Abstract
Tobacco endogenous pararetroviruses (TEPRVs) represent the first virus-derived repetitive sequence family found in plants. The sequence conservation of TEPRVs and the lack of an exogenous form of the virus suggest that TEPRVs serve a beneficial function, perhaps by furnishing virus resistance via homologous sequence interactions. This hypothesis is supported by the observation that TEPRVs are methylated and negligibly transcribed. Moreover, transgenes driven by the TEPRV enhancer are silenced and methylated when introduced into tobacco, but remain active and unmethylated in non-host species devoid of sequences homologous to TEPRVs. In transgenic Arabidopsis, the TEPRV enhancer is active primarily in shoot meristems. This suggests that the virus giving rise to TEPRVs could infect germ cell precursors, a prerequisite for meiotically heritable insertions into host chromosomes. The copy number, organization and methylation of TEPRVs in tetraploid tobacco and one of its diploid ancestors, Nicotiana sylvestris, the presumed original host for the virus, have remained constant since polyploid formation. The remarkable conservation of these features in two independently evolving species further supports a role for TEPRVs in viral immunity.
Keywords: DNA methylation/endogenous virus/genome evolution/homology-dependent virus resistance/pararetrovirus
Introduction
During the past few years, the belief that plant viruses, unlike many of their bacterial and animal counterparts, do not invade host chromosomes has been overturned by several independent findings of endogenous viral sequences in diverse plant species (Hull et al., 2000). Sequences derived from both groups of plant DNA viruses, the single-stranded DNA geminiviruses (Kenton et al., 1995; Bejarano et al., 1996; Ashby et al., 1997) and the double-stranded DNA pararetroviruses (Harper et al., 1999; Jakowitsch et al., 1999; Ndowora et al., 1999; Budiman et al., 2000; Lockhart et al., 2000; Mao et al., 2000), have been found inserted into plant chromosomes. Integration of viral sequences has apparently occurred by illegitimate recombination because the viruses involved do not encode an integrase function. Three cases of endogenous pararetroviruses (EPRVs) are particularly interesting because they illustrate the different possible fates of integrated viral sequences.
Although some endogenous viruses could simply be benign components of plant genomes, others could potentially be pathogenic or, conversely, provide viral immunity. For endogenous viruses to be pathogenic, they must encode functional proteins and therefore cannot be significantly mutated. The pathogenic potential of EPRVs has been demonstrated for banana streak virus (BSV; Harper et al., 1999; Ndowora et al., 1999) and tobacco vein clearing virus (TVCV; Lockhart et al., 2000). Normally silent endogenous copies of these viruses were activated by in vitro propagation (BSV) or in hybrid plants cultivated under sub-optimal environmental conditions (TVCV), resulting in episomal virus infections (Hull et al., 2000).
Tobacco (Nicotiana tabacum) endogenous pararetroviruses (TEPRVs) provide an example of integrated viral sequences that might supply resistance to the cognate virus. All of the TEPRV copies for which sequence information is available contain defective open reading frames (ORFs), indicating that they do not encode functional proteins and are negligibly transcribed (Jakowitsch et al., 1999). Moreover, tobacco shows no signs of infection under a variety of growth conditions and the corresponding free virus, which would represent a previously unknown pararetrovirus, has not yet been detected. Despite the presence of imperfect ORFs, the TEPRVs analyzed so far display 91–98% DNA sequence similarity (Jakowitsch et al., 1999). This notable sequence conservation and the inability to recover the exogenous form of the virus suggested that TEPRVs serve a beneficial function, perhaps conferring viral resistance to tobacco by means of homologous sequence interactions (Jakowitsch et al., 1999).
In plants, a type of homology-dependent virus resistance that is not associated with endogenous viruses and that acts post-transcriptionally to degrade viral RNAs has been discovered recently (Vance and Vaucheret, 2001; Voinnet, 2001; Waterhouse et al., 2001). Homology-dependent post-transcriptional gene silencing (PTGS) is able to counteract RNA viruses, which can be both inducers and targets of PTGS, and it can also eliminate transcripts of DNA viruses (Covey and Al-Kaff, 2000). Many plant viruses encode suppressors of PTGS (Voinnet et al., 1999). Consistent with a role in viral defense, plants defective in this silencing mechanism display an increased susceptibility to some RNA viruses (Mourrain et al., 2000).
PTGS-based virus resistance is transient, inducible by aberrant or double-stranded RNA in the cytoplasm, and is not associated with stable changes in host chromosomes. In contrast, the presence of endogenous viruses opens up the possibility of a different, heritable type of homology-dependent resistance that is manifested in the nucleus at the genome level. Here we describe studies aimed at determining whether TEPRVs exhibit features that would be compatible with this potentially new type of homology-dependent virus resistance.
Results
The considerable sequence conservation of TEPRVs and the inability to detect the corresponding free virus led to the proposal that TEPRVs contribute to pathogen resistance (Jakowitsch et al., 1999). A possible mechanism of resistance involves silencing of free viral genomes by DNA methylation that is imposed by homologous sequence interactions with methylated TEPRVs. Conceivably, multiple dispersed TEPRVs could become coordinately methylated as a result of DNA–DNA pairing or RNA–DNA associations. These TEPRV sequence-specific methylation signals could in turn act on invading homologous viruses, triggering methylation and silencing of viral promoters. The outcome would be repression of virus replication (Jakowitsch et al., 1999). According to this hypothesis, TEPRVs should be stably methylated. To test this, methylation of TEPRVs was examined using isoschizomer pairs of methylation-sensitive and -insensitive restriction enzymes for which there are recognition sites in the consensus TEPRV sequence. This consensus sequence was compiled from partially overlapping λ genomic clones comprising 22 distinct, incomplete copies of TEPRV (Jakowitsch et al., 1999). Although there is no guarantee that these restriction enzyme sites are present in all 500–1000 copies of TEPRV, which display some sequence heterogeneity, differences in the degree of digestion by members of an isoschizomer pair nevertheless provide a reliable measure of the overall methylation of the TEPRV population.
