Abstract
Viroids, small single-stranded circular RNAs (246–401 nucleotides), do not have mRNA capacity and must recruit host proteins to assist in the steps of their biological cycle. The nature of these cellular factors is poorly understood due to a lack of reliable experimental approaches. Here, to screen for host proteins interacting with viroid RNAs in vivo, we UV-irradiated avocado leaves infected with avocado sunblotch viroid (ASBVd), the type member of chloroplast viroids containing hammerhead ribozymes. This resulted in the detection of several ASBVd–host protein adducts. Tandem mass spectrometry analysis of the most abundant cross-linked species identified the protein component as two closely related chloroplast RNA-binding proteins (PARBP33 and PARBP35) of a family whose members previously have been shown to be involved in stabilization, maturation and editing of chloroplast transcripts. PARBP33 behaves as an RNA chaperone that stimulates in vitro the hammerhead-mediated self-cleavage of the multimeric ASBVd transcripts that result from rolling circle replication, indicating that this reaction, despite its RNA-based mechanism, is facilitated by proteins. The structural and functional parallelism between PARBP33 and PARBP35, and some proteins involved in viral RNA replication, indicates that viroids and RNA viruses recruit similar host proteins for their replication.
Keywords: avocado sunblotch viroid/catalytic RNAs/hammerhead ribozymes/RNA-binding proteins/RNA chaperones
Introduction
There is considerable experimental data supporting the view that due to the limited information content of their genomes, bacterial, plant and animal RNA viruses rely on different host proteins which are diverted from their normal functions to assist in the replication and transcription of the RNA from these pathogens (for reviews see Blumenthal and Carmichael, 1979; Buck, 1996; De and Banerjee, 1997; Lai, 1998). The most convincing data behind this view come from a yeast-derived system for studying replication of brome mosaic virus, with which many yeast mutants that affect viral RNA replication or transcription have been obtained (Janda and Ahlquist, 1993). The situation is even more extreme in viroids, whose genome is constituted exclusively of a small circular RNA of 246–401 nucleotides without any apparent coding capacity, and which are able to replicate autonomously in certain plants (for reviews see Diener, 1999; Flores et al., 2000a,b). Therefore, viroid-based systems offer a simplified context for identifying host proteins assisting the replication of an RNA without interference by switchings between replication and transcription, or between transcription and translation, as when dealing with viral RNAs.
The ∼30 characterized viroid species have been classified into two different families (Flores et al., 2000a). Most viroids, which contain in their molecules a typical central conserved region (CCR), belong to the family Pospiviroidae, whereas three viroids that lack the CCR but are able to adopt self-cleaving hammerhead ribozymes are grouped in the family Avsunviroidae (for a review see Flores et al., 2000b). Moreover, in those viroid species studied to date, it has been shown that while members of the family Pospiviroidae replicate and accumulate in the nucleus (Diener, 1971; Spiesmacher et al., 1983; Harders et al., 1989; Bonfiglioli et al., 1996; Woo et al., 1999), members of the family Avsunviroidae replicate and accumulate in the chloroplast (Bonfiglioli et al., 1994; Lima et al., 1994; Bussière et al., 1999; Navarro et al., 1999).
Viroids replicate through an RNA-based rolling circle mechanism for which two different pathways, symmetric and asymmetric, have been proposed (Branch and Robertson, 1984). Avocado sunblotch viroid (ASBVd), a circular RNA of 247 nucleotides (Hutchins et al., 1986), is the type species of the family Avsunviroidae. ASBVd and possibly the other members of its family replicate through the symmetric pathway (Bruening et al., 1982; Hutchins et al., 1985; Daròs et al., 1994; Bussière et al., 1999), in contrast to potato spindle tuber viroid (PSTVd) (Diener, 1972; Gross et al., 1978) the type species of the family Pospiviroidae, and possibly the other members of this family that follow the asymmetric pathway (Branch et al., 1988; Feldstein et al., 1998). In ASBVd, the monomeric circular RNAs of both polarities serve as templates for synthesis of their complementary oligomeric RNAs, in a process apparently catalyzed by a nuclear-encoded chloroplast RNA polymerase (Navarro et al., 2000) that starts transcription in a defined (A + U)-rich region in each polarity strand (Navarro and Flores, 2000). The resulting (+) and (–) oligomers, the (+) polarity being assigned arbitrarily to the strand that accumulates more abundantly in infected tissue, self-cleave through the activity of hammerhead ribozymes embedded in these RNAs (Hutchins et al., 1986; Prody et al., 1986; Forster and Symons, 1987; for a review see Flores et al., 2001), to yield monomeric linear forms that presumably are circularized by an RNA ligase. This latter step, however, has been proposed to occur autocatalytically at least in peach latent mosaic viroid (Côté et al., 2001).
The search for cellular factors which assist steps in the biological cycle of viroids, particularly replication, has been based on indirect assays, mainly in vitro binding between viroid RNAs and host proteins extracted from the infected tissue (Wolff et al., 1985; Hadidi, 1988) or expressed from cDNA libraries (Sagesser et al., 1997), or in vitro analysis of subcellular fractions (Hadidi, 1988; Klaff et al., 1989). These approaches have met with only partial success and, most importantly, they leave unanswered the questions of whether the observed interactions also occur in vivo and their possible functional significance. In an attempt to circumvent these limitations, we have focused on more direct techniques. UV cross-linking is a powerful methodology for characterization of RNA–protein interactions in ribonucleoprotein complexes. UV light is a ‘zero-length’ cross-linking agent able to induce formation of covalent bonds between nucleic acids and proteins at their contact points, thereby ‘freezing’ the interaction between the two molecules (Hockensmith et al., 1991; Pashev et al., 1991). In the present work, we have applied a strategy of this kind by UV-irradiating ASBVd-infected avocado leaves to screen for host proteins directly interacting with viroid RNAs in vivo. This has led to the identification of several ASBVd–host protein adducts and to the characterization of the protein component of the most abundant cross-linked species as two closely related chloroplast RNA-binding proteins belonging to a family whose members have been shown previously to be involved in different steps of RNA metabolism in this organelle. At least one of these avocado proteins behaves as an RNA chaperone and stimulates in vitro, and possibly in vivo, the hammerhead-mediated self-cleavage of multimeric ASBVd transcripts.
Results
Formation of ASBVd–protein cross-linked adducts by UV irradiation of ASBVd-infected leaves
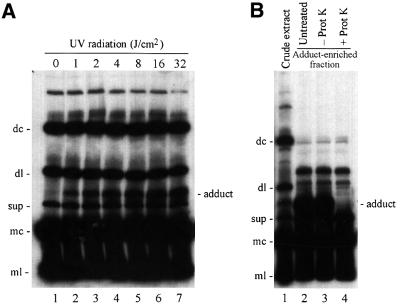
To search for host factors that interact directly in vivo with viroid RNAs, sections of ASBVd-infected avocado leaves were irradiated with different doses of UV light and immediately homogenized in an aqueous buffer containing 5 M urea. The extracts were separated by denaturing PAGE and ASBVd RNAs were revealed by northern blot hybridization with probes complementary to (+) and (–) viroid strands. Analysis with the probe for detecting the (+) viroid RNAs showed a series of prominent bands, whose intensity remained unchanged with the radiation dose (Figure 1A) corresponding to a series of ASBVd RNAs identified and characterized in previous studies (Hutchins et al., 1985; Daròs et al., 1994). Prominent among them are the dimeric circular (dc) and linear (dl), the supragenomic (sup), and the monomeric circular (mc) and linear (ml) ASBVd (+) RNAs. However, a previously undescribed additional band was also detected in a position between the dl- and sup-ASBVd (+) RNAs in the irradiated extracts (Figure 1A, lanes 2–7). Interestingly, the intensity of this band grew with the radiation dose from 1 to 32 J/cm2, suggesting that UV promoted the formation of a covalent adduct by cross-linking an ASBVd RNA to another molecule of host origin. However, the possibility of an intra- or intermolecular viroid RNA cross-link could not be discarded from these results. No band changing its intensity with the UV irradiation dose was observed when a probe for detecting (–) viroid strands was used in parallel northern blot hybridizations, and no ASBVd-specific signals were detected with extracts from non-infected avocado leaves (data not shown).
