Abstract
The biological activity of the soluble form of the Notch ligand (sNL) and requirement of the intracellular domain (ICD) of the Notch ligand have been debated. Here we show that soluble Delta1 (sD1) activates Notch2 (N2), but much more weakly than full-length Delta1 (fD1). Furthermore, tracing the N2 molecule after sD1 stimulation revealed that sD1 has a defect in the cleavage releasing ICD of N2 (intracellular cleavage), although it triggers cleavage in the extracellular domain of N2. This represents the molecular basis of the lower activity of sD1 and suggests the presence of an unknown mechanism regulating activation of the intracellular cleavage. The fact that Delta1 lacking its ICD (D1ΔICD) exhibits the phenotype similar to that exhibited by sD1 indicates that the ICD of D1 (D1ICD) is involved in such an as yet unknown mechanism. Furthermore, the findings that D1ΔICD acts in a dominant-negative fashion against fD1 and that the signal-transducing activity of sD1 is enhanced by antibody-mediated cross-linking suggest that the multi merization of Delta1 mediated by D1ICD may be required for activation of the N2 intracellular cleavage.
Keywords: extracellular cleavage/intracellular cleavage/multimerization/Notch/Notch ligand
Introduction
The Notch family of genes encodes transmembrane receptors that are involved in the cell fate decision in vertebrates and invertebrates (Weinmaster, 1997; Greenwald, 1998; Artavanis-Tsakonas, 1999). In mammals, multiple Notch homologs have been identified, including Notch1 to Notch4 (Ellisen et al., 1991; Weinmaster et al., 1991, 1992; Kopan and Weitraud, 1993; Lardelli et al., 1994; Uyttendaele et al., 1996). The extracellular region comprises 29–36 epidermal growth factor (EGF)-like repeats and three copies of a Lin-12/Notch/Glp motif. The intracellular region contains cdc10/Ankyrin repeats and a PEST-containing domain. The Notch receptors are initially synthesized as ∼300 kDa proteins, which are then proteolytically processed in the Golgi apparatus into an extracellular subunit (NEC) containing multiple EGF repeats and lin-12/Notch repeats (Blaumueller et al., 1997; Logeat et al., 1998), and a single-pass transmembrane subunit (NTM) containing a short extracellular tail and an intracellular domain (ICD; NICD). These subunits are reassembled in the trans-Golgi network and are presented as a heterodimeric, mature receptor at the cell surface (Blaumueller et al., 1997). The lin-12/Notch repeats and Ca2+ ion are involved in maintaining the heterodimeric complex of NEC and NTM (Rand et al., 2000).
Binding of a Notch ligand (NL) to NEC triggers cleavage of Notch, releasing NICD from the cell membrane, which is then translocated into the nucleus to activate transcription of target genes in cooperation with RBP-Jκ (Kopan et al., 1996; Chan and Jan, 1998; Jarriault et al., 1998; Schroeter et al., 1998; Struhl and Adachi, 1998; Shimizu et al., 2000). This cleavage is mediated by a presenilin-containing complex and occurs within the transmembrane domain of Notch (intracellular cleavage) (De Strooper et al., 1999; Struhl and Greenwald, 1999; Ye et al., 1999). It has recently been proposed that prior to this cleavage, an additional cleavage at the extracellular domain of NTM occurs in a ligand-dependent manner (Brou et al., 2000; Mumm et al., 2000) and that the extracellular cleavage autonomously promotes intracellular cleavage (Mumm et al., 2000). However, these proposals were based on experiments using Notch1 (N1) proteins with most of the extracellular domain truncated, or experiments using a partial peptide of N1. Therefore, the relationships between ligand stimulation and cleavage of the extracellular region of the native Notch protein, and between ligand-induced extracellular cleavage and subsequent intracellular cleavage, have not been fully addressed.
Delta and Serrate (Jagged), comprising a Delta/Serrate/Lag-2 motif, tandem EGF repeats and a short ICD, are known to be ligands for the Notch receptor. As a natural protein in vivo, Drosophila Delta exists in both the transmembrane and soluble forms (Klueg et al., 1998). It has recently been proposed that the soluble form of Delta is generated by Kuzbanian, a metalloprotease of the ADAM family (Qi et al., 1999). Results of examinations of the biological activity of the soluble Notch ligands have been controversial. Whereas all experiments using cell-culture systems have shown that they behave as agonists (Li et al., 1998; Qi et al., 1999; Han et al., 2000; Karanu et al., 2000; Morrison et al., 2000), in vivo experiments have demonstrated that soluble Delta and Serrate act as antagonists (Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997). It remains to be elucidated why such contradictory conclusions are drawn. To explain the discrepancy, the difference in the activity between the soluble and full-length forms should be clarified.
In the present study, we show that the signal-transducing activity of the soluble form of Delta(-like-)1 (sD1) for Notch2 (N2) is obviously lower than that of full-length Delta1 (fD1), and that in coexistence with fD1 it inhibits the fD1-triggered N2 signal. This implies that sD1 is a partial agonist, while fD1 is a full agonist. Furthermore, we demonstrate the molecular basis of the impaired signal-transducing activity of sD1; it triggers cleavage of the extracellular domain of N2TM, but promotes the cleavage step that releases N2ICD only very little. This indicates that, although the extracellular domain of NL alone is sufficient for extracellular cleavage, intracellular cleavage requires some other domain of NL, and that extracellular cleavage does not autonomously promote intracellular cleavage, suggesting the existence of an unknown mechanism that regulates the activation of the intracellular cleavage. Experiments using Delta1 without the ICD (D1ΔICD) demonstrate that NLICD is important for the intracellular cleavage. Furthermore, the findings that D1ΔICD acts as a dominant-negative molecule against fD1 when they coexist and that the signal-transducing activity of sD1–Fc (sD1 fused to hIgG Fc portion) is enhanced by the addition of anti-Fc antibody suggest that oligomerization of NL is involved in Notch signaling.