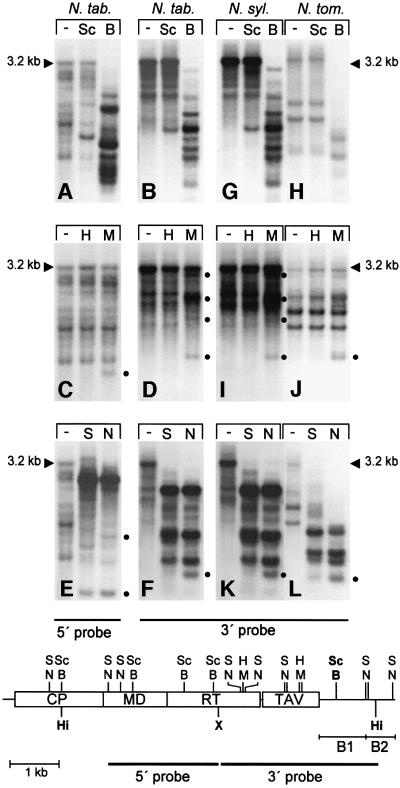
Tobacco DNA was digested with HindIII–XbaI, which, according to the TEPRV consensus sequence, should produce two fragments, each ∼3.2 kb in size, contained within the 5′ and 3′ halves of a full-length TEPRV (Figure 1, map). In addition to this standard double digest, an enzyme from one of three different isoschizomer pairs, which reveal cytosine (C) methylation in symmetrical CG dinucleotides (HpaII–MspI: CmCGG), C(A/T)G trinucleotides (ScrFI–BstNI: CmCWGG, where W = A or T) and in potentially non-symmetrical Cs (Sau3AI–NdeII: GATmC), was added. Following gel electrophoresis to separate DNA fragments and transfer to nitrocellulose, blots were hybridized to probes originating from either the 5′ or 3′ half of the TEPRV sequence. Complex hybridization patterns were obtained, as expected for a dispersed repetitive sequence. Bands in addition to the predicted 3.2 kb HindIII–XbaI fragments were observed with both 5′ (Figure 1A, C and E, lanes ‘–’) and 3′ (Figure 1B, D and F, lanes ‘–’) probes, attesting to the aforementioned partial sequence heterogeneity and/or incompleteness of some TEPRV copies.
Fig. 1. Methylation of TEPRVs. Total DNA isolated from tetraploid tobacco (N. tab.) and its two diploid ancestors, N.sylvestris (N. syl.) and N.tomentosiformis (N. tom.), was digested with XbaI (X) and HindIII (Hi; ‘–’ lane of each panel; arrowheads indicate 3.2 kb bands produced by this digest) or with X, Hi and one member of three isoschizomer pairs of methylation-sensitive/insensitive restriction enzymes: ScrFI–BstNI (Sc and B; blots A, B, G and H); HpaII–MspI (H and M; blots C, D, I and J) or Sau3AI–NdeII (S and N; blots E, F, K and L). These enzymes have recognition sequences of CCWGG, CCGG and GATC, respectively (methylation at bold Cs inhibits digestion by the first enzyme in each pair). Dots to the right of H/M and S/N blots indicate positions of new fragments following digestion with M or N. The map shows the ORFs of the putative tobacco pararetrovirus (Jakowitsch et al., 1999), the positions of restriction enzyme sites (twin parallel bars indicate two closely spaced sites for the indicated enzyme pair) and the 5′ and 3′ probes used. Blots (A), (C) and (E) were probed with the 5′ region; the remaining blots with the 3′ region. CP, coat protein; MD, movement domain; RT, reverse transcriptase; TAV, trans-activating domain.
Striking differences in digestion were obtained with ScrFI–BstNI, for which there are five sites in the TEPRV consensus sequence (Figure 1, map). While little cleavage beyond that obtained with the standard HindIII–XbaI double digest was observed with methylation-sensitive ScrFI (Figure 1A and B, compare lanes ‘–’ and Sc), significant digestion was obtained when methylation-insensitive BstNI was used. Enhanced cleavage, which led to the disappearance of the 3.2 kb HindIII–XbaI bands and the appearance of many new ones, was observed with both 5′ (Figure 1A, lane B) and 3′ (Figure 1B, lane B) probes. Although the partial sequence heterogeneity of TEPRVs makes it difficult to assign methylation to specific sites in the consensus TEPRV sequence, these results nevertheless demonstrate numerous recognition sites for this enzyme pair in the TEPRV population and considerable methylation of CWGs throughout the entire TEPRV sequence.
There are three HpaII–MspI sites in the 3′ portion of the consensus sequence (Figure 1, map). Methylation at either C prevents digestion by HpaII, whereas MspI can cut when the internal C is methylated but is inhibited by methylation in the external C. Increased digestion with MspI as compared with HpaII therefore indicates CG methylation (CmCGG); failure of both enzymes to cut despite the presence of the recognition sequence demonstrates methylation in the outer C (mCCGG), which may or may not be associated with adjacent CG methylation. With both 3′ and 5′ TERPV probes, the HpaII and MspI digests appeared similar to the standard HindIII–XbaI digest, particularly with respect to the persistence of the 3.2 kb HindIII–XbaI fragments (Figure 1C and D, compare lanes ‘–’, H and M). Nevertheless, at least four new fragments produced by MspI digestion were detected with the 3′ probe (Figure 1D, lane M, dots) and one with the 5′ probe (Figure 1C, lane M, dot). The relatively poor cleavage obtained with both enzymes, together with the substantial CWG methylation observed at the ScrFI–BstNI sites, suggests a generally high level of CNG methylation. The additional digestion obtained with MspI indicates a moderate number of sites with methylation in the internal but not external C (CmCGG), particularly in the 3′ region that contains three recognition sites in the consensus sequence. Even though there are no HpaII–MspI sites in the 5′ portion of the TEPRV consensus sequence, the appearance of one smaller fragment following digestion with MspI indicates that the recognition site for this enzyme pair is indeed present in the 5′ region of some TEPRV copies, which is again in agreement with the partial sequence divergence of TEPRV family members.
There are 10 sites for Sau3AI–NdeII (GATC) distributed throughout the TEPRV consensus sequence (Figure 1, map). Eight of these contain the C residue in a CNN context and two in a CNG context. Considerable digestion in addition to that observed with the standard HindIII–XbaI digest was seen when methylation-sensitive Sau3AI was added, as indicated by complete disappearance of the 3.2 kb HindIII–XbaI fragments with both 5′ and 3′ probes (Figure 1E and F, compare lanes ‘–’ and S). The only change in this pattern following addition of methylation-insensitive NdeII was the appearance of several new fragments with both probes (Figure 1E and F, lanes N, dots). The similar extent of digestion obtained with Sau3AI and NdeII suggests in general a minor degree of methylation at non-symmetrical CNNs within the TEPRV repeat family.
