Abstract
The RING finger protein CNOT4 is a component of the CCR4–NOT complex. This complex is implicated in repression of RNA polymerase II transcription. Here we demonstrate that CNOT4 functions as a ubiquitin–protein ligase (E3). We show that the unique C4C4 RING domain of CNOT4 interacts with a subset of ubiquitin-conjugating enzymes (E2s). Using NMR spectroscopy, we detail the interaction of CNOT4 with UbcH5B and characterize RING residues that are critical for this interaction. CNOT4 acts as a potent E3 ligase in vitro. Mutations that destabilize the E2–E3 interface abolish this activity. Based on these results, we present a model of how E3 ligase function within the CCR4–NOT complex relates to transcriptional regulation.
Keywords: CCR4–NOT complex/RING finger/TFIID/transcription regulation/ubiquitin–protein ligase
Introduction
The recognition of core promoter elements by basal transcription factors is an essential step in the assembly of a functional RNA polymerase II (pol II) transcription complex. Key to this process is the basal transcription factor TFIID, an assemblage of TATA-binding protein (TBP) and TBP-associated factors (TAFIIs). TBP recognizes and binds TATA elements present in promoters of many genes transcribed by pol II. Several TAFIIs establish TATA-independent DNA contacts with core promoter elements such as the initiator or the downstream promoter element, and thereby contribute to the recognition and binding of promoters that lack a canonical TATA sequence (reviewed in Albright and Tjian, 2000).
Several general negative regulators of pol II-directed transcription have been identified (Hampsey, 1998). The CCR4–NOT complex is one of them. The NOT genes were isolated in a genetic screen in the yeast (y) Saccharomyces cerevisiae as repressors of pol II transcription from the HIS3 core promoter (Collart and Struhl, 1993, 1994; Oberholzer and Collart, 1998). The five NOT proteins assemble with CCR4 and CAF1 into the CCR4–NOT complex (Liu et al., 1998). With the exception of yNOT1, none of the other subunits is essential for vegetative growth of yeast cells. However, double null mutations of non-essential yCCR4–NOT genes yield synthetic lethality in various combinations, indicating a vital function for the complex (Maillet et al., 2000). NOT1 interacts with most of the other subunits and provides a scaffold for the assembly of the CCR4–NOT complex (Bai et al., 1999).
Little is known about the molecular basis of CCR4–NOT repressor function. Experiments in yeast suggest a link between yCCR4–NOT and yTFIID. First, yNOT1 interacts with yTBP (Lee et al., 1998) as well as yTAFII130 (M.A.Collart, unpublished observations). Secondly, yNOT2 and yNOT5 co-immunoprecipitate with yTBP, and yTAFIIs 60, 61 and 90 are pulled down by GST–yNOT5 (Badarinarayana et al., 2000). Thirdly, yNOT4 and yNOT5 interact with yTAFII19 genetically and biochemically (Lemaire and Collart, 2000). Recently, the view of CCR4–NOT proteins acting on the level of pol II transcription has been challenged by the characterization of yCCR4 and yCAF1 as nucleases involved in mRNA deadenylation (Daugeron et al., 2001; Tucker et al., 2001).
We have identified the human counterpart of the yCCR4–NOT complex (Albert et al., 2000). The human CCR4–NOT proteins are now denoted as CNOT subunits by the HUGO Gene Nomenclature Committee (http://www.gene.ucl.ac.uk/nomenclature/), indicating their association within the CCR4–NOT complex, and we will use this nomenclature. For a further understanding of CCR4–NOT, we focused on the CNOT4 subunit. A CNOT4 cDNA can complement a ynot4 null mutation, indicating functional conservation (Albert et al., 2000). The N-terminus of CNOT4 harbors a conserved domain with eight cysteine residues. We recently determined the structure of this domain and showed that it adopts a RING finger fold of the novel C4C4 type (Hanzawa et al., 2001). RING fingers are a specialized type of zinc fingers, which bind two atoms of zinc with a defined octet of cysteine and histidine residues in a ‘cross-brace’ manner. Two different RING variants, the ‘classical’ C3HC4 type (RING-HC) and the C3H2C3 type (RING-H2), have been described (Borden and Freemont, 1996). The atypical C4C4 RING of CNOT4 is the first shown to utilize exclusively cysteines for metal binding. Besides the CNOT4 RING, five C3HC4 RING structures have been determined: from the viral IEEHV protein (Barlow et al., 1994), promyelocytic leukemia protein PML (Borden et al., 1995), recombination protein RAG1 (Bellon et al., 1997), tyrosine kinase adaptor c-Cbl (Zheng et al., 2000) and TFIIH subunit MAT1 (Gervais et al., 2001). Zinc ligation is required for proper folding of RING domains and for their subsequent biological activities. To date, nearly 700 different RING proteins, including 119 in humans, have been identified (Pfam database, August 2001; Bateman et al., 1999). One unifying theme is utilization of the RING as a versatile protein–protein interaction module in disparate cellular processes (Borden, 2000).
Several RING proteins function as ubiquitin–protein ligases (Joazeiro and Weissman, 2000). Ubiquitylation is accomplished via a hierarchical enzyme cascade involving a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin–protein ligase (E3) (Hershko and Ciechanover, 1998). All known E3 ligases contain either a RING or HECT domain (Pickart, 2001). The main function of E3 ligases is to catalyze isopeptide bond formation between ubiquitin and a lysine residue in the substrate. Repeated cycles of this process lead to the assembly of polyubiquitin chains on the substrate, destining it for recognition and subsequent proteolytic degradation by the 26S proteasome, an abundant cellular protease complex. By bridging between the E2-ubiquitin thioester and the substrate, E3 ligases contribute to the specificity of ubiquitylation.
Recently, the co-crystal structure of the RING E3 ligase c-Cbl and the E2 enzyme UbcH7 has been reported (Zheng et al., 2000). c-Cbl functions as an E3 for activated receptor tyrosine kinases (RTKs) (Thien and Langdon, 2001). The crystal structure provided insights into how coupling of substrate and E2 binding in c-Cbl are accomplished (Zheng et al., 2000). As in other single-subunit E3s, separate c-Cbl domains are used for E2 and substrate binding: the RING establishes E2 contacts and an SH2 domain binds to RTKs. Alternatively, RING proteins are components of E3 ligase complexes in which E2 and substrate binding are delegated to different subunits (Jackson et al., 2000). For example, the Rbx1 subunit of SCF (Skp1–Cullin–F-box) E3 ligase complexes binds to E2 via its RING, and an F-box adaptor protein binds the substrate.
Ubiquitylation is also involved in regulation of transcription activation (Salghetti et al., 2001) and in modification of components of the basal transcription machinery, including pol II (Beaudenon et al., 1999; Mitsui and Sharp, 1999) and TFIID (Perletti et al., 2001). Mechanisms and proteins involved in TFIID ubiquitylation are unknown.
Here we report that the CNOT4 subunit of the CCR4–NOT complex functions as an E3 ligase. We show that an essential function of CNOT4 resides in its RING. CNOT4 interacts via this domain with a subset of E2s, including UbcH5B. Using nuclear magnetic resonance (NMR) spectroscopy, we detail the CNOT4–UbcH5B interaction on a molecular level. By mutagenesis, we identify conserved RING residues that are critical for E2 interaction. We also demonstrate that CNOT4 is a potent E3 ligase in vitro. Residues involved in E2 interaction also play a crucial role for the intrinsic E3 ligase activity of CNOT4. Based on our results, we propose how this activity is integrated to balance CCR4–NOT functions in transcription.
