealed by the strong phosphotyrosine signal in the cytoplasmic extract (Figure 2B). In gel shift assays with oligonucleotides harboring a single GAS element, it was found that the mutant displayed no DNA-binding activity in the nuclear fraction, while DNA binding in cytoplasmic extracts was normal as compared with wild-type STAT1 (Figure 2C). We also tried to trap the mutant LL407,409AA with anti-STAT1 antibodies microinjected into the nucleus of IFN-γ-stimulated cells, but even then the protein did not accumulate in the nucleus (Figure 2D). These data strongly indicate that the mutant LL407,409AA is excluded from the nucleus as a tyrosine-phosphorylated molecule.
It had been demonstrated before that cytokine-induced nuclear import of STAT1 requires the import receptor NPI-1 (Sekimoto et al., 1997), which is known to bind to clusters of basic residues (Weis et al., 1995; Moroianu et al., 1996). Several conserved basic side chains are found around residue 410 of STAT1 (Figure 1). Replacement of the Arg405–406 His motif with alanine–asparagine did not influence nuclear accumulation (not shown). However, mutation of the residues Lys410 and Lys413 to glutamic acid produced a mutant protein that no longer responded with nuclear accumulation to stimulation of cells with IFN-γ (Figure 2A, panel F). We examined extracts from U3A cells expressing the KK410,413EE mutant to check for nucleocytoplasmic distribution, tyrosine phosphorylation and DNA binding in response to IFN-γ. As is shown in Figure 2E, STAT1-KK410,413EE was found in both cytoplasmic and nuclear extracts irrespective of IFN-γ. Faint tyrosine phosphorylation of the mutant STAT1 was detectable in cytoplasmic extracts already prior to stimulation with IFN-γ, which was strongly boosted after the addition of IFN-γ to the cells. However, the signal remained confined largely to the cytoplasmic extracts, and only a very weak band indicating the presence of tyrosine-phosphorylated STAT1 was observable in the nuclear extracts (Figure 2E). STAT1-KK410,413EE proved incapable of binding specifically to oligonucleotides harboring a STAT recognition site, since no gel shift activity was detectable (not shown).
We therefore examined whether the observed phenotype could be explained by an inability to dimerize. HeLa cells were transiently co-transfected with cDNAs encoding GFP- and Flag-tagged STAT1 variants of either KK410,413EE, wild-type or Tyr701Phe, a tyrosine phosphorylation-incompetent mutant (Shuai et al., 1993a). Expressed proteins were precipitated with anti-Flag antibodies and the collected proteins were probed for co-precipitated GFP-tagged variants. No GFP-tagged STAT1 was precipitated by the Flag-tagged Tyr701Phe mutant, while both wild-type STAT1–GFP and STAT1-KK410,413EE–GFP co-precipitated with the respective Flag-tagged variants, thus indicating the ability of these molecules to dimerize (Figure 2F).
Taken together, the nuclear presence of unphosphorylated STAT1 did not require the integrity of the NLS in the DNA-binding domain. In the following, we will use the term dimer-specific (ds) NLS to stress the fact that this signal is only relevant for the nuclear import of tyrosine-phosphorylated dimers of STAT1.
STAT1 is constitutively shuttling between cytoplasm and nucleus prior to cytokine stimulation
We furthered these observations of nuclear translocation of unphosphorylated STAT1 with a novel in vivo assay, which directly reveals nucleocytoplasmic transport. The assay is based on the co-microinjection of fluorescein isothiocyanate (FITC)-labeled bovine serum albumin (BSA) (injection marker) and STAT1 antibodies (injection antibody) for intracellular binding to STAT1 in order to immobilize the protein in the respective compartment (nuclear or cytosolic). One hour later, the cells were fixed and STAT1 was detected by incubation with a STAT1-specific antibody derived from a different species (detection antibody), followed by treatment with a species-specific Cy3-coupled secondary antibody. Accumulation of STAT1 in the microinjected compartment is indicative of ongoing nucleocytoplasmic trafficking. A related approach has been used before to inactivate proteins that are involved in nucleocytoplasmic transport, such as NTF-2 or ran (Hieda et al., 1999; Steggerda et al., 2000). We microinjected Hek cells, reconstituted U3A cells, HeLa cells and HeLa-S3 cells with identical outcome. The results shown here were obtained with HeLa-S3 cells, which displayed significant levels of nuclear STAT1 already prior to cytokine stimulation (Figure 3A). It is of note that the constitutive nuclear presence of STAT1 in these cells does not require tyrosine phosphorylation (Meyer et al., 2002).