Fig. 1. Northern blot analysis of extracts from ASBVd-infected avocado leaves fractionated by denaturing PAGE and hybridized with a probe for detecting (+) viroid strands. (A) Extracts from leaves irradiated with different UV doses. Lane 1, non-irradiated leaves; lanes 2–7, leaves irradiated with 1, 2, 4, 8, 16 and 32 J/cm2 of 254 nm UV light, respectively. (B) Proteinase K digestion of a preparation from irradiated tissue. Lane 1, control of crude extract; lanes 2, 3 and 4, adduct-enriched fraction untreated, incubated in buffer without proteinase K, and digested with proteinase K, respectively. The adduct-enriched fraction was obtained by cutting a segment of a non-denaturing polyacrylamide gel and eluting its contents. The positions of the dimeric circular (dc) and linear (dl), supragenomic (sup), and monomeric circular (mc) and linear (ml) ASBVd (+) RNAs, are indicated on the left of both panels. The position of an ASBVd–protein adduct is indicated on the right.
To investigate further the nature of the ASBVd cross-linked species, an RNA fraction from irradiated tissue enriched in this species was treated with proteinase K and the digestion products were analyzed by denaturing PAGE and northern blot hybridization with a probe for detecting (+) viroid strands. The band corresponding to the ASBVd cross-linked species completely disappeared as a consequence of the treatment, whereas none of the other bands corresponding to major ASBVd RNAs suffered any alteration (Figure 1B, compare lanes 3 and 4). This result showed that the ASBVd cross-linked species resulted from the formation of a covalent adduct between an ASBVd (+) RNA and a protein from the host.
Purification of the ASBVd–protein adduct
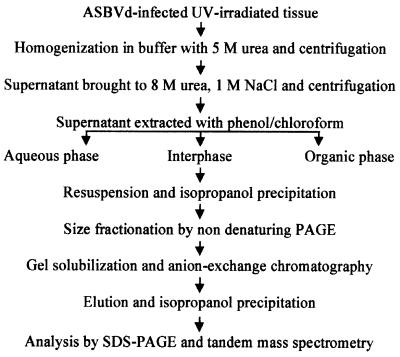
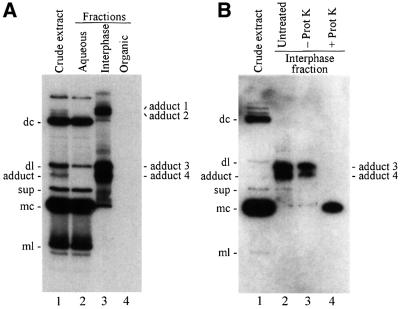
To identify the host protein, which together with the ASBVd RNA forms the adduct, a protocol to purify it from UV-irradiated tissue was developed (Figure 2). First, the tissue was homogenized in the presence of an aqueous buffer, but the adduct was only recovered in the supernatant when the homogenization was made in partially denaturing conditions with urea (data not shown). The extract, typically obtained in 5 M urea, was clarified further by increasing the urea concentration up to 8 M and adding NaCl up to 1 M, and discarding by centrifugation the material sedimenting under these conditions. The supernatant was extracted with a buffer-saturated mix of phenol and chloroform, and the aqueous and organic phases, as well as the interphase, were recovered. The ASBVd–protein adduct, composed of two moieties which behave differently in this kind of partition, was observed accumulating in the interphase. This purification step was particularly useful, as it enabled the easy recovery of a fraction enriched in the adduct and its separation from other viroid RNAs, as revealed by analysis of the three fractions by denaturing PAGE and northern blot hybridization analysis (Figure 3A). Viroid RNAs remained in the aqueous phase during the partition: bands arising from dc-, sup-, mc- and ml-ASBVd (+) RNAs exhibited approximately the same intensity in the initial crude extract and the aqueous phase fraction (Figure 3A, lanes 1 and 2), while they were not detected in the interphase fraction or only observed as weak bands in the case of the two most abundant dc- and mc-ASBVd (+) RNAs (Figure 3A, lane 3). In contrast, the band generated by the ASBVd–protein adduct was prominent in the interphase and absent in the aqueous phase (Figure 3A, lanes 2 and 3). Furthermore, the analysis of the interphase fraction revealed the existence of three additional bands with a similar behavior to the adduct during partition (Figure 3A, lane 3). These three new species remained unnoticed in the initial analysis, possibly because the two of higher molecular weight accumulated at lower concentrations and could only be detected in the 20-fold concentrated interphase fraction, and because the most concentrated species, with a slightly slower mobility than the previously identified adduct, co-migrated with the dl-ASBVd (+) RNA in the denaturing PAGE and could only be observed clearly after separating the two species in two different fractions (Figure 3A, compare lanes 1, 2 and 3). As expected, no viroid RNAs were detected in the organic phase (Figure 3A, lane 4).
Fig. 2. Purification scheme of ASBVd–protein adducts from ASBVd-infected UV-irradiated avocado tissue. Following phenol/chloroform extraction, ASBVd–protein adducts accumulated in the interphase fraction, which was used subsequently for further purification (see Materials and methods for details).
Fig. 3. Northern blot analysis of extracts from ASBVd-infected UV-irradiated avocado leaves. Extracts were fractionated by denaturing PAGE and hybridized with a probe for detecting (+) viroid strands. (A) Partition after treatment with phenol/chloroform. Lane 1, crude extract; lanes 2, 3 and 4, fractions corresponding to the aqueous phase, interphase and organic phase, respectively. The interphase and organic phase fractions were concentrated 20-fold by alcohol precipitation with respect to the crude extract and the aqueous phase fraction. (B) Proteinase K digestion of a preparation enriched in adducts 3 and 4 derived from the concentrated interphase fraction. Lane 1, control of crude extract; lanes 2, 3 and 4, preparation untreated, incubated in buffer without proteinase K, and digested with proteinase K, respectively. The preparation enriched in adducts 3 and 4 was obtained by cutting a segment of a non-denaturing polyacrylamide gel and eluting its contents. The positions of the different ASBVd (+) RNAs and of the four ASBVd–protein cross-linked adducts are indicated on the left and right sides of both panels. Other details as in the legend to Figure 1.
The nature of the three new putative ASBVd cross-linked species was investigated by treating the concentrated interphase fraction with proteinase K, and analyzing the products by denaturing PAGE and northern blot hybridization with a probe for detecting (+) viroid strands. The three new species behaved like the ASBVd–protein adduct previously investigated and completely disappeared as a consequence of the digestion (data not shown). Therefore, the four bands corresponded to four different ASBVd–protein adducts that were named 1, 2, 3 and 4 according to their position from the origin of the gel (Figure 3A), with the ASBVd–protein cross-linked species adduct 4 being detected initially. Digestion with proteinase K of the two closely migrating adducts 3 and 4 also allowed the identification of the ASBVd RNA cross-linked to host proteins as mc-ASBVd (+) RNA (Figure 3B, compare lanes 3 and 4).
At this point, purification efforts were focused on the most abundant ASBVd–protein adducts 3 and 4. Components of the concentrated interphase fraction were separated by non-denaturing PAGE using solubilizable disulfide gels, and the gel segment containing adducts 3 and 4 was excised and solubilized. Adducts were recovered by anion-exchange chromatography, concentrated by alcohol precipitation and separated in duplicate SDS–polyacrylamide gels for preparative and analytical purposes. RNAs in the first gel were stained sequentially with ethidium bromide and silver, while RNAs in the second gel were transferred to a nylon membrane and hybridized with a probe for detecting ASBVd (+) strands. Three prominent bands that were observed in the preparative gel by ethidium bromide (Figure 4A) and silver staining (data not shown) were also recognized by the viroid-specific probe (Figure 4B). The two slowest migrating species corresponded to ASBVd–protein adducts 3 and 4, and the fastest migrating species to mc-ASBVd RNA according to the mobility of a purified standard in this type of gel (data not shown).