Results
The C4C4 RING of CNOT4 harbors a crucial function
The N-terminus of CNOT4 is highly conserved, with 44% identity to yNOT4 in the first 230 residues. CNOT4 harbors a unique combination of a C4C4 RING (residues 14–63), a RRM RNA recognition motif (residues 111–194) and a C3H-type zinc finger (residues 197–227) in this region (Figure 1A). We previously determined the solution structure of the C4C4 RING by NMR (Hanzawa et al., 2001). This analysis revealed overall similarities to the solved structures of other RING-HC domains.
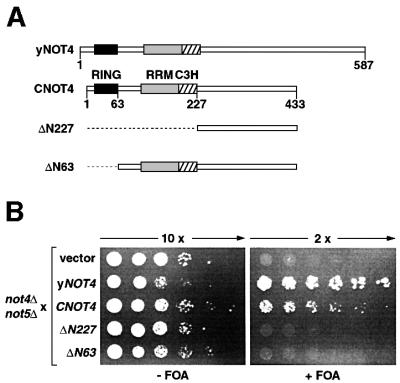
Fig. 1. Complementation analysis of yeast strain YOU637 (not4Δ not5Δ). (A) Schematic representation of yNOT4, CNOT4 and CNOT4 derivatives. The RING, RRM and C3H zinc finger domains are shown as black, gray and hatched boxes, respectively. Truncations of ΔN227 and ΔN63 are indicated by stippled lines. (B) YOU637 cells were transformed with empty vector, yNOT4 and CNOT4 full-length and partial cDNAs as indicated. Ten- and 2-fold serial dilutions of transformants were spotted on selective medium in either the absence (–) or presence (+) of 0.1% FOA, and colonies were grown for 4 days at 30°C.
We analyzed the physiological importance of the N-terminus of CNOT4 by hetero-species complementation. Full-length and partial CNOT4 cDNAs (Figure 1A) were tested for their ability to suppress the synthetic lethal phenotype of a yeast strain harboring a not4 not5 double null mutation. Western blot analysis of yeast extracts confirmed the presence of full-length CNOT4, ΔN227 and ΔN63, ruling out that lack of protein expression would account for lack of complementation (unpublished data). The not4Δ not5Δ strain carried the yNOT5 gene on a URA3 plasmid, which allows plasmid shuffling using 5-fluoro-orotic acid (FOA). As expected, episomal expression of yNOT4 suppressed synthetic lethality of not4Δ not5Δ cells on FOA media (Figure 1B). Full-length CNOT4 also allowed survival of not4Δ not5Δ cells, albeit that it was slightly less efficient (Figure 1B; Albert et al., 2000). In contrast, transformants that expressed ΔN227 lacking the conserved N-terminus did not survive on FOA media. Apparently, an essential function resides within this region. To narrow it down further, not4Δ not5Δ cells were transformed with ΔN63 lacking only the RING. Expression of ΔN63 could not suppress the lethality of not4Δ not5Δ cells (Figure 1B), indicating that the essential function of CNOT4 resides within its RING.
The non-conserved C-terminus of CNOT4 mediates CNOT1 association
Since RING domains are described as protein–protein interaction modules, we reasoned that this domain of CNOT4 mediates association with other CCR4–NOT proteins, in particular with CNOT1. We therefore assayed CNOT4 deletion mutants lacking parts from either the N- or C-terminus (Figure 2A) in directed yeast two-hybrid experiments for interaction with full-length yNOT1 and a partial CNOT1 derivative, C1270 (Figure 2A). The latter encompasses the C-terminal 1270 residues of full-length CNOT1. This region can establish a positive two-hybrid interaction with full-length CNOT4 (Albert et al., 2000; Figure 2B). From the tested CNOT4 deletion mutants, only N-terminal truncations ΔN63 and ΔN227 displayed positive interactions with C1270, whereas C-terminal truncations N227, N190, N78 and N63 did not (Figure 2B). Despite the low degree of conservation in this region, this interaction is evolutionarily conserved, since we also detected interactions of CNOT1-C1270 with yNOT4 as fusions to both LexA and B42 (Figure 2B). Also, analysis of truncated versions of yNOT4 for interaction with yNOT1 indicates that similar domains are responsible for interaction between the yeast orthologs (Figure 2C). Taken together, these data provide a coherent picture of the non-conserved C-terminal part of the (C)NOT4 proteins interacting with (C)NOT1 proteins.
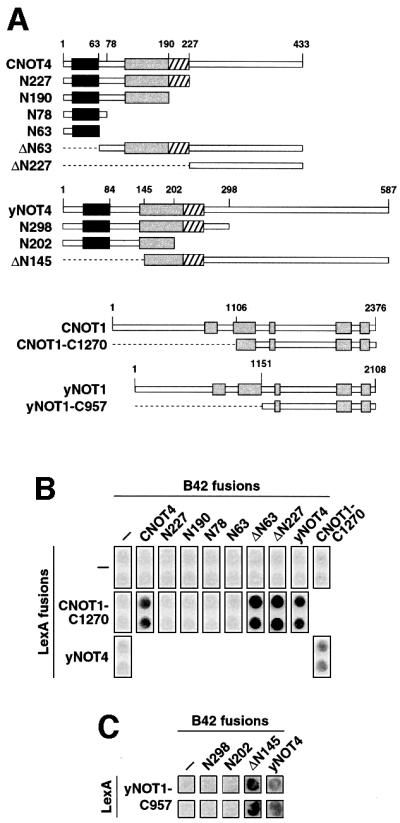
Fig. 2. CNOT4 interacts with CNOT1. (A) Survey of CNOT4, yNOT4, CNOT1 and yNOT1 derivatives used in two-hybrid assays. Conserved motifs within NOT1 proteins are indicated by gray shading. (B) Two-hybrid interactions of CNOT4 with CNOT1. EGY48 yeast cells containing a LexA-responsive lacZ reporter gene were transformed with the indicated LexA and B42 fusion plasmids. Empty vector controls (–) were included. Transformants were assayed on X-gal plates for blue staining. (C) Two-hybrid interactions of yNOT4 with yNOT1. Analysis was performed as in (B).
CNOT4 interacts with a subset of ubiquitin-conjugating enzymes via its RING
To obtain insight into the essential function of the conserved N-terminal part of CNOT4, we decided to identify interacting proteins by yeast two-hybrid cDNA screening methods. We used a fusion of the N-terminal 227 residues of CNOT4 to the LexA DNA-binding domain, LexA-N227 (Figure 3A), as bait to screen a HeLa cell cDNA library. From this screen, we isolated 10 clones. Five of them encoded closely related ubiquitin-conjugating enzymes. Three were partial cDNAs encoding UbcH6 starting at codon 21, one was a partial cDNA for UbcH9 starting at codon 39, and one cDNA encoded full-length UbcH5B (Figure 3A). All three E2 enzymes share extensive sequence homology to the highly related yUBC4 and yUBC5 proteins, with 90% similarity to yUBC4 for UbcH5B, and 80% similarity for both UbcH6 and UbcH9 (unpublished data). We obtained full-length cDNAs for UbcH6 and UbcH9 and used them in all subsequent experiments.