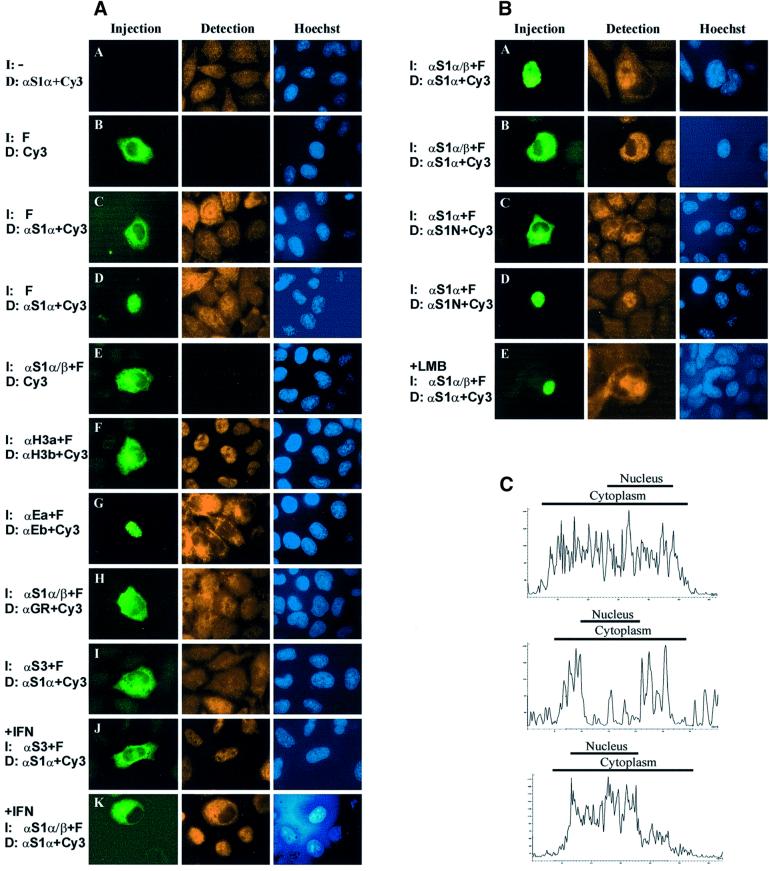
Fig. 3. Antibody microinjections reveal the nucleocytoplasmic shuttling of STAT1 in unstimulated cells. (A) Fluorescence micrographs of control experiments characterizing the microinjection assay employed. One hour after injection (I) of the indicated reagents, the HeLa-S3 cells were fixed, nuclei stained with Hoechst dye and the indicated proteins detected (D) by immunocytochemistry. The following specific antibodies were used: STAT1α (αS1α), STAT1α/β (αS1α/β), STAT1 N-terminus (αS1N), histone H3 antibodies a and b (αH3a and αH3b), Eps15 antibodies a and b (αEa and αEb) and glucocorticoid receptor (αGR); in addition, FITC–BSA (F) and the appropriate Cy3-coupled secondary antibodies (Cy3) were used. Shown are the injection site (Injection), the immunocytochemically detected protein (Detection) and the corresponding nuclei (Hoechst). In panel A, uninjected cells were stained for STAT1 showing the pancellular STAT1 distribution in the resting cells. Panels B–D demonstrate the absence of cross-over fluorescence (B) and the undisturbed STAT1 distribution after cytoplasmic (C) and nuclear (D) microinjection of FITC–BSA. Panel E depicts the absence of cross-species reactivity of the Cy3-coupled secondary antibody with the injection antibody. Panel F shows the persistent nuclear localization of histone H3 after cytoplasmic microinjection of histone H3 antibodies. The unaltered cytoplasmic localization of the cytoplasmic marker protein Eps15 after nuclear microinjection of anti-Eps15 is depicted in panel G. Panel H shows the persistent predominantly nuclear localization of glucocorticoid receptor after cytoplasmic microinjection of STAT1 antibodies. Panel I shows the undisturbed STAT1 distribution after cytoplasmic microinjection of STAT3 antibodies. In panel J, treatment of cells was identical to I, except that IFN-γ was present during the 1 h incubation with the microinjected STAT3 antibody, resulting in STAT1 nuclear accumulation. Exchange of the microinjected STAT3 antibody with an antibody against STAT1 prevented the IFN-γ-induced nuclear accumulation of STAT1 (K), demonstrating that STAT1 immobilization resulted specifically from STAT1 antibody injection. (B) Nucleocytoplasmic shuttling of unphosphorylated STAT1 as revealed by microinjection of STAT1 antibodies into the nucleus (A, D and E) or cytoplasm (B and C) of HeLa-S3 cells in the absence (A–D) or presence (E) of LMB. Two different combinations of injection/detection antibodies were used in (A, B and E) and (C and D), respectively (see Materials and methods for details). In (A) and (E), the nuclear microinjection was performed into multinucleated cells. Note the accumulation of STAT1 in the injected nucleus and the absence of STAT1 immunoreactivity in the uninjected nuclei of the same cell. (C) Laser scanning topographic sections of STAT1 immunofluorescence intensities before microinjection (top) and after STAT1 antibody injection into the cytoplasm (middle) or nucleus (bottom). Data were generated by confocal microscopy using a Leica TCS-SP2 microscope. The focus was adjusted through the center of the nucleus. Fluorescence intensity was quantified using Leica Confocal Software and plotted versus the longitudinal cell axis. Horizontal axis (µm); vertical axis (relative fluorescence units). Bars indicate the dimensions of nuclear and cytoplasmic compartments.