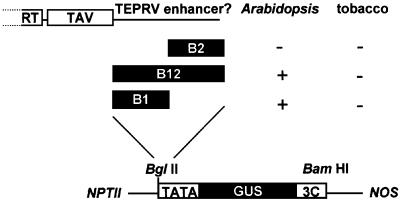
These experiments demonstrated that TEPRVs were significantly methylated, most heavily at symmetrical CNGs and CGs. To test whether introduced transgenes controlled by TEPRV transcriptional regulatory sequences would become methylated, as might be expected if TEPRVs provided virus resistance by inducing methylation and silencing of homologous sequences, we first sought to identify the enhancer–promoter (enh–pro) region of the putative tobacco pararetrovirus. Based on the location of the enh–pro in other pararetroviruses, we tested fragments downstream of the last major ORF of the putative virus for the ability to drive expression of a β-glucuronidase (GUS) reporter gene fused to a minimal promoter in various plant species. The B1 and B12 fragments, but not the B2 fragment alone (Figure 2), activated GUS expression in stably transformed Arabidopsis (Figure 3, top and data not shown), which does not contain TEPRV-like sequences (Jakowitsch et al., 1999). The B1 fragment also activated green fluorescent protein (GFP) expression in bombarded onion epidermal cells (Figure 4). Therefore, TEPRV enhancer activity is contained within the B1 fragment. Despite being active in non-host plants, the B1–GUS and B12–GUS transgenes were silent in stably transformed tobacco cells (Figure 2) and in regenerated plants (Figure 3, bottom). The intact transfer of the B1–GUS and B12–GUS expression cassettes in these plants was confirmed by DNA blotting analysis and sequencing of a PCR product spanning the B1 enhancer region (data not shown). Although not active in transgenic tobacco plants, the B1 enhancer was active in transient GFP expression assays in bombarded flower petal cells of Nicotiana sylvestris, which displays a TEPRV hybridization pattern identical to tobacco (discussed below). This demonstrates that the factors necessary for B1 enhancer activity are present in host plants (defined as those harboring TEPRVs), suggesting that silencing of integrated B1–GUS transgenes in tobacco is not due to lack of appropriate factors but requires a stable chromatin context for initiation and maintenance.
Fig. 2. Identification of the TEPRV enhancer. To identify the enhancer element of the putative tobacco pararetrovirus giving rise to TEPRVs, three fragments (B1, B12 and B2) that are downstream of the last major ORF (TAV) of the putative viral genome were placed upstream of a minimal promoter (TATA), which is negligibly active in plants, adjacent to a GUS reporter gene. GUS expression was tested in stably transformed Arabidopsis plants and in transformed tobacco calli, all of which were pre-screened for kanamycin resistance and NOP, demonstrating that both flanking marker genes (NPTII and NOS) were present and expressed. Plus signs indicate fragments displaying enhancer activity in the plant material indicated. The actual numbers of GUS-positive transformants obtained with each construct over the total number of KanR NOP+ transformants tested were as follows. Arabidopsis: TATA, 0/15; B2, 0/19; B12, 20/24; B1, 16/19. Tobacco: TATA, 0/4; B2, n.d.; B12, 0/11; B1, 0/12. TAV, trans-activating domain; NPTII, neomycinphosphotransferase; NOS, nopaline synthase.
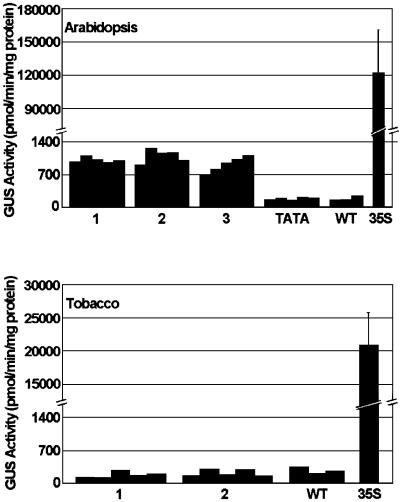
Fig. 3. Quantification of GUS activity in B1–GUS transgenic Arabidopsis and tobacco seedlings. GUS activity was measured in whole seedling extracts from three (Arabidopsis) or two (tobacco) independent transgenic lines containing the B1–GUS construct (data for five seedlings/line are shown). Segregation analysis of kanamycin resistance demonstrated that all lines contained a single transgene locus; DNA blotting using a GUS probe detected only a single band of the size expected for an intact B1–GUS expression cassette in all lines (data not shown). Seedlings containing a 35S–GUS transgene were used as positive controls (average of three separate determinations; bars indicate standard deviations). Three seedlings from lines not containing a GUS gene (WT) were used as a negative controls; for Arabidopsis, an additional control included five seedlings from a line containing only a minimal promoter (TATA)–GUS chimeric transgene. Lack of activity of this construct has been demonstrated previously in tobacco seedlings (Matzke et al., 2001). Leaves of adult tobacco plants transformed with B1–GUS or B12–GUS transgenes (several independent lines tested for each construct) were negative for GUS activity (data not shown).
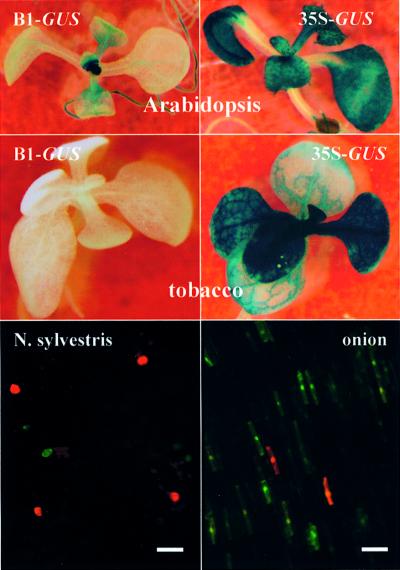
Fig. 4. Shoot meristem activity of the B1 enhancer in Arabidopsis and transient expression in onion and N.sylvestris. Top left: shoot meristem activity of B1–GUS transgenes in Arabidopsis seedlings. Middle left: B1–GUS tobacco seedlings did not exhibit GUS expression in any tissue. 35S–GUS transgenes display fairly constitutive expression in Arabidopsis (top right) and tobacco (middle right). Bottom: transient expression of smRS-GFP and DsRed1 expression in onion epidermal cells (right) and N.sylvestris petal cells (left) co-bombarded with B1–smRS-GFP (green) and 35S–DsRed1 (red) constructs. Bar = 0.1 mm.