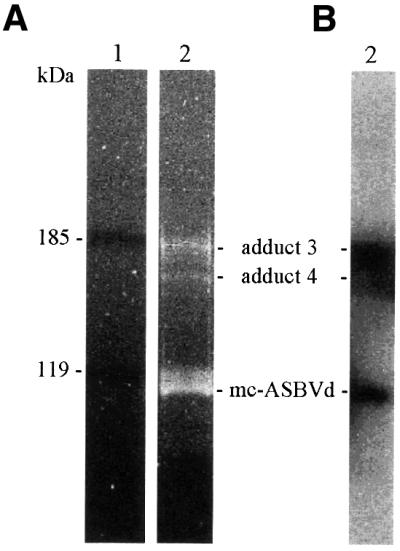
Fig. 4. Purified ASBVd–protein adducts 3 and 4 separated by SDS–PAGE in 7.5% gels and revealed by ethidium bromide staining (A), or transferred to a membrane and hybridized with a probe for detecting (+) viroid strands (B). Lane 1, pre-stained protein standards; lane 2, ASBVd–protein adducts. Molecular masses of protein standards (in kDa) are indicated on the left, and the positions of the ASBVd–protein adducts 3 and 4, and of the mc-ASBVd RNA are indicated between the two panels.
Identification of host proteins cross-linked to ASBVd RNA
The minimal amounts of purified ASBVd–protein adducts impeded application of classical N-terminal sequencing using Edman degradation. Therefore, we focused on the identification of the protein component of the most abundant adduct 3 (Figure 4), using a more sensitive analysis by mass spectrometry (Mann et al., 2001). The band corresponding to adduct 3 was cut from the silver-stained gel, subjected to in-gel trypsin digestion, the resulting peptides eluted and four of them sequenced by tandem mass spectrometry. When peptide amino acid sequence information was used to search for homologs in protein sequence databases, eight entries from six different plant species were retrieved (Figure 5), all belonging to a family of nuclear-encoded RNA-binding proteins localized to the chloroplast, more specifically to group II of this family (Schuster and Gruissem, 1991; Ye et al., 1991; Bar-Zvi et al., 1992; Ohta et al., 1995). These results strongly suggested that the protein cross-linked to ASBVd RNA was an avocado homolog of these proteins.
Fig. 5. Identification of the protein moiety cross-linked to mc-ASBVd (+) forming adduct 3. (A) Peptide sequence tags derived from tandem mass spectrometry analysis and entries retrieved from protein sequence databases using the program PeptideSearch. The sequence tags, which correspond to Y′′-type ions and are written from the C- to the N-terminal end following an established convention, are composed of a string of amino acid residues flanked by the mass of the lowest and the largest ion of the series in parentheses (Mann and Wilm, 1994). Accession numbers correspond to Swiss-Prot (S) or TrEMBL sequence databases (E). (B) Multiple sequence alignment of the retrieved proteins from databases using the program PeptideSearch. Regions matching the four sequence tags are boxed, with non-identical residues indicated by italics. The sequences of avocado proteins PARBP33 and PARBP35 (see Results) have also been included in the alignment.
To acquire more direct evidence of this, a pair of degenerate primers derived from the partial sequence of two of the tryptic peptides was used to amplify by RT–PCR a fragment of the cDNA from avocado corresponding to the cross-linked protein. From the sequence of the 71 bp amplified fragment, two pairs of nested primers were designed in upstream and downstream orientation to allow 5′ and 3′ RACE (rapid amplification of cDNA ends). Analysis of the sequence of the resulting clones showed the existence of two different but closely related cDNAs. The sequence of the complete first mRNA (1253 nucleotides) consisted of a 5′-untranslated region (UTR) of 67 nucleotides, followed by an open reading frame (ORF) of 900 nucleotides encoding a protein of 300 amino acids with a molecular mass of 32.9 kDa and a 3′-UTR of 286 nucleotides with a poly(A)+ tail. The second mRNA (1244 nucleotides) had a 5′-UTR of 71 nucleotides, an ORF of 945 nucleotides encoding a protein of 315 amino acids with a molecular mass of 34.7 kDa and a 3′-UTR of 228 nucleotides with a poly(A)+ tail. These sequence data have been deposited in DDBJ/EMBL/GenBank (accession Nos AJ421780 and AJ421781).
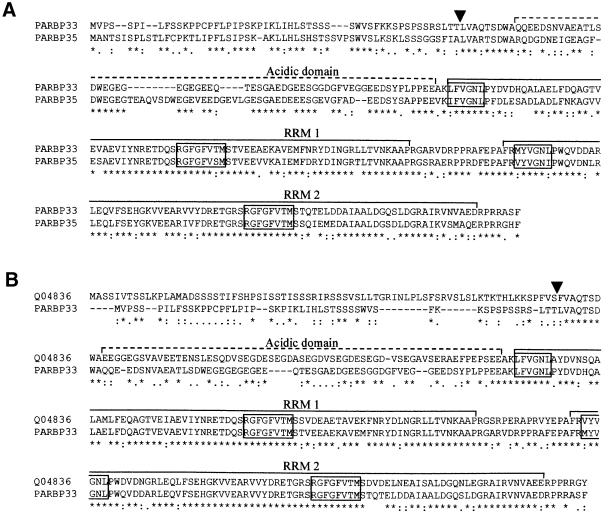
Inspection of the amino acid sequence of these two proteins showed that two of the tryptic peptides sequenced by mass spectrometry corresponded to the 32.9 kDa protein and the other two tryptic peptides to the 34.7 kDa protein, thus indicating that adduct 3 was a mixture of two species. The amino acid sequences of the two avocado proteins revealed a high identity between them and with members of group II of the family of chloroplast RNA-binding proteins (Ohta et al., 1995). Figure 6 shows the sequence alignments between the two avocado proteins (65% identity) and between one of them and a representative member of this group II, the precursor of the chloroplastic RNA-binding protein (329 amino acids) from Arabidopsis thaliana (ATRBP33, Swiss-Prot database accession No. Q04836) (Bar-Zvi et al., 1992; Cheng et al., 1994; Ohta et al., 1995): the two latter proteins showed 57% sequence identity and the same domain distribution (see below). Consequently, the avocado proteins were named precursors of the chloroplast RNA-binding proteins from Persea americana (PARBP33 and PARBP35). Considering the close similarity existing between these two proteins, hereafter we concentrated our attention on PARBP33.
Fig. 6. Sequence alignments between the avocado chloroplast RNA-binding proteins PARBP33 and PARBP35 (A) and between PARBP33 and ATRBP33 from A.thaliana (Swiss-Prot database accession No. Q04836) (B). The alignments were obtained using the Clustal W program with opening gap and end gap penalties of 10, and extending gap and separation gap penalties of 0.05. Arrowheads point to the processing site that in ATRBP33 separates the transit peptide from the mature protein (Ohta et al., 1995) and the putative processing sites in PARBP33 and PARBP35. Dashed and continuous lines delimit the acidic domain and the two RNA recognition motifs (RRMs), respectively. Conserved RNP-1 octapeptide and RNP-2 hexapeptide in both RRMs are boxed. ‘*’ indicates identical residues in the alignment, and ‘:’ and ‘.’ indicate conserved and semi-conserved substitutions, respectively.