We performed a number of control experiments to establish the validity of this novel assay. The STAT1 distribution in uninjected control cells is shown in panel A of Figure 3A. First, we injected FITC-labeled BSA in the cytoplasm of HeLa-S3 cells. The cells were fixed and viewed without additional manipulations (Figure 3A, panel B), or STAT1 distribution was visualized with a detection antibody and a Cy3-coupled secondary antibody (Figure 3A, panels C and D). No cross-over fluorescence was detectable. Also, we microinjected the injection antibody, but omitted the detection antibody and directly stained the cells with the Cy3-coupled secondary antibody (Figure 3A, panel E). This experiment showed that no cross-species recognition was observable with the secondary antibody. Moreover, cytoplasmic microinjections of histone H3 antibodies did not result in cytoplasmic accumulation of this nuclear protein (Figure 3A, panel F). Conversely, nuclear microinjection of antibodies against the cytoplasmic marker protein Eps15 (Tebar et al., 1996) did not result in its nuclear relocation (Figure 3A, panel G). Further, microinjection of STAT1 antibodies did not interfere with the constitutive nucleocytoplasmic shuttling of glucocorticoid receptor (Madan and DeFranco, 1993) (Figure 3A, panel H). In addition, cytoplasmic microinjection of STAT3 antibodies did not interfere with the resting distribution or the IFN-γ-induced nuclear accumulation of STAT1 (Figure 3A, panels I and J). The latter two experiments confirmed the target specificity of the approach. The potential of this technique to immobilize STAT1 was demonstrated directly (Figure 3A, panel K) when STAT1 antibodies were delivered into the cytoplasm followed by IFN-γ stimulation for 1 h. The protein now remained in the cytoplasm and no nuclear accumulation was observable.
We then injected antibodies directed against STAT1 into unstimulated cells. Strikingly, nuclear microinjection of STAT1 antibodies was followed by nuclear accumulation of STAT1 in the injected cell (Figure 3B, panel A). Conversely, cytoplasmic microinjections of STAT1 antibodies depleted the nucleus of STAT1 immunoreactivity during a 1 h incubation (Figure 3B, panel B). Thus, immobilization of STAT1 by antibody microinjection leads to accumulation of this protein in the respective compartment. Two different combinations of STAT1 injection/detection antibodies resulted in accumulation of STAT1 in the microinjected compartment (Figure 3B). We and others have shown previously that treatment of cells with LMB, an inhibitor of nuclear export processes dependent upon the exportin CRM-1 and leucine-rich export signals, did not influence the steady-state distribution of STAT1 in resting cells (Begitt et al., 2000; McBride et al., 2000). Therefore, we microinjected a single nucleus of a HeLa polykaryon with STAT1 antibodies after the addition of LMB. As is shown in panel E of Figure 3B, nucleocytoplasmic shuttling proceeded uninhibited. Figure 3C shows a quantitative confocal microscopic analysis of the STAT1 distribution for typical cells before (top) and after cytoplasmic (middle) or nuclear (bottom) STAT1 antibody injection.
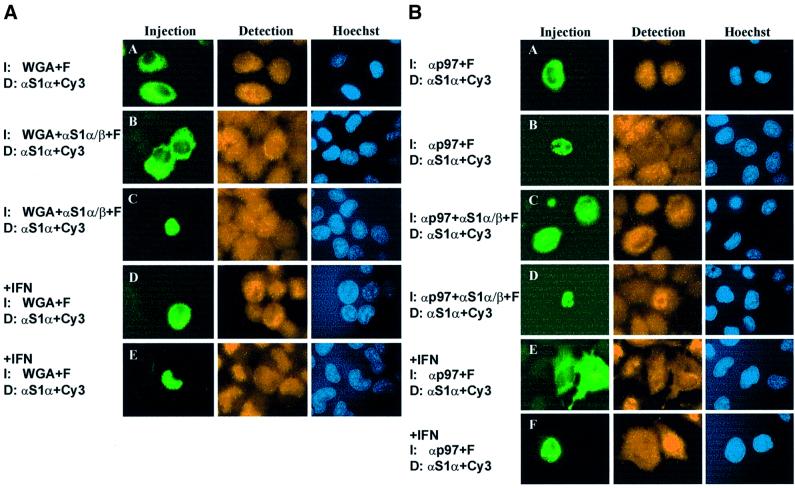
We examined next whether inactivating nuclear pore proteins by wheat germ agglutinin (WGA) (Finlay et al., 1987) inhibited nucleocytoplasmic shuttling of unphosphorylated STAT1. Cytoplasmic microinjection of WGA was without observable effects on the resting distribution of STAT1 (Figure 4A, panel A), but cytoplasmic co-microinjection of STAT1 antibodies and WGA prevented nuclear export, as the nucleus now could not be depleted of STAT1 (Figure 4A, panel B). Nuclear co-injections of WGA and STAT1 antibodies were performed in order to test the effects of WGA on nuclear import of unphosphorylated STAT1. Again, WGA clearly reduced the antibody-induced accumulation, albeit less strongly than after injection into the cytoplasm (Figure 4A, panel C). To demonstrate the negative effects of nuclear and cytoplasmic WGA on active nuclear import, cells were microinjected with WGA before stimulation with IFN-γ. Clearly, both cytoplasmic (Figure 4A, panel D) and nuclear (panel E) WGA were equally potent in blocking IFN-γ-induced STAT1 nuclear accumulation. Potentially damaging effects of WGA on the antibody reactivity were excluded by treating the STAT1 antibody used for microinjection with 0.5 mg/ml WGA, since no differences as compared with untreated antibody could be detected in subsequent western blot experiments (not shown).
