Abstract
Here we show that Dictyostelium discoideum dynamin A is a fast GTPase, binds to negatively charged lipids, and self-assembles into rings and helices in a nucleotide-dependent manner, similar to human dynamin-1. Chemical modification of two cysteine residues, positioned in the middle domain and GTPase effector domain (GED), leads to altered assembly properties and the stabilization of a highly regular ring complex. Single particle analysis of this dynamin A* ring complex led to a three-dimensional map, which shows that the nucleotide-free complex consists of two layers with 11-fold symmetry. Our results reveal the molecular organization of the complex and indicate the importance of the middle domain and GED for the assembly of dynamin family proteins. Nucleotide-dependent changes observed with the unmodified and modified protein support a mechanochemical action of dynamin, in which tightening and stretching of a helix contribute to membrane fission.
Keywords: conformational changes/protease inhibitor/protein assembly/ring complex
Introduction
The high molecular weight GTPases of the dynamin family are structurally related, but functionally diverse (reviewed in van der Bliek, 1999; McNiven et al., 2000). They share a highly conserved N-terminal GTPase domain and a conserved C-terminal GTPase effector domain (GED), separated by a less conserved ‘middle domain’. In addition, a pleckstrin homology (PH) domain and a proline-rich domain (PRD) are found in various dynamins (reviewed in van der Bliek, 1999). The functions of dynamins include vesicle formation events along the endocytotic and the secretory pathways, vesicular transport, mitochondrial morphogenesis and contribution to viral resistance (reviewed in McNiven et al., 2000).
Human dynamin-1 plays an essential role in membrane fission during receptor-mediated endocytosis and synaptic vesicle recycling (reviewed in Schmid et al., 1998). Dynamin-1 was reported to self-assemble into rings and stacks in vitro (Hinshaw and Schmid, 1995) and into helices or ‘spirals’ around the necks of clathrin-coated pits in vivo (Takei et al., 1995). These rings and helices have the same dimensions as the electron-dense collars around the neck of coated pits accumulated at the neuromuscular junction of shibire mutants of Drosophila melanogaster expressing a temperature-sensitive dynamin (Kosaka and Ikeda, 1983). Assembly of dynamin-1 is favoured by low ionic strength, GTP analogues, GDP in combination with γ-phosphate analogues and acidic lipid membranes (Hinshaw and Schmid, 1995; Takei et al., 1995, 1998, 1999; Carr and Hinshaw, 1997; Sweitzer and Hinshaw, 1998; Stowell et al., 1999). Once dynamin has assem bled around a lipid tube, membrane fission occurs upon GTP hydrolysis (Sweitzer and Hinshaw, 1998). Mechanochemical models for the action of dynamins are based either on constriction (Sweitzer and Hinshaw, 1998; Smirnova et al., 1999) or stretching of the helix (Kozlov, 1999; Stowell et al., 1999). The function of dynamin as a mechanoenzyme has been challenged by the suggestion that GTP-bound dynamin activates downstream effectors responsible for the fission event rather than actively causing membrane fission upon GTP hydrolysis (Sever et al., 1999, 2000). However, recent studies show that GTP hydrolysis and an associated conformational change are required for endocytosis, supporting a mechanochemical function of dynamin-1 (Hill et al., 2001; Jeong et al., 2001; Marks et al., 2001).
The lower eukaryote Dictyostelium discoideum has at least three dynamins, dynamin A, B and C. Dynamin A is a 96 kDa cytosolic protein which functions in membrane severing events (Wienke et al., 1999). The dynamin A GTPase domain (residues 1–304), the atomic structure of which has recently been solved (Niemann et al., 2001), shares 62 and 61% sequence identity with the GTPase domain of human dynamin-1 and human DLP1, respectively. The middle domain of dynamin A (residues 305–511) shows the highest degree of sequence similarity to DLP1 (49%). The region from residue 512 to 734 shows no similarity to the sequence of other members of the dynamin family in that it contains a high proportion of glutamine (25%), asparagine (23%) and serine (14%) residues in long stretches of up to 13 amino acids. Long repeats of Gln, Asn or Ser residues are frequently found in Dictyostelium proteins, but their structure and function are unknown (Subirana and Palau, 1999; Katti et al., 2000). The central part of this Gln, Asn and Ser rich region (residues 573–624) comes closest to the Pro-rich domain observed in other dynamin family members. The C-terminal domain of dynamin A (residues 735–853) shares 51 and 43% sequence identity with the GED of DLP1 and dynamin-1, respectively (Wienke et al., 1999).
Here we show that dynamin A, similar to human dynamin-1, forms ring-like structures and helical assemblies in a nucleotide-dependent fashion. A covalently modified form of the protein, obtained in the presence of the protease inhibitor N-CBZ-l-phenylalanine chloromethyl ketone (ZPCK), displayed normal GTPase activity but altered assembly properties. The modified protein, dynamin A*, formed highly regular and stable ring complexes in the absence of nucleotides. Using single particle analysis, we reconstructed the three-dimensional structure of the nucleotide-free dynamin A* ring complex, revealing details of its molecular organization. In addition, the ring complex was analysed in the presence of the ‘non-hydrolysable’ GTP analogue GppNHp to determine nucleotide-dependent structural changes. Based on these data, we propose a model for the mechanochemical action of dynamins in membrane fission.
Results
Dynamin A purification
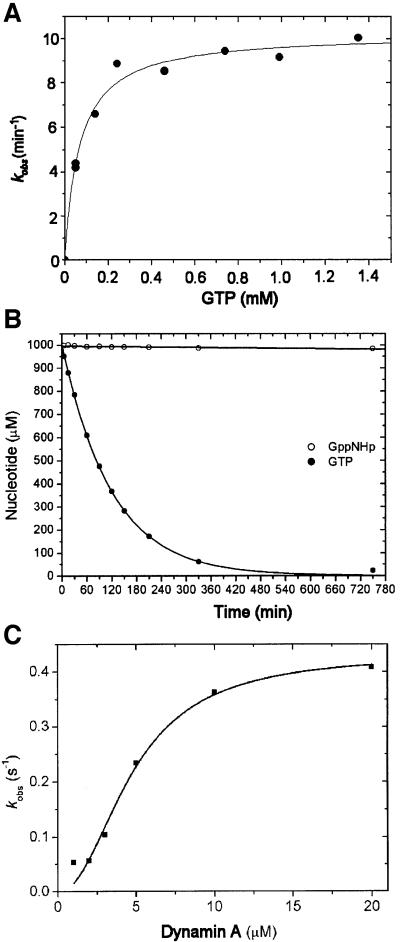
We have shown previously that disruption of the gene encoding dynamin A disturbs endosomal trafficking in D.discoideum (Wienke et al., 1999). In complementation experiments to rescue this phenotype, we observed that dynamin A can be overproduced up to 20-fold compared with wild-type levels in D.discoideum without affecting the viability or growth of the cells. Here, we used these overproducing cells to study the biochemical and structural properties of dynamin A in detail. In the first step of the purification, dynamin A is separated from soluble proteins in the whole-cell lysate by sedimentation at 30 000 g. Dynamin A could be effectively extracted from the resulting pellet fraction by homogenizing it in buffers containing 300 mM NaCl and 5 mM Mg2+-GTP. Other nucleotides, including GDP, ATP and ADP, were tested but had no significant effect on the solubility of dynamin A (data not shown). Purification of dynamin A was greatly facilitated by the combined centrifugation and GTP extraction step, and the protein could subsequently be purified to homogeneity by an additional DEAE chromatography step. The purified protein displayed cooperative GTPase activity. The apparent kcat increased ∼10-fold over a dynamin A concentration range of 1–20 µM (Figure 1C).