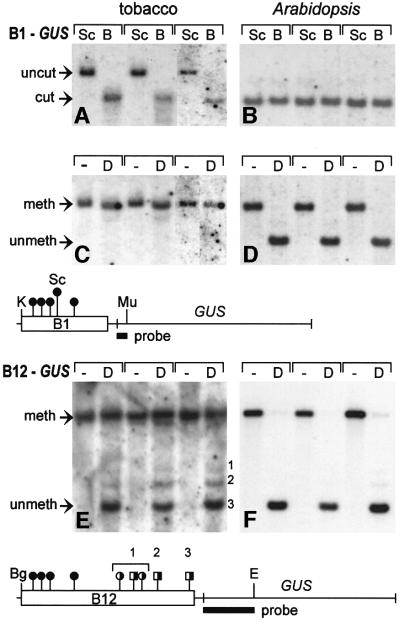
To test whether inactivity of integrated B1–GUS and B12–GUS transgenes in tobacco was associated with methylation of the B1 and B12 enhancer-containing fragments, ScrFI–BstNI (CmCWGG) and DdeI (mCTNAG) digests were performed on total DNA isolated from GUS-positive Arabidopsis transgenic plants and GUS-negative transgenic tobacco plants. The ScrFI–BstNI pair had also been used to assess methylation in the TEPRV population; however, DdeI, for which there are four recognition sites in the B1 fragment and an additional five in the B2 region, is not suitable for analyzing TEPRV methylation because of the lack of a methylation-insensitive isoschizomer. Therefore, it would not be possible to ascertain whether a failure of DdeI to digest is due to methylation or the absence of the recognition sequence. Of the two other enzyme pairs used to analyze methylation of TEPRVs (HpaII–MspI: CmCGG and Sau3AI–NdeII: GATmC), no internal recognition sites for the former are present in the B12 sequence. Although three Sau3AI–NdeII sites flank the B2 fragment in the consensus sequence (Figure 1, map), this enzyme pair is not informative for studying transgene methylation because both the B1 and B2 fragments were flanked by artificial GATC sites during cloning procedures.
In Arabidopsis, little or no methylation could be detected at the single ScrFI site in the B1 enhancer-containing fragment, as demonstrated by equal digestion with ScrFI and BstNI (Figure 5B), or in any of the DdeI sites in the B1 (Figure 5D) or the B2 portion (Figure 5F), as indicated by virtually complete conversion to the smallest expected fragment. In contrast, the ScrFI site in the B1 fragment was completely methylated in tobacco, as demonstrated by digestion and conversion to the smaller fragment only when the methylation-insensitive isoschizomer BstNI was used (Figure 5A, lanes B) and the DdeI sites were completely or partially methylated as indicated, respectively, by either no conversion (B1–GUS; Figure 5C, lanes D) or partial conversion (B12–GUS; Figure 5E, lanes D) to the smallest expected fragment. The more complete methylation in the B1 fragment might be due to the fact that all four DdeI sites in this region contain CTG trinucleotides (Figure 5, maps, circles), whereas three of the five DdeI sites in the B2 portion contain CTN trinucleotides, where N is not G (Figure 5, B12–GUS map, squares). The more complete methylation of the B12 sequence at CNGs than at CNNs is consistent with the results on TEPRVs, which are highly methylated in CNGs and more lightly methylated in CNNs. Methylation at CWG in the B1 enhancer fragment in transgenic tobacco was also demonstrated by the ScrFI–BstNI isoschizomer pair. Collectively, these results demonstrate that the methylation patterns of the B12 enhancer-containing fragments in transgenic tobacco mirror those of TEPRVs.
Fig. 5. Methylation analysis of B1–GUS and B12–GUS transgenes in tobacco and Arabidopsis. Methylation was analyzed in the B1 and B12 enhancer regions of GUS transgenes using ScrFI–BstNI (CCWGG) and DdeI (CTNAG; methylation at the bold Cs inhibits enzyme activity). There is one site for ScrFI–BstNI in the B1 fragment (circle labeled ‘Sc’, B1–GUS map) and none in the B2 fragment. There are four sites for DdeI in the B1 fragment (unlabeled filled circles, B1–GUS map) and an additional five in the B2 fragment (unlabeled partially filled circles and squares, B12–GUS map). Digests were performed on total DNA from three independent lines of tobacco or Arabidopsis containing single copies of the B1–GUS (blots A–D) or B12–GUS (blots E and F) constructs (determined by segregation of kanamycin resistance and Southern blot analysis of the GUS expression cassette; data not shown). (A and B) Double digestion with KpnI (K)–MunI (Mu) and digestion with either ScrFI (Sc; methylation sensitive) or BstNI (B; methylation insensitive). (C and D) Double digestion with K–Mu and either minus (‘–’ lanes) or plus DdeI (D lanes). The faint band slightly below the major band in (C), lanes D, suggests incomplete methylation at one of the upstream DdeI sites. (E and F) Double digestion with BglII (Bg)–EcoRV (E) and either minus (‘–’ lanes) or plus DdeI (D lanes). Blots were probed with GUS coding sequences (black bars). Little or no methylation was detected in Arabidopsis. The filled and partially filled lollipops indicate methylation and partial methylation at the respective sites in tobacco. For DdeI, circles denote sites in which the ‘N’ in the recognition sequence is G; squares indicate sites in which N is not G. The numbered bands in (E) correspond to partial methylation at the correspondingly numbered DdeI sites on the map. The fragments produced by cutting at sites under the number 1 bracket are not resolved on the gel system used.
Even though the B1–GUS transgene was expressed in Arabidopsis, GUS enzyme activity was low in extracts of seedlings compared with that driven by the cauliflower mosaic virus 35S promoter (Figure 3, top). To determine whether the weak activity resulted from tissue-specific expression of the B1 enhancer, histochemical GUS assays were performed on B1–GUS transgenic Arabidopsis seedlings. In three independent lines tested, GUS activity was concentrated in shoot apical meristems (Figure 4, B1–GUS, Arabidopsis). Histochemical GUS assays with progeny of several independent lines of B1–GUS transgenic tobacco plants revealed no comparable GUS activity in the shoot meristem or in any other portion of the seedling (Figure 4, B1–GUS, tobacco).
To attempt to reactivate TEPRVs and B1–GUS transgenes in tobacco, two approaches were used: treatment with 5-azacytidine (5-azaC) and crosses to Nicotiana clevelandii. If TEPRVs are inactivated by methylation, then treatment of B1–GUS seedlings with 5-azaC, an inhibitor of DNA methylation, might stimulate GUS expression. However, histochemical staining revealed no GUS activity after incubating seedlings for 10 days on concentrations of 5-azaC that have been used successfully in other systems (Wang and Waterhouse, 2000). These results do not rule out a role for methylation in silencing of B1–GUS transgenes or TEPRVs, which contain substantial CWG methylation, because 5-azaC has been reported to affect methylation at CG dinucleotides but not CNG trinucleotides (Kovarik et al., 1994).