Fig. 4. Influence of wheat germ agglutinin (WGA) and p97 on STAT1 shuttling. Shown are fluorescence micrographs of fixed cells immunostained for STAT1. (A) WGA inhibits the nucleocytoplasmic translocation of STAT1 in unstimulated (A–C) and IFN-γ-treated cells (D and E). WGA alone (A, D and E) or in combination with STAT1 antibodies (B and C) was injected in the cytoplasm (A, B and D) or nucleus (C and E) of HeLa-S3 cells. (B) p97 functions in the nucleocytoplasmic shuttling of phosphorylated, but not unphosphorylated STAT1. Antibodies directed against p97 alone (A, B, E and F) or in combination with anti-STAT1 (C and D) were microinjected in the cytoplasm (A, C and E) or nucleus (B, D and F) of unstimulated HeLa-S3 cells. Subsequently, cells were either left untreated (A–D) or stimulated with IFN-γ for 1 h (E and F).
To characterize further the nuclear import pathway of unphosphorylated STAT1, we extended the antibody microinjection assay by injecting antibodies against p97 into the cytoplasm or nucleus of HeLa-S3 cells (Figure 4B). No differences in the nucleocytoplasmic STAT1 distribution between uninjected and anti-p97-injected cells were observed (Figure 4B, panels A and B). To demonstrate unambiguously the ongoing nucleocytoplasmic translocation in the absence of active p97, we co-microinjected STAT1 and p97 antibodies into the nuclei or cytoplasm of resting HeLa-S3 cells (Figure 4B, panels C and D). This treatment did not slow down the antibody-induced accumulation of unphosphorylated STAT1 measurably, as compared with anti-STAT1 microinjections alone (Figure 3B). We additionally tested the influence of p97 precipitation on IFN-γ-induced nuclear import of STAT1. Injection of p97 antibodies into either compartment blocked IFN-γ-inducible nuclear accumulation of STAT1 (Figure 4B, panels E and F), thus confirming the central role of this import receptor for nuclear import of activated STAT1 (Sekimoto et al., 1997). These data also demonstrate that the pool of p97 available for STAT1 nuclear import is significantly depleted after antibody injection. Thus, the constant nucleocytoplasmic shuttling of unphosphorylated STAT1 is likely to occur independently of the import receptor p97.
Inactivation of the dsNLS selectively blocks cytokine-induced gene induction
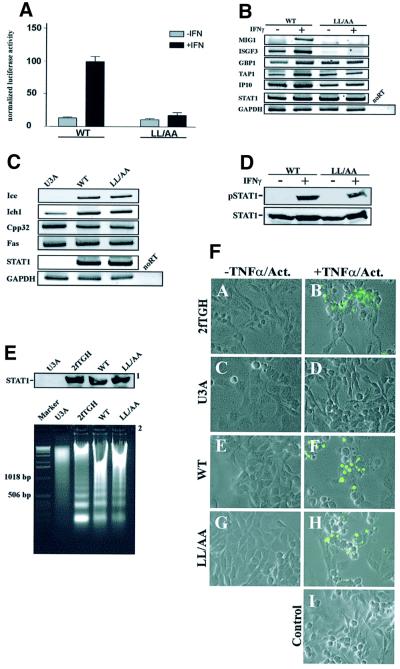
The results presented thus far show that at least two parallel import pathways allow for the independent nuclear uptake of unphosphorylated and activated STAT1, with activated STAT1 depending upon dsNLS for nuclear access. This prompted us to investigate the consequences of specifically blocking the nuclear uptake of tyrosine-phosphorylated STAT1 on both constitutive and IFN-γ-inducible STAT1 target genes. For transcriptional analysis, the DNA binding-competent double mutant LL407,409AA was used, which was constitutively phosphorylated on residue Ser727, as was wild-type STAT1 (not shown). This modification significantly increases IFN-γ-induced transcriptional activation (Wen et al., 1995). We first employed a reporter gene assay in U3A cells to compare the transcription potentials of the mutant LL407,409AA and wild-type STAT1. As is shown in Figure 5A, cells expressing wild-type STAT1 reacted strongly to IFN-γ with an ∼10-fold increase in luciferase activity. In contrast, the dsNLS mutant was incapable of inducing the GAS-driven luciferase reporter gene in response to IFN-γ stimulation. We then used RT–PCR assays to probe the induction of endogenous IFN-γ-responsive genes in stably transfected U3A cells. Again, the mutant failed to respond to interferon treatment with gene induction, while IFN-γ stimulation of control cells expressing wild-type STAT1 resulted in the increased transcription of all IFN-γ-responsive genes tested (Figure 5B).