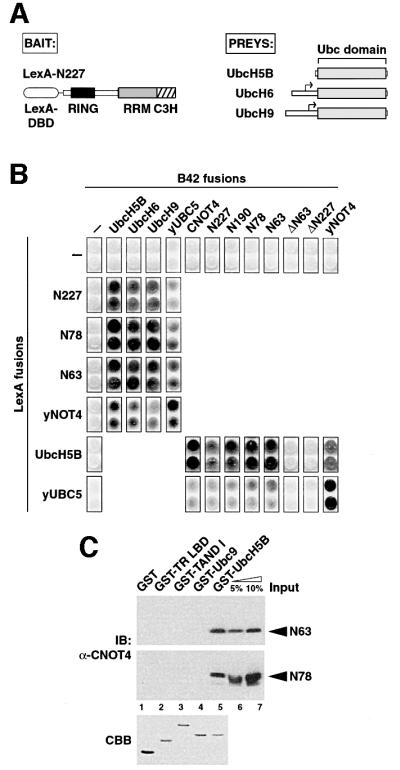
Fig. 3. Specific interactions of CNOT4 with ubiquitin-conjugating E2 enzymes. (A) Schematic illustrations of two-hybrid ‘bait’ LexA-N227 (left) and of E2s isolated as ‘preys’ (right). Small arrows denote start codons of the partial UbcH6 and UbcH9 library clones. (B) (C)NOT4 interactions with human and yeast Ubc proteins in the two-hybrid system. (C) GST pull-down analysis of His6-N63 and His6-N78 RING finger proteins using different GST fusion proteins: lane 1, GST alone; lane 2, GST fusions with the ligand-binding domain of chicken thyroid receptor α (GST-TR LBD); lane 3, Drosphila TAFII230 N-terminal domain (GST-TAND I); lane 4, SUMO-1-conjugating enzyme Ubc9 (GST-Ubc9); and lane 5, GST–UbcH5B. The upper and middle panels show immunoblots (IB) using anti-CNOT4 antibody 19A12. Five and ten percent of recombinant N63 and N78 used in the pull-down reactions were loaded as input controls (lanes 6 and 7). The lower panel shows a Coomassie Blue (CBB) stain of an SDS–gel with 2 µg of each GST protein loaded.
To determine the CNOT4 domain involved in E2 interactions, additional truncation mutants of CNOT4 were assayed in two-hybrid experiments. This analysis revealed that the N-terminal 63 residues spanning the RING (N63) are sufficient for binding UbcH5B, UbcH6 and UbcH9 (Figure 3B). This domain was absolutely required for E2 binding, since deleting it (ΔN63) resulted in a complete loss of the interaction with UbcH5B. LexA and B42 fusions of yNOT4 were also capable of interactions with these human Ubc proteins, indicating an evolutionary conservation of E2 binding (Figure 3B). Further support for this notion is the strong interaction of yNOT4 with yUBC5 and the (weak) capacity of the CNOT4 RING domain for interactions with yUBC5 (Figure 3B).
To test whether a CNOT4–UbcH5B interaction is direct, we performed GST pull-down assays with purified recombinant proteins. In these assays, His6-tagged CNOT4 RING derivatives N63 and N78 were retained by immobilized GST–UbcH5B as revealed by western blots using anti-CNOT4 antibodies (Figure 3C, lane 5). Quantitation of this result indicates a Kd in the range of 10–6–10–7 M. The CNOT4 proteins did not bind to GST or to various GST fusion proteins, including SUMO-1-conjugating factor Ubc9 (Figure 3D, lane 4), which is involved not in ubiquitylation but in sumoylation (Johnson and Blobel, 1997). This further supports the specific nature of the CNOT4–UbcH5B interaction.
In summary, these experiments establish that (i) CNOT4 interacts via its RING finger with a subset of E2s, namely UbcH5B, UbcH6 and UbcH9, and that (ii) the RING is necessary and sufficient for these interactions.
Mapping the CNOT4–UbcH5B interaction surface
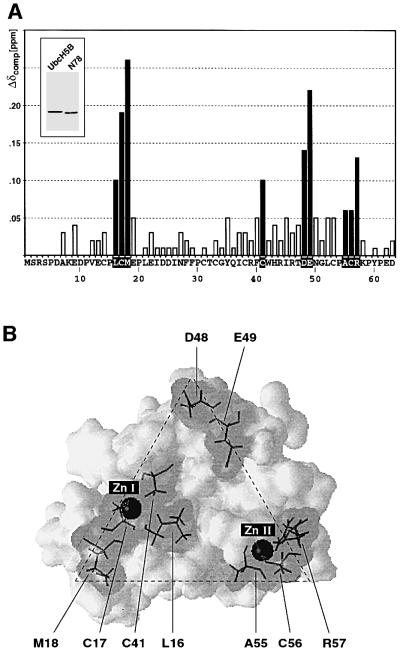
NMR spectroscopy yielded the structure for the C4C4 RING of CNOT4 (Hanzawa et al., 2001). Assignment of the N-terminal 78 residues allows us to identify RING residues potentially involved in E2 interactions. To this end, we expressed UbcH5B as a fusion with GST in bacteria and purified it to homogeneity. After proteolytic removal of GST, UbcH5B was titrated to 15N-labeled CNOT4 RING derivative His6-N78, and 15N-HSQC spectra were recorded. Comparison of the two-dimensional NMR spectra of His6-N78 in the absence and presence of UbcH5B revealed chemical shift changes for a number of RING residues. The shifts were of the fast-exchange type, which is indicative of an interaction in the Kd range of 10–5–10–6 M and consistent with the GST pull-down result of Figure 3C. The most prominent chemical shifts (Δδcomp >0.1 p.p.m.) were observed for residues C17, M18, D48, E49 and R57, and minor shifts (Δδcomp 0.05–0.1 p.p.m.) for residues L16, C41, A55 and C56 (Figure 4A). Strikingly, in the three-dimensional structure, these residues map to one side of the molecule and their side chains are solvent-exposed and accessible (Figure 4B). These residues probably delineate the UbcH5B interaction surface of the CNOT4 RING. The overall shape of this interface is triangular. Residues M18, D48 and R57 form the corners of this triangle, which is centered around a shallow hydrophobic groove comprising L16, I45 and P54. L16, C17 and M18 form part of loop region L1, C41 and D48 are located in a short α-helix, and the remaining residues, i.e. E49, A55, C56 and R57, reside within loop region L3 of the C4C4 RING (Hanzawa et al., 2001). Besides their structural role, two cysteines from zinc-binding site I, C17 and C41, and one from zinc-binding site II, C56, also seem to be involved in E2 interactions.
Fig. 4. Titration of UbcH5B with CNOT4 RING. (A) Compound chemical shifts (Δδcomp) of 15N-labeled His6-N78 upon titration of UbcH5B. Residues showing chemical shift changes >0.05 p.p.m. are shown as black bars. The inset shows a Coomassie Blue-stained gel of UbcH5B (lane 1) and His6-N78 (lane 2). Only shifts for residues 7–63 could be detected in 15N-HSQC spectra. (B) Three-dimensional surface representation of CNOT4 RING structure. Residues showing shifts >0.05 p.p.m. are indicated by gray shading. Stippled lines outline the triangular shape of the E2 interface.