The second strategy to revive TEPRVs and silent B1–GUS transgenes was based on previous experiments with TVCV, an example of a potentially pathogenic EPRV (Lockhart et al., 2000). Endogenous TVCV was activated and produced symptoms of episomal virus infection after formation of a hybrid between the host plant, Nicotiana glutinosa and N.clevelandii, which lacks endogenous TVCV-like sequences. This finding suggested either that N.clevelandii encodes an EPRV-activating protein or that formation of the hybrid N.glutinosa–N.clevelandii genome triggered loss of DNA methylation or chromatin structural alterations, which reactivated normally cryptic endogenous TVCVs in the N.glutinosa sub-genome (Lockhart et al., 2000). However, neither dormant TEPRVs nor silent B1–GUS transgenes were stimulated in N.tabacum–N.clevelandii hybrid plants, as indicated by no obvious symptoms of viral infection or GUS activity in histochemical assays, respectively (data not shown). Moreover, no methylation changes were observed in TEPRVs or in the B1 enhancer upstream of the GUS coding region in hybrid plants (data not shown). While the failure to reactivate TEPRVs could be due to the absence of functional copies, the B1–GUS transgene was shown to be intact by DNA blotting and sequence analysis.
Tobacco is an allotetraploid derived from two diploid progenitors: N.sylvestris (S sub-genome) and Nicotiana tomentosiformis (T sub-genome). The three Nicotiana species have been evolving independently since polyploid formation. Previous work suggested that TEPRVs were present primarily in the S sub-genome of tobacco (Jakowitsch et al., 1999). To determine the genome organization and extent of methylation of TEPRVs in N.sylvestris and N.tomentosiformis, DNA from the two diploid species was digested with isoschizomer pairs of methylation-sensitive and -insensitive restriction enzymes used for tobacco and probed with a probe from the 3′ portion of the TEPRV sequence. Normally, the hybridization pattern of a dispersed repeat in tobacco is the superimposition of the patterns from the two diploid parents. With all enzymes tested, however, tobacco (Figure 1B, D and F) appeared identical to N.sylvestris (Figure 1G, I and K), whereas N.tomentosiformis hybridized to the TEPRV probe but displayed a distinct, fainter pattern (Figure 1H, J and L). This indicates that TEPRV sequences integrated initially into the genome of diploid N.sylvestris (or a progenitor) and attained a fixed copy number prior to polyploid tobacco formation. TEPRV sequences have subsequently been maintained in a similar state in both N.sylvestris and tobacco. Sequences from a similar virus apparently invaded the genome of diploid N.tomentosiformis after polyploid formation, as the hybridization pattern of this species comprises bands that do not have counterparts in N.tabacum.
Discussion
This work has assessed features of TEPRVs that might be associated with resistance to the exogenous virus by a mechanism involving homology-dependent gene silencing and DNA methylation. Earlier observations supporting this proposal included the inability to detect the (previously unidentified) pararetrovirus in tobacco and the sequence similarity of TEPRVs, which suggested selection for a function based on nucleic acid sequence homology (Jakowitsch et al., 1999). The data reported in this paper support this hypothesis in several ways. First, TEPRVs have been found to be substantially and persistently methylated, particularly in CNG and CG nucleotide groups, which probably accounts for their negligible expression. Secondly, stably integrated GUS reporter genes driven by the TEPRV enhancer in tobacco were readily silenced and methylated in a pattern similar to TEPRVs, suggesting involvement of the same methylation signal, whereas the same reporter gene constructs were active and unmethylated in a non-host plant lacking TEPRV-homologous sequences. Finally, the copy number, genome organization and degree of methylation of TEPRVs have remained unchanged since the formation of polyploid tobacco, as demonstrated by the identical TEPRV hybridization patterns of tobacco and one of its diploid ancestors, N.sylvestris, which was apparently the original host for the virus. Taken together, the results are consistent with a scenario in which stably methylated TEPRVs have supplied long-term viral immunity, perhaps accompanied by attenuation or extinction of the cognate exogenous virus. A possible mechanism for virus resistance that is compatible with our data invokes enhancer methylation and transcriptional silencing induced by homologous sequence interactions among TEPRVs and free viral genomes (Figure 6). Unlike the transient and inducible nature of PTGS-based virus resistance (Vance and Vaucheret, 2001; Voinnet, 2001; Waterhouse et al., 2001), this type of homology-dependent resistance would be associated with permanent changes in host chromosomes and continuous protection that is inherited from one generation to the next.
Fig. 6. Model for heritable homology-dependent virus resistance conferred by EPRVs. Originally, no EPRVs are present in host chromosomes (left). The virus can infect the plant and free virions and virus DNA are detectable. Over evolutionary time, viral sequences integrate randomly by illegitimate recombination into host chromosomes. Once a threshold copy number is reached (∼500–1000), EPRVs become methylated (filled circles) and silenced through a homology-dependent gene silencing mechanism. Methylated EPRVs can trigger methylation and silencing of free viral genomes through a homology-dependent process involving DNA–DNA or RNA–DNA interactions. Consequently, virus replication is blocked and the host has gained long-term viral immunity.
Direct tests of the TEPRV virus resistance hypothesis are not possible due to the absence of detectable exogenous virus. Although TEPRVs show a relatively high degree of sequence similarity, they are still too heterogeneous to reconstruct the original infectious virus exactly. However, the fossil viruses, as revealed in the integration patterns of TEPRVs and related sequences in N.tomentosiformis, support the idea that multicopy EPRVs can confer virus resistance. These patterns also help to illuminate the chronological order of insertion events. Judging from the copy number of TEPRVs in tetraploid tobacco and diploid N.sylvestris, integration of viral sequences (taken as a measure of active virus in the nucleus) can occur until a copy number of 500–1000 is attained. The fact that additional insertions do not seem to have taken place to a significant extent in either N.sylvestris or tobacco after polyploid formation is consistent with the notion that this level of TEPRVs subdued the virus in both species. Tetraploid tobacco is believed to have arisen not more than 6 million years ago (Okamuro and Goldberg, 1985). Since that time, significant alterations in the polyploid genome have taken place. These alterations include the elimination of the N.sylvestris rDNA repeats (Volkov et al., 1999), the occurrence of at least four intergenomic translocations (Kenton et al., 1993; Moscone et al., 1996) and intergenomic exchange between endo-1,3-β-glucosidase genes (Sperisen et al., 1991). Against this background of obvious change in the polyploid genome, minimal, if any, alterations appear to have occurred in copy number, organization and methylation of TEPRVs in either tobacco or N.sylvestris. The faithful reproduction of methylation patterns in both species suggests not only preservation of the recognition sequences for the respective restriction enzymes, but also non-random re-establishment and/or maintenance of the epigenetic state over evolutionary time. The extraordinary conservation of these features in a repetitive sequence family that is nominally ‘junk’ in two species, which have been evolving independently since polyploid formation, strongly supports the idea that methylated TEPRVs have been subjected to the same selection pressures and have contributed advantageous traits to tobacco and N.sylvestris.