Fig. 5. Mutations of the dimer-specific STAT1 NLS prevent cytokine-induced gene activation, while constitutive functions of STAT continue. (A) U3A cells were transiently co-transfected with expression vectors for wild-type STAT1 or the LL407,409AA import mutant (LL/AA) and an IFN-γ-responsive luciferase reporter gene as described. (B) RT–PCR analysis of IFN-γ-inducible STAT1 target genes. The assay was performed with stable U3A-derived cell lines complemented with wild-type STAT1 or STAT1-LL407,409AA. Cells were grown for 15 h in DMEM with reduced serum (1%), followed by the addition of serum to 10% and treatment with or without IFN-γ for 4 h, RNA extraction and reverse transcription. PCR amplification of the resulting cDNAs was performed with primer pairs representing the mRNAs indicated on the left. The products were resolved by gel electrophoresis and stained with ethidium bromide. (C) RT–PCR analysis of constitutively expressed STAT1-dependent genes. Gene expression in the absence of IFN-γ stimulation was monitored in growth medium with U3A cells and U3A-derived stable cell lines. (D) Whole-cell extracts from similar numbers of cells were immunoblotted to verify STAT1 expression (STAT1) and tyrosine phosphorylation (pSTAT1) of the stable lines used in (B) and (C). (E) DNA laddering assay with U3A cells, the parental line 2fTGH and U3A cells transiently expressing wild-type STAT1 or the LL407,409AA mutant. Apoptosis was detected after 18 h exposure to TNF-α and actinomycin D by assaying oligonucleosomal DNA fragmentation. Shown are the levels of STAT1 expression as detected by western blotting (1) and the DNA fragmentation patterns as revealed by agarose gel electrophoresis (2). (F) Transiently transfected cells from (E) were treated with TNF-α/actinomycin or left untreated. Subsequently, the cells were fixed and subjected to the TUNEL procedure. After treatment with TNF-α/actinomycin, there was an increase in the number of cells labeled with fluoresceinated dUTP in the cell populations expressing wild-type STAT1 or the dsNLS mutant, suggestive of apoptosis. Non-apoptotic cells were devoid of fluorescence. A control reaction with 2fTGH cells omitting the terminal deoxytransferase is included (Control).
However, when we turned to the expression of genes that require unphosphorylated nuclear STAT1 for their constitutive expression, we could no longer discriminate between the dsNLS mutant and wild-type STAT1. Expression of the apoptosis-associated proteins Fas, Cpp32, Ich1 and Ice was produced to similar levels by both STAT1 variants (Figure 5C). In addition, when we looked at the induction of apoptosis not on the single gene level but on the cellular level, we again were unable to detect differences between wild-type STAT1 and the dsNLS mutant. The induction of apoptosis by tumor necrosis factor-α (TNF-α) was equally efficient in U3A cells expressing wild-type STAT1 or the dsNLS mutant, as judged by oligonucleosomal DNA laddering and the TUNEL procedure (Figure 5D–F). Taken together, these observations demonstrate that the dsNLS in the STAT DNA-binding domain is necessary for the nuclear import and subsequent transcriptional activity of tyrosine-phosphorylated STAT1. On the other hand, this signal is dispensable for constitutive nuclear functions of STAT1.
Discussion
It becomes increasingly clear that nuclear transport plays a crucial role in regulating the activity of transcription factors (Kaffman and O’Shea, 1999; Turpin et al., 1999). In this study, we have begun to distinguish between different pathways used by STAT1 to afford nuclear translocation, and it was discovered that tyrosine-phosphorylated and unphosphorylated STAT1 use separate mechanisms to enter the nucleus. Here, we describe the identification of a nuclear import signal that specifically regulates nuclear entry of tyrosine-phosphorylated dimeric STAT1, named dsNLS. The signal is situated in the DNA-binding domain, which is the most frequent site of NLS localization in DNA-binding proteins (LaCasse and Lefebvre, 1995). The crucial residues identified are well conserved among the STAT proteins, hinting that this signal is of general importance for the cytokine-inducible functions of these transcription factors.
The STAT1 dsNLS differs from conventional NLSs in several ways. The experimentally best characterized examples of NLS sequences are distinguished by a single or double cluster of basic residues, often lysines (Mattaj and Englmeier, 1998). The STAT1 dsNLS does not resemble this consensus (Figure 1), but positively charged residues are important. Two lysines in position 410 and 413 are dispensable for STAT1 dimerization (Figure 2), yet they are critical for nuclear import of the tyrosine-phosphorylated molecule. Nevertheless, an extended cluster of positive charges in the dsNLS is missing, and neighboring basic residues in positions 405 and 406 do not participate in forming the NLS. These data are in line with a recent report that implicated this region in nucleocytoplasmic translocation of STAT1 (Melen et al., 2001). Remarkably, hydrophobic residues are also important for nuclear uptake of phosphorylated STAT1. While conservative mutations (leucine to isoleucine or valine) that preserved the hydrophobic nature of the side chains were without effects on import, introduction of alanines or threonines in positions 407 and 409 precluded nuclear entry of phosphorylated STAT1. Contributions of hydrophobic residues to NLS function have been reported before for several proteins (Makkerh et al., 1996; Mahalingam et al., 2001; Sherman et al., 2001). In accordance with these findings, the mutant phenotype displayed a complete loss of IFN-γ-induced transcriptional responses (Figure 5A and B). The dsNLS mutant STAT1-LL407,409AA, despite normal DNA-binding capability, was unable to transactivate a GAS-driven reporter construct and also failed to activate STAT1 target genes in their native chromosomal environment.
An interesting feature of the STAT1 NLS is its non-transferability. An isolated peptide spanning the entire dsNLS region (residues 376–427) was inactive in nuclear import upon cytoplasmic microinjection, and suppression of the intrinsic export activity of this fragment by incubation with LMB did not induce nuclear translocation (see Supplementary data). Therefore, the dsNLS is functional only in its native protein environment, which is the STAT1 dimer. This arrangement directly couples phosphorylation and dimerization to immediate nuclear import and thus the rapid onset of cytokine-induced gene induction. On the other hand, tyrosine dephosphorylation will, at the same time, inactivate the dimer-specific NLS and thus contribute to the restoration of the predominantly cytoplasmic localization in unstimulated cells.