Like ATRBP33, PARBP33 contains a putative N-terminal transit peptide rich in serine (∼30% of the residues) that could potentially mediate the import of the protein to the chloroplast. The exact length of the putative transit peptide in PARBP33 could not be inferred from sequence comparisons with ATRBP33 or other members of group II, but, from the alignment (Figure 6B), an approximate size of 53 residues was estimated, 17% of them identical to the putative transit peptide of ATRBP33. Following the putative transit peptide, PARBP33 has an acidic domain showing a 33% identity with the corresponding acidic domain of ATRBP33. In both proteins, ∼40% of the residues of this domain are aspartic or glutamic acid. Within the acidic domain, ATRBP33 contains six tandem repeats of SEGDX (residues 108–137) that are not present in PARBP33, which, in contrast, has four EG repeats (residues 79–86). After the acidic domain, PARBP33 has two consecutive RNA-binding domains of the RNA recognition motif (RRM) class, each with the two conserved boxes RNP-1 and RNP-2 (Burd and Dreyfuss, 1994). The first and second PARBP33 RRMs have 77 and 79 amino acids, and are 86 and 82% identical to those of ATRBP33, respectively. RNP-1 and RNP-2 boxes of the first PARBP33 RRM are RGFGFVTM and LFVGNL, respectively, whereas the second PARBP33 RMM contains RGFGFVTM and MYVGNL, respectively (Figure 6B). Both RRMs are separated by a spacer region of 15 amino acids 60% identical to the spacer region of ATRBP33. Finally, in the C-terminal end of PARBP33, there is a small positively charged region of seven amino acids 43% identical to that of ATRBP33.
Immunodetection of the ASBVd–PARBP33 adduct
Mass spectrometry analysis resulted in identification of PARBP33 (and PARBP35) as the avocado protein cross-linked to mc-ASBVd (+) RNA forming adduct 3. To corroborate this finding by an independent approach, PARBP33, tagged at its C-terminal end with the octapeptide VEH6, was expressed in Escherichia coli, purified by immobilized Ni affinity chromatography and used to obtain a rabbit antiserum. Once this antiserum had been shown to react specifically with the purified recombinant protein and with the native protein present in extracts from avocado leaves (data not shown), two aliquots of the concentrated interphase fraction resulting from phenol extraction of UV-irradiated ASBVd-infected leaves (see Figure 3) were separated by PAGE in urea gels. Material from one lane was transferred to a PVDF membrane and analyzed by western blot with the PARBP33-specific antiserum, whereas material from the other lane was transferred to a nylon membrane and analyzed by northern blot hybridization with a probe for detecting (+) viroid strands. Antibodies reacted with a species migrating at exactly the same position as the ASBVd–PARBP33 adduct revealed by northern blot hybridization (Figure 7). This result confirmed that the ASBVd–PARBP33 adduct (adduct 3) was the result of the UV-induced cross-link between mc-ASBVd (+) RNA and the chloroplast RNA-binding protein PARBP33, and also showed that the protein moiety of adduct 4 is distinct from PARBP33. The strong signal in the lower part of the western blot is due to free PARBP33 (data not shown) and the signal in the upper part of the blot is presumably generated by PARBP33 cross-linked to chloroplastic RNAs. This intense background precluded the clear distinction of a signal in the region of the blot corresponding to adducts 1 and 2.
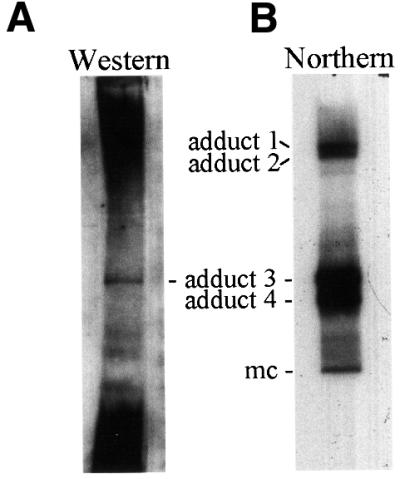
Fig. 7. ASBVd–protein adduct 3 as revealed with a PARBP33-specific antiserum (A) and with a probe specific for (+) viroid strands (B). Aliquots of the concentrated interphase fraction from ASBVd-infected UV-irradiated leaves were separated by denaturing PAGE in urea gels and transferred to PVDF and nylon membranes, respectively. The positions of the ASBVd–protein adducts and of the mc-ASBVd RNA are indicated between both panels.
Assisting effect of PARBP33 protein in self-cleavage of dimeric ASBVd (+) transcripts
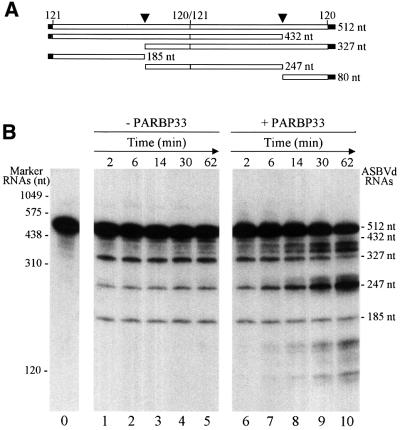
Some RNA-binding proteins of viral and cellular origin stimulate catalysis by hammerhead ribozymes in vitro, a property that has been proposed to result from the RNA chaperone activity of these proteins (Herschlag et al., 1994; Huang and Wu, 1998). This prompted us to check whether PARBP33 could influence self-cleavage of ASBVd RNAs in vitro. To examine this possibility, radioactively labeled dimeric ASBVd (+) transcripts were incubated with and without recombinant PARBP33. We chose this RNA as a representative of the oligomeric intermediates generated in the rolling circle mechanism of replication of this viroid (Bruening et al., 1982; Hutchins et al., 1986; Daròs et al., 1994) and also because hammerhead structures in ASBVd appear to act as double, and not as single, self-cleaving domains (Forster et al., 1988). When aliquots of both reactions taken at different times were analyzed by denaturing PAGE, the primary transcript of 512 nucleotides was observed to self-cleave as early as 2 min later, yielding two products of 432 and 327 nucleotides that resulted from the action of one or the other hammerhead structure and three products of 247, 185 and 80 nucleotides (the latter, not visible because of its small size, migrated out of the gel) that resulted from the action of both hammerhead structures (Figure 8). Comparative analysis of the products from both self-cleavage reactions showed that PARBP33 accelerated RNA self-cleavage: the dimeric ASBVd (+) transcript of 512 nucleotides and its derivatives of 432 and 327 nucleotides resulting from processing at one of the two self-cleavage sites disappeared more quickly during incubation, along with the accumulation of the ml-ASBVd (+) RNA of 247 nucleotides which resulted from processing at both self-cleavage sites (Figure 8B). Some other bands that increased their intensity with the incubation time, two between the ASBVd RNAs of 432 and 327 nucleotides, and two which migrated faster than the ASBVd RNA of 185 nucleotides, most probably resulted from RNA sites particularly prone to non-enzymatic degradation (some of the bands were visible in the protein-free incubation) or from RNase traces contaminating PARBP33.
Fig. 8. Effect of the avocado PARBP33 protein on hammerhead-mediated self-cleavage of dimeric ASBVd (+) transcripts. (A) Schematic representation of a dimeric ASBVd (+) transcript and of the self-cleavage products thereof. Vector and viroid sequences are denoted by black and white bars, respectively, and the self-cleavage sites are marked by arrowheads. Numbers on top of the bars refer to ASBVd positions (Hutchins et al., 1986) and numbers on the right indicate the size in nucleotides. (B) Self-cleavage kinetics of a radioactively labeled dimeric ASBVd (+) transcript incubated under standard self-cleavage conditions in the absence (lanes 1–5) or presence of PARBP33 (lanes 6–10). Reaction aliquots were taken at different times and analyzed by denaturing PAGE. Lane 0, control aliquot without incubation; lanes 1–5 and 6–10, aliquots taken after incubation for 2, 6, 14, 30 and 62 min, respectively. The positions of marker RNAs (with their sizes in nucleotides) are indicated on the left, and those of the uncleaved transcript and of the resulting self-cleavage fragments thereof on the right.