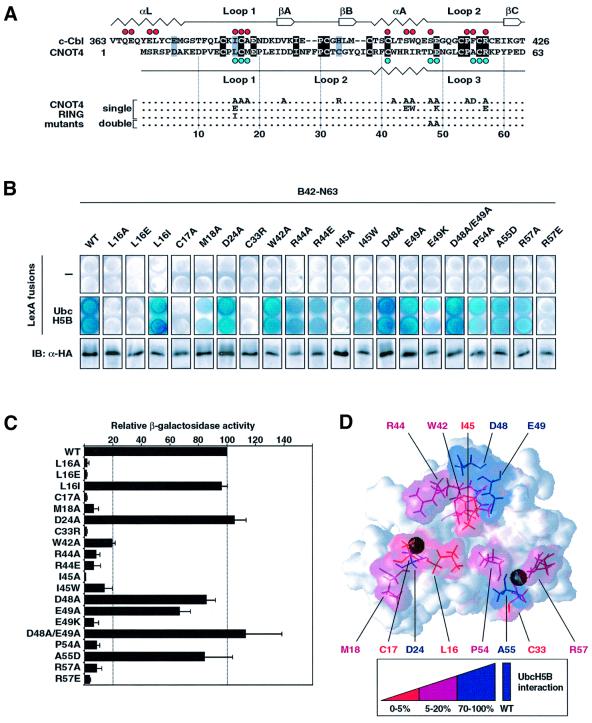
The crystal structure of c-Cbl in complex with UbcH7 revealed the architecture of the E2-binding site on c-Cbl (Zheng et al., 2000). UbcH7 mainly packs in a groove on the RING, but makes additional contacts with a proximal α-helical linker domain. Comparison of the E2 interfaces of CNOT4 and c-Cbl reveals some interesting structural similarities, despite the fact that the two RING proteins share only weak homology (Figure 5A). In both cases, the E2-binding sites are provided by a shallow groove, which is formed by an α-helix and two extended zinc-chelating loops. Within the first loop region, hydrophobic residues L16, C17 and M18 of CNOT4, corresponding to I383, C384 and A385 of c-Cbl, are part of the respective E2 interfaces (Figure 5A). The same applies for a cysteine in the α-helix, C41 in CNOT4 and C404 in c-Cbl. Finally, the C-terminal loop region accommodates further E2 contacts in both CNOT4 and c-Cbl, and again this is reflected by the conservation of R57 in CNOT4 and of R420 in c-Cbl. It is important to note that despite these structural correlations, the chemical nature of many side chains is not conserved. In particular, c-Cbl α-helix residues S407, W408 and S411, which are part of the UbcH7-binding site, are replaced by R44, I45 and D48 in CNOT4, and only D48 shows a major chemical shift upon UbcH5B binding (Figures 4A and 5A). Moreover, the RING is located at the N-terminus in CNOT4 (residues 14–63) but is centrally placed in c-Cbl (residues 381–426). c-Cbl uses an N-terminally adjacent linker helix (residues 363–374) for additional UbcH7 contacts. This helix is absent in CNOT4.
Fig. 5. Effect of CNOT4 RING mutations on UbcH5B interaction. (A) Survey of CNOT4 RING mutations. An alignment of CNOT4 and the c-Cbl RING sequences is shown on the top, with secondary structure elements displayed below and above the sequences, respectively. Red dots indicate contacts of the c-Cbl–UbcH7 interface (Zheng et al., 2000), those in blue indicate CNOT4 chemical shifts >0.05 p.p.m. (B) Yeast strains expressing empty (–) vector (upper panels) or LexA-UbcH5B (middle panels) were transformed with HA-tagged wild type (WT) and mutant B42-N63 as indicated. Equal expression levels of all B42 fusion proteins were confirmed by anti-HA immunoblot analysis (lower panels). Individual clones were assayed on X-gal plates (B), and by quantitative determination of the means and SEM of relative β-galactosidase activities in yeast cell extracts, with B42-N63 (WT) set to 100% (C). (D) Three-dimensional map illustrating the effects of mutations on UbcH5B binding by color coding: abolishment, red; strongly affected, magenta; not affected, blue.
Identification of CNOT4 RING residues that are critical for UbcH5B interaction
Our NMR approach detects an altered chemical environment for amide groups. The observed chemical shifts do not reveal direct contacts per se, but rather indicate dynamic conformational changes at a particular position or its close proximity. For this reason, it is essential to complement the NMR data by site-directed mutagenesis. In this, our strategy was guided by structural information on the E2 interfaces of the CNOT4 and c-Cbl RING domains. Effects of mutations were monitored in two-hybrid experiments with LexA-UbcH5B, comparing the relative strength of E2 interactions of wild-type and mutant B42-N63. To avoid debilitating effects of cysteine mutations that disrupt zinc ligation and overall folding, we preferentially targeted non-metal-chelating residues supported as potential UbcH5B contact points by NMR, including L16, M18, D48, E49, A55 and R57. Also, R44 and I45 in the structurally conserved α-helix were mutated. The corresponding c-Cbl residues S407 and W408 are involved in UbcH7 contacts. All but one of these CNOT4 residues were replaced by alanine, and A55 was replaced by aspartate to yield mutant A55D (Figure 5A). As controls, we used two single point mutations hitting zinc-binding site I, C17A, or zinc-binding site II, C33R, as well as mutation D24A. The latter is supposed to have no effect on UbcH5B interaction, as this residue is located opposite the proposed E2 interface.
The result of this analysis is shown in Figure 5B and its quantification in Figure 5C. In summary, we observed three kinds of effects: (i) RING mutations that completely abolished UbcH5B interaction; (ii) mutations with intermediary, yet pronounced effects; and (iii) mutations with very mild or no effects. As expected, the first class included the two cysteine mutations, C17A and C33R. Notably, mutations of two non-zinc-ligating residues, L16A and I45A, had the same deleterious effect. These residues map to the center of the hydrophobic groove, which defines the proposed UbcH5B-binding site (Figure 5D). A structural neighbor of L16 and I45 is P54. Mutation of this residue results in a 10-fold drop in interaction (Figure 5C), which is consistent with P54 being part of the Ubc5B interaction surface. However, in the case of P54, one should be cautious as this residue also orientates the C-terminal part of loop 3. Mutations M18A, R44A and R57A resulted in similarly weakened E2 interactions (Figure 5B and C). In the structure, their solvent-exposed side chains demarcate the outer borders of the hydrophobic groove. Finally, some mutations did not yield pronounced effects. Mutation D24A was completely neutral, supporting the specificity of our interaction analysis. Mutation A55D also resulted in an inert phenotype. Surprisingly, and in contrast to their pronounced chemical shift changes, D48 and E49 seem to play no crucial role for UbcH5B interaction. Neither single mutations D48A and E49A nor double mutation D48A/E49A had major effects on UbcH5B interaction. An explanation for this might be that the NMR experiment detects not only shifts of contacted residues but also distortions of structural neighbors. The mutagenesis approach directly scores interaction capacities.
Thus far, these data point to an important role for a hydrophobic groove in the RING domain for the CNOT4–UbcH5B interaction. Notably, the three central residues within this area, L16, I45 and P54, are absolutely conserved in CNOT4 proteins from various species (Albert et al., 2000). Chemically related or identical amino acids are found at corresponding positions in many other RING fingers, suggesting that a common theme of E2 interaction is realized in different RINGs. In c-Cbl, these residues are I383, W408 and P417. We therefore tested two more CNOT4 mutants, L16I and I45W, in order to mimic corresponding c-Cbl residues. Mutation L16I had a dramatic effect: it completely reverted E2 binding from undetectable back to wild-type levels (Figure 5B and C). A weaker phenotype was seen for mutant I45W, which displayed ∼15% of wild-type binding (Figure 5C). Yet, compared with I45A, it clearly regained a capacity for E2 interaction.