The results presented here establish that residues that previously have been implicated in the nuclear export of unphosphorylated STAT1 (McBride et al., 2000) instead constitute part of a bona fide nuclear import signal for the activated protein. Our data confirm the observation that an isolated STAT1 peptide from the DNA-binding domain confers LMB-sensitive export activity on heterologous fusion proteins (see Supplementary data). Unexpectedly, the respective mutations of full-length STAT1 did not result in impaired nuclear export, since single point mutations affecting positions 404, 407 and 409 did not alter resting distribution nor nuclear clearance after IFN-γ stimulation. Most surprisingly, a double mutation in positions Leu407 and Leu409 did not lead to nuclear accumulation of STAT1 in resting cells despite continued nuclear presence (Figure 2). Inactivation of leucine-rich NESs by treatment of cells with LMB also did not block nucleocytoplasmic shuttling of the unphosphorylated protein (Figure 3B). Thus, we conclude that the segment between residues 400 and 410 in the STAT DNA-binding domain is not crucial for nuclear export of unphosphorylated STAT1. The possibility remains that this motif is involved in clearance of tyrosine-phosphorylated STAT1 from the nucleus.
Although tyrosine-phosphorylated STAT1 is blocked from nuclear entry after destruction of the dsNLS, the unphosphorylated mutant STAT1 nevertheless is found in the nuclei of unstimulated as well as IFN-γ-treated cells (Figure 2), indicating the ability of these mutants to enter the nuclear compartment in resting cells. This fact is underscored by the ability of the dsNLS import mutant STAT1-LL407,409AA to induce constitutive genes and to promote apoptosis (Figure 5C–F). In order to test directly for nucleocytoplasmic translocation of unphosphorylated STAT1, an antibody microinjection assay to immobilize the protein was developed. We reasoned that antibody cross-linking of a shuttling protein should lead to its precipitation and accumulation in the microinjected compartment. The localization of the targeted protein was determined subsequently by immunocytochemical decoration. Critical for the success of this assay are the target specificity of the microinjected antibody and the absence of cross-reactivity between secondary antibody and injection antibody. To ensure specificity, affinity-purified monoclonal and polyclonal injection antibodies were used and their specificity confirmed by western blotting (not shown). The lack of both cross-species fluorescence between the Cy3-labeled secondary antibody and the injection antibody as well as of cross-over fluorescence of the fluorescent reagents is illustrated in Figure 3A. Importantly, our results reveal the potential of this assay specifically to target and precipitate a translocating molecule. Only the STAT1 antibody prevented the IFN-γ-induced nuclear translocation of STAT1, while a STAT3 antibody did not. In contrast, non-shuttling proteins stayed in place after microinjection of specific antibodies, and microinjected STAT1 antibodies did not perturb the subcellular localization of a shuttling protein such as the glucocorticoid receptor (Figure 3A). STAT1 nonetheless reacted to STAT1 antibody microinjections in unstimulated cells with a readily observable translocation into the microinjected compartment (Figure 3B and C). Notably, this translocation is shown to be inhibited by WGA (Figure 4A), which is known to bind with high affinity to O-linked glycoproteins of the nuclear pore, including the p62 complex at the central pore channel (Finlay et al., 1987). For this reason, the observed sensitivity towards WGA demonstrates exclusive passage through the nuclear pore.
Nuclear import of tyrosine-phoshorylated STAT1 was determined to require importin β p97 (Sekimoto et al., 1997). Confirming these data, we find that nuclear or cytoplasmic microinjection of cells with p97 antibodies resulted in the blocking of IFN-γ-induced nuclear accumulation (Figure 4B). However, importin p97 does not seem to be required for the import of unphosphorylated STAT1. Co-microinjection of antibodies directed against p97 and STAT1 did not prevent trapping of STAT1 in the nucleus. Almost all cases of facilitated nuclear transport that have been characterized thus far require a member of the importin superfamily together with the small GTPase ran (Mattaj and Englmeier, 1998). Therefore, at present, we cannot rule out the possibility that STAT1 contains yet another NLS that mediates import of the unphosphorylated protein.
The above results indicate that tyrosine phosphorylation determines the choice of pathway in nucleocytoplasmic shuttling of STAT1. This allows for fast and independent modulation of both cytokine-inducible and constitutive nuclear functions, and stresses the autonomous nature of the distinct biological roles of STAT1.
Materials and methods
Cell culture
Cells were grown at 37°C in a humidified 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS; Biochrom, Berlin) and antibiotics. Transient transfections were carried out using lipofectamine-plus (Life Technologies). Stimulation with IFN-γ was performed in growth medium supplemented with 5 ng/ml IFN-γ (Life Technologies).
Plasmids
The plasmid expressing full-length human STAT1 (amino acids 1–746) fused to the N-terminus of GFP (pSTAT1-GFP) was described previously (Begitt et al., 2000). A double-stranded oligonucleotide coding for the Flag sequence (DYKDDDDK) followed by a stop codon was cloned into the BamHI site of pSTAT1-GFP to yield STAT1 with a Flag tag fused to residue 746. Untagged STAT1 was expressed in U3A cells from the vector pcDNA3 (Invitrogen). Recombinant proteins for microinjection were produced from plasmid pGST-GFP (Begitt et al., 2000). Mutagenesis was performed with the Quik-Change kit (Stratagene) and confirmed by DNA sequencing.