Fig. 1. GTPase activity of dynamin A. (A) GTPase activity of 1 µM dynamin A at 20°C as a function of GTP concentration. Values of 68 µM for KM and 0.17/s for kcat were determined with freshly purified dynamin A. (B) Hydrolysis of GTP and GppNHp by dynamin A*, the modified form of the enzyme. Complete hydrolysis of 1 mM GTP by 1 µM dynamin A* occurred with τ1/2 = 70 min at 20°C. No significant hydrolysis of 1 mM GppNHp could be detected following a 12 h incubation with 1 µM dynamin A at 20°C. (C) Cooperativity of the GTPase activity of dynamin A. The apparent kcat increased significantly with increasing dynamin A concentration. A fit of the data to the equation kobs = Vmax × [dynamin A]n/([dynamin A]n + kn) is shown, with Vmax = 0.43 ± 0.04/s, k = 4.75 ± 0.6 µM and n = 2.1 ± 0.4. The specific activity of dynamin A decreased upon storage at 80°C and up to 3-fold higher values for Vmax were measured with freshly purified protein.
Lipid binding by dynamin A
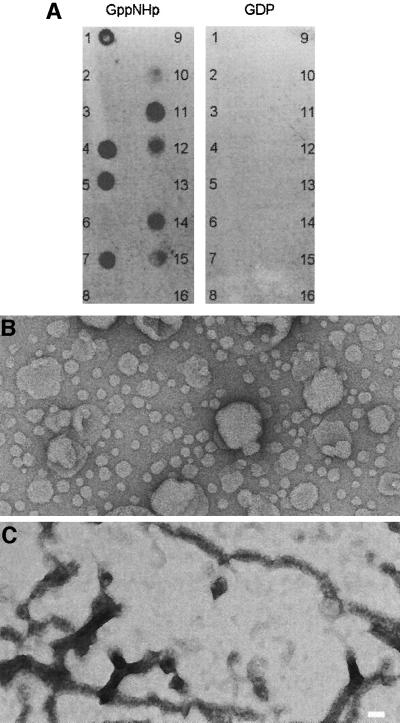
Co-sedimentation studies showed that dynamin A remained soluble in the absence of lipid vesicles and when mixed with phosphatidylethanolamine (PtdEth) vesicles, but was found partially in the pellet when incubated with phosphatidylserine (PtdSer) vesicles (data not shown). Since neutral PtdEth differs from negatively charged PtdSer only by the lack of a charged carboxyl group, these results argue for the preferred binding of dynamin A to negatively charged phospholipid membranes. The lipid binding properties of dynamin A were further examined by testing the protein’s ability to interact with 12 different lipids that were spotted on a nitrocellulose membrane. Figure 2A shows that the GDP-bound form of dynamin A did not interact with any of the lipids. However, in the presence of GppNHp, binding to phosphatidic acid and several phosphatidylinositol phosphates was observed (Figure 2A). Electron microscopy (EM) of negatively stained samples revealed spherical liposomes of various sizes in the absence of dynamin A (Figure 2B). Addition of 2 µM dynamin A and 1 mM GTP-γS led to the formation of branched networks, never observed in the absence of lipids (Figure 2C). Similar results were obtained when GppNHp was used instead of GTP-γS (data not shown).
Fig. 2. Interaction of dynamin A with lipids. (A) Membranes with immobilized lipids were incubated with 20 µg/ml dynamin A in the presence of 1 mM GppNHp or 1 mM GDP at 4°C overnight. Antibody PAD1 was used for detection (Wienke et al., 1999). Only in the presence of GppNHp did dynamin A bind to PtdIns, PtdInsP, PtdInsP2, PtdInsP3 and phosphatidic acid. The following lipids (100 pmol) were spotted on the membrane: (1) d-myo-phosphatidylinositol (PtdIns); (2) PtdIns (animal); (3) PtdIns (soy); (4) d-myo-phosphatidylinositol 3-phosphate [PtdIns(3)P]; (5) PtdIns(4)P; (6) PtdIns(4)P (animal); (7) PtdIns(5)P; (8) PtdEth (natural); (9) phosphatidylcholine (natural); (10) d-myo-phosphatidylinositol 3,4-bisphosphate [PtdIns(3,4)P2]; (11) PtdIns(3,5)P2; (12) PtdIns(4,5)P2; (13) PtdIns(4,5)P2 (animal); (14) d-myo-phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3]; (15) phosphatidic acid (natural); (16) PtdSer (natural). All synthetic phosphoinositides are d(+)-sn-1,2-di-O-hexadecanoylglyceryl, 3-O-phospho linked. (B) Spherical liposomes were observed in samples containing extruded lipids without added dynamin A. (C) Networks of tubular structure were observed after addition of dynamin A and GTP-γS to the mixture of extruded lipids. Magnification in (B) and (C) is the same; the scale bar represents 50 nm.
Nucleotide-dependent oligomerization of dynamin A
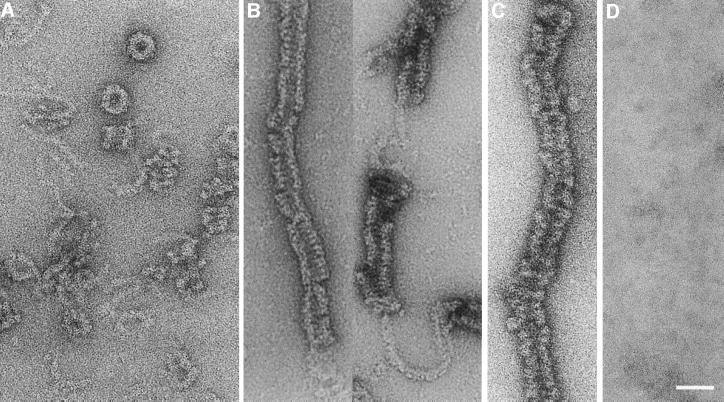
EM was used to study the morphology of negatively stained dynamin A in different nucleotide states. In the absence of guanine nucleotides, dynamin A formed round structures with a diameter of 32 nm and irregular assemblies that consist of short helices with a diameter of ∼30 nm connected to strings of density ∼6 nm in diameter (Figure 3A).
Fig. 3. Micrographs showing the nucleotide-dependent assembly behaviour of purified dynamin A. (A) In the absence of nucleotide, dynamin A forms round structures and loose filamentous structures that are frequently connected to the round structures or short helical assemblies. These structures are stable in 150 mM NaCl. (B) Addition of GppNHp induces the formation of long filamentous assemblies. The irregular nature of the assemblies makes it difficult to identify them unambiguously as either helices or stacks of closed rings; however, a helical nature is suggested by the frequent observation of ∼6 nm diameter density connecting adjacent regions of filamentous assemblies. (C) Long filamentous assemblies were also observed in the presence of GDP–AlFx. (D) Ring-like structures and other forms of assembled dynamin A were rarely observed in the presence of GDP, where unassembled protein appears to dominate. The scale bar represents 50 nm.