In contrast to the acquisition of TEPRVs prior to polyploid formation in N.sylvestris, the EPRVs in diploid N.tomentosiformis must have integrated after the polyploidization event because the major portion does not seem to be present in the T sub-genome of tobacco. We are currently isolating and sequencing EPRVs from the N.tomentosiformis genome. Preliminary data indicate that they are similar but not identical to TEPRVs (W.Gregor and A.J.M.Matzke, unpublished results), suggesting that they are derived from a related virus. Moreover, the EPRV hybridization pattern of N.tomentosiformis is identical to that of a close relative, Nicotiana otophora (M.F.Mette and W.Gregor, unpublished results), indicating that the endogenous viruses were acquired before divergence of the two species.
Experiments to identify TEPRV transcriptional regulatory regions revealed two features that are relevant for the TEPRV virus resistance hypothesis. The first concerns the unusual inactivity of the B1 enhancer in tobacco. Indeed, this enhancer is the only pararetroviral transcriptional regulatory sequence identified so far that is inactive when used to drive expression of stably integrated reporter genes in transgenic tobacco plants. In addition to the 35S and 19S promoters of cauliflower mosaic virus, all pararetroviral transcriptional regulatory regions tested to date are active in transgenic tobacco, including those from commelina yellow mottle virus (Medberry et al., 1992), mirabilis mosaic virus (Dey and Maiti, 1999), BSV (Schenk et al., 2001), figwort mosaic virus (Sanger et al., 1990), peanut chlorotic streak virus (Maiti et al., 1998), rice tungro bacilliform virus (Petruccelli et al., 2001) and cassava vein mosaic virus (Verdaguer et al., 1998), which is most related to the putative virus giving rise to TEPRVs (Jakowitsch et al., 1999). The unprecedented inactivity of the functional B1 enhancer in tobacco argues for targeted silencing and methylation of this transcriptional regulatory region specifically in a genome harboring homologous TEPRV sequences. Silencing appears to require a chromatin context, as transient expression of a B1–GFP reporter gene was observed in N.sylvestris. Transient activity is not inconsistent with the possibility that methylated TEPRVs trigger silencing of free viruses, as pararetroviral RNA is normally transcribed in the nucleus from an episomal mini-chromosome containing supercoiled viral DNA associated with histones, a template that is similar to chromatin (Covey et al., 1990). In principle, free viruses and homologous genomic DNA can be coordinately regulated, presumably via sequence interactions, as demonstrated by the parallel transcriptional inactivation of cauliflower mosaic virus and stably integrated transgenes driven by the 35S promoter (Covey and Al-Kaff, 2000).
The second feature of the B1 enhancer that is pertinent to the TEPRV virus resistance hypothesis is the concentration of activity in shoot meristems of Arabidopsis. Other pararetroviral transcriptional regulatory regions studied so far are expressed constitutively and/or in vascular tissue. Although there have been no previous reports of plant DNA viruses infecting meristematic cells (Hull et al., 2000), the activity of the TEPRV B1 enhancer in shoot apical meristems of Arabidopsis suggests that the virus giving rise to TEPRVs could indeed replicate in this tissue in the host. This would provide a plausible explanation for how integration of viral sequences occurred in cells contributing to the germ line, which is required for these sequences to become meiotically heritable components of the host genome. Analyses of the tissue specificities of other EPRV transcriptional regulatory regions will establish whether they are also active in cells of shoot meristems.
Although animal retroviruses and plant pararetroviruses have different types of genome and modes of replication, there are striking similarities in their endogenous forms. For example, there are a number of animal endogenous retroviruses (ERVs) for which no exogenous counterpart is known. Similar to the TEPRV example, this suggests that successful pathogen control has been achieved through endogenization (Löwer et al., 1996; Best et al., 1997; Löwer, 1999). Although resistance to ERVs can be brought about by several different mechanisms, it has been postulated recently that ‘co-suppression’ (equivalent to PTGS) might be involved in some cases (Löwer, 1999). Silencing through different types of homologous sequence interactions might turn out to be a common theme in virus resistance conferred by endogenous viruses in plants and animals.
It is likely that the EPRVs identified so far represent only the tip of an iceberg. EPRVs have been detected by PCR analysis in many other plants species (P.Heslop-Harrison, personal communication), although none seems to be present in the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000). EPRVs thus appear to be a significant new type of repetitive DNA in plants. The presence of different but related EPRVs in N.sylvestris and N.tomentosiformis, and similar skewed distributions of endogenous viruses in progenitors of other allopolyploids (Hull et al., 2000) illustrate their contribution to genome divergence between related species. In addition to the insight they provide about host–pathogen interactions, EPRVs can potentially be useful for analyzing plant phylogenies and dating polyploidization events.
Materials and methods
TEPRV sequences
A consensus sequence of the putative virus giving rise to TEPRVs (Jakowitsch et al., 1999) has the DDBJ/EMBL/GenBank accession No. AJ238747. The 12 longest contiguous sequences for individual clones [nomenclature as in Jakowitsch et al. (1999)] have been submitted to the database and have the following DDBJ/EMBL/GenBank accession Nos: AJ414165 (clone V2), AJ414164 (clone V3), AJ414166 (clone V4), AJ413172 (clone V6), AJ414168 (clone V9), AJ414167 (clone V14), AJ414169 (clone E15), AJ414170 (clone E16), AJ414174 (clone E17), AJ414171 (clone E18), AJ414172 (clone E21) and AJ414173 (clone E22). Sequences of all 22 TEPRV clones reported previously are available on our laboratory website (http://www.imolbio.oeaw.ac.at/pmg) as supplementary data to the Jakowitsch et al. (1999) reference.