CNOT4 harbors E3 ligase activity
Several RING proteins display E3 ligase activity (Joazeiro and Weissman, 2000). Having demonstrated that its RING finger interacts with E2 proteins, we subsequently asked whether CNOT4 harbors E3 ligase activity. Therefore, we established a CNOT4-dependent, substrate-independent in vitro ubiquitylation assay, which used recombinant GST–ubiquitin, UbcH5B, His6-CNOT4 and mammalian E1 (Figure 6A). Accumulation of high molecular weight polyubiquitin adducts (Ubn) was detected in western blots using anti-ubiquitin antibodies. When the reactions were performed in the absence of ATP, no Ubn species were observed (Figure 6B, lane 1). In the presence of ATP, however, His6-CNOT4 strongly stimulated Ubn accumulation (Figure 6B, lane 3). Similar results were obtained when free ubiquitin was used instead of GST–ubiquitin (data not shown). Consistent with formation of thioester intermediates in the E1:E2:E3 cascade, reducing agents such as β-mercaptoethanol interfered with ubiquitylation as expected (Figure 6B, lane 2). Omission of any component from the reactions had the same effect (Figure 6B, lanes 4–7). Addition of N-ethylmaleimide (NEM), which modifies the active site cysteine of E2s, abolished the formation of Ubn chains (Figure 6C). Next, we assayed RING point mutants L16A and C33R in the context of full-length His6-CNOT4 for their ability to promote ubiquitin–protein ligation. In contrast to wild-type CNOT4, the mutant proteins showed no E3 ligase activity (Figure 6C, lanes 1–3). Thus, mutations that interfere with UbcH5B binding in vivo prevent polyubiquitin chain assembly in vitro. We also assayed CNOT4 deletion mutants His6-N78, His6-N63 and GST–ΔN227 in this assay. Whereas both N78 and N63 promoted Ubn formation, ΔN227 did not (Figure 6C, lanes 4–6). We conclude that the RING of CNOT4 is required and sufficient for ubiquitylation in vitro. Notably, neither a GST fusion of c-Cbl residues 47–447 nor GST alone displayed E3 activity in these assays (Figure 6C, lanes 7 and 8). As c-Cbl does not cooperate with UbcH5B in these assays, our in vitro ubiquitylation system displays E2–E3 specificity.
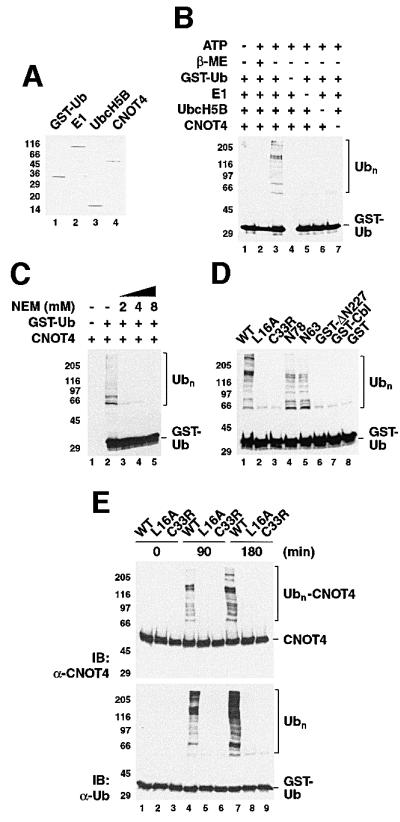
Fig. 6. E3 ligase activity of CNOT4 in vitro. (A) Coomassie Blue-stained gel showing components of the in vitro ubiquitylation system: lane 1, GST–ubiquitin; lane 2, ubiquitin-activating enzyme E1; lane 3, UbcH5B; and lane 4, His6-CNOT4. (B) Ubiquitylation reactions were performed for 90 min in the absence (–) or presence (+) of 2 mM ATP with the indicated components. In lane 2, β-mercaptoethanol (β-ME) was included to 2 M. The western blot used anti-ubiquitin antibody. Polyubiquitin chains (Ubn) are bracketed. (C) Effect of the thiol-specific reagent N-ethylmaleimide (NEM) on Ubn formation. (D) Ubiquitylation reactions used wild-type (WT) and mutant His6-CNOT4, N78 and N63 proteins, or GST fusion proteins as indicated. GST–Cbl denotes a fusion of c-Cbl residues 47–447. The reactions were performed for 90 min (left panel), and the reaction products probed with anti-ubiquitin antibody. (E) Ubiquitylation reactions using wild-type (WT) and mutant (L16A, C33R) His6-CNOT4 were first analyzed with anti-CNOT4 (upper panel) and reprobed with anti-ubiquitin antibody (lower panel).
Since all proteins were purified to homogeneity (Figure 6A), Ubn accumulation in these substrate-independent reactions most probably reflects E3 autoubiquitylation. This in vitro phenomenon has been described previously for heterologous GST–RING proteins, including c-Cbl (Joazeiro et al., 1999) and AO7 (Lorick et al., 1999). We employed His6-tagged CNOT4 proteins, which contain only lysines within CNOT4, as targets for the E2-ubiquitin thioester. To assay whether internal lysines are acceptors of polyubiquitin chains, we reacted wild-type and mutant His6-CNOT4 for up to 3 h and probed the reactions products with anti-CNOT4 and anti-ubiquitin antibodies (Figure 6D). This analysis showed that Ubn chains are attached to wild-type but not to mutant CNOT4, as revealed by the appearance of high molecular weight species in the CNOT4 western blot (Figure 6D, upper panel, lanes 4 and 7). The complete lack of Ubn–CNOT4 adducts in the mutant L16A even after 3 h reaction time underscores the absolute requirement of L16 for UbcH5B interaction and subsequent ubiquitylation. In contrast, wild-type CNOT4 became autoubiquitylated efficiently, with an estimated 10% of CNOT4 attached to Ubn. Taken together, our biochemical analysis establishes that CNOT4 is a potent E3 ligase that cooperates with UbcH5B in RING-dependent polyubiquitylation.
Discussion
Our findings indicate that the CNOT4 subunit of the CCR4–NOT transcription repressor complex is an E2-dependent RING E3 ligase. CNOT4 preferentially interacts with the UBC4/5 subfamily of closely related human E2 enzymes UbcH5B, UbcH6 and UbcH9, both in vivo and in vitro. This interaction is mediated by its atypical C4C4 RING, which harbors an essential CNOT4 function as revealed by hetero-species complementation. We mapped the E2–RING interface on a molecular level. This analysis allowed us to make a detailed comparison of the UbcH5B–CNOT4 RING interface with that of UbcH7–c-Cbl (Zheng et al., 2000), which is the only available atomic resolution structure of a RING E3 ligase bound to its cognate E2. As in c-Cbl, E2 contacts of the CNOT4 RING are restricted to a shallow hydrophobic groove. Our mutagenesis revealed an important role for conserved residues within this groove for UbcH5B interaction. Finally, we showed in biochemical assays that CNOT4 is an E3 ligase capable of catalyzing the assembly of polyubiquitin chains in a UbcH5B-dependent manner.