Addition of GppNHp to apo-dynamin A induced formation of long filamentous assemblies with a diameter of ∼30 nm (Figure 3B). The irregular nature of the assemblies makes it difficult to unambiguously identify them as either helices or stacks of closed rings; however, a helical nature is suggested by the frequent observation of ∼6 nm diameter density connecting adjacent regions of filamentous assemblies. A similar shift from round structures to filamentous assemblies was also observed after addition of GDP–AlFx and GTP-γS. However, the filamentous assemblies formed in the presence of GDP–AlFx were less regular in appearance (Figure 3C). GDP appeared to destabilize higher oligomeric structures. Ring-like structures and larger assemblies were rarely observed after addition of GDP or extended incubation with GTP (Figure 3D).
Assembly of dynamin A*
Different assembly behaviour was observed when the protein was purified in the presence of ZPCK, which leads to the modification of two cysteine residues. Modified dynamin A (dynamin A*) appeared exclusively to form ring complexes in the absence of nucleotides (Figure 4A). These ring complexes are highly stable and remained the dominant form even in the presence of GDP or GTP (data not shown). Nevertheless, dynamin A* was able to bind and hydrolyse GTP (Figure 1B).
Fig. 4. Micrographs showing nucleotide-free dynamin A* and dynamin A* complexed with GppNHp. Addition of ZPCK in the presence of GTP leads to the modification of cysteine residues C375 and C831. (A) The modified protein exclusively forms ring-like structures in the absence of nucleotide. (B) Ring-like structures and short, regular filamentous assemblies were observed in the presence of GppNHp.
Binding of GppNHp led to the formation of short filamentous assemblies in addition to ring-like structures (Figure 4B), some of which resemble the nucleotide-free ring complexes. To confirm that the GTP analogue GppNHp is not hydrolysed during the incubation prior to fixation, we measured the rate of GppNHp turnover by dynamin A*. After 12 h at 20°C, no hydrolysis of 1 mM GppNHp by 1 µM dynamin A* was detectable, whereas 1 mM GTP was completely hydrolysed with a half-life of 70 min (Figure 1B). Normal dynamin A could be converted to the modified form by incubation with ZPCK and GTP, as determined by electron microscopic examination. Incubation with ZPCK alone had no effect. Mass spectroscopy analysis revealed that the modifications occur at residues C375 and C831 (A.Schlosser, B.Klockow, D.J.Manstein and W.Lehmann, unpublished results).
Structure of the dynamin A* ring complex
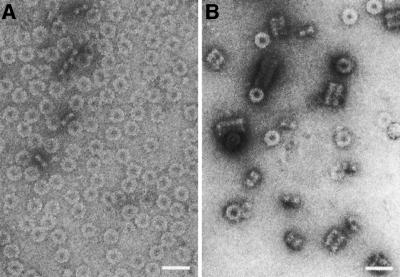
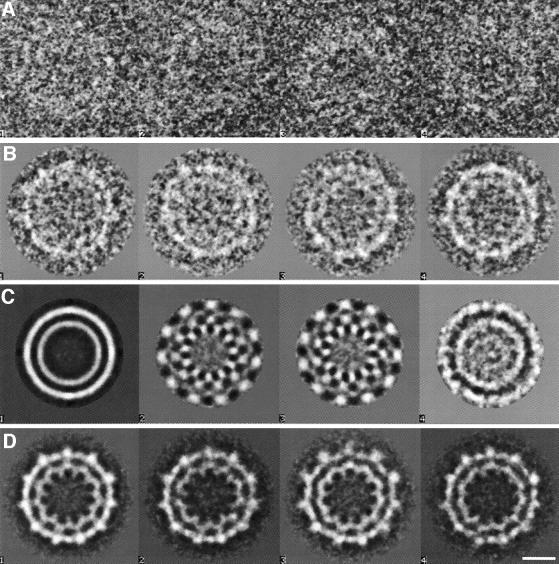
The highly uniform rings formed by dynamin A* were investigated to elucidate the structure of the nucleotide-free ring complex. Eight hundred and ninety-eight top views were subjected to bias-free symmetry analysis (Dube et al., 1993), as described in Materials and methods and shown in Figure 5. Briefly, following translational alignment, rotational symmetry was detected by multivariate statistical analysis (MSA) and eigenvector analysis (van Heel and Frank, 1981). The eigenvectors of a dataset define the main differences between the individual images. As the eigenvectors are themselves images (‘eigenimages’; van Heel 1984, 1989), they reflect these differences, including the symmetry properties of the analysed structure (Dube et al., 1993). The eigenimages obtained for the top views of the dynamin A* ring complex reveal an 11-fold rotational symmetry (Figure 5C). The absence of eigenimages with other rotational symmetries indicates that the composition of the ring complexes is highly uniform. Rotational alignment and classification yielded nine highly similar classes of ring complexes, of which one was picked as a representative reference image. All individual images were aligned onto this reference image and its mirror image by multi-reference analysis. Sub sequent classification led to indistinguishable superpositions (Figure 5D), confirming the uniform composition of the ring complexes and allowing the superimposition of all 898 top views. The resulting averaged image shows the ring complex in projection (Figure 6A) and has a resolution of 25 Å as determined by Fourier ring correlation. The image shows the complex to be composed of two concentric rings, each with 11-fold symmetry. On the outer ring, globular masses are visible with low-density connections to the inner ring. The inner ring forms spike-like structures pointing towards the centre. The maximum diameter of the ring complex is 32 nm and the minimum diameter (at the tip of the spike-like structures) is 12 nm. The diameters of the two concentric rings determined from the rotationally averaged image are 26 nm for the outer and 18 nm for the inner ring. The 11-fold symmetry of both rings suggests that the complex consists of 11n asymmetric units, each containing a segment of the outer and the inner ring.
Fig. 5. Symmetry analysis of 898 top views of the dynamin A* ring complex. (A) Four original images from the data set as picked from the digitized micrograph are shown. (B) Four of the 90 classes obtained after multivariate statistical analysis classification of the centred top views are shown. The two concentric rings and their 11-fold symmetry become noticeable. (C) The first four eigenimages of the analysis are shown. The first one resembles the total average of the data set (van Heel, 1989), the two subsequent eigenimages (identical symmetric wave patterns 90° out of phase) illustrate the most important difference between the individual images, that is a rotational misalignment of an 11-fold symmetric component. No further symmetry is obvious from the following eigenimages, indicating a uniform composition of the ring complexes. (D) The four final classes of aligned images are shown. The lack of significant differences allows all images to be superimposed (see Figure 6A). Same scale in (A)–(D); the scale bar represents 10 nm.
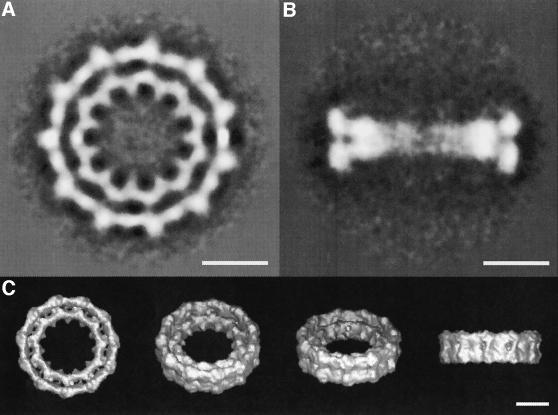
Fig. 6. Averaged images of top and side views lead to the three-dimensional map of the nucleotide-free dynamin A* ring complex. (A) Averaged image of all 898 top views of nucleotide-free dynamin A* ring complexes at 25 Å resolution. The two concentric rings have an 11-fold symmetry. The inner and outer rings have averaged diameters of 18 and 26 nm, respectively. The inner ring forms spike-like structures towards the centre. (B) Averaged image of all 310 side views of nucleotide-free dynamin A* ring complexes. The side view has approximate 2-fold and mirror symmetry, and shows that the ring complex consists of two layers. At the periphery, the outer and inner ring are visible. (C) Three-dimensional map of the nucleotide-free dynamin A* ring complex reconstructed from top and side views. The scale bars in (A)–(C) all represent 10 nm.