Cloning of TEPRV enhancer and vector construction
PCR products from the putative enhancer region of the putative tobacco pararetrovirus genome were designed to be BglII–BamHI fragments and sequenced to confirm that they contained the desired B1, B12 or B2 regions. The B1 region extended from nucleotides 6398 to 7402 and the B2 region from 7398 to 198 in the consensus sequence of the putative tobacco pararetrovirus circular genome (Jakowitsch et al., 1999). These fragments were then cloned upstream of a minimal promoter (TATA), which lacks significant activity (Benfey et al., 1989), joined to a GUS coding region and a 3C terminator (Benfey et al., 1989) in a plasmid vector. The resulting plasmids were linearized with BglII and introduced into a BglII site between a selection marker gene encoding neomycin phosphotransferase (NPTII) and a screening marker gene encoding nopaline synthase (NOS) positioned between left and right T-DNA borders in the binary vector BV4, then introduced into Agrobacterium tumefaciens by triparental mating (Matzke and Matzke, 1986).
Production of transgenic plants
Transgenic tobacco plants containing the GUS chimeric transgenes were made by Agrobacterium-mediated leaf disk transformation. Selection was on kanamycin-containing medium; nopaline (NOP) was used as a biochemical screening marker for transformation. Transgenic Arabidopsis plants were obtained by the floral dip method (Clough and Bent, 1998). Kanamycin-resistant (KanR) T1 Arabidopsis plants and KanR tobacco calli emerging on leaf disks were screened for NOP, and KanR NOP+ material was further screened for GUS enzyme activity (Jefferson, 1987).
DNA blotting analysis
Isolation of total DNA from leaves of greenhouse-grown tobacco plants, hybrid progeny and Arabidopsis plants was performed using the DNeasy Plant Maxi Kit (Qiagen, Hilden). Restriction enzyme digests were carried out according to the manufacturer’s instructions. In our hands, EcoRII (CmCWGG) was not a suitable methylation-sensitive isoschizomer for BstNI (CCWGG); therefore, we used ScrFI (CmCNGG). DNA blotting hybridization and probing with 32P-labled RNA probes containing the 5′ part of the GUS coding region were carried out as described previously (Mette et al., 1999). DNA blotting hybridization using DNA probes containing TEPRV sequences was performed as described by Jakowitsch et al. (1999).
GUS reporter gene assays
GUS enzyme activity was quantified in tobacco and Arabidopsis seedling extracts according to the procedures of Jefferson (1987). Standard deviations were calculated using Microsoft Excel. GUS histochemical staining was performed on Arabidopsis and tobacco seedlings 15 days after imbibition. Plantlets were pre-treated for 15 min on ice in 90% acetone followed by vacuum infiltration for 2 h in GUS staining solution [1.9 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc), 0.5 mM potassium ferricyanate/ferrocyanate, 0.3% Triton X-100, 20% methanol, 100 mM sodium phosphate buffer pH 7.2; Jefferson, 1987]. The enzyme reaction was performed overnight at 37°C in the dark. Tissues were cleared and stored in 70% ethanol. As a positive control for 35S–GUS activity in tobacco, line 17 (Matzke et al., 2001) was used. In histochemical assays, homozygous seedlings of this line have relatively uniform GUS activity with the exception of the cotyledons, which show GUS staining primarily in vascular tissue.
Biolistic transformation
Transient expression of the B1 enhancer-containing fragment was analyzed in different plant species by introducing the B1–GUS or B1–soluble modified red-shifted (smRS)-GFP (Davis and Vierstra, 1998) constructs by particle bombardment. As control constructs, we used 35S–smRS-GFP, 35S–DsRed1 (Matz et al., 1999) and 35S–GUS. The plant material that was bombarded included onion epidermal cells, and young leaves and flower petals of tobacco and N.sylvestris. Onion epidermis was the most suitable tissue for the biolistic assays, followed by flower petals of N.sylvestris, which are non-pigmented and fleshy. Tobacco and N.sylvestris leaves were unsuitable for analyzing transient GFP expression because of high background fluorescence. Because of the lower activity of the B1 enhancer compared with the 35S enhancer, we were unable to detect B1–GUS activity in bombarded leaves or petals, although B1–GUS activity was observed in bombarded onion cells. In contrast, B1–smRS-GFP activity could be visualized in both bombarded onion and flower petal cells of N.sylvestris. A 35S–smRS-GFP–NOSter plasmid was obtained as clone CD3-327 from the Arabidopsis Biological Resource Center of Ohio State University. In this clone, the 35S promoter was replaced with a fragment containing B1–TATA. Conversely, the smRS-GFP coding region was replaced with DsRed1 (Clontech, Becton Dickinson, Belgium). Bombardment was performed with a Bio-Rad Gene Gun using 900 p.s.i. rupture disks (Bio-Rad, Vienna, Austria). Microcarrier gold particles (1 µm) were coated with plasmid DNA of the reporter gene constructs according to the manufacturer’s instructions. Red and green images were acquired using a Leica MZ FLIII fluorescence stereomicroscope (Leica, Vienna, Austria) equipped with a Color-Cool-View CD camera (Photonic Science, UK). Red and green images were merged using Image-Pro-plus Version 3.0 (Media Cybernetics, MD).
5-azaC treatment
In accordance with a published procedure (Wang and Waterhouse, 2000), tobacco seeds were sterilized, placed on filter paper soaked with MS medium containing either 0, 5, 10, 20, 50 or 100 µM 5-azaC (Sigma-Aldrich) and incubated for 10–12 days at 25°C in constant light. To ensure a constant drug concentration, the medium was replaced daily. With increasing 5-azaC, seedlings were increasingly retarded in growth, but they still developed a shoot apical meristem and primary leaves at all concentrations used. GUS activity in these seedlings was tested by histochemical GUS staining as described above.
Acknowledgments
Acknowledgements
We thank Dr N.-H.Chua for the TATA–GUS–3c chimeric gene, Dr R.Vierstra for the smRS-GFP construct, the IPK seed collection (Gatersleben) for N.clevelandii seeds and Dr W.Gregor for helpful comments on the manuscript. Our work has been supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung, Grant nr. Z21-MED.