Antibody microinjections and fluorescence analysis
The following mouse monoclonal (mAb), rabbit polyclonal (rAb) and goat polyclonal (gAb) primary antibodies were used (all from Santa Cruz unless stated otherwise): gAb anti-H3a (C16) and rAb anti-H3b (06599, Upstate) against histone H3; rAb anti-Ea (C20) and gAb anti-Eb (D15) against Eps15; mAb anti-S1α/β (C136) against STAT1α/β; rAb anti-S1α (C24) against STAT1α; mAb anti-S1N (1, Transduction) against the STAT1 N-terminus; rAb anti-S3 (C20) against STAT3; rAb anti-GR (H300) against the glucocorticoid receptor; and mAb anti-p97 (3E9, Affinity Bioreagents). Antibodies were microinjected at a concentration of 150 µg/ml in phosphate-buffered saline (PBS) into ∼50 HeLa-S3 cells per coverslip in a 15 min period using the Transjector 5246 (Eppendorf). Co-injected FITC–BSA or tetramethylrhodamine isothiocyanate (TRITC)–BSA (0.2 mg/ml, Sigma) indicated the injection site. WGA (Sigma) was injected at a concentration of 0.5 mg/ml in PBS. After finishing the injections, the cells were incubated for 1 h at 37°C. Cells were then fixed with methanol at –20°C, washed in PBS and blocked in 25% FCS/PBS. Samples were incubated for 45 min with detection antibodies diluted 1:400–1:800 in 25% FCS/PBS. After three washes in PBS, the cells were incubated with the appropriate species-specific and affinity-purified secondary immunoglobulins conjugated with Cy3 (Jackson Research), and the nuclei were counterstained with 5 µg/ml Hoechst 33258 (Sigma). Fluorescence analysis was performed as described (Begitt et al., 2000).
Fractionated cell extraction, western blotting and electrophoretic mobility shift assay (EMSA)
Extractions were performed with U3A cells that were stably or transiently transfected with wild-type or mutant STAT1 constructs. Cells were left to recover from transfection for 24 h before pools of transfectants were plated onto 10 cm Petri dishes. Twelve hours later, the cells were lysed in 300 µl of cytosolic extraction buffer [10 mM KCl, 20 mM HEPES, 0.2% NP-40, 0.1 mM EDTA, 10% glycerol, 0.1 mM vanadate, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), complete protease inhibitors (Roche), pH 7.4] for 5 min on ice. Then the extracts were spun at 16 000 g for 10 s at 4°C, the supernatants were collected, spun again as before for 10 min, and used as cytosolic extracts for EMSA and western blotting. The pellets were washed in PBS and suspended in 300 µl of nuclear extraction buffer (420 mM KCl, 20 mM HEPES, 0.1 mM vanadate, 20% glycerol, 1 mM EDTA, 0.1 mM PMSF, 1 mM DTT, complete protease inhibitors, pH 7.6) and left on ice for 30 min with occasional gentle agitation. The extract were spun at 16 000 g for 20 min at 4°C, and the supernatants were used as nuclear extracts. The methods described by Begitt et al. (2000) were used for 7% SDS–PAGE, transfer onto nitrocellulose membranes, probing with antibodies and development of blots. The purity and protein content of fractionated extracts were assessed with antibodies against Eps15 and Sp1 (Santa Cruz). EMSA analysis was performed as described (Begitt et al., 2000).
Immunoprecipitation
A 10 cm dish with HeLa-S3 cells transiently expressing STAT1–GFP and STAT1-Flag was incubated with IFN-γ for 45 min and then for another 15 min in the additional presence of 0.8 mM sodium vanadate and 0.2 mM H2O2. Cells were lysed in 400 µl of buffer A (50 mM Tris–HCl pH 8, 1% NP-40, 5 mM EDTA, 2 mM EGTA, 10% glycerol, 50 mM NaF, 10 mM glycerol phosphate, 0.1 mM vanadate, 400 mM NaCl, 3 mM DTT, 0.1 mM PMSF and complete protease inhibitors) for 30 min on ice. Insoluble material was separated by centrifugation (16 000 g, 5 min), and 1.5 ml of diluted lysate [1:15 (v/v) with buffer A] was incubated with 20 µl packed volume of agarose-bound monoclonal anti-Flag antibodies (M2 affinity-gel; Sigma) for 2 h in the cold. The precipitate was washed three times with buffer A, and bound proteins were eluted by boiling in SDS sample buffer and separated by 7% SDS–PAGE. STAT1 was detected as described; Flag-tagged STAT1 was detected with antibody M2 (Sigma).
RT–PCR and luciferase assay
Total RNA was extracted using the StrataPrep Total Miniprep Kit (Stratagene) from 5 × 106 U3A cells stably expressing either wild-type STAT1 or the dsNLS import mutant LL407,409AA. First-strand cDNA synthesis was performed with 100 ng of each RNA sample using the ProStar HF Single-Tube RT–PCR System (Stratagene). All PCRs were performed in 50 µl reaction volume containing 100 ng of each upstream and downstream primer, 2.5 U of Taq DNA polymerase (Qiagen) and 10 mM of each deoxynucleotide in a buffer mix provided by the RT–PCR system. A primer list is provided in the Supplementary data. PCR products were analyzed by 4–20% polyacrylamide gel electrophoresis (Invitrogen) and stained with ethidium bromide (0.5 µg/ml). Luciferase assays were performed as described (Begitt et al., 2000).