Discussion
Viroids, in spite of their minimal genome size, manage to independently complete a complex infectious cycle in host plants that, in addition to replication, includes intracellular, intercellular and long-distance movement of the genomic RNA (Palukaitis, 1987; Woo et al., 1999; Zhu et al., 2001), activation of host defense mechanisms (Itaya et al., 2001; Papaefthimiou et al., 2001) and, in most cases, induction of a pathogenic effect. However, because the available evidence strongly suggests that viroids do not code for any protein (for a review see Diener, 1999), these RNAs must interact with cellular proteins and recruit them to mediate different steps of their biological cycle. Apart from some data on the host RNA polymerase involved in replication of certain members of the family Pospiviroidae (Mühlbach and Sänger, 1979; Flores and Semancik, 1982; Schindler and Mühlbach, 1992; Warrilow and Symons, 1999), and on the possible participation of a phloem protein in movement of a viroid of this family (Gómez and Pallás, 2001; Owens et al., 2001), very little is known about other biologically relevant cellular proteins. This is particularly true for members of the other family, Avsunviroidae. Only some information about the nature of the chloroplastic RNA polymerase presumably catalyzing their replication has been obtained recently for ASBVd using an avocado system (Navarro et al., 2000) and for PLMVd with a bacterial system (Pelchat et al., 2001). However, additional host proteins are most likely to be involved in this and in other replication steps including self-cleavage which, despite being an RNA-based reaction, presumably is assisted in vivo by one or more cellular proteins.
To search for such proteins, we applied a strategy of UV irradiation of viroid-infected leaves to induce in situ cross-linking between viroid RNAs and host proteins physically associated with them in vivo. With this approach, we first detected by northern blot hybridization one putative ASBVd–host protein adduct, whose nature was confirmed by digestion with proteinase K. Disclosure of this interaction in intact tissue strongly suggests that it is biologically significant. Moreover, the adduct was detected even with very low UV doses, and samples subjected to considerably higher doses showed no degradation of the avocado RNAs (data not shown), indicating that no disorganization occurred that could have promoted spurious interactions between ASBVd RNAs and host proteins. Following extraction with phenol/chloroform, the characteristic viroid RNAs remained in the aqueous phase, whereas the adduct accumulated in the interphase, probably due to its covalently attached protein moiety. Northern blot analysis of the concentrated interphase fraction showed the existence of at least three additional ASBVd RNA–host protein adducts, and proteinase K digestion revealed that mc-ASBVd (+) RNA was the RNA moiety in the two most abundant adducts (3 and 4, Figure 3). Similarly to adducts 3 and 4, which exhibited slightly slower mobilities than that of mc-ASBVd (+) RNA, adducts 1 and 2 showed a parallel retardation with respect to dc-ASBVd (+) RNA, suggesting that they are the counterparts of adducts 3 and 4 in which the cross-linked RNA moiety is the dc- instead of the mc-ASBVd (+) RNA.
We anticipated two properties for host protein candidates to interact in vivo with ASBVd RNAs: chloroplastic localization and RNA-binding activity. Tandem mass spectrometry analysis of the protein component of the most abundant adduct 3 led to peptide sequence tags matching the sequences of some members of a family of RNA-binding proteins from different plant species (Figure 5). This family of nuclear-encoded chloroplastic proteins has been divided into three groups on the basis of phylogenetic comparisons (Ohta et al., 1995). One member of each group has been identified in A.thaliana (Ohta et al., 1995) although recent data indicate the existence of a second member of group II (Sato et al., 2000). In other species with a more complex genome, such as Nicotiana sylvestris, the family is composed of two proteins of group I, two of group II and one of group III (Ye et al., 1991). Other members of this family have been described in spinach (Schuster and Gruissem, 1991), maize (Cook and Walker, 1992) and barley (Churin et al., 1999). The 28RNP protein from spinach, which belongs to group II, has been involved in maturation of some chloroplastic mRNAs, more specifically in the formation of their 3′ ends (Schuster and Gruissem, 1991). Further studies with this tobacco family of proteins have shown that they are surprisingly abundant in the chloroplastic stroma, most of them forming complexes with mRNAs contributing to increase their stability (Nakamura et al., 2001), whereas others, especially those of group II, are found associated with unspliced pre-tRNAs, pointing to their involvement in the splicing or stability of intron-containing pre-tRNAs (Nakamura et al., 1999). Furthermore, group II protein cp31 has been implicated recently in RNA editing of tobacco chloroplast RNAs (Hirose and Sugiura, 2001). Collectively, these observations indicate that members of this protein family mediate different steps of the post-transcriptional metabolism of chloroplast RNAs, including stabilization, maturation and editing, thus contributing to the control of gene expression in this organelle.
Cloning and sequencing of the complete cDNAs of the protein component of the most abundant viroid adduct revealed the existence of two proteins, and confirmed that they were the avocado homologs of group II RNA-binding proteins; consequently, they were termed PARBP33 and PARBP35. These proteins contain the same domain distribution as the other members of the group, including an N-terminal putative transit peptide potentially directing the protein into the chloroplast, an acidic domain that forms the N-terminal region in the mature protein and two consecutive RNA-binding domains of the RMM class (Burd and Dreyfuss, 1994; Varani and Nagai, 1998). Independent confirmation that PARBP33 (and most probably PARBP35) was actually the protein cross-linked to ASBVd RNA, and not an accompanying contaminant (a remote possibility considering that adduct 3 was excised from an SDS–gel region close to a protein standard of 185 kDa, a size significantly larger than that of the mature PARBP33 and PARBP35) was obtained by co-localization in parallel blots of adduct 3 with an ASBVd RNA-specific probe and a PARBP33-specific polyclonal antiserum.
Once the bona fide nature of the ASBVd RNA–PARBP33 adduct had been established, we wondered about the role(s) of this protein in the viroid infection cycle. As already indicated, members of the family of chloroplast RNA-binding proteins to which PARBP33 belongs are involved in multiple steps of the post-transcriptional metabolism of chloroplast RNA transcripts, including maturation. We reasoned that hammerhead-mediated self-cleavage of oligomeric ASBVd RNAs, a key step of the rolling circle mechanism of replication, could be favored by PARBP33 protein. In vitro experiments using radioactively labeled dimeric ASBVd (+) transcripts demonstrated that this was indeed the case (Figure 8). Although a previous work has shown the RNA chaperone activity of certain non-specific RNA-binding proteins in hammerhead ribozyme catalysis (Herschlag et al., 1994), these proteins had a heterologous viral origin. Therefore, the situation reported here is, to our knowledge, the first example in which an assisting effect on hammerhead-mediated RNA self-cleavage is demonstrated for a protein found to be physically associated with the RNA in vivo, providing a more direct support for the idea that, in the cellular habitat, catalysis by hammerhead ribozymes is a protein-facilitated process. On the other hand, considering that the family of proteins to which PARBP33 and PARBP35 belong is involved in binding ribosome-free chloroplastic transcripts contributing to their stabilization, the observed interaction of these proteins with mc-ASBVd (+) RNA, the final product of the replication cycle, may also lend a protective effect to the viroid progeny resembling that provided by viral coat proteins to their cognate RNAs. It is interesting to note here that mc-ASBVd (+) RNA may reach very high accumulation levels in vivo, suggesting the existence of some mechanism protecting this RNA from degradation.