References
- Ashby M.K., Warry,A., Bejarano,E., Khashoggi,A., Burrell,M. and Lichtenstein,C.P. (1997) Analysis of multiple copies of geminiviral DNA in the genome of four closely related Nicotiana species suggests a unique integration event. Plant Mol. Biol., 35, 313–321. [DOI] [PubMed] [Google Scholar]
- Bejarano E.R., Khashoggi,A., Witty,M. and Lichtenstein,C. (1996) Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc. Natl Acad. Sci. USA, 93, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P.N., Ren,L. and Chua,N.H. (1989) The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J., 8, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S., le Tissier,P.R. and Stoye,J.P. (1997) Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol., 5, 313–339. [DOI] [PubMed] [Google Scholar]
- Budiman M.A., Mao,L., Wood,T.C. and Wing,R.A. (2000) A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res., 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Covey S.N. and Al-Kaff,N.S. (2000) Plant DNA viruses and gene silencing. Plant Mol. Biol., 43, 307–322. [DOI] [PubMed] [Google Scholar]
- Covey S.N., Turner,D.S., Lucy,A.P. and Saunders,K. (1990) Host regulation of the cauliflower mosaic virus multiplication cycle. Proc. Natl Acad. Sci. USA, 87, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J. and Vierstra,R.D. (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol., 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Dey N. and Maiti,I.B. (1999) Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol., 40, 771–782. [DOI] [PubMed] [Google Scholar]
- Harper G., Osuji,J.O., Heslop-Harrison,J.S. and Hull,R. (1999) Integration of banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology, 255, 207–213. [DOI] [PubMed] [Google Scholar]
- Hull R., Harper,G. and Lockhart,B. (2000) Viral sequences integrated into plant genomes. Trends Plant Sci., 5, 362–365. [DOI] [PubMed] [Google Scholar]
- Jakowitsch J., Mette,M.F., van der Winden,J., Matzke,M.A. and Matzke,A.J.M. (1999) Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl Acad. Sci. USA, 96, 13241–13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep., 5, 387–405. [Google Scholar]
- Kenton A., Parokonny,A.S., Gleba,Y.Y. and Bennett,M.D. (1993) Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol. Gen. Genet., 240, 159–169. [DOI] [PubMed] [Google Scholar]
- Kenton A., Khashoggi,A., Parokonny,A. and Bennett,M.D. (1995) Chromosomal location of endogenous geminivirus-related DNA sequences in Nicotiana tabacum L. Chromosome Res., 3, 346–350. [DOI] [PubMed] [Google Scholar]
- Kovarik A., Koukalova,B., Holy,A. and Bezdek,M. (1994) Sequence-specific hypomethylation of the tobacco genome induced with dihydroxypropyladenine, ethionine and 5-azacytidine. FEBS Lett., 353, 309–311. [DOI] [PubMed] [Google Scholar]
- Lockhart B., Menke,J., Dahal,G. and Olszewski,N.E. (2000) Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol., 81, 1579–1585. [DOI] [PubMed] [Google Scholar]
- Löwer R. (1999) The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol., 7, 350–356. [DOI] [PubMed] [Google Scholar]
- Löwer R., Löwer,J. and Kurth,R. (1996) The viruses in all of us: characteristics and biological significance of endogenous retrovirus sequences. Proc. Natl Acad. Sci. USA, 93, 5177–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti I.B., Richins,R.D. and Shepard,R.J. (1998) Gene expression regulated by gene VI of caulimovirus: transactivation of downstream genes of transcripts by gene VI of peanut chlorotic streak virus in transgenic tobacco. Virus Res., 57, 113–124. [DOI] [PubMed] [Google Scholar]
- Mao L. et al. (2000) Rice transposable elements: a survey of 73 000 sequence-tagged-connectors. Genome Res., 10, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M.V., Fradkov,A.F., Labas,Y., Savitsky,A.P., Zaraisky,A.G., Markelov,M.L. and Lukyanov,S.A. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotechnol., 17, 969–973. [DOI] [PubMed] [Google Scholar]
- Matzke A.J.M. and Matzke,M.A. (1986) A set of novel Ti plasmid-derived vectors for the production of transgenic plants. Plant Mol. Biol., 7, 357–365. [DOI] [PubMed] [Google Scholar]
- Matzke M., Mette,M.F., Jakowitsch,J., Kanno,T., Moscone,E.A., van der Winden,J. and Matzke,A.J.M. (2001) A test for transvection in plants: DNA pairing may lead to trans-activation or silencing of complex heteroalleles in tobacco. Genetics, 158, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medberry S.L., Lockhart,B.E. and Olszewski,N.E. (1992) The commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell, 4, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M.F., van der Winden,J., Matzke,M.A. and Matzke,A.J.M. (1999) Production of aberrant promoter transcripts contributes to methylation and silencing of homologous promoters in trans. EMBO J., 18, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscone E.A., Matzke,M.A. and Matzke,A.J.M. (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma, 105, 231–236. [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Ndowora T., Dahai,G., LaFleur,D., Harper,G., Hull,R., Olszewski,N. and Lockhart,B. (1999) Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology, 255, 214–220. [DOI] [PubMed] [Google Scholar]
- Okamuro J.K. and Goldberg,R.B. (1985) Tobacco single-copy DNA is highly homologous to sequences present in the genomes of its diploid progenitors. Mol. Gen. Genet., 198, 290–298. [Google Scholar]
- Petruccelli S., Shunhong,D., Carcamo,R., Yin,Y., Chen,S. and Beachy,R.N. (2001) Transcriptional factor RF2a alters expression of the rice tungro bacilliform virus promoter in transgenic tobacco plants. Proc. Natl Acad. Sci. USA, 98, 7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger M., Daubert,S. and Goodman,R.M. (1990) Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol. Biol., 14, 433–443. [DOI] [PubMed] [Google Scholar]
- Schenk P.M., Remans,T., Sági,L., Elliott,A.R., Dietzgen,R.G., Swennen,R., Ebert,P.R., Grof,C.P. and Manners,J.M. (2001) Promoters for pregenomic RNA of banana streak badnavirus are active for transgene expression in monocot and dicot plants. Plant Mol. Biol., 47, 399–412. [DOI] [PubMed] [Google Scholar]
- Sperisen C., Ryals,J. and Meins,F. (1991) Comparison of cloned genes provides evidence for intergenomic exchange of DNA in the evolution of a tobacco glucan endo-1,3-β-glucosidase gene family. Proc. Natl Acad. Sci. USA, 88, 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Vance V.B. and Vaucheret,H. (2001) RNA silencing—defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Verdaguer B., de Kochko,A., Fux,C.I., Beachy,R.N. and Fauquet,C. (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol., 37, 1055–1067. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto,Y. and Baulcombe,D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov R.A., Borisjuk,N.V., Panchuk,I.I., Schweizer,D. and Hemleben,V. (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol., 16, 311–320. [DOI] [PubMed] [Google Scholar]
- Wang M.B. and Waterhouse,P.M. (2000) High-efficiency silencing of a β-glucuronidase gene in rice is correlated with a repetitive transgene structure but is independent of DNA methylation. Plant Mol. Biol., 43, 67–82. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]