Specificity in E2–RING E3 interactions
Several lines of evidence argue for the specific nature of the CNOT4 RING interaction with a limited number of E2 enzymes of the UBC4/5 subfamily. First, UbcH5B, UbcH6 and UbcH9 were the only E2s isolated from the HeLa cDNA library screen. Secondly, CNOT4 did not interact with other, more distantly related E2s in various assays (T.K.Albert et al., in preparation). Thirdly, we demonstrated that the UbcH5B–CNOT4 interaction is functional, since UbcH5B supported autoubiquitylation of CNOT4 in vitro. Fourthly, the interaction is specific, since UbcH5B did not support c-Cbl E3 activity. Several E2s, among them UbcH5B and UbcH7, bind different RINGs as well as HECT E3s (Pickart, 2001). This promiscuity is structurally reflected in the limited number of E2 contacts to either HECT or RING E3s. For UbcH7, those contacts involve F63, P97 and A98 in complexes with both HECT E3 E6-AP and RING E3 c-Cbl (Huang et al., 1999; Zheng et al., 2000). These residues are completely conserved in UbcH5B, UbcH6 and UbcH9 (unpublished data). Why does UbcH5B cooperate with CNOT4 but not with c-Cbl, and how is this discrimination achieved? Obviously, other E2 residues are also relevant for specificity. In fact, preliminary results indicate that this is the case for UbcH5B–CNOT4 (T.K.Albert et al., in preparation). In this context, it is worth noting that c-Cbl establishes additional E2 contacts with its linker helix, which is absent in CNOT4. These involve hydrogen bond contacts with UbcH7 residues R5 and R15 (Zheng et al., 2000), which are not conserved in UbcH5B. This indicates that non-conserved E2 residues contribute to the specificity of E2–E3 interactions.
Structural aspects of the CNOT4–UbcH5B interaction
Direct support for the UbcH5B interaction with CNOT4 comes from our NMR study. This approach allowed us to map the E2-binding site at the level of individual CNOT4 RING residues by monitoring chemical shift changes of N78 residues upon titration with UbcH5B (Figure 4). Affected amino acids belong to distinct structural elements within the RING. They provide a shallow hydrophobic groove as the E2-binding site. A parallel mutagenesis revealed a crucial role in UbcH5B interaction for two conserved non-zinc-ligating residues, L16 and I45, which form the center of the hydrophobic groove. Mutations L16A and I45A completely abolished E2 binding (Figure 5). In addition, a L35A mutation in yNOT4 (corresponding to L16 of CNOT4) debilitates its complementation capacity when assayed as in Figure 1B (K.W.Mulder, T.K.Albert and H.Th.M.Timmers, in preparation). L16 and I45 map to loop L1 and the α-helix of the CNOT4 RING, respectively (Hanzawa et al., 2001). The corresponding amino acids I383 and W408 of c-Cbl make contacts with UbcH7 side chains (Zheng et al., 2000). As proposed by the authors, residues at those positions within the characteristic RING groove may have a crucial role for various E2–E3 interactions. By employing a structure-guided mutagenesis of CNOT4, we add strong support to this hypothesis. It is noteworthy that c-Cbl mutation W408A abrogates E2 binding and E3 autoubiquitylation (Joazeiro et al., 1999), as does the corresponding mutation L51A in the RING E3 ligase BRCA1 (Ruffner et al., 2001). CNOT4 mutation L16I, which mimics c-Cbl residue I383 and BRCA1 residue I26, displays wild-type UbcH5B binding. At this proximal position of zinc-binding site I, RING E3 ligases almost exclusively use amino acids with hydrophobic side chains, including isoleucine, leucine and valine (unpublished data). This preference explains the wild-type phenotype of L16I. Taken together, our data underscore the importance of RING residues at structurally defined positions for E2 interactions. Thus, general principles of E2 interactions are realized in different RINGs, including the atypical C4C4 RING of CNOT4 and the classical C4HC3 RING of c-Cbl.
Functional implications of CNOT4 E3 ligase activity
The discovery of the yeast NOT proteins in a genetic screen led to their classification as transcriptional repressors (Collart and Struhl, 1993, 1994; Oberholzer and Collart, 1998). Specifically, the screen was set up to isolate genes that repress transcription of the TC core promoter of the HIS3 gene. As mentioned above, several observations link NOT genes to TFIID. By characterizing CNOT4 as an E3 ligase, our study adds a novel aspect to CCR4–NOT functions. The E3 activity is confined to the N-terminus of CNOT4, whereas its C-terminal region mediates association with CNOT1, the scaffolding platform of the CCR4–NOT complex. Important questions remain. First, what are the physiological targets of CNOT4 E3 ligase activity? Secondly, what are the functional consequences of this activity?
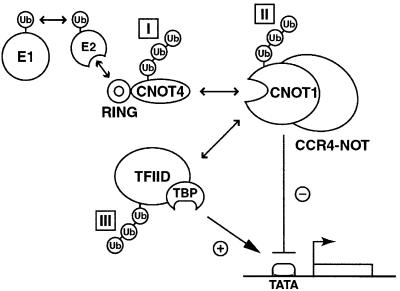
Candidates for substrates include (i) CNOT4 itself; (ii) other components of the CCR4–NOT complex; and (iii) proteins that transiently associate with either CNOT4 or the CCR4–NOT complex (Figure 7). Regulation of E3 ligases by their own ubiquitin-dependent degradation is not unprecedented. Examples include autoubiquitylation of HECT protein E6-AP, and RING proteins Mdm2 and inhibitors of apoptosis, c-IAP1 and XIAP (for references see Pickart, 2001). We observed autoubiquitylation of CNOT4 in reconstituted in vitro ubiquitylation assays. It is unclear whether this phenomenon is of physiological relevance to CNOT4 function in vivo. One prediction of an autoregulation mode for CNOT4 is that other essential functions reside outside of its RING, which are regulated by ubiquitin-mediated, RING-dependent CNOT4 degradation. Such functions await their discovery; they might be harbored in the conserved RRM and C3H zinc finger domains. However, two observations argue for targets other than CNOT4 itself. First, the RING contains an essential CNOT4 function, as revealed by complementation. Secondly, and more directly, inhibition of proteasome activity does not stabilize exogenously expressed CNOT4 in mammalian cells (unpublished data).
Fig. 7. Model illustrating potential targets for RING-dependent CNOT4 E3 ligase activity: I, CNOT4 itself; II, CNOT1 or other CCR4–NOT subunits; and III, TFIID. Arrows indicate physical and functional connections. See text for details.
The CNOT1 protein presents a putative target for CNOT4 action. In this scenario, and in analogy to other single-subunit E3s such as c-Cbl, CNOT4 uses two separate domains for E2 (UbcH5B) and substrate (CNOT1) binding. As predicted by this model, expression of the substrate-binding domain without the RING would elicit a dominant-negative effect on substrate turnover, leading to a stabilization of CNOT1. As CNOT1 is essential for the assembly and integrity of the CCR4–NOT repressor complex, one would expect stabilization of the complex and thereby enhancement of repression as the final outcome. Likewise, overexpression of the RING without the substrate-binding domain might result in a dominant-negative effect by titrating limiting E2s. In support of this model, overexpression of the conserved N-terminal 298 residues of yNOT4 containing the RING has a dominant-negative effect (M.A.Collart, unpublished observations). A caveat of this model is the fact that yNOT4 and yNOT1 display similar mutant phenotypes (Collart and Struhl, 1994). If the major function of CNOT4 is to decrease CNOT1 protein levels, the two genes should have opposite mutant phenotypes.