Cellular proteins participating in viral RNA-dependent RNA synthesis have been classified into those forming part of the RNA-dependent RNA polymerase (RdRp) holoenzyme and those binding to RNA template and directing the RdRp to the template (Lai, 1998), with a third class binding to primary transcripts and mediating their processing, although these modes are not mutually exclusive. Typical representative proteins that do not form part of the RdRp holoenzyme are the polypyrimidine tract-binding (PTB) protein and the heterogeneous nuclear ribonucleoprotein (hnRNP) A1, which in uninfected cells are involved in pre-mRNA splicing in the nucleus (with the second additionally involved in translational regulation in the cytoplasm). In virus-infected cells they also participate in the transcription and replication of viral RNA, probably mediating 5′–3′ cross-talk (Shi et al., 2000; Huang and Lai, 2001). In the choroplast context, group II RNA-binding proteins to which PARBP33 and PARBP35 belong apparently play a similar role to that of hnRNP A1 and PTB proteins that also have RNA-binding domains of the RRM class. Consistent with this role, PARBP33 and PARBP35 were found in close association with mc-ASBVd (+) RNA in ASBVd-infected cells. Moreover, in vitro, and possibly in vivo, PARBP33 assists the hammerhead-mediated self-cleavage of multimeric ASBVd RNAs, although other additional functions cannot be excluded; for example, PARBP33 and PARBP35 might escort the viroid into the chloroplast. Incidentally, these results also suggest a possible involvement of proteins such as hnRNP A1 and PTB in the replication of nuclear viroids of the family Pospiviroidae. In summary, viroids and RNA viruses seem to recruit similar cellular proteins into replication complexes in which they may provide template specificity for RNA-dependent RNA synthesis (a process catalyzed in viroids by DNA-dependent RNA polymerases that are forced to accept RNA templates) and facilitate appropriate processing.
Materials and methods
Tissue irradiation and homogenization
Leaf sections from ASBVd-infected avocado plants (Persea americana Mill.) were irradiated for different times using a UV cross-linker (Hoefer) equipped with 254 nm UV lamps and an internal dosimeter. Leaves were set on ice at ∼10 cm from the radiation source. Following irradiation, crude extracts were obtained by homogenizing the tissue in buffer B (0.1 M Tris–HCl pH 9.0, 0.1 M NaCl, 0.1 M 2-mercaptoethanol, 10 mM EDTA) containing 5 M urea, at a ratio of 5 ml of buffer per gram of fresh weight. Extracts were clarified by low-speed centrifugation.
RNA analysis by denaturing PAGE and northern blot hybridization
RNAs were separated by denaturing 5% PAGE using TBE (89 mM Tris, 89 mM boric acid, 2.5 mM EDTA) for the electrode buffer, and the same plus 8 M urea for the gel buffer. After ethidium bromide staining, RNAs were electroblotted to nylon membranes (Hybond-N, Amersham Pharmacia Biotech) and fixed by UV irradiation. Membranes were hybridized at 65°C in the presence of 50% formamide with 32P-labeled RNA probes specific for both ASBVd polarity strands (Daròs et al., 1994).
Proteinase K digestion
Preparations containing ASBVd–protein adducts were mixed with 20 µg of proteinase K in a volume of 100 µl of 10 mM Tris–HCl pH 8.0, 5 mM EDTA, 1 mM 2-mercaptoethanol and 0.5% SDS, and incubated first at 42°C for 30 min and then at 55°C for another 30 min. After digestion, RNAs were recovered by ethanol precipitation.
Purification of the RNA–protein adduct
ASBVd-infected avocado leaves (15 g) were irradiated with 60 J/cm2 of UV light (see above) and homogenized with 75 ml of buffer B containing 5 M urea. The extract was clarified by low-speed centrifugation, and solid urea and NaCl were added while stirring to final concentrations of 8 and 1 M, respectively. Precipitated material was discarded by centrifugation and the supernatant (∼60 ml) was extracted with 30 ml of phenol/chloroform (1:1) equilibrated and saturated with 0.1 M Tris–HCl pH 9.0. After centrifuging at 8000 g for 1 h, the aqueous and the organic phases, as well as the interphase, were recovered. No further treatment was applied to the aqueous phase. The organic phase was mixed with 1 vol. of isopropanol, kept for 1 h at –20°C, and the precipitated material was recovered by centrifugation, washed with 70% ethanol, dried and resuspended in buffer C (25 mM Tris–HCl pH 9.0, 25 mM NaCl, 10 mM 2-mercaptoethanol, 1 mM EDTA, 8 M urea and 1% NP-40). The interphase was re-extracted with 60 ml of buffer B (containing 8 M urea and 1 M NaCl) and 30 ml of buffer-saturated phenol/chloroform, recovered by centrifugation and dispersed by stirring in 15 ml of buffer B (containing 8 M urea, 1 M NaCl and 1% SDS). After adding 1 vol. of isopropanol and stirring for 15 min, the mixture was kept for 1 h at –20°C, and the precipitated material was recovered by centrifugation, washed with 70% ethanol, dried and resuspended in buffer C.
ASBVd–protein adducts present in the interphase fraction were purified further by non-denaturing 5% PAGE in TAE buffer (40 mM Tris, 20 mM sodium acetate, 1 mM EDTA pH 7.2) using a polymerization mixture of acrylamide:N,N′-bis(acryloyl)cystamine (29:1) that resulted in a gel solubilizable in an excess of a reducing agent (Hansen et al., 1980). After staining with ethidium bromide, the segment delimited by the DNA markers of 300 and 400 bp was sliced and solubilized with 0.1 vol. of 2-mercaptoethanol. Then, 1 vol. of buffer RW (0.1 M Tris–HCl pH 7.0, 0.25 M NaCl, 10 mM EDTA, 10 mM 2-mercaptoethanol, 8 M urea and 1% NP-40) was added and the sample was loaded on a Qiagen tip-100 anion-exchange column previously equilibrated with 10 ml of RW buffer. The column was washed three times with 3.3 ml of RW buffer and eluted three times with 3.3 ml of buffer RE (0.1 M Tris–HCl pH 9.0, 1.5 M NaCl, 10 mM EDTA, 10 mM 2-mercaptoethanol, 8 M urea and 1% NP-40). Adducts in the eluted fractions were precipitated with isopropanol and resuspended in buffer C.
Mass spectrometry protein sequencing and database search
Purified ASBVd–protein adducts were separated by SDS–PAGE in 7.5% gels that were stained with silver. Protein analysis by tandem mass spectrometry was performed by Protana A/S (Odense, Denmark). To this end, the band corresponding to adduct 3 was sliced and subjected to in-gel digestion with trypsin (Shevchenko et al., 1996). Tryptic peptides were micropurified and sequenced on a Sciex QSTAR (quadruple-TOF mass spectrometer) equipped with a Protana A/S nanoelectrospray source. The resulting peptide amino acid sequences were queried against a non-redundant protein database using the WWW version of the PeptideSearch tool from the EMBL Protein and Peptide Group (Heidelberg, Germany).
RT–PCR amplification of cDNA, cloning and sequencing
Nucleic acids from avocado leaves were extracted with buffer-saturated phenol/chloroform and the polysaccharides were removed with methoxyethanol (Daròs et al., 1994). Poly(A)+ mRNA was purified from the fraction insoluble in 2 M LiCl using the Oligotex mRNA kit (Qiagen). A fragment of 71 bp of the PARBP33 cDNA was RT–PCR amplified with the Superscript II RNase H– reverse transcriptase (Life Technologies) and Taq DNA polymerase (Roche Molecular Biochemicals) using the degenerated primers PI [5′-GA(A/G)(A/T) (C/G)NGCNGA(A/G)GTNAT-3′] and PII [5′-GTN(C/G)(A/T) CATNGTNAC(A/G)AA-3′] homologous and complementary, respectively, to the cDNA regions coding for peptides ESAEV(L/I) and FVTMST identified by mass spectrometry (alternative residues in partially degenerated positions are given in parentheses; N refers to completely degenerated positions). From the sequence of the 71 bp fragment, two pairs of nested primers were designed: PIII (5′-CGAGGTTATTTACAATAGAGAAACAG-3′) and PIV (5′-GATCAGAGTCGCGGGTTTGG-3′) homologous to the cDNA fragment, and PV (5′-ACTCTGATCTGTTTCTCTATTG-3′) and PVI (5′-CACAAAACCAAACCCGCG-3′) complementary to the same sequence. These primers were used to RT–PCR amplify the 5′ and 3′ halves of the cDNA with the Marathon cDNA amplification kit (Clontech), that were cloned in plasmid pGEM-T Easy (Promega). The inserts from 12 recombinant plasmids (six for the 5′ half and six for the 3′ half) were sequenced in both orientations with an ABI PRISM DNA-377 apparatus (Perkin-Elmer). Three additional clones from the 3′ half exhibited a slightly different sequence corresponding to a closely related protein (PARBP35), the 5′ half cDNA of which was amplified by RT–PCR with primer PVII (5′-TCATAGCGGTCAAACATCTCAATC-3′).
