Abstract
Mitochondria are one of the hallmarks of eukaryotic cells, exporting ATP in exchange for cytosolic ADP using ADP/ATP carriers (AAC) located in the inner mitochondrial membrane. In contrast, several evolutionarily important anaerobic eukaryotes lack mitochondria but contain hydrogenosomes, peculiar organelles of controversial ancestry that also supply ATP but, like some fermentative bacteria, make molecular hydrogen in the process. We have now identified genes from two species of the hydrogenosome-containing fungus Neocallimastix that have three-fold sequence repeats and signature motifs that, along with phylogenetic analysis, identify them as AACs. When expressed in a mitochondrial AAC- deficient yeast strain, the hydrogenosomal protein was correctly targeted to the yeast mitochondria inner membrane and yielded mitochondria able to perform ADP/ATP exchange. Characteristic inhibitors of mitochondrial AACs blocked adenine nucleotide exchange by the Neocallimastix protein. Thus, our data demonstrate that fungal hydrogenosomes and yeast mitochondria use the same pathway for ADP/ATP exchange. These experiments provide some of the strongest evidence yet that yeast mitochondria and Neocallimastix hydrogenosomes are but two manifestations of the same fundamental organelle.
Keywords: ADP/ATP carrier/hydrogenosomes/mitochondria/Neocallimastix
Introduction
A variety of microbial eukaryotes found in diverse anaerobic habitats lack mitochondria (Fenchel and Finlay, 1995). Some of these anaerobes, such as Tricho monas, ciliates, and chytrid fungi like Neocallimastix, possess ATP-producing organelles called hydrogenosomes, so called because they produce hydrogen (Müller, 1993). Hydrogenosomes generate ATP by substrate-level phosphorylation, and they lack an electron transport chain and, with one possible exception, an associated organelle genome (Embley et al., 1997; Akhmanova et al., 1998; Dyall et al., 2000). Hydrogenosome-containing eukaryotes do not form a coherent phylogenetic group, but appear throughout the evolutionary tree (Embley and Hirt, 1998). Hydrogenosomes in contemporary eukaryotes have thus originated independently, revealing an extraordinary capacity of eukaryotes to repeatedly evolve hydrogen-producing organelles. To understand how eukaryotes achieve this, we need to determine both the origins of the unusual prokaryotic-like anaerobic biochemistry used to make hydrogen (Horner et al., 1999, 2000), and the origin(s) of the intracellular compartment to which the enzymes for making hydrogen are sorted.
Recent work on the (oxygen-sensitive) hallmark enzymes of hydrogenosomes, iron hydrogenase and pyruvate:ferredoxin oxidoreductase (PFO), suggests an early evolutionary origin of their genes with unexpectedly widespread retention among eukaryotic lineages (Horner et al., 1999, 2000; Rotte et al., 2001). The complex gene history of these enzymes involves possible gene duplications, gene fusions, and lineage specific retention and loss. Moreover, in at least one case, that of ciliates, a relatively late horizontal gene transfer from a prokaryote may have provided the hydrogenase gene needed for hydrogenosome function (Akhmanova et al., 1998; Horner et al., 2000). The eukaryotes shown to contain hydrogenase and/or PFO have targeted these enzymes to different cellular compartments including the cytosol, plastids, hydrogenosomes and mitochondria (Horner et al., 1999, 2000; Florin et al., 2001; Rotte et al., 2001).
The unfolding phylogeny for hydrogenase and PFO contributes to illuminating the apparent facility of eukaryotes to make hydrogen, but cannot explain the origins of the hydrogenosome. Early theories of a separate endosymbiotic origin for the hydrogenosome from an anaerobic bacterium (Whatley et al., 1979) have given way to hypotheses positing endogenous origins. Thus, for ciliates and Trichomonas, the weight of evidence is consistent with them having changed the biochemistry of one-time mitochondria to produce hydrogen, as an adaptation to anaerobic environments (reviewed in Embley et al., 1997; Akhmanova et al., 1998; Embley and Martin, 1998; Dyall et al., 2000). The origin of hydrogenosomes in rumen chytrid fungi like Neocallimastix is potentially exceptional, rekindling the question of their evolutionary origin. Protein import into fungal hydrogenosomes has been reported to have features of both mitochondrial (van der Giezen et al., 1998) and peroxisomal (Marvin-Sikkema et al., 1993a) import systems. Moreover, ultrastructural data have been interpreted to support each of these organelles as the progenitor of chytrid hydrogenosomes (Marvin-Sikkema et al., 1993b; Benchimol et al., 1997; Hackstein and Vogels, 1997; van der Giezen et al., 1997b; Akhmanova et al., 1998). The debate is complicated further by the lack of an associated organelle genome, which could provide the most incisive evidence regarding the identity of fungal hydrogenosomes (van der Giezen et al., 1997b).