Another intriguing potential target for CNOT4 E3 activity is the basal transcription factor TFIID. In this scenario, the CCR4–NOT complex would resemble an SCF-like holo-E3 complex, delegating E2 binding to its RING subunit CNOT4 and substrate recognition, i.e. TFIID binding, to a different component(s). Data obtained in yeast reveal a multitude of genetic and physical connections between the CCR4–NOT and TFIID subunits (see Introduction). Most strikingly in this context, not4 mutants show a synthetic phenotype when combined with a temperature-sensitive (ts) taf19-1 allele (Lemaire and Collart, 2000). Depletion of yTAFII19 preferentially affects transcription from promoters without a consensus TATA sequence, including TC of HIS3 (Moqtaderi et al., 1996). The essential yTAFII19 is needed for the stability of TFIID and, in support of this, yeast cells harboring the ts taf19-1 allele show decreased steady-state levels of yTAFII145, yTAFII40, yTAFII25 and yTBP at the non-permissive temperature (Lemaire and Collart, 2000). Thus, targeting TAFII19 for degradation has deleterious effects on the integrity of yTFIID. As both CCR4–NOT and TFIID complexes are conserved, a similar functional relationship in higher eukaryotes is plausible. Notably, murine TFIID subunits TAFII135 and TBP are depleted in extracts from differentiated mouse cell lines in a proteasome-mediated manner (Perletti et al., 2001), indicating that TFIID subunits can be targeted selectively by ubiquitin-dependent degradation in higher eukaryotes. How could CNOT4 E3 activity be integrated in this network of CCR4–NOT and TFIID functions? As summarized in Figure 7, a model of CCR4–NOT targeting TFIID would be based on various genetic, physical and functional links between the two complexes. At a subset of CCR4–NOT-regulated core promoters, the net result of CNOT4-dependent TFIID degradation would be transcription repression. In this, the model prediction meets the results of genetic CCR4–NOT analyses.
Finally, we propose that either CNOT4 association with the CCR4–NOT complex, or its E3 activity within the complex is tightly controlled, as such activity lies at the heart of gene regulation. This could occur by compartmentalization away from the complex, e.g. by regulated subcellular localization. A similar regulation mode has been shown for the p53-specific RING E3 Mdm2. Upon interaction with p14ARF, nuclear Mdm2 relocalizes to the nucleolus where it is inhibited for degradation of p53 (Lohrum et al., 2000). Alternatively, E3 activity of the CCR4–NOT complex might be regulated by post-translational modification. Upon stress or glucose depletion, yNOT3 and yNOT5 become phosphorylated and subsequently degraded (M.A.Collart, unpublished observations). Future studies in our groups aim at elucidating these processes in more detail.
The characterization of CNOT4 as E3 ligase, its cooperation with UBC4/5-type E2 enzymes in in vitro-based assays and the identification of CNOT4 mutations that interfere with E2 binding allow testing for target functions in a biochemically defined manner. Revealing those targets should add substantial insights into CCR4–NOT functions and provide a better understanding of pol II transcription regulation.
Materials and methods
Yeast culture and assays
Culture of yeast cells and assays were as described (Albert et al., 2000). For the two-hybrid screen, bait LexA-N227 was used to screen a B42 fusion library of HeLa cDNAs with a complexity of 1 × 107 clones. Using 30 µg of library DNA, we obtained ∼1.5 × 106 primary yeast transformants. Positive clones were selected on leucine-deficient media. Library plasmids were recovered, sequenced and tested with the original bait and control plasmids.
DNA, plasmids and mutagenesis
Full-length IMAGE cDNA clones for UbcH6 and UbcH9 were obtained from RZPD, Berlin. CNOT4 N- and C-terminal truncations were created by standard PCR procedures, digested with appropriate restriction enzymes and introduced into polylinkers of yeast expression vector pGEN for complemetation analysis, and pEG202 (LexA-DBD fusion) or pJG4-5 (B42-AD fusion) for two-hybrid analysis (Albert et al., 2000). GST–UbcH5B was created by inserting an EcoRI–NotI UbcH5B fragment of library plasmid pJG-UbcH5B into EcoRI–NotI-digested pGEX-2T derivative pRP265NB. Site-directed mutagenesis on template B42-N63 was carried out by standard overlap PCR procedures. Mutagenic primers contained the following codons: L16A/E/I, CTT→GCA/GAA/ATT; C17A, TGC→GCC; M18A, ATG→GCG; D24A, GAT→GCG; C33R, TGT→CGT; W42A, TGG→GCG; R44A/E, CGA→GCG/GAG; I45A/W, ATT→GCC/TGG; D48A, GAT→GCT; E49A/K, GAA→GCA/AAA; P54A, CCT→GCT; A55D, GCA→GAC; and R57A/E, AGA→GCA/GAA. All constructs created by PCR were analyzed by DNA sequencing. Cloning details are available upon request.
Western blotting and antibodies
Western blotting was performed as described (Albert et al., 2000). Primary antibodies used were mouse monoclonal antibodies anti-HA 12CA5, anti-ubiquitin P4D1 (Santa Cruz) and anti-CNOT4 19A12.
Recombinant proteins
Expression and purification of His6-CNOT4 derivatives were as described (Hanzawa et al., 2001). GST fusion proteins were expressed in BL21(DE3) cells grown to mid-log phase, induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 30°C, and lysed in buffer LB (300 mM KCl, 50 mM Tris–HCl pH 8.0, 2 mM EDTA 0.1% Triton X-100, 20% sucrose) containing 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail (Roche) and 250 µg/ml lysozyme. After freeze–thawing and sonification, lysates were centrifuged at 50 000 r.p.m. for 1 h at 4°C. Supernatants were bound to glutathione–agarose in LB buffer without sucrose, washed with buffer WB (150 mM KCl, 20 mM potassium phosphate pH 7.0) and eluted with buffer EB (WB plus 10 mM glutathione). After dialysis against buffer DB (EB without glutathione) containing 10% glycerol, eluted proteins were stored at –80°C. For NMR analysis, GST–UbcH5B was purified further on a Superdex 75 size exclusion column (Pharmacia). The GST tag was removed by cleavage with 1 U of thrombin (Sigma) per mg of protein at 37°C for 4 h. Thrombin was inactivated by addition of 0.5 mM PMSF. After centrifugation (15 000 r.p.m., 1 h, 4°C), the cleared supernatant was dialyzed against NMR buffer (150 mM KCl, 20 mM potassium phosphate pH 7.0, 10 µM ZnCl2) and concentrated using a stirred Amicon ultrafiltration cell (3 kDa cut-off; Millipore).
GST pull-down
Pull-down experiments were carried out with 4 µg of GST–Ubc fusion proteins immobilized on 20 µl (dry volume) of glutathione–agarose (Sigma). A 4 µg aliquot of His6-N63/N78 was bound in 500 µl of G/A buffer (50 mM potassium phosphate pH 6.6, 50 mM KCl, 0.1% NP-40, 10 µM ZnCl2) containing 0.5 mM PMSF, 1 mM DTT, 1 µg/ml aprotinin, leupeptin and pepstatin (all Sigma), with tumbling for 1 h at 4°C. Beads were washed three times with G/A buffer before elution by boiling in 2× SDS sample buffer. After SDS–gel electrophoresis, eluted material was analyzed by western blot analysis.