Protein expression and purification
With the previous sequence information, a cDNA corresponding to the complete PARBP33 ORF was RT–PCR amplified from avocado poly(A)+ mRNA with Superscript II RNase H– reverse transcriptase and Pfu DNA polymerase (Stratagene), using the homologous primer PVIII (5′-CGTACGCATATGGTACCCTCCTCCCCCATC-3′) with an NdeI site, and the complementary primer PIX (5′-CGTACGGTCGACAA-AGGAGGCACGCCTTGGTCG-3′) with a SalI site (both restriction sites underlined). NdeI–SalI-digested cDNA was cloned in NdeI–XhoI-linearized plasmid pET-23a(+) (Novagen). The resulting plasmid directed, under the control of the T7 RNA polymerase promoter, the expression of a recombinant version of PARBP33 from avocado containing the C-terminal octapeptide tag VEH6. Escherichia coli cells of the strain BL21(DE3)pLysS (Novagen) were transformed with this plasmid, and a 250 ml culture was grown at 37°C to ∼0.6 OD600. The recombinant protein, whose expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 25°C, was purified from the cell pellet in denaturing conditions (for antiserum production) and non-denaturing conditions (for RNA self-cleavage) using the Ni-NTA resin (Qiagen) and following the manufacturer’s instructions.
Antiserum production and western blot analysis
Approximately 100 µg of the recombinant avocado protein PARBP33 were emulsified with an equal volume of complete Freund’s adjuvant and injected intradermaly into a rabbit. Immunizations were repeated twice more at 1 week intervals but using incomplete adjuvant. The rabbit was bled 1 week after the last immunization and the serum recovered by centrifugation. Western blot analysis was carried out using a 1:2000 dilution of the antiserum and the luminescent detection system ECL+Plus (Amersham Pharmacia Biotech).
In vitro self-cleavage of RNA
Radioactively labeled dimeric ASBVd (+) RNA was synthesized by in vitro transcription, purified by denaturing PAGE and resuspended in 1 mM EDTA pH 8.0 (Hutchins et al., 1986). Aliquots (500 ng) were boiled for 2 min and snap-cooled on ice. After adding the self-cleavage buffer (final concentration 50 mM Tris–HCl pH 8.0, 10 mM MgCl2 and 0.5 mM EDTA) and 500 ng of recombinant PARBP33, or the same volume of protein buffer, the reaction was incubated at 37°C. Samples were taken at different times and the reaction was stopped by mixing with 1 vol. of 20 mM EDTA pH 8.0 in 96% formamide, boiling for 2 min and snap-cooling on ice. RNAs were separated by denaturing PAGE and the extent of the self-cleavage reaction was monitored by autoradiography.
Acknowledgments
Acknowledgements
We thank M.Bordás, V.Moncholí and A.Ahuir for excellent technical assistance, Drs V.Pallás, C.Hernández and A.E.Martínez for critical reading and suggestions, and Barraclough-Donnellan for English revision. R.F. was partially supported by grant PB98-0500 of the DGES de España. J.A.D. was the recipient of a postdoctoral contract from the Ministerio de Ciencia y Tecnología de España.
References
- Bar-Zvi D., Shagan,T., Schindler,U. and Cashmore,A.R. (1992) RNP-T, a ribonucleoprotein from Arabidopsis thaliana, contains two RNP-80 motifs and a novel acidic repeat arranged in an α-helix conformation. Plant Mol. Biol., 20, 833–838. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. and Carmichael,G.G. (1979) RNA replication: function and structure of Qβ-replicase. Annu. Rev. Biochem., 48, 525–548. [DOI] [PubMed] [Google Scholar]
- Bonfiglioli R.G., McFadden,G.I. and Symons,R.H. (1994) In situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J., 6, 99–103. [Google Scholar]
- Bonfiglioli R.G., Webb,D.R. and Symons,R.H. (1996) Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J., 9, 457–465. [Google Scholar]
- Branch A.D. and Robertson,H.D. (1984) A replication cycle for viroids and other small infectious RNAs. Science, 223, 450–455. [DOI] [PubMed] [Google Scholar]
- Branch A.D., Benenfeld,B.J. and Robertson.H.D. (1988) Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl Acad. Sci. USA, 85, 9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening G., Gould,A.R., Murphy,P.J. and Symons,R.H. (1982) Oligomers of avocado sunblotch viroid are found in infected avocado leaves. FEBS Lett., 148, 71–78. [Google Scholar]
- Buck K.W. (1996) Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res., 47, 159–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Bussière F., Lehoux,J., Thompson,D.A., Skrzeczkowski,L.J. and Perreault,J.P. (1999) Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J. Virol., 73, 6353–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.H., Cline,K. and DeLisle,A.J. (1994) An Arabidopsis chloroplast RNA-binding protein gene encodes multiple mRNAs with different 5′ ends. Plant Physiol., 106, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churin Y., Hess,W.R. and Borner,T. (1999) Cloning and characterization of three cDNAs encoding chloroplast RNA-binding proteins from barley (Hordeum vulgare L.): differential regulation of expression by light and plastid development. Curr. Genet., 36, 173–181. [DOI] [PubMed] [Google Scholar]
- Cook W.B. and Walker,J.C. (1992) Identification of a maize nucleic acid-binding protein (NBP) belonging to a family of nuclear-encoded chloroplast proteins. Nucleic Acids Res., 20, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté F., Lévesque,D. and Perreault,J.P. (2001) Natural 2′,5′-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J. Virol., 75, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs J.A., Marcos,J.F., Hernández,C. and Flores,R. (1994) Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl Acad. Sci. USA, 91, 12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De B.P. and Banerjee,A.K. (1997) Role of host proteins in gene expression of nonsegmented negative strand RNA viruses. Adv. Virus Res., 48, 169–204. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1971) Potato spindle tuber ‘virus’: a plant virus with properties of a free nucleic acid. III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ.Virology, 43, 75–89. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1972) Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology, 50, 606–609. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1999) Viroids and the nature of viroid diseases. Arch. Virol., Suppl. 15, 203–220. [DOI] [PubMed] [Google Scholar]
- Feldstein P.A., Hu,Y. and Owens,R.A. (1998) Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl Acad. Sci. USA, 95, 6560–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R. and Semancik,J.S. (1982) Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl Acad. Sci. USA, 79, 6285–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Randles,J.W., Bar-Joseph,M. and Diener,T.O. (2000a) Viroids. In van Regenmortel,M.H.V. et al. (eds), Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA, pp. 1009–1024.