Discriminating evidence for the ancestry of chytrid hydrogenosomes should be preserved in the membrane proteins that are essential for its key function, that of ATP export to supply the cytosol with metabolic energy. Here we report the identification and characterization of an ADP/ATP carrier in the hydrogenosomes of the anaerobic fungi Neocallimastix frontalis and Neocallimastix patriciarum. The hydrogenosomal ADP/ATP carrier has similar properties in primary structure, mode of transport and sensitivity towards inhibitors as its mitochondrial counterparts. Moreover, phylogenetic analyses demonstrate its common ancestry with mitochondrial AAC from aerobic fungi. Expressed in AAC-deficient yeast, the hydrogenosomal protein is correctly imported to mitochondria and able to exchange ATP for ADP.
Results
Isolation of AAC genes from Neocallimastix species
PCR primers designed against conserved regions of ADP/ATP carriers (AAC) were used to amplify a fragment of 450 bp from N.frontalis genomic DNA. The amino acid sequence revealed a strong similarity to AAC genes from aerobic fungi. The fragment was used as a heterologous probe to isolate several full-length clones from a N.patriciarum cDNA library. PCR primers were designed to amplify the entire genomic coding sequence for both species. Comparison of the genomic and cDNA sequences established that no introns were present in the Neocallimastix genes.
Southern blotting of genomic DNA from N.frontalis digested with EcoRI and VspI demonstrated three hybridising bands when the original 450 bp AAC fragment was used as a probe (data not shown). The full-length N.frontalis AAC cloned in this study does not contain restriction sites for either of these enzymes, suggesting that there are two or three copies of this gene present on the Neocallimastix genome. There are three, very similar copies of AAC present in most organisms (Fiore et al., 1998). Sequencing of several N.patriciarum cDNA clones did not reveal any sequence variation. Since it is known that expression of different AAC is not constitutive (Lawson and Douglas, 1988), it is possible that the cDNA library does not contain all of the AAC isoforms.
Sequence features of the Neocallimastix AAC sequences
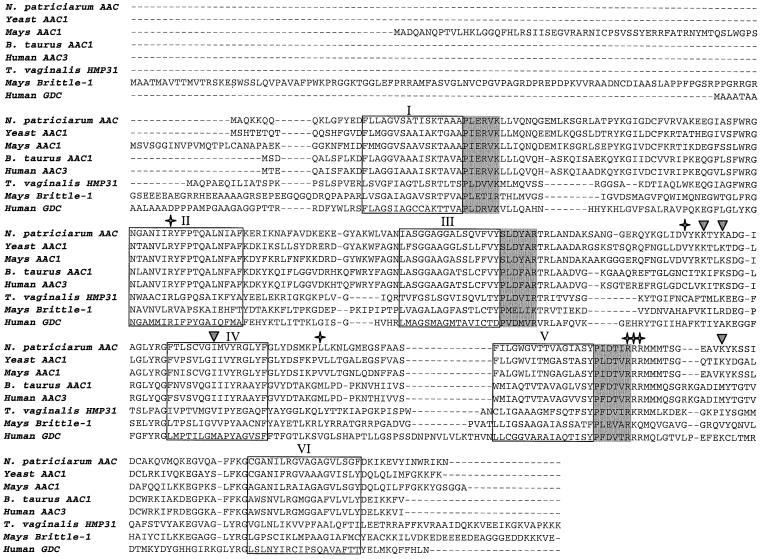
The fungal genes show a high level of sequence similarity to AAC sequences from aerobic mitochondria-containing eukaryotes and contain all of the features that are characteristic for mitochondrial AAC (Figure 1). The two Neocallimastix genes encode identical proteins of 308 amino acids with calculated masses of 33.7 kDa. Hydrophobicity profiles identify six transmembrane spanning regions in the Neocallimastix sequences (data not shown). The amino acid sequence shows the characteristic tripartite repeats of mitochondrial carriers and the conserved (D/E)-X-hydrophobic-(K/R) motif, which follows each odd transmembrane spanning helix (Saraste and Walker, 1982). Most of the amino acids important for the function of yeast and bovine AAC are conserved in the Neocallimastix sequences (Boulay et al., 1983; Dalbon et al., 1988; Klingenberg, 1989; Gawaz et al., 1990; Lawson et al., 1990; Kuan and Saier, 1993). Like other transporters belonging to the mitochondrial carrier family (MCF) (Walker, 1992; Sirrenberg et al., 1996), the inferred amino acid sequences of the Neocallimastix genes reported here do not appear to encode any N-terminal mitochondrial targeting pre-sequences (von Heijne et al., 1989), nor known peroxisomal targeting signals (Rachubinski and Subramani, 1995).
Fig. 1. Alignment of the novel Neocallimastix AAC protein reported here with typical members of the AAC subfamily, T.vaginalis hydrogenosomal HMP31, the maize brittle-1 protein and the human Graves’ disease carrier (GDC). Stars denote amino acids implicated in nucleotide binding or atractylate reactions in Bos taurus (Boulay et al., 1983; Dalbon et al., 1988); inverted triangles denote amino acids implicated in carrier function in S.cerevisiae (Gawaz et al., 1990; Lawson et al., 1990); shaded areas are signature motifs following each odd membrane spanning domain (Saraste and Walker, 1982); and boxed areas denote the six transmembrane helices.
Phylogenetic analysis of the Neocallimastix AAC