To gain three-dimensional structural information, we also analysed 310 side views of single dynamin A* ring complexes, which are orthogonal to the top views described above. The resulting averaged image has low density within the ring, presumably due to conformational flexibility, but at the periphery distinct masses form the outer and inner rings (Figure 6B). The apparent 2-fold symmetry of the side view implies that the entire complex consists of two layers. This is not noticeable in the top view, indicating that the two layers superimpose well when the ring complex is observed along the central axis. To obtain a more detailed view of the molecular organization of the complex, a three-dimensional map of the dynamin A* ring complex was calculated by angular reconstitution, using the top view and 10 classes of side views (Figure 6C). As suggested by the projection structures, the ring complex is organized as a double layer, with each layer containing an 11-fold symmetric inner and outer ring. The entire complex is thus composed of 22 asymmetric units, each forming a segment of the outer and inner ring of one layer. The asymmetric units facing each other in the two layers form spike-like structures and thus appear to form a functional dimer, which may represent the building block of the dynamin A ring complex. When contoured assuming that each asymmetric unit corresponds to a single dynamin A monomer (total mol. wt 22 × 96 kDa = 2.1 MDa), the molecular envelope of the complex exhibits a reasonable packing density (Figure 6C).
Dynamin A* in the presence of GppNHp
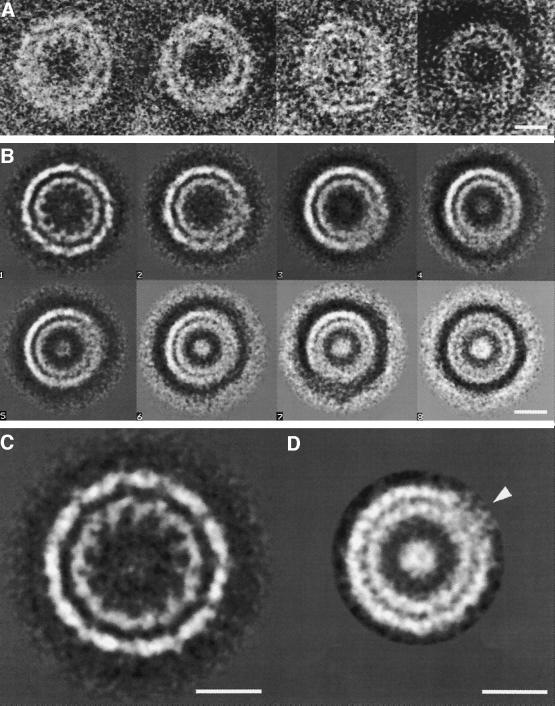
Addition of GppNHp to nucleotide-free dynamin A* induced changes in the diameter of some of the ring complexes and formation of short, tightly packed filamentous assemblies with an inter-ring spacing of ∼10 nm (Figure 4B). To elucidate the conformational changes associated with binding of GppNHp to dynamin A*, 954 top views of ring-like structures were selected for single particle analysis as described above. Even at the level of individual images the ring-like structures showed noticeable diameter variation (Figure 7A). The eigenimage analysis confirmed diameter variations to be the main inter-image variations. After classification, four ring-like structures of different diameters were used together with their mirror images as references to align all images, which were then grouped into eight classes (Figure 7B). The structural images thus obtained differ not only in diameter, but also in the level of structural detail displayed. The ring complex with the largest diameter (class 1) is well defined throughout, but with decreasing diameter (from class 1 to class 8) the distinction between the inner and outer ring diminishes and becomes blurred over an increasing angular segment of the smaller ring-like structures. These gradual changes suggest that the large ring complexes (class 1) can reorganize to form smaller ring-like structures (classes 2–8), or vice versa. As nucleotide-free dynamin A* ring complexes more closely resemble the structure in class 1 than that in class 8, binding of GppNHp appears to induce the formation of small ring-like structures.
Fig. 7. Diameter variation of the dynamin A ring complex in the presence of GppNHp. (A) Four individual images as picked from the micrograph are shown. Note the varying diameters of the displayed ring-like structures. (B) Nine hundred and fifty-four aligned top views of the dynamin A ring complex in the presence of GppNHp are grouped into eight classes by multivariate statistical analysis and classification to visualize the diameter variations. Note that a segment in which inner and outer rings are blurred increases as the diameter decreases. (C) Averaged image of 184 top views of ring complexes with large diameter (Figure 7B, class 1). The inner and outer rings have averaged diameters of 18 and 26 nm, respectively. An 11-fold rotational symmetry was detected by eigenvector analysis. (D) Averaged image of 188 top views of ring complexes with small diameter (Figure 7B, classes 6–8), presumably short, constricted helices (see text). The inner and outer rings have averaged diameters of 15 and 21 nm, respectively. No rotational symmetry was detected by eigenvector analysis, and structural details such as the spike-like structures are not visible. A small segment remains blurry (arrowhead). The scale bars in (A)–(D) all represent 10 nm.
We addressed this hypothesis by separate analysis of large and small ring complexes, using either the 184 images in the class with the largest diameter (Figure 7B, class 1) or the 188 images of ring complexes in the three similar classes with the smallest diameters (Figure 7B, classes 6–8). The resulting averaged image of large diameter ring complexes (Figure 7C) has inner and outer diameters of 26 and 18 nm, respectively, identical to those of the nucleotide-free ring complex. The averaged image of small diameter ring-like structures (Figure 7D) has diameters of 21 and 15 nm, respectively, demonstrating that these assemblies have constricted ∼20% upon GppNHp binding. Neither the averaged image (Figure 7D) nor the eigenvector analysis (data not shown) revealed any rotational symmetry for the small ring-like structures, whereas eigenvector analysis revealed 11-fold symmetry for the large ring complexes (Figure 7C). Moreover, as expected from the small-diameter class averages, in the averaged image of the small ring-like structures the distinction between the inner and outer rings is blurred over a small angular segment of the otherwise well defined averaged image (Figure 7D, arrowhead). These observations indicate that the two averaged images represent differently organized structures and suggest that the large ring complex represents a single well-defined structure, whereas the small ring-like structures are likely to be averaged images of top views through short helices that are structurally heterogeneous. As a result, in the small ring-like structures, rotational symmetry could be blurred and structural details lost during alignment due to imperfect superposition. In addition, the end of a helix might be conformationally labile and thus contribute to blurring upon image averaging. Such conformational variability is indeed observed in some individual images, where the end of the protein strand becomes visible (Figure 7A). Taken together, the analysis of the top views shows a clear reorganization and associated constriction of the dynamin A* ring complex upon binding of GppNHp.