NMR spectroscopy
For NMR titration experiments, UbcH5B was added gradually to 0.5 mM 15N-labeled His6-N78 in NMR buffer to equimolar amounts. Recordings of the 15N-HSQC spectra were as described (Hanzawa et al., 2001). Compound chemical shift changes were calculated as in Mulder et al. (1999).
In vitro ubiquitylation assay
A 50 ng aliquot of porcine heart E1 (Calbiochem), 100 ng of UbcH5B, 500 ng of His6-CNOT4 and 3 µg of GST–ubiquitin were used in a 20 µl reaction containing 50 mM Tris–HCl pH 7.5, 50 mM KCl, 2.5 mM MgCl2, 0.5 mM EDTA pH 8.0, 0.25 mM DTT, 2 mM ATP, 10 mM creatine phosphate and 10 U of creatine phosphokinase (Calbiochem). Reaction mixtures were incubated at 30°C for various times as indicated.
Acknowledgments
Acknowledgements
We thank members of the Timmers laboratory for comments, F.Sauer for the GST–ubiquitin plasmid, R.Bernards for Ubc9 expression plasmids, N.Pavletich for a GST–Cbl construct, H.Stunnenberg for GST–cTRα, and V.Bardwell, L.Maillet and K.W.Mulder for sharing unpublished reagents and results. Grants from the European Community to H.Th.M.T., M.A.C. (ERB-FRX-CT96-0064) and R.B. (‘Access to Large Scale Facilities’), from The Netherlands Organization for Scientific Research (NWO) to H.Th.M.T. and R.B., and from the Dutch Cancer Foundation to M.J.de R. and F.A.J.v.d.H. supported this work. T.K.A. acknowledges support by an NWO fellowship.
References
- Albert T.K., Lemaire,M., van Berkum,N.L., Gentz,R., Collart,M.A. and Timmers,H.T. (2000) Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res., 28, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- Badarinarayana V., Chiang,Y.C. and Denis,C.L. (2000) Functional interaction of CCR4–NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics, 155, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Salvadore,C., Chiang,Y.-C., Collart,M.A., Liu,H.Y. and Denis,C.L. (1999) The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol., 19, 6642–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow P.N., Luisi,B., Milner,A., Elliott,M. and Everett,R. (1994) Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol., 237, 201–211. [DOI] [PubMed] [Google Scholar]
- Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L. (1999) Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res., 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudenon S.L., Huacani,M.R., Wang,G., McDonnell,D.P. and Huibregtse,J.M. (1999) Rsp5 ubiquitin–protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6972–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon S.F., Rodgers,K.K., Schatz,D.G., Coleman,J.E. and Steitz,T.A. (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nature Struct. Biol., 4, 586–591. [DOI] [PubMed] [Google Scholar]
- Borden K.L. (2000) RING domains: master builders of molecular scaffolds? J. Mol. Biol., 295, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Borden K.L. and Freemont,P.S. (1996) The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol., 6, 395–401. [DOI] [PubMed] [Google Scholar]
- Borden K.L., Boddy,M.N., Lally,J., O’Reilly,N.J., Martin,S., Howe,K., Solomon,E. and Freemont,P.S. (1995) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J., 14, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J., 12, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1994) NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev., 8, 525–537. [DOI] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion,F. and Seraphin,B. (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res., 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais V., Busso,D., Wasielewski,E., Poterszman,A., Egly,J.M., Thierry,J.C. and Kieffer,B. (2001) Solution structure of the N-terminal domain of the human TFIIH MAT1 subunit: new insights into the RING finger family. J. Biol. Chem., 276, 7457–7464. [DOI] [PubMed] [Google Scholar]
- Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa H., de Ruwe,M.J., Albert,T.K., van der Vliet,P.C., Timmers,H.T. and Boelens,R. (2001) The structure of the C4C4 RING finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem., 276, 10185–10190. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Huang L., Kinnucan,E., Wang,G., Beaudenon,S., Howley,P.M., Huibregtse,J.M. and Pavletich,N.P. (1999) Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science, 286, 1321–1326. [DOI] [PubMed] [Google Scholar]
- Jackson P.K., Eldridge,A.G., Freed,E., Furstenthal,L., Hsu,J.Y., Kaiser,B.K. and Reimann,J.D. (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol., 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman,A.M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell, 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A., Wing,S.S., Huang,H., Leverson,J.D., Hunter,T. and Liu,Y.C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin–protein ligase. Science, 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Johnson E.S. and Blobel,G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem., 272, 26799–26802. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Wyrick,J.J., Koh,S.S., Jennings,E.G., Gadbois,E.L. and Young,R.A. (1998) Interplay of positive and negative regulators in transcription initiation by RNA pol II holoenzyme. Mol. Cell. Biol., 18, 4455–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M. and Collart,M.A. (2000) The TATA-binding protein-associated factor yTAFII19p fuctionally interacts with components of the global transcriptional regulator Ccr4–Not complex and physically interacts with the Not5p subunit. J. Biol. Chem., 275, 26925–26934. [DOI] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana,V., Audino,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum M.A., Ashcroft,M., Kubbutat,M.H. and Vousden,K.H. (2000) Contribution of two independent MDM2-binding domains in p14(ARF) to p53 stabilization. Curr. Biol., 10, 539–542. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Tu,C., Hong,Y.K., Shuster,E.O. and Collart,M.A. (2000) The essential function of Not1 lies within the Ccr4–Not complex. J. Mol. Biol., 303, 131–143. [DOI] [PubMed] [Google Scholar]
- Mitsui A. and Sharp,P.A. (1999) Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl Acad. Sci. USA, 96, 6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai,Y., Poon,D., Weil,A.P. and Struhl,K. (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature, 383, 188–191. [DOI] [PubMed] [Google Scholar]
- Mulder F.A., Schipper,D., Bott,R. and Boelens,R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol., 292, 111–123. [DOI] [PubMed] [Google Scholar]
- Oberholzer U. and Collart,M.A. (1998) Characterization of NOT5 that encodes a new component of the Not protein complex. Gene, 207, 61–69. [DOI] [PubMed] [Google Scholar]
- Perletti L., Kopf,E., Carre,L. and Davidson,I. (2001) Coordinate regulation of RARγ2, TBP, and TAFII135 by targeted proteolysis during retinoic-acid induced differentiation of F9 embryonal carcinom cells. BMC Mol. Biol., 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Ruffner H., Joazeiro,C.A., Hemmati,D., Hunter,T. and Verma,I.M. (2001) Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl Acad. Sci. USA, 98, 5134–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy,A.A., Chenoweth,J.G. and Tansey,W.P. (2001) Regulation of transcriptional activation domain function by ubiquitin. Science, 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- Thien C.B. and Langdon,W.Y. (2001) Cbl: many adaptations to regulate protein tyrosine kinases. Nature Rev. Mol. Cell. Biol., 2, 294–307. [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang,P., Jeffrey,P.D. and Pavletich,N.P. (2000) Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin–protein ligases. Cell, 102, 533–539. [DOI] [PubMed] [Google Scholar]