Results
Lower signal-transducing activity of sD1
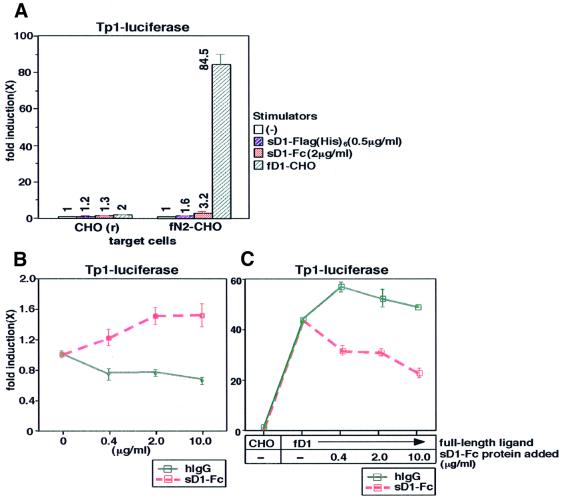
To define a biological activity of a soluble form of Notch ligand (sNL), we assessed the signal-transducing activity of mouse sD1 encompassing the entire extracellular region by comparing it with that of the full-length form in a transient reporter assay with CHO(r) cells overexpressing mouse full-length N2 (fN2-CHO), which is a highly sensitive assessment system for N2 signaling. Results showed that both Fc-fused and Flag(His)6-tagged sD1 proteins [sD1–Fc and sD1-Flag(His)6] activated the transcription of a reporter gene driven by the RBP-Jκ-responsive promoter, TP-1 (Figure 1A and B), but the transcriptional activity was obviously lower than that of fD1 [represented by the stimulation with CHO(r) expressing fD1 (fD1-CHO)] (Figure 1A). On the other hand, in coexistence with fD1, sD1 inhibited the fD1-mediated N2 activation compared with control hIgG (Figure 1C). These data indicate that sNL is a partial agonist, while full-length NL (fNL) is a full agonist.
Fig. 1. Lower signal-transducing activity of soluble Delta1 protein. (A) Comparison of signal-transducing activity of sD1–Fc, sD1-Flag(His)6 and fD1. A transient reporter assay with a TP1-luciferase reporter plasmid, pGa981-6, was performed using fN2-CHO cells. Following transfection of pGa981-6 into fN2-CHO, sD1–Fc, sD1-Flag(His)6 or fD1-CHO was added to the transfected cells. Fold induction of the luciferase activity for each sample (mean of triplet measurements with standard deviation) was calculated against the control. The values are also shown in the graph. (B) N2-mediated transcriptional activation by sD1–Fc at increasing concentrations. Various concentrations of sD1–Fc were added to fN2-CHO cells transfected with pGa981-6. (C) The inhibitory effect of sD1–Fc on fD1-induced N2 signaling. fD1-CHO cells and sD1–Fc proteins at various concentrations were added simultaneously to the pGa981-6-transfected fN2-CHO cells. The same concentration of hIgG was added as a control.
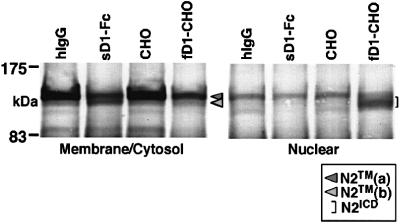
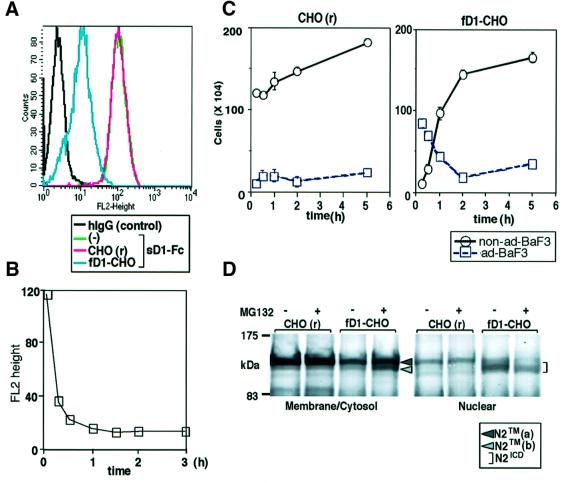
We further evaluated the difference in the signal-transducing activity between the two molecules from another viewpoint, i.e. the nuclear accumulation of N2ICD. It is generally accepted that the nuclear accumulation of NICD generated by cleavage within the transmembrane domain of the Notch receptor after fNL stimulation is associated with activation of the transcription of downstream genes in Notch signaling. To evaluate the cleavage and nuclear accumulation in a serial manner, we used BaF3 cells capable of displaying these two events following stimulation with fD1. As previously reported, stimulation with fD1 decreased the amount of N2TM in the membrane/cytosol fraction [designated N2TM(a); Figure 2] and, instead, N2-derived fragments representing N2ICD were accumulated in the nuclear-rich fraction (Shimizu et al., 2000). In contrast, the stimulation with sD1–Fc did not result in detectable N2ICD in the nuclear-rich fraction, although it also reduced the amount of N2TM(a) in the membrane/cytosol fraction. Instead, a new band representing a protein smaller than N2TM(a) emerged in the membrane/cytosol fraction [designated N2TM(b); Figure 2]. The fact that hardly any N2ICD was generated after stimulation with sD1–Fc was compatible with the results of reporter assays (Figure 1).
Fig. 2. N2 molecule traced after sD1–Fc and fD1 stimulations. BaF3 was stimulated for 1.5 h under the conditions indicated in the figure and then separated into membrane/cytosol-rich and nucleus-rich fractions. In each fraction, N2 fragments containing an ICD were analyzed by western blot analysis using the bhN6 antibody after immunoprecipitation with an anti-N2 polyclonal antibody.
sD1 has a defect in the cleavage required for release of N2ICD, although it can trigger the extracellular cleavage of N2
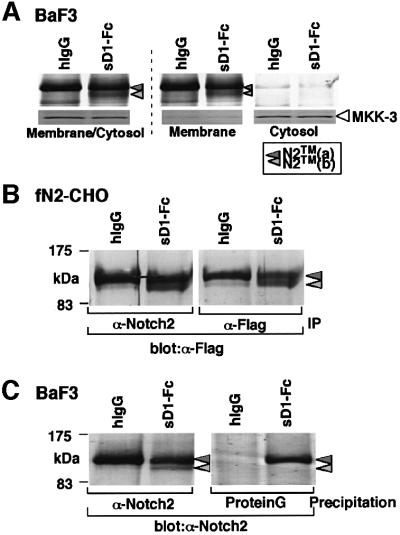
To understand better the lower signal-transducing activity of sD1, we then characterized N2TM(b) generated by sD1 stimulation, which was scarcely seen after fD1 stimulation (Figure 2). The decrease in the amount of N2TM(a) and the appearance of N2TM(b) in the membrane/cytosol fraction after stimulation with sD1–Fc (Figure 2) indicated that N2TM(b) represented a molecule derived from N2TM(a). A further fractionation of the membrane/cytosol fraction demonstrated that N2TM(b) and N2TM(a) were present in the membrane but not in the cytosol fraction (Figure 3A), suggesting that N2TM(b) was a membrane-associated molecule lacking either the N- or the C-terminal tail of N2TM(a). To determine which side of N2TM(a) was cleaved to generate the N2TM(b) fragment, we performed the two experiments. In the first, we used fN2-CHO(r), [CHO(r) with exogenous fN2 tagged with a Flag sequence at the C-terminus] to investigate whether the Flag tag remained in N2TM(b) generated after sD1–Fc stimulation. The result was that the anti-Flag antibody detected N2TM(b) (Figure 3B), indicating that N2TM(b) lacks the N-terminus but not the C-terminus of N2TM(a). In the second experiment, we assessed whether N2TM(b) was coprecipitated with sD1–Fc. In a previous report, we described that N2TM(a) is precipitated with sD1–Fc (Shimizu et al., 2000). If the cleavage after sD1 stimulation occurs within the short extracellular domain of N2TM(a), sD1–Fc-bound N2EC probably loses the association with N2TM(b), and thus N2TM(b) is not coprecipitated with sD1–Fc. As expected, sD1–Fc coprecipitated only N2TM(a) and not N2TM(b) (Figure 3C). This result also suggests that N2TM(b) was generated from N2TM(a) by the cleavage in the juxtamembrane portion of the extracellular region (see Figure 7).
Fig. 3. Characterization of the N2TM(b) fragment induced by sD1–Fc-stimulation. (A) To determine whether N2TM(b) is a transmembrane protein, membrane/cytosol-rich fraction prepared from BaF3 after the sD1–Fc stimulation was then separated into membrane and cytosol fractions. N2 proteins in each fraction was subjected to western blot after immunoprecipitation with an anti-N2 polyclonal antibody. As a control for correct fractionation of membrane and cytosol franctions, an antibody against MKK3, MAP kinase, was used for each fraction in western blot analysis. (B) Generation of N2TM(b) fragment containing a Flag(His)6 tag at the C-terminus. fN2-CHO [CHO(r) with exogenous N2 with a Flag(His)6 tag at the C-terminus] was incubated in the presence of either sD1–Fc or hIgG at 6.7 nM. After 1.5 h, the stimulated cells were collected and solubilized in a TNE buffer. The cell lysates were precipitated with an anti-Flag monoclonal (M2) or an anti-N2 polyclonal antibody. The precipitates were analyzed by western blot with the M2 antibody. IP, immunoprecipitation. (C) Co-precipitation analysis. BaF3 was incubated in RPMI medium containing sD1–Fc or hIgG at 6.7 nM for 1.5 h, then subjected to a cross-linking reaction to form the binding complex of sD1–Fc and N2. Following the reaction, the BaF3 lysates were divided into two aliquots. One was precipitated with an anti-N2 polyclonal antibody to identify N2 protein fragments. To precipitate sD1–Fc-containing complex, protein G beads were added directly to the other. These precipitates were analyzed by western blot with the bhN6 antibody.
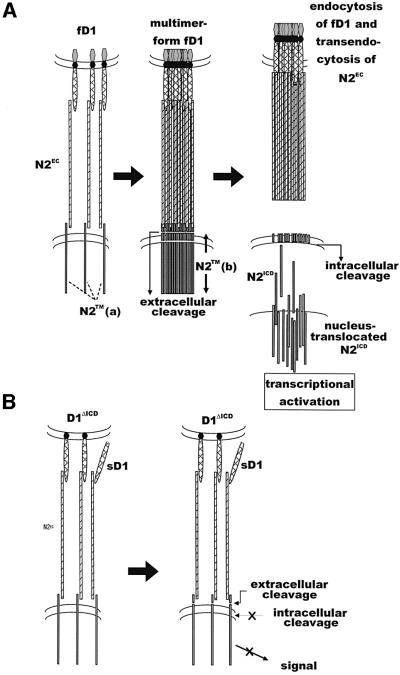
Fig. 7. Scheme of the ligand-induced cleavage of Notch receptor. (A) Upon binding to the Notch receptor, fNL forms appropriate homomultimers, leading to the intracellular cleavage of Notch in addition to the extracellular cleavage. Cleaved NICD translocates into the nucleus. Endocytosis of NL and NEC occurs at some time point during the series of process. (B) NLΔICD or sNL can bind to the Notch receptor and cleave Notch at the extracellular site, but do not form appropriate multimers or excute the intracellular cleavage of Notch, resulting in the failure of the Notch activation.
We then investigated whether the same cleavage occurred during the process of fD1-mediated N2 signaling, to verify that N2TM(b) generated by sD1 was not an artifact. The amount of sD1–Fc binding to BaF3 was significantly reduced when the binding assay was performed after the co-culture of BaF3 with fD1-CHO, as compared with the co-culture with control CHO(r) (Figure 4A). A time-course analysis showed that the reduction in sD1–Fc binding started within 15 min and reached a plateau 1.5 h from the initiation of the co-culture (Figure 4B). During the co-culture, we observed that BaF3 cells, which previously adhered to fD1-CHO within 10 min, were detached from it in a time-dependent fashion (Figure 4C). One possible explanation for these phenomena is that the fD1-induced N2 extracellular cleavage results in the dissociation of N2EC together with the bound fD1 molecule from N2TM, which then results in the reduction in fD1-bindable N2 receptors on BaF3 cell surface (see Figure 7).
Fig. 4. Involvement of extracellular cleavage in fD1-mediated N2 activation. (A) Reduction in the amount of sD1–Fc binding to BaF3 after co-culture with fD1-CHO. BaF3 cells were collected at 1.5 h after co-culture with either CHO(r) or fD1-CHO. Cell-binding assay using sD1–Fc at 6.7 nM was performed for BaF3 cells recovered from the co-culture. (B) Time-course analysis of binding of sD1–Fc to BaF3 after co-culture with fD1-CHO. BaF3 cells co-cultured for the times indicated in the figure were subjected to cell-binding assays. The extent of fluorescence brightness giving the highest frequency (y-axis) was plotted against time (x-axis). (C) Time-dependent detachment of BaF3 from fD1-CHO. The time-course of the number of detached BaF3 cells was recorded in a cell–cell association assay. ad-BaF3, BaF3 that adhered to CHO cells; non-ad-BaF3, BaF3 that did not adhere to CHO cells. (D) Relationship between extracellular cleavage and nuclear transport of N2ICD. MG-132, an inhibitor of cleavage for release of NICD, was added to a co-culture system of BaF3 and fD1-CHO at a final concentration of 25 µM. After 1.5 h of co-culture, the BaF3 cells were collected and separated into membrane/cytosol-rich and nucleus-rich fractions. In each fraction, N2 fragments containing an ICD were analyzed by western blot using the bhN6 antibody after immunoprecipitation.
To obtain more direct evidence of the extracellular cleavage of N2TM(a) by fD1 and to determine the relationship between this extracellular cleavage and the cleavage following it, we added MG-132, a known inhibitor of the intracellular cleavage that results in the release of NICD (De Strooper et al., 1999; Mumm et al., 2000), into the co-culture system of BaF3 and fD1-CHO. The addition of MG-132 in fact reduced the amount of fD1-induced N2ICD in the nucleus-rich fraction (Figure 4D), implying that it prevented fD1-induced intracellular cleavage. In addition, N2TM(b) was detected in the membrane/cytosol fraction when MG-132 was added (Figure 4D). This indicated that extracellular cleavage also occurred during the process of fD1-mediated N2 signaling, as in the case of sD1, and that stimulation with fD1 induced cleavage of N2TM(a) in the extracellular region, prior to cleavage in the transmembrane region. The above findings lead to the conclusions that the extracellular cleavage does not autonomously trigger the N2 intracellular cleavage and that sD1 has a defect in the cleavage required for release of N2ICD, although it can trigger extracellular cleavage of N2 (see Figure 7).
Requirement of NLICD for full activation of N2
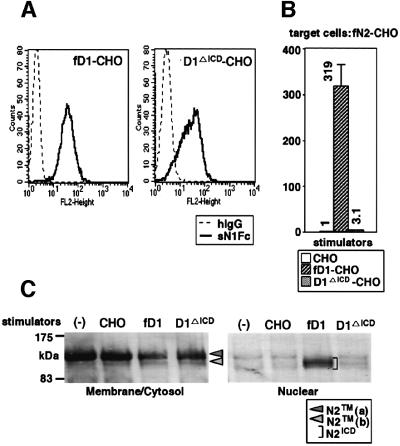
To determine which region of Delta1 is involved in progression of the intracellular cleavage, we generated a CHO(r) cell line expressing D1ΔICD (D1ΔICD-CHO) and investigated its signal-transducing activity. Using cell-binding assays with sN1, we first confirmed that sN1 bound to fD1-CHO and D1ΔICD-CHO in an indistinguishable manner (Figure 5A), indicating that fD1 and D1ΔICD were approximately equally expressed on the cell surface. We also observed that the amount of sD1–Fc binding to BaF3 after co-culture with D1ΔICD-CHO was reduced to a degree similar to that after the co-culture with fD1-CHO (data not shown), and that once D1ΔICD-CHO-adhered BaF3 cells were detached from it exactly like BaF3 cells co-cultured with fD1-CHO (data not shown). In contrast, the reporter assays using these cell lines showed that the signal-transducing activity of D1ΔICD was obviously lower than that of fD1 (Figure 5B). Correspondingly, N2ICD was hardly detected in the nucleus-rich fraction after stimulation with D1ΔICD, unlike after stimulation with fD1, while D1ΔICD reduced the amount of N2TM(a) in the membrane/cytosol fraction (Figure 5C). These observations indicate that D1ΔICD can bind to N2 and induce its extracellular cleavage, but cannot facilitate the ensuing intracellular cleavage, being similar to the phenotype exhibited by sD1, although emergence of N2TM(b) was less clear when stimulated with D1ΔICD than that with sD1. Therefore, it was concluded that the ICD of D1 (D1ICD) is essential for D1-induced N2 intracellular cleavage and full activation of N2, and that the lower signal-transducing activity of sD1 is a consequence of the lack of the ICD rather than the lack of the membrane anchorage.
Fig. 5. Requirement of the intracellular domain of Delta1 for full activation of N2. (A) Generation of D1ΔICD-CHO [CHO(r) cells expressing the truncated Delta1 lacking its intracellular domain]. To investigate the expression of fD1 and D1ΔICD, a cell-binding assay using sN1-Fc (6.7 nM) was performed against the fD1-CHO and D1ΔICD-CHO cells. (B) Comparison of signal-transducing activity of fD1 and D1ΔICD. To examine activity of the two molecules, a transient reporter assay with a TP1-luciferase reporter plasmid was performed using fN2-CHO cells. Fold-induction of luciferase activity for fD1-CHO and D1ΔICD-CHO (mean of triplicate measurements with standard deviation) was calculated against luciferase activity when parental CHO(r) was used as stimulator. (C) N2 fragments after fD1 and D1ΔICD stimulations. BaF3 was stimulated for 1.5 h under the conditions indicated in the figure and then separated into two fractions, membrane/cytosol-rich and nucleus-rich. In each fraction, N2 fragments containing an intracellular domain were analyzed by western blot using the bhN6 antibody after immunoprecipitation.
Importance of multimerization of NL for full activation of N2
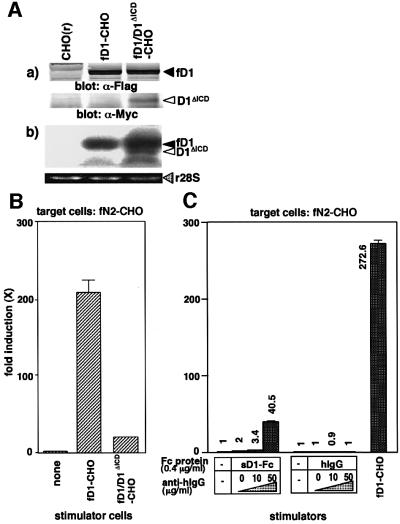
To see an effect of D1ΔICD on fD1-triggered N2 activation in the coexistence of the two molecules, we generated the fD1-CHO cell line expressing D1ΔICD (fD1/D1ΔICD-CHO) (Figure 6A) and investigated its signal-transducing activity. The result showed that the intensity of the N2 signal transduction by fD1/D1ΔICD-CHO was about one-tenth of that by fD1-CHO, indicating that the activity of fD1 was reduced to about one-tenth in the presence of D1ΔICD (Figure 6B). This suggests that D1ΔICD acts in a dominant-negative fashion against fD1, in agreement with previous report indicating that the Delta proteins lacking the ICD act as dominant-negative proteins in Drosophila and vertebrates (Chitnis et al., 1995; Sun and Artavanis-Tsakonas, 1996; Jen et al., 1997). Since the expression level of D1ΔICD was less than that of fD1 in the fD1/ D1ΔICD-CHO cells [Figure 6A, (b)], the strong dominant-negative activity of D1ΔICD was likely to occur via interaction with fD1 rather than simple binding competition, raising the possibility that NL molecules interact with each other. Therefore, we investigated whether the signal-transducing activity of sD1–Fc was enhanced by addition of an anti-Fc polyclonal antibody, which can cross-link the sD1–Fc molecules. Results showed that addition of the anti-Fc antibody significantly increased the signal-transducing activity of sD1–Fc, but not of control IgG, in a dose-dependent manner (Figure 6C). We also note that the same phenomenon also occurred using soluble Jagged1 protein fused to Fc (data not shown). These results suggest that multimerization of the Notch ligand plays an important role in full activation of N2 (see Figure 7).
Fig. 6. Involvement of multimerization of Delta1 in the N2 activation. (A) Generation of fD1-CHO cells expressing Myc-tagged D1ΔICD (fD1/D1ΔICD-CHO). (a) Expression of Myc-tagged D1ΔICD and Flag-tagged fD1 proteins in fD1/D1ΔICD-CHO cells were examined by western blot analysis with an anti-Flag or an anti-Myc antibody. (b) To compare the expression levels of mRNA of fD1 with D1ΔICD in the fD1/D1ΔICD-CHO cells, total RNA (10 µg) extracted from the cells was subjected to northern blot using the 5′-end fragment of mouse Delta1 cDNA as a probe. The lower panel shows ethidium bromide-stained 28S ribosomal RNA (r28S) in each lane. (B) Enhancement of the signal-transducing activity of sD1–Fc by addition of an anti-Fc antibody. A transient reporter assay was performed using pGa981-6 plasmid-transfected fN2-CHO cells in the presence of sD1–Fc and the anti-Fc antibody at various concentrations. hIgG was added as a control for sD1–Fc. The relative induction of luciferase activity in each sample (mean of triplicate measurements with standard deviation) was calculated against luciferase activity in the presence of hIgG alone. (C) A dominant-negative effect of D1ΔICD on fD1-triggered N2 activation. fD1/D1ΔICD-CHO [CHO(r) cells co-expressing fD1 and D1ΔICD] was generated and its signal-transducing activity was examined by a transient reporter assay with pGa981-6 plasmid-transfected fN2-CHO cells.
Discussion
In this study, we compared the signal-transducing activity of sD1 and fD1 and used the resultant information to analyze the activation process of N2 after ligand stimulation. We found that sD1 functions as a partial agonist and that the mechanism of such a function could stem from the incomplete activity of sD1 in the intracellular cleavage required to release N2ICD. Furthermore, experiments using sD1 and D1ΔICD demonstrated that the extracellular cleavage at the extracellular domain of N2 does not autonomously induce intracellular cleavage, which takes place in the transmembrane domain, and that D1ICD is involved in some unidentified mechanisms that exist between the two cleavage processes. Given that the signal-transducing activity of sD1–Fc was enhanced by the addition of an anti-Fc antibody and that D1ΔICD acted in a dominant-negative fashion against fD1 for N2 signaling, we suggest that multimerization of NL would be important for intracellular cleavage (Figure 7).
A soluble form of Drosophila Delta does exist in vivo (Klueg et al., 1998), possibly generated by Kuzbanian (Qi et al., 1999). Therefore, it is important to understand the exact physiological function of sNL, although the precise C-terminal sequence of naturally occurring soluble Delta is unknown. However, conclusions regarding the biological activity of sNL to date have been discordant, as manoeuvered mammalian sD1 and soluble Jagged1 have been characterized as having an agonistic activity in in vitro experiments (Li et al., 1998; Qi et al., 1999; Han et al., 2000; Karanu et al., 2000; Morrison et al., 2000) and an antagonistic activity has been proposed for soluble Delta and Serrate in Drosophila in vivo assessments (Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997). Examination of this issue was enabled by transcriptional activation assay using CHO(r) cells overexpressing mouse fN2 (fN2-CHO) (Shimizu et al., 2000), which is a very sensitive system. We have demonstrated that sD1 comprising entire extracellular domain induces N2 activation, but the activity is markedly lower than that induced by fD1 (Figure 1A). This indicates that sD1 is a partial agonist, while fD1 is a full activator for Notch signaling. The fact that the coexistence of sD1 led to inhibition of fD1-induced N2 activation (Figure 1C) further supports the nature of sD1 as a partial agonist. We also note that the signal-transducing activity of soluble Jagged1-Fc is lower than that of full-length Jagged1 (data not shown). Hence, a partial activity in the soluble form may be common to all kinds of NL, including naturally existing sNL, since they do not harbor NLICD that is essential for full activation of Notch signaling (Figure 5); this scenario explains the contradiction concerning the biological activity of sNL. Regarding the physiological function of sNL in vivo, we speculate that it is associated with the strict control of the Notch signaling, which is known to be critical for the exact cell fate decision, because the abnormal phenotype is seen in patients with haploinsufficiency of Jagged1 (Li et al., 1997; Oda et al., 1997).
In an analysis tracing the N2 fragments, we ascertained the molecular basis of the incomplete function of sD1. Consistent with the low transcriptional activity of sD1 (Figure 1A), the stimulation with sD1–Fc did not result in nuclear accumulation of N2ICD (Figure 2), whose amount is considered to determine the level of subsequent RBP-Jκ-mediated transcriptional activation (Jarriault et al., 1995; Schroeter et al., 1998). Furthermore, the experiments using sD1–Fc incidentally unveiled the mechanism of the cleavage of N2TM in the extracellular region (Figures 2 and 3), which was recently demonstrated by different approaches (Brou et al., 2000; Mumm et al., 2000). This cleavage was also evident after the binding of fD1 (Figure 4), but only when the intracellular cleavage that takes place in the transmembrane portion of N2TM was blocked by an inhibitor (Figure 4D). Taken together, these data imply that sD1 induces the extracellular cleavage of N2, but fails to sufficiently promote the intracellular cleavage that releases N2ICD, while fD1 efficiently triggers both cleavages. This indicates that the extracellular domain of Notch ligand alone is sufficient for extracellular cleavage of N2, and that extracellular cleavage is not necessarily followed by progression to intracellular cleavage, suggesting the existence of an as yet unknown mechanism regulating activation of the intracellular cleavage.
However, this notion was controversial, being in contrast to the recent report on extracellular cleavage of N1 in which the intracellular cleavage and subsequent signal transduction are described as autonomous events after extracellular cleavage (Mumm et al., 2000), which was drawn from experiments using the truncated N1 protein lacking NEC. Regarding this discrepancy, we raise the possibility that the intracellular cleavage in the truncated Notch protein lacking NEC progresses through a mechanism different from the intracellular cleavage induced by a ligand in the natural Notch protein. Indeed, the difference in intracellular cleavage between the two molecules was reported; the intracellular cleavage in the natural Notch protein occurred within 15 min of ligand binding (Shimizu et al., 2000), whereas that in the truncated Notch protein required >60 min (Schroeter et al., 1998) after protein synthesis. In addition, we also found that the amino acid sequence surrounding the extracellular cleavage site identified using truncated N1 (Brou et al., 2000; Mumm et al., 2000) is not conserved in N2. Consistent with this, addition of 1,10-o-phenanthroline, a reagent identified as an inhibitor of the extracellular cleavage in truncated Notch1 (Mumm et al., 2000), did not prevent sD1–Fc-induced extracellular cleavage of N2 (data not shown). Therefore, the extracellular cleavage site and the activation mechanism required for intracellular cleavage in the mutant Notch protein lacking NEC may be somewhat diverse from those in the natural Notch proteins. Alternatively, the discrepancy may result from an unidentified difference between N1 and N2.
One clue to the mechanism regulating activation of the intracellular cleavage was found in the data from experiments using the truncated Dl protein lacking its ICD, D1ΔICD (Figure 5). The characteristics of D1ΔICD are similar to those of sD1 rather than those of fD1, i.e. a low level of transcriptional activity from the TP-1 promoter, sufficiency in cleaving the extracellular domain of N2TM, and insufficiency in cleaving N2TM within the transmembrane portion and in releasing N2ICD, although there was a slight difference in the emergence of N2TM(b) (Figure 5B and C). These indicate that ICD of Delta1 is indispensable for the cleavage of N2TM within its transmembrane portion, which is essential for full activation of N2 (Figure 7).
Another clue to the mechanism lies in the data from experiments using sD1–Fc plus an anti-Fc antibody. Addition of the anti-Fc antibody resulted in enhancement of the signal-transducing activity of sD1–Fc (Figure 6C), suggesting that multimerization of the ligand is associated with the full activation of N2 (Figure 7). Dimerization of NL is inefficient, since the signal-transducing activities of sD1–Fc (dimer) and sD1-Flag(His6) (monomer) were the same (Figure 1). It is possible that an assembly of a large number of NL molecules is required for fN activation, since a monoclonal anti-Fc antibody did not work, unlike a polyclonal antibody (data not shown). Putting together the inevitable role of D1ICD for D1-induced N2 intracellular cleavage and subsequent N2 activation, as we discussed above, NLICD may be the region that is used for the multimerization of fNL. As for the strong dominant-negative effect of D1ΔICD against fD1 (Figure 6), we raise the possibility that D1ΔICD can interact with fD1 using its transmembrane and/or extracellular domains to participate in the fD1 assembly, which interfere with the appropriate fD1 multimerization suitable for the intracellular cleavage of N2.
As is well known with regard to cytokines and their receptor systems (reviewed in Heldin, 1995), the multimerization of NL may be associated with the assembly of the Notch receptor, which may result in its conformational change and allow the transmembrane domain of Notch to be subjected to the action of the presenilin-containing protease complex and subsequent cleavage. Upon fD1 binding, N2 extracellular cleavage must be preceeded by fD1 multimerization and N2 assembly, although these steps are not necessary for N2 extracellular cleavage itself (Figure 7).
It was reported recently that transendocytosis by the ligand-expressing cells of the ligand-bound Notch extracellular domain together with the ligand appears to be necessary for efficient Notch processing and signal transduction (Parks et al., 2000). Integrating this into our model of Notch receptor activation, the endocytosis process may be positioned after the ligand multimerization or Notch assembly. However, we are not certain whether the transendocytosis is always necessary for the Notch signaling, since cell-free ligands can activate Notch signaling, particularly when the antibody-mediated ligand cross-linking or ligand-coating technique is used (Figure 6; Morrison et al., 2000; Varnum-Finney et al., 2000). Given the well-established notion in the G-coupled receptor and the cytokine receptor systems that endocytosis is associated with receptor/ligand degradation rather than directly associated with signal activation, the possibility may remain that transendocytosis in the Notch signaling pathway participates in degradation of Notch and NL.
Comparing the effect of D1ΔICD with that of sD1, emergence of N2TM(b) was less clear when stimulated with the former (Figures 2 and 3) than that with the latter (Figure 5C), despite our assumption that N2TM(b) was indeed generated after D1ΔICD stimulation, since the amount of N2TM(a) was obviously reduced. As an explanation of this phenomenon, we speculate that some unknown degradation process for N2TM(b) is accelerated by stimulation with D1ΔICD but not with sD1.
In the present study, we demonstrated the biological activities of sNL and the existence of a novel mechanism involved in the activation of the Notch receptor by its ligand. Although many investigators are using sNL to determine the involvement of the Notch signal in the regulation of cell differentiation in various experimental approaches, we have to be careful of the interpretation of results, since the demonstrated phenotype could be caused by the inhibition, rather than activation, of the Notch signal. We believe that the findings described here will facilitate understanding of the complexities of Notch signaling in higher vertebrates.
Materials and methods
Plasmid construction
To generate D1ΔICD, mouse Delta1 cDNA (a gift from A.Gossler; Bettenhausen et al., 1995) was truncated at the codon CGG corresponding to arginine (amino acid 570). The resulting D1DICD was constructed in an expression vector pTraserCMV (Clontech) after addition of a Flag or a Myc tag.
Soluble fusion proteins
sD1 proteins [sD1–Fc and sD1-Flag(His)6] and sN1 protein (sN1-Fc) were prepared as described previously (Shimizu et al., 1999, 2000).
Cell culture
BaF3 cells were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 0.5 ng/ml recombinant mouse interleukin-3 (a gift from Kirin Brewery, Japan). CHO(r) (a gift from S.Shirahata, Kyushu University), fN2-CHO (Shimizu et al., 2000), fD1-CHO (Shimizu et al., 2000), D1ΔICD-CHO and fD1/D1ΔICD-CHO cells were maintained in alpha-minimal essential medium containing 10% FBS.
Antibody
An anti-human IgG goat polyclonal antibody used for multimerization of sD1–Fc was purchased from DAKO Japan Co., Ltd.
Cell-binding assay
Binding of sD1–Fc to the pro-B cell line BaF3 was performed as described previously (Shimizu et al., 1999), with the minor modification that the binding reaction was terminated at 5 min.
Co-precipitation using sD1–Fc
Co-precipitation using sD1–Fc has been described elsewhere (Shimizu et al., 2000). Disuccinimidyl glutarate (Pierce) was used to cross-link sD1–Fc and the bound Notch receptor.
Cell–cell association assay
Cell–cell association assay was performed as described previously (Shimizu et al., 2000). Briefly, CHO(r) and fD1-CHO cells were inoculated at 1 × 106 into a 6 cm plate. After overnight culture, 1 × 106 BaF3 cells were spread over the monolayer of cells. Following co-culture at 37°C for the time indicated in Figure 4C, BaF3 cells that did not adhere to the cell layer were collected by swirling the plate very gently and washing the wells gently once with RPMI medium. The population obtained through these procedures was defined as non-adhered BaF3. Next, phosphate-buffered saline containing 2 mM EGTA was added to the wells and the BaF3 cells adhering to the cell layer were allowed to dissociate by tapping the plate. These BaF3 cells, together with additional cells collected by washing with RPMI medium, were defined as adhered BaF3. The cells in each fraction were then counted.
Transient transcription assay
A total of 4 × 104 fN2-CHO was inoculated into a 24-well plate and transfected with a TP1-luciferase reporter plasmid (Minoguchi et al., 1997), pGa981-6, by a liposome-based method (SuperFect, Qiagen). Following transfection, sD1–Fc and sD1-Flag(His)6 were added at the respective concentrations shown in Figure 1A and cultured for 30–40 h. When fD1 or D1ΔICD were used as a stimulator, these cells were added at 5 × 104 to the pGa981-6-transfected fN2-CHO and co-cultured for 30–40 h. The mixture of cells was then used for luciferase assay.
Subcellular fractionation of BaF3
After 1.5 h co-culture with fD1-CHO or D1ΔICD-CHO, BaF3 was collected and membrane/cytosol-rich and nucleus-rich fractions were prepared as described elsewhere (Shimizu et al., 2000). The membrane/cytosol-rich fraction was further centrifuged at 105 000 g for 30 min at 4°C to separate the membrane (pellet) and the cytosol (supernatant) fractions. These fractions were used for immunoprecipitation with an anti-N2 polyclonal antibody.
Acknowledgments
Acknowledgements
We thank Dr S.Artavanis-Tsakonas for providing us with the bhN6 anti-N2 antibody, Dr A.Gossler for mouse Delta1 cDNA, Dr T.Honjo for pGa981-6 plasmid and Dr S.Shirahata for CHO ras clone-I cells. We also wish to thank Mr Chris W.P.Reynolds for his review of the manuscript. This work was supported by grants-in-aid from the Ministry of Education, Science, Sport, Culture and Technology of Japan, and the Ministry of Health, Welfare and Labor of Japan.
References
- Artavanis-Tsakonas S., Rand,M.D. and Lake,R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis,M., Simon,D., Guenet,J. and Gossler,A. (1995) Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development, 121, 2407–2418. [DOI] [PubMed] [Google Scholar]
- Blaumueller C.M., Qi,H., Zagouras,P. and Artavanis-Tsakonas,S. (1997) Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell, 90, 281–291. [DOI] [PubMed] [Google Scholar]
- Brou C. et al. (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell, 5, 207–216. [DOI] [PubMed] [Google Scholar]
- Chan Y.M. and Jan,Y.N. (1998) Roles for proteolysis and trafficking in notch maturation and signal transduction. Cell, 94, 423–426. [DOI] [PubMed] [Google Scholar]
- Chitnis A., Henrique,D., Lewis,J., Ish-Horowicz,D. and Kintner,C. (1995) Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature, 375, 761–766. [DOI] [PubMed] [Google Scholar]
- De Strooper B. et al. (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature, 398, 518–522. [DOI] [PubMed] [Google Scholar]
- Ellisen L.W., Bird,J., West,D.C., Soreng,A.L., Reynolds,T.C., Smith,S.D. and Sklar,J. (1991) TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell, 66, 649–661. [DOI] [PubMed] [Google Scholar]
- Greenwald I. (1998) LIN-12/Notch signaling: lessons from worms and flies. Genes Dev., 12, 1751–1762. [DOI] [PubMed] [Google Scholar]
- Han W., Ye,Q. and Moore,M.A. (2000) A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic progenitor cells. Blood, 95, 1616–1625. [PubMed] [Google Scholar]
- Heldin C.H. (1995) Dimerization of cell surface receptors in signal transduction. Cell, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- Hukriede N.A., Gu,Y. and Fleming,R.J. (1997) A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development, 124, 3427–3437. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou,C., Logeat,F., Schroeter,E.H., Kopan,R. and Israel,A. (1995) Signalling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Le Bail,O., Hirsinger,E., Pourquie,O., Logeat,F., Strong,C.F., Brou,C., Seidah,N.G. and Israel,A. (1998) Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol., 18, 7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen W.C., Wettstein,D., Turner,D., Chitnis,A. and Kintner,C. (1997) The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development, 124, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Karanu F.N., Murdoch,B., Gallacher,L., Wu,D.M., Koremoto,M., Sakano,S. and Bhatia,M. (2000) The notch ligand jagged-1 represents a novel growth factor of human hematopoietic stem cells. J. Exp. Med., 192, 1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klueg K.M., Parody,T.R. and Muskavitch,M.A. (1998) Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol. Biol. Cell, 9, 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R. and Weintraub,H. (1993) Mouse notch: expression in hair follicles correlates with cell fate determination. J. Cell Biol., 3, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Schroeter,E.H., Weintraub,H. and Nye,J.S. (1996) Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl Acad. Sci. USA, 93, 1683–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli M., Dahlstrand,J. and Lendahl,U. (1994) The novel Notch homologue mouse Notch3 lacks specific epidermal growth factor repeats and is expressed in proliferating neuroepithelium. Mech. Dev., 46, 123–136. [DOI] [PubMed] [Google Scholar]
- Li L. et al. (1997) Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nature Genet., 16, 243–251. [DOI] [PubMed] [Google Scholar]
- Li L. et al. (1998). The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity, 8, 43–55. [DOI] [PubMed] [Google Scholar]
- Logeat F., Bessia,C., Brou,C., Le Bail,O., Jarriault,S., Seidah,N.G. and Israel,A. (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl Acad. Sci. USA, 95, 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoguchi S., Taniguchi,Y., Kato,H., Okazaki,T., Strobl,L.J., Zimber-Strobl,U., Bornkamm,G.W. and Honjo,T. (1997) RBP-L, a transcription factor related to RBP-Jkappa. Mol. Cell. Biol., 17, 2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Perez,S.E., Qiao,Z., Verdi,J.M., Hicks,C., Weinmaster,G. and Anderson,D.J. (2000) Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell, 101, 499–510. [DOI] [PubMed] [Google Scholar]
- Mumm J.S., Schroeter,E.H., Saxena,M.T., Griesemer,A., Tian,X., Pan,D.J., Ray,W.J. and Kopan,R. (2000) A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell, 5, 197–206. [DOI] [PubMed] [Google Scholar]
- Oda T. et al. (1997) Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature Genet., 16, 235–242. [DOI] [PubMed] [Google Scholar]
- Parks A.L., Klueg,K.M., Stout,J.R. and Muskavitch,M.A. (2000) Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development, 127, 1373–1385. [DOI] [PubMed] [Google Scholar]
- Qi H., Rand,M.D., Wu,X., Sestan,N., Wang,W., Rakic,P., Xu,T. and Artavanis-Tsakonas,S. (1999) Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science, 283, 91–94. [DOI] [PubMed] [Google Scholar]
- Rand M.D., Grimm,L.M., Artavanis-Tsakonas,S., Patriub,V., Blacklow,S.C., Sklar,J. and Aster,J.C. (2000) Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol., 20, 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Chiba,S., Kumano,K., Hosoya,N., Takahashi,T., Kanda,Y., Hamada,Y., Yazaki,Y. and Hirai,H. (1999) Mouse Jagged1 physically interacts with Notch2 and other Notch receptors: Assessment by quantitative methods. J. Biol. Chem., 274, 32961–32969. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Chiba,S., Hosoya,N., Kumano,K., Saito,T., Kurokawa,M., Kanda,Y., Hamada,Y. and Hirai,H. (2000) Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol. Cell. Biol., 20, 6913–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. and Adachi,A. (1998) Nuclear access and action of notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- Struhl G. and Greenwald,I. (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature, 398, 522–525. [DOI] [PubMed] [Google Scholar]
- Sun X. and Artavanis-Tsakonas,S. (1996) The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Development, 122, 2465–2474. [DOI] [PubMed] [Google Scholar]
- Sun X. and Artavanis-Tsakonas,S. (1997) Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development, 124, 3439–3448. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H., Marazzi,G., Wu,G., Yan,Q., Sassoon,D. and Kitajewski,J. (1996) Notch/Int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development, 122, 2251–2259. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B., Wu,L., Yu,M., Brashem-Stein,C., Staats,S., Flowers,D., Griffin,J.D. and Bernstein,I.D. (2000) Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci., 113, 4313–4318. [DOI] [PubMed] [Google Scholar]
- Weinmaster G. (1997) The ins and outs of notch signaling. Mol. Cell. Neurosci., 9, 91–102. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts,V. and Lemke,G. (1991) A homolog of Drosophila Notch expressed during mammalian development. Development, 113, 199–205. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts,V. and Lemke,G. (1992) Notch2: a second mammalian Notch gene. Development, 116, 931–941. [DOI] [PubMed] [Google Scholar]
- Ye Y., Lukinova,N. and Fortini,M.E. (1999) Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature, 398, 525–529. [DOI] [PubMed] [Google Scholar]