Discussion
Here we provide biochemical and morphological evidence that dynamin A is a fast GTPase, binds to membranes and forms macromolecular assemblies in a nucleotide-dependent fashion similar to those observed for dynamin-1, although dynamin A assemblies display a somewhat higher stability in the presence of salt. In good agreement with work on other dynamin-related proteins, our results support the notion that the PH domain and the PRD are not essential for oligomerization and lipid binding, or can be functionally replaced by other domains. Based on sedimentation experiments with DLP1 (also referred to as DRP1, DVLP, Dymple and HdynIV), Shin et al. (1999) first suggested that dynamin-related proteins lacking both a PH domain and a PRD form structures similar to those formed by dynamin-1. DLP1 and ADL2 were recently shown to assemble in macromolecular structures in a nucleotide-dependent manner (Kim et al., 2001; Smirnova et al., 2001). In addition, DLP1 was shown to bind and tubulate lipid membranes (Yoon et al., 2001). Finally, in the presence of PtdSer, purified dynamin-1 lacking the PRD was shown to form helical tubes that are better ordered than those formed by the full-length protein (Zhang and Hinshaw, 2001).
Detailed analysis shows that the rings and filamentous assemblies formed by dynamin A and dynamin A* resemble those formed by dynamin-1 (Hinshaw and Schmid, 1995; Hinshaw, 1999). The periphery of the side views of the dynamin A* ring complex is strikingly similar to the T-shaped repeat of dynamin-1 bound to lipid (Hinshaw, 1999; Zhang and Hinshaw, 2001). The three-dimensional structure of the complexes of both dynamin A* and dynamin-1 support the idea that the complexes are assembled from dimers. The formation of the macromolecular assemblies from dimers is also in agreement with biochemical studies showing that dynamins form dimers and tetramers in solution (Carr and Hinshaw, 1997; Muhlberg et al., 1997; Binns et al., 1999; Shin et al., 1999). Oligomerization appears to be a general feature of the GTPases of the dynamin family: the Mx proteins exist predominantly as trimers in solution (Melén et al., 1992) and the more distantly related hGBP1 can form monomers, dimers and tetramers depending on its nucleotide state (Prakash et al., 2000). Furthermore, oligomerization to helical arrays has been observed in vitro for murine Mx proteins and phragmoplastin from soybean (Nakayama et al., 1993; Zhang et al., 2000).
The effects of the modification of dynamin A at residues C375 and C831 by the protease inhibitor ZPCK were first noticed when electron micrographs of negatively stained protein that was purified in the presence and absence of ZPCK were compared. Residues C375 and C831 are located in the middle domain and GED, respectively. This suggests a role for these domains in dynamin assembly, consistent with results obtained for the assembly of dynamin-1 (Muhlberg et al., 1997; Okamoto et al., 1999; Smirnova et al., 1999) and DLP1 (Shin et al., 1999). The modifications at these sites appear to favour assembly of rings with zero pitch. As a result, the variable ring-like structures formed by unmodified protein are stabilized as homogeneous, 11-fold symmetric ring complexes of dynamin A* that are well suited to structural analysis.
In general, dynamin A* appears to offer a promising model for structural studies of dynamin function, given the homogeneity of the various molecular assemblies observed and its ability to reproduce important conformational changes of the native protein upon binding of nucleotide triphosphates. The similarity in the diameter and gross morphology of the dynamin A and dynamin A* ring-like structures suggests that the chemical modification only moderately affects the pitch and curvature of the radial packing interactions, permitting formation of a closed, symmetric ring complex. Furthermore, the GTP-dependent conformational changes of dynamin A* (increase in pitch, constriction, tendency to stack) correspond qualitatively to the changes observed in unmodified protein. Consistent with this, dynamin A* displayed a fast rate of GTP-turnover. However, GDP-binding affinity appeared to be greatly reduced by the covalent modification, and electron microscopic visualization showed that the dynamin A* ring complexes persist in the presence of GDP or after complete hydrolysis of GTP (data not shown), in contrast to dynamin A complexes (Figure 3D). Presumably, GDP binding is coupled to an obligate conformational change that disrupts the radial subunit packing interactions in the native protein (Figure 8D). In the dynamin A* ring complex, these packing interactions appear to be more energetically favourable than the GDP binding interaction, rendering the complex resistant to GDP-driven disassembly.
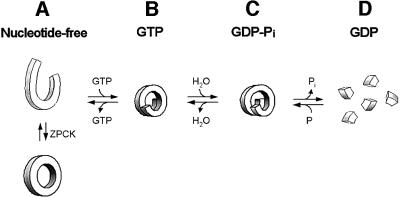
Fig. 8. Model of the mechanochemical cycle of dynamin A. The equilibrium between the different assembly states is highly nucleotide dependent. (A) In the absence of nucleotide, dynamin A forms strings of density ∼6 nm in diameter and round structures with a diameter of 32 nm. Modification by ZPCK stabilizes a uniform ring complex suitable for structural analysis. (B) Addition of GppNHp or GTP induces helix formation. (C) GTP hydrolysis is associated with a conformational change that leads to the disruption of contacts within the dynamin helix and ultimately to the dissociation into dimers in the GDP-bound state (D). The conformational changes upon GTP binding and GTP hydrolysis could cause constriction and stretching of the helix. Phosphate release appears to initiate helix disassembly. An increase in helical pitch has been predicted to occur, based on theoretical considerations (Kozlov, 1999) and was directly observed in the case of dynamin-1 (Stowell et al., 1999; Marks et al., 2001).
Upon addition of GppNHp to dynamin A*, some of the ring complexes undergo conformational changes that appear to involve both ring constriction and an increase in helical pitch, leading to the formation of structurally heterogeneous ring-like structures. This tightening of the ring diameter and/or helix formation requires a reorganization of the ring complex that leads to the loss of its 11-fold symmetry. A similar reorganization is observed for the assembly of tobacco mosaic virus (reviewed in Klug, 1999), in which a ring complex of two concentric rings in two layers, each comprising 17 monomers, reorganizes upon viral RNA binding into a helix of 16.33 monomers per turn. Reorganization of the tobacco mosaic virus assembly is strongly pH- and ionic strength-dependent, leading to the presence of ring complexes, stacks of ring complexes and short helices.
The fact that not all dynamin A* ring complexes undergo the conformational change upon addition of saturating concentrations of GppNHp suggests that the closed ring, zero pitch structure is highly stable and therefore kinetically trapped. This interpretation is consistent with the observation that the rings are not disrupted by concentrations of NaCl (300 mM) known to disrupt dynamin-1 assemblies (Shpetner and Vallee, 1989).
Binding of GppNHp to dynamin A leads to the formation of long but irregular filamentous assemblies. This is in agreement with the reported self-assembly of dynamins in the presence of GTP analogues (Takei et al., 1995, 1998; Carr and Hinshaw, 1997). Addition of GppNHp to dynamin A* also leads to the formation of filamentous structures, indicating that nucleotide binding induces a further conformational change that favours axial packing interactions. The inter-ring spacing is larger than in other nucleotide states (see below). The filament diameters of ∼30 nm suggests that they might be formed, at least in part, from unconstricted ring complexes assembled into stacks. Such stacking could reflect the high stability of the dynamin A* ring complex: the fact that the ring complexes are preformed in the absence of nucleotide could kinetically favour stack formation over helix formation upon addition of GppNHp. These stacks might be a constrained state stabilized by both intra- and inter-ring interaction, whereas single ring complexes are subject to only the intra-ring interaction. The absence of inter-ring interactions allows the single rings to reorganize and constrict, as seen in the top views (Figure 7B). Constriction is also seen in side views of the unmodified protein (Figure 3B). The conformational changes that follow GppNHp binding to dynamin A and dynamin A* appear to stabilize additional dynamin–dynamin interactions, leading to stack or helix formation and reorganization to a more constricted helix. These conformational changes may be reflected in nucleotide-dependent differences in the susceptibility of dynamin A* to proteolysis by trypsin and our observation that the ZPCK-induced modification occurred only in the presence of GTP (data not shown).
Our results add new insight into the mechanism of dynamin action. They show that both mechanisms proposed for mechanochemical action of dynamins, i.e. tightening and stretching of the helix, do occur as part of the dynamin A GTPase cycle and may contribute to membrane fission (Figure 8). Previously, Sweitzer and Hinshaw (1998) demonstrated in vitro that human dynamin-1 can deform lipids to tubes in the absence of nucleotide by self-assembling around the lipid membrane into a helix 50 nm in diameter. Addition of GTP transformed most of those tubes into coated vesicles, but several tubes, constricted to 40 nm, remained. These constricted tubes were interpreted to have undergone GTP hydrolysis and to be precursors of vesicles (Sweitzer and Hinshaw, 1998). In contrast, our results provide evidence that helix tightening may already be triggered by the binding of GTP, i.e. in the absence of hydrolysis (Figure 7).
Since membrane fission is unlikely to be achieved by the 20% constriction we observe upon GppNHp binding, subsequent conformational changes are required (Kozlov, 1999). Indeed, experimental data show that helix formation around lipid tubes is not sufficient for fission, but that GTP hydrolysis is required as well (Takei et al., 1995, 1998; Sweitzer and Hinshaw, 1998; Marks et al., 2001). Consequently, dynamin is expected to sever the bound lipid tube during the conformational change associated with GTP hydrolysis. Theoretical considerations suggest an increase in pitch to be responsible for disruption of the membrane (Kozlov, 1999) (Figure 8C). Helix stretching upon GTP hydrolysis was experimentally observed in vitro for dynamin-1 on artificial lipid nanotubes (Stowell et al., 1999). The use of preformed, rigid nanotubes precludes the observation of constriction and vesiculation, but permitted measurement of a 9 nm increase in pitch (from 11 to 20 nm). For dynamin A*, a distinct increase in inter-ring spacing (from 10 nm to an irregular spacing of ∼12 nm) was observed when the tight stacks in the GppNHp-bound state were compared with the less regular filamentous assemblies in other nucleotide states (data not shown). Similar but structurally less well defined transitions were also observed with the unmodified protein (Figure 3).
Most likely, further details will have to be considered to understand fully the in vivo mechanism of membrane fission, such as changes in lipid composition (Schmidt et al., 1999; Weigert et al., 1999) and lipid–protein interaction. The lipid contact is likely to be mediated by the spike-like structures at the inside of the dynamin A* ring complex, ideally situated to penetrate and perforate the lipid layer upon ring assembly. Experimental data show that dynamin-1 indeed inserts into a lipid layer (Burger et al., 2000). The degree of penetration is most pronounced in the presence of negatively charged lipids (Burger et al., 2000), adding another level of regulation for dynamin action. Taken together, the mechanism of membrane disruption by dynamins appears to combine lipid constriction and even penetration prior to GTP hydrolysis, followed by major conformational changes upon GTP hydrolysis that lead to membrane disruption. This model agrees with a recent theoretical approach suggesting that the combination of tightening and stretching of the dynamin helix plus a change in the spontaneous curvature of the neck membrane leads to the collapse of the neck membrane and subsequent fission (Kozlov, 2001). Dynamin disassembly is expected to occur in the GDP-bound state (Figure 8D), as suggested by the absence of large assemblies in negatively stained electron microscope preparations (Figure 3D). Spontaneous GDP dissociation and subsequent GTP binding reactivates dynamin to actively contribute to membrane fission.
The proposed model can now be tested in D.discoideum. Cell lines in which the dymA gene was deleted by homologous recombination have been generated (Wienke et al., 1999). These cell lines will be used for complementation studies and the production of mutant forms of the protein for further functional and structural analysis in vitro. Elucidation of the molecular contacts in dynamin assemblies will be crucial for understanding the role of the protein in vesicle formation. We are now working towards this goal by combining a three-dimensional reconstruction of the highly ordered dynamin A* ring complex with the X-ray crystallographic structure of the dynamin A monomer. A 2.3 Å resolution structure of the dynamin A GTPase domain has recently been determined (Niemann et al., 2001).
Materials and methods
Reagents
Nucleotides and buffer reagents were purchased from Sigma (Deisenhofen, Germany), lipids from Waco (Osaka, Japan) or Avanti Polar Lipids (Alabaster), and PIP-strips™ from Echelon Research Laboratories (Salt Lake City, UT).
Protein expression and purification
Dynamin A overproducing cell line HDM21 was grown in 5 l flasks containing 2.5 l HL-5C. The flasks were incubated on gyratory shakers at 180 rpm at 21°C. Cells were harvested at a density of ∼4–6 × 106 cells/ml by centrifugation for 7 min at 2700 rpm in a Beckman J-6 centrifuge and washed once in phosphate buffered saline. The wet weight of the resulting cell pellet was determined. Typically, 30–35 g were obtained from a 15 l shaking culture. The cells were resuspended in 15 vols Tris–EDTA buffer (TEB) containing 50 mM Tris–HCl pH 8, 2.5 mM EDTA, 1 mM DTT, 5 mM benzamidine, 2 mM N-CBZ-Pro-Ala, 2 µg/ml pepstatin A, 100 µg/ml N-tosyl-l-lysine chloromethyl ketone (TLCK), 80 µg/ml N-tosyl-l-argininemethylester (TAME), 5 µg/ml leupeptin, 0.1 mM phenylmethylsulfonyl fluoride. Cell lysis was achieved by the addition of 15 vols of TEB containing 1% Triton X-100 and incubation on ice for 30 min. After centrifugation (30 000 g, 1 h), the recombinant protein remained in the pellet. The pellet was washed with TEB and centrifuged as before. The recombinant protein was released into the supernatant by extraction of the resulting pellet with 1.5 vols TEB buffer containing 5 mM GTP, 7 mM MgCl2 and 300 mM NaCl. After centrifugation (20 000 g, 20 min), the supernatant was dialysed against TEB containing 50 mM NaCl and purified by use of a column packed with Fractogel TSK DEAE-650(S) (Merck, Darmstadt, Germany) connected to an FPLC system. The protein had a purity of >95% judged from Coomassie-stained SDS polyacrylamide gels. Dynamin A* was purified using the same protocol, except for the addition of 100 µg/ml ZPCK to the lysis and GTP extraction buffers and the absence of DTT in these buffers.
Preparation of protein-free liposomes
Small unilamellar liposomes were prepared as described by Otter-Nilsson et al. (1999). The lipid mixture consisted of 41 mol% phosphatidyl choline, 18 mol% phosphatidyl ethanolamine, 13.5 mol% phosphatidylinositol, 13.5 mol% phosphatidic acid, 5 mol% PtdSer, 4.5 mol% phosphatidylinositol phosphate (PtdInsP) and 4.5 mol% phosphatidylinositol bisphosphate (PtdInsP2). Lipid solutions were transferred to 10 ml glass vials and dried under a gentle stream of nitrogen. Lipid films were resuspended by vortexing in buffer (20 mM HEPES–KOH pH 7.2, 100 mM KCl, 1 mM MgCl2) to a final concentration of 1.0 mM. After 10 rapid freeze–thaw cycles (30 s in liquid nitrogen, 30 s in a 37°C water bath and 5 s vortexing), liposomes with an average size of 400 nm were formed by extrusion with a handheld extrusion apparatus (Lipofast, Avestin) through 400 nm polycarbonate filters (21 passages). Freshly prepared liposomes were stored at 4°C and used within 24 h.
Transmission electron microscopy
One micromolar (∼0.1 mg/ml) nucleotide-free dynamin A in buffer EM150 (50 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM MgCl2) was incubated with either EM150, 1 mM GppNHp, or 1 mM GDP + 1 mM AlCl3 + 10 mM NaF (for generation of AlFx) or 1 mM GDP for 10 min at room temperature or 20 h on ice. Alternatively, 1 µM dynamin A in buffer EM150 was incubated with 1 mM GTP until GTP was completely hydrolysed to GDP and was subsequently incubated with buffer or 1 mM AlCl3 + 10 mM NaF. One millimolar nucleotide is >10-fold higher than KM for GTP (68 µM). We applied a 5 µl sample to freshly glow-discharged carbon-coated grids, stained with 1% uranyl acetate, and observed the protein at 120 kV at 51 200-fold magnification in a Philips Tecnai F20 or at 80 kV at 46 400-fold magnification on a Philips EM 400T. Electron micrographs were recorded under low-dose conditions at an underfocus of ∼0.6 µm and their quality was assessed by optical diffraction.
Image analysis
Micrographs were digitized on a Zeiss SCAI scanner with a 7 µm pixel size. Groups of 2 × 2 pixels were averaged, corresponding to a pixel size of 2.7 and 3.0 Å on the specimen. Images were analysed using the IMAGIC-5 software system (Imagic Science Software GmbH, Berlin, Germany) (van Heel et al., 1996).
Well-defined particles were selected interactively, band-pass filtered and aligned using a reference-free alignment as described by Dube et al. (1993). The images were centred by translational alignment relative to a rotationally averaged sum and subjected to multivariate statistical analysis (MSA) classification with 10 images/class to check homogeneity and discard bad images (to increase the quality of the symmetry analysis, the datasets were enlarged by adding nine rotationally randomized images per original image). The eigenimages obtained during MSA classification were analysed to detect rotational symmetry. The class averages were rotationally aligned against each other and classified by MSA into four to 20 classes to obtain the references. When 11-fold rotational symmetry was present, the reference was 11-fold rotational averaged. All individual images were aligned onto the references and their mirror images by multi-reference alignment. This step was followed by MSA classification. When the obtained classes were similar, the total sum was used as the final averaged image, without imposing rotational symmetry.
The three-dimensional structure was calculated using the averaged top view and 10 classes of side views and imposing 11-fold rotational symmetry. Euler angle assignment was done by angular reconstitution (Schatz et al., 1995; Serysheva et al., 1995; van Heel et al., 1996) using the IMAGIC-5 software system (Imagic Science Software GmbH).
Sedimentation assay
Lipid vesicles were made by evaporating chloroform in a speedvac and resuspending the dry lipids in water at 1 mg/ml by extensive sonication. Two micromolar (∼0.2 mg/ml) dynamin A in EM150 buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM MgCl2) was incubated with either 1 mM nucleotide (GDP, GDP–AlFx, GTP, GppNHp or GTP-γS) or 0.1 mg/ml lipid vesicles (PtdSer or PtdEth) for 10 min at room temperature. Samples were centrifuged at 100 000 g (48 000 rpm in a Beckman RP100 AT3 rotor) for 10 min at 4°C. Supernatant (25 µl) and pellet (resuspended in 25 µl EM150) were analysed by SDS–PAGE and stained with Coomassie Blue.
GTPase assays
All GTPase assays were performed at 20°C. In a quenched flow apparatus (Kintec) 2–80 µM dynamin A in buffer EM150 was rapidly mixed with 800 µM GTP including 20 nCi/ml [γ-32P]GTP (Amersham). After reaching steady state, the reaction was stopped at different time points by quenching with 1 M HClO4. Following neutralization with 8 M KOAc, samples were centrifuged in a table centrifuge for 2 min at maximum speed and 1.5 µl of the supernatant was applied to a polyethyleneimine cellulose thin-layer chromatography layer (Cellulose MN 300 PEI, Macherey-Nagel). Separation of [γ-32P]phosphate and [γ-32P]GTP was achieved using 350 mM KH2PO4 pH 3.4, as mobile phase. [γ-32P]phosphate and [γ-32P]GTP were quantified by a phosphoimager (BAS 650, Fuji). In addition, analytical separation of nucleotides was carried out by reversed-phase HPLC on a C18-column in 100 mM K2HPO4/KH2PO4 pH 6.5, 10 mM tetrabutylammonium bromide and 7.5% acetonitrile as described previously (Tucker et al., 1986), using a UV absorption detector at 254 nm.
Acknowledgments
Acknowledgements
We thank Helga Clasen for expert technical assistance, Marin van Heel for helpful advice with single particle analysis, B.Brügger and F.T.Wieland for help and advice with liposome preparation and lipid interaction assays, A.Schlosser and W.Lehmann for mass spectrometry, F.Jon Kull for stimulating discussion and Kenneth C.Holmes for support. The Boehringer Ingelheim Fonds supported B.K. and H.H.N.
References
- Binns D.D., Barylko,B., Grichine,N., Atkinson,M.A., Helms,M.K., Jameson,D.M., Eccleston,J.F. and Albanesi,J.P. (1999) Correlation between self-association modes and GTPase activation of dynamin. J. Protein Chem., 18, 277–290. [DOI] [PubMed] [Google Scholar]
- Burger K.N.J., Demel,R.A., Schmid,S.L. and de Kruijff,B. (2000) Dynamin is membrane-active: lipid insertion is induced by phosphoinositides and phosphatidic acid. Biochemistry, 39, 12485–12493. [DOI] [PubMed] [Google Scholar]
- Carr J.F. and Hinshaw,J.E. (1997) Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and γ-phosphate analogues. J. Biol. Chem., 272, 28030–28035. [DOI] [PubMed] [Google Scholar]
- Dube P., Tavares,P., Lurz,R. and van Heel,M. (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J., 12, 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E., van der Kaay,J., Downes,C.P. and Smythe,E. (2001) The role of dynamin and its binding partners in coated pit invagination and scission. J. Cell Biol., 152, 309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E. (1999) Dynamin spirals. Curr. Opin. Struct. Biol., 9, 260–267. [DOI] [PubMed] [Google Scholar]
- Hinshaw J.E. and Schmid,S.L. (1995) Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature, 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Jeong M.J., Yoo,J., Lee,S.S., Lee,K.I., Cho,A., Kwon,B.M., Moon,M.J., Park,Y.M. and Han,M.Y. (2001) Increased GTP-binding to dynamin II does not stimulate receptor-mediated endocytosis. Biochem. Biophys. Res. Commun., 283, 136–142. [DOI] [PubMed] [Google Scholar]
- Katti M.V., Sami-Subbu,R., Ranjekar,P.K. and Gupta,V.S. (2000) Amino acid repeat patterns in protein sequences: their diversity and structural–functional implications. Protein Sci., 9, 1203–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.W., Park,D.S., Park,S.C., Kim,S.H., Cheong,G.W. and Hwang,I. (2001) Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol., 127, 1243–1255. [PMC free article] [PubMed] [Google Scholar]
- Klug A. (1999) The tobacco mosaic virus particle: structure and assembly. Philos. Trans. R. Soc. Lond. B Biol. Sci., 354, 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T. and Ikeda,K. (1983) Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J. Cell Biol., 97, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M.M. (1999) Dynamin: possible mechanism of “Pinchase” action. Biophys. J., 77, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M.M. (2001) Fission of biological membranes: interplay between dynamin and lipids. Traffic, 2, 51–65. [DOI] [PubMed] [Google Scholar]
- Marks B., Stowell,M.H.B., Vallis,Y., Mills,I.G., Gibson,A., Hopkins,C.R. and McMahon,H.T. (2001) GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature, 410, 231–235. [DOI] [PubMed] [Google Scholar]
- McNiven M.A., Cao,H., Pitts,K.R. and Yoon,Y. (2000) The dynamin family of mechanoenzymes: pinching in new places. Trends Biochem. Sci., 25, 115–120. [DOI] [PubMed] [Google Scholar]
- Melén K., Ronni,T., Broni,B., Krug,R.M., von Bonsdorff,C.-H. and Julkunen,L. (1992) Interferon-induced Mx proteins form oligomers and contain a putative leucine zipper. J. Biol. Chem., 267, 25898–25907. [PubMed] [Google Scholar]
- Muhlberg A.B., Warnock,D.E. and Schmid,S.L. (1997) Domain structure and intramolecular regulation of dynamin GTPase. EMBO J., 16, 6676–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Yazaki,K., Kusano,A., Nagata,K., Hanai,N. and Ishihama,A. (1993) Structure of mouse Mx1 protein. Molecular assembly and GTP-dependent conformational change. J. Biol. Chem., 268, 15033–15038. [PubMed] [Google Scholar]
- Niemann H.H., Knetsch,M.L.W., Scherer,A., Manstein,D.J. and Kull,F.J. (2001) Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J., 20, 5813–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto P.M., Tripet,B., Litowski,J., Hodges,R.S. and Vallee,R.B. (1999) Multiple distinct coiled-coils are involved in dynamin self-assembly. J. Biol. Chem., 274, 10277–10286. [DOI] [PubMed] [Google Scholar]
- Otter-Nilsson M., Hendriks,R., Pecheur-Huet,E.I., Hoekstra,D. and Nilsson,T. (1999) Cytosolic ATPases, p97 and NSF, are sufficient to mediate rapid membrane fusion. EMBO J., 18, 2074–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash B., Praefcke,G.J., Renault,L., Wittinghofer,A. and Herrmann,C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature, 403, 567–571. [DOI] [PubMed] [Google Scholar]
- Schatz M., Orlova,E.V., Dube,P., Jager,J. and van Heel,M. (1995) Structure of Lumbricus terrestris hemoglobin at 30 Å resolution determined using angular reconstitution. J. Struct. Biol., 114, 28–40. [DOI] [PubMed] [Google Scholar]
- Schmid S.L., McNiven,M.A. and De Camilli,P. (1998) Dynamin and its partners: a progress report. Curr. Opin. Cell Biol., 10, 504–512. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wolde,M., Thiele,C., Fest,W., Kratzin,H., Podtelejnikov,A.V., Witke,W., Huttner,W.B. and Soling,H.D. (1999) Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature, 401, 133–141. [DOI] [PubMed] [Google Scholar]
- Serysheva I.I., Orlova,E.V., Chiu,W., Sherman,M.B., Hamilton,S.L. and van Heel,M. (1995) Electron cryomicroscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel. Nature Struct. Biol., 2, 18–24. [DOI] [PubMed] [Google Scholar]
- Sever S., Muhlberg,A.B. and Schmid,S.L. (1999) Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature, 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Sever S., Damke,H. and Schmid,S.L. (2000) Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol., 150, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.-W., Takatsu,H., Mukai,H., Munekata,E., Murakami,K. and Nakayama,K. (1999) Intermolecular and interdomain interactions of a dynamin-related GTP-binding protein, Dnm1p/Vps1p-like protein. J. Biol. Chem., 274, 2780–2785. [DOI] [PubMed] [Google Scholar]
- Shpetner H.S. and Vallee,R.B. (1989) Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell, 59, 421–432. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Shurland,D.L., Newman-Smith,E.D., Pishvaee,B. and van der Bliek,A.M. (1999) A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem., 274, 14942–14947. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic,L., Shurland,D.L. and van der Bliek,A.M. (2001) Dynamin-related protein DRP1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell, 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell M.H.B., Marks,B., Wigge,P. and McMahon,H.T. (1999) Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nature Cell Biol., 1, 27–32. [DOI] [PubMed] [Google Scholar]
- Subirana J.A. and Palau,J. (1999) Structural features of single amino acid repeats in proteins. FEBS Lett., 448, 1–3. [DOI] [PubMed] [Google Scholar]
- Sweitzer S.M. and Hinshaw,J.E. (1998) Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell, 93, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Takei K., McPherson,P.S., Schmid,S.L. and De Camilli,P. (1995) Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature, 374, 186–190. [DOI] [PubMed] [Google Scholar]
- Takei K., Haucke,V., Slepnev,V., Farsad,K., Salazar,M., Chen,H. and De Camilli,P. (1998) Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell, 94, 131–141. [DOI] [PubMed] [Google Scholar]
- Takei K., Slepnev,V., Haucke,V., Farsad,K. and De Camilli,P. (1999) Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nature Cell Biol., 1, 33–39. [DOI] [PubMed] [Google Scholar]
- Tucker J., Sczakiel,G., Feuerstein,J., John,J., Goody,R.S. and Wittinghofer,A. (1986) Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J., 5, 1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A.M. (1999) Functional diversity in the dynamin family. Trends Cell Biol., 9, 96–102. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1984) Multivariate statistical classification of noisy images (randomly oriented biological macromolecules). Ultramicroscopy, 13, 165–183. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1989) Classification of very large electron microscopical image data sets. Optik, 82, 114–126. [Google Scholar]
- van Heel M. and Frank,J. (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy, 6, 187–194. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz,G. and Orlova,E.V. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Weigert R. et al. (1999) CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature, 402, 429–433. [DOI] [PubMed] [Google Scholar]
- Wienke D.C., Knetsch,M.L.W., Neuhaus,E.M., Reedy,M.C. and Manstein,D.J. (1999) Disruption of a dynamin homologue affects endocytosis, organelle morphology and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell, 10, 225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Pitts,K.R. and McNiven,M.A. (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell, 12, 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. and Hinshaw,J.E. (2001) Three-dimensional reconstruction of dynamin in the constricted state. Nature Cell Biol., 3, 922–926. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hong,Z. and Verma,D.P. (2000) Phragmoplastin polymerizes into spiral coiled structures via intermolecular interaction of two self-assembly domains. J. Biol. Chem., 275, 8779–8784. [DOI] [PubMed] [Google Scholar]