- Flores R., Daròs,J.A. and Hernández,C. (2000b) The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res., 55, 271–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Hernández,C., De la Peña,M., Vera,A. and Daròs,J.A. (2001). Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol., 341, 540–552. [DOI] [PubMed] [Google Scholar]
- Forster A.C. and Symons,R.H. (1987) Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell, 49, 211–220. [DOI] [PubMed] [Google Scholar]
- Forster A.C., Davies,C., Sheldon,C.C., Jeffries,A.C. and Symons,R.H. (1988) Self-cleaving viroid and newt RNAs may only be active as dimers. Nature, 334, 265–267. [DOI] [PubMed] [Google Scholar]
- Gómez G. and Pallás,V. (2001) Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant–Microbe Interact., 14, 910–913. [DOI] [PubMed] [Google Scholar]
- Gross H.J., Domdey,H., Lossow,C., Jank,P., Raba,M., Alberty,H. and Sänger,H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Hadidi A. (1988) Synthesis of disease-associated proteins in viroid-infected tomato leaves and binding of viroids to host proteins. Phytopathology, 78, 575–578. [Google Scholar]
- Hansen J.N., Pheiffer,B.H. and Boehnert,J.A. (1980) Chemical and electrophoretic properties of solubilizable disulfide gels. Anal. Biochem., 105, 192–201. [DOI] [PubMed] [Google Scholar]
- Harders J., Lukacs,N., Robert-Nicoud,M., Jovin,J.M. and Riesner,D. (1989) Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J., 8, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D., Khosla,M., Tsuchihashi,Z. and Karpel,R.L. (1994) An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J., 13, 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T. and Sugiura,M. (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J., 20, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockensmith J.W., Kubasek,W.L., Vorachek,W.R., Evertsz,E.M. and von Hippel,P.H. (1991) Laser cross-linking of protein–nucleic acid complexes. Methods Enzymol., 208, 211–236. [DOI] [PubMed] [Google Scholar]
- Huang P. and Lai,M.M.C. (2001) Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′–3′-end cross talks of mouse hepatitis virus RNA. J. Virol., 75, 5009–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.S. and Wu,H.N. (1998) Identification and characterization of the RNA chaperone activity of hepatitis δ antigen peptides. J. Biol. Chem., 273, 26455–26461. [DOI] [PubMed] [Google Scholar]
- Hutchins C.J., Keese,P., Visvader,J.E., Rathjen,P.D., McInnes,J.L. and Symons,R.H. (1985) Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol., 4, 293–304. [DOI] [PubMed] [Google Scholar]
- Hutchins C., Rathjen,P.D., Forster,A.C. and Symons,R.H. (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res., 14, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya A., Folimonov,A., Matsuda,Y., Nelsom,R. and Ding,B. (2001) Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant–Microbe Interact., 14, 1332–1334. [DOI] [PubMed] [Google Scholar]
- Janda M. and Ahlquist,P. (1993) RNA-dependent replication, transcription and persistence of brome mosaic virus RNA replicons. Cell, 72, 961–970. [DOI] [PubMed] [Google Scholar]
- Klaff P., Gruner,R., Hecker,R., Sättler,A., Theissen,G. and Riesner,D. (1989) Reconstituted and cellular viroid–protein complexes. J. Gen. Virol., 70, 2257–2270. [Google Scholar]
- Lai M.M.C. (1998) Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology, 244, 1–12. [DOI] [PubMed] [Google Scholar]
- Lima M.I., Fonseca,M.E.N., Flores,R. and Kitajima,E.W. (1994) Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch. Virol., 138, 385–390. [DOI] [PubMed] [Google Scholar]
- Mann M. and Wilm,M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem., 66, 4390–4399. [DOI] [PubMed] [Google Scholar]
- Mann M., Hendrickson,R.C. and Pandey,A. (2001) Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem., 70, 437–473. [DOI] [PubMed] [Google Scholar]
- Mühlbach H.P. and Sänger,H.L. (1979) Viroid replication is inhibited by α-amanitin. Nature, 278, 185–188. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ohta,M., Sugiura,M. and Sugita,M. (1999) Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett., 460, 437–441. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ohta,M., Sugiura,M. and Sugita,M. (2001) Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem., 276, 147–152. [DOI] [PubMed] [Google Scholar]
- Navarro J.A., Daròs,J.A. and Flores,R. (1999) Complexes containing both polarity strands of avocado sunblotch viroid: identification in chloroplasts and characterization. Virology, 253, 77–85. [DOI] [PubMed] [Google Scholar]
- Navarro J.A. and Flores,R. (2000) Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J., 19, 2662–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J.A., Vera,A. and Flores,R. (2000) A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology, 268, 218–225. [DOI] [PubMed] [Google Scholar]
- Ohta M., Sugita,M. and Sugiura,M. (1995) Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol. Biol., 27, 529–539. [DOI] [PubMed] [Google Scholar]
- Owens R.A., Blackburn,M. and Ding,B. (2001) Possible involvement of phloem protein 2 in long-distance viroid movement. Mol. Plant–Microbe Interact., 14, 905–909. [DOI] [PubMed] [Google Scholar]
- Palukaitis P. (1987) Potato spindle tuber viroid: investigation of long-distance intra-plant transport route. Virology, 158, 239–241. [DOI] [PubMed] [Google Scholar]
- Papaefthimiou I., Hamilton,A.J., Denti,M.A., Baulcombe,D.C., Tsagris,M. and Tabler,M. (2001) Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res., 29, 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashev I.G., Dimitrov,S.I. and Angelov,D. (1991) Crosslinking proteins to nucleic acids by ultraviolet laser irradiation. Trends Biochem. Sci., 16, 323–326. [DOI] [PubMed] [Google Scholar]
- Pelchat M., Côté,F. and Perreault,J.P. (2001) Study of the polymerization step of the rolling circle replication of peach latent mosaic viroid. Arch. Virol., 146, 1753–1763. [DOI] [PubMed] [Google Scholar]
- Prody G.A., Bakos,J.T., Buzayan,J.M., Schneider,I.R. and Bruening,G. (1986) Autolytic processing of dimeric plant virus satellite RNA. Science, 231, 1577–1580. [DOI] [PubMed] [Google Scholar]
- Sagesser R., Martínez,E., Tsagris,M. and Tabler,M. (1997) Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res., 25, 3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura,Y., Kaneko,T., Katoh,T., Asamizu,E., Kotani,H. and Tabata,S. (2000) Structural analysis of Arabidopsis thaliana chromosome 5. X. Sequence features of the regions of 3.076.755 bp covered by sixty P1 and TAC clones. DNA Res., 7, 31–63 [DOI] [PubMed] [Google Scholar]
- Schindler I.M. and Mühlbach,H.P. (1992) Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci., 84, 221–229. [Google Scholar]
- Schuster G. and Gruissem,W. (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J., 10, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. Mann,M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shi S.T., Huang,P., Li,H.P. and Lai,M.M. (2000) Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J., 19, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiesmacher E., Mühlbach,H.P., Schnölzer,M., Haas,B. and Sänger,H.L. (1983) Oligomeric forms of potato spindle tuber viroid (PSTV) and of its complementary RNA are present in nuclei isolated from viroid-infected potato cells. Biosci. Rep., 3, 767–774. [DOI] [PubMed] [Google Scholar]
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- Warrilow D. and Symons,R.H. (1999) Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo.Arch. Virol., 144, 2367–2375. [DOI] [PubMed] [Google Scholar]
- Wolff P., Gilz,R., Schumacher,J. and Riesner,D. (1985) Complexes of viroids with histones and other proteins. Nucleic Acids Res., 13, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo Y.-M., Itaya,A., Owens,R.A., Tang,L., Hammons,R.W., Chou,H.-C., Lai,M.M.C. and Ding,B. (1999) Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplasts. Plant J., 17, 627–635. [Google Scholar]
- Ye L., Li,Y., Fukami-Kobayashi,K., Go,M., Konishi,T., Watanabe,A. and Sugiura,M. (1991) Diversity of a ribonucleoprotein family in tobacco chloroplasts: two new chloroplast ribonucleoproteins and a phylogenetic tree of ten chloroplast RNA-binding domains. Nucleic Acids Res., 19, 6485–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Green,L., Woo,Y.-M., Owens,R.A. and Ding,B. (2001) Cellular basis of potato spindle tuber viroid systemic movement. Virology, 279, 69–77. [DOI] [PubMed] [Google Scholar]