Assays for apoptosis
Oligonucleosomal DNA laddering and the TUNEL procedure were carried out according to standard techniques (for details see Supplementary data).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Petra Striedelmeyer for excellent technical assistance, and Gloria Brujula-Sanchez who participated in the initial characterization of the export activity. Assistance with confocal microscopy by Dr Gerd Kempermann, Max-Delbrück-Centrum, Berlin, is gratefully acknowledged. This work was supported by a grant from the Bundesministerium für Bildung und Forschung (0311872 to U.V.).
References
- Begitt A., Meyer,T., van Rossum,M. and Vinkemeier,U. (2000) Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl Acad. Sci. USA, 97, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., Wright,K.L., Ting,J.P. and Stark,G.R. (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J., 19, 4111–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr (1997a) Phosphotyrosine signaling and the single cell:metazoan boundary. Proc. Natl Acad. Sci. USA, 94, 11767–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr (1997b) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- Decker T., Kovarik,P. and Meinke,A. (1997) GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res., 17, 121–134. [DOI] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer,D.D., Price,T.M. and Forbes,D.J. (1987) Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol., 104, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J., Rui,L., Luo,G., Yu-Lee,L.Y. and Carter-Su,C. (1999) A functional DNA binding domain is required for growth hormone-induced nuclear accumulation of Stat5B. J. Biol. Chem., 274, 5138–5145. [DOI] [PubMed] [Google Scholar]
- Hieda M., Tachibana,T., Yokoya,F., Kose,S., Imamoto,N. and Yoneda,Y. (1999) A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol., 144, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T. (1997) A new player in cell death. Science, 278, 1578–1579. [DOI] [PubMed] [Google Scholar]
- Ihle J.N. (1995) The Janus protein tyrosine kinase family and its role in cytokine signaling. Adv. Immunol., 60, 1–35. [DOI] [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kudo N., Matsumori,N., Taoka,H., Fujiwara,D., Schreiner,E.P., Wolff,B., Yoshida,M. and Horinouchi,S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA, 96, 9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Commane,M., Flickinger,T.W., Horvath,C.M. and Stark,G.R. (1997) Defective TNF-α-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science, 278, 1630–1632. [DOI] [PubMed] [Google Scholar]
- LaCasse E.C. and Lefebvre,Y.A. (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res., 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.J. (2001) Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol., 73, 271–277. [DOI] [PubMed] [Google Scholar]
- Madan A.P. and DeFranco,D.B. (1993) Bidirectional transport of glucocorticoid receptors across the nuclear envelope. Proc. Natl Acad. Sci. USA, 90, 3588–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam S., Van Tine,B., Santiago,M.L., Gao,F., Shaw,G.M. and Hahn,B.H. (2001) Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J. Virol., 75, 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkerh J.P., Dingwall,C. and Laskey,R.A. (1996) Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr. Biol., 6, 1025–1027. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- McBride K.M., McDonald,C. and Reich,N.C. (2000) Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J., 19, 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K., Kinnunen,L. and Julkunen,I. (2001) Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem., 276, 16447–16455. [DOI] [PubMed] [Google Scholar]
- Meyer T., Gavenis,K. and Vinkemeier,U. (2002) Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res., 272, 45–55. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel,G. and Radu,A. (1996) The binding site of karyopherin α for karyopherin β overlaps with a nuclear localization sequence. Proc. Natl Acad. Sci. USA, 93, 6572–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K. and David,M. (2000) Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol., 20, 7273–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C.V., Chatterjee-Kishore,M., Nguyen,H. and Stark,G.R. (2000) Complex roles of Stat1 in regulating gene expression. Oncogene, 19, 2619–2627. [DOI] [PubMed] [Google Scholar]
- Schindler C. and Darnell,J.E.,Jr (1995) Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Imamoto,N., Nakajima,K., Hirano,T. and Yoneda,Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J., 16, 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.P., de Noronha,C.M., Heusch,M.I., Greene,S. and Greene,W.C. (2001) Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol., 75, 1522–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Stark,G.R., Kerr,I.M. and Darnell,J.E.,Jr (1993a) A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science, 261, 1744–1746. [DOI] [PubMed] [Google Scholar]
- Shuai K., Ziemiecki,A., Wilks,A.F., Harpur,A.G., Sadowski,H.B., Gilman,M.Z., Darnell,J.E. (1993b) Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature, 366, 580–583. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Steggerda S.M., Black,B.E. and Paschal,B.M. (2000) Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol. Biol. Cell, 11, 703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebar F., Sorkina.T., SorkinA., Ericsson,M. and Kirchhausen,T. (1996) Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J. Biol. Chem., 271, 28727–28730. [DOI] [PubMed] [Google Scholar]
- Turpin P., Ossareh-Nazari,B. and Dargemont,C. (1999) Nuclear transport and transcriptional regulation. FEBS Lett., 452, 82–86. [DOI] [PubMed] [Google Scholar]
- Weis K., Mattaj,I.W. and Lamond,A.I. (1995) Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science, 268, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Wen Z., Zhong,Z. and Darnell,J.E.,Jr (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell, 82, 241–250. [DOI] [PubMed] [Google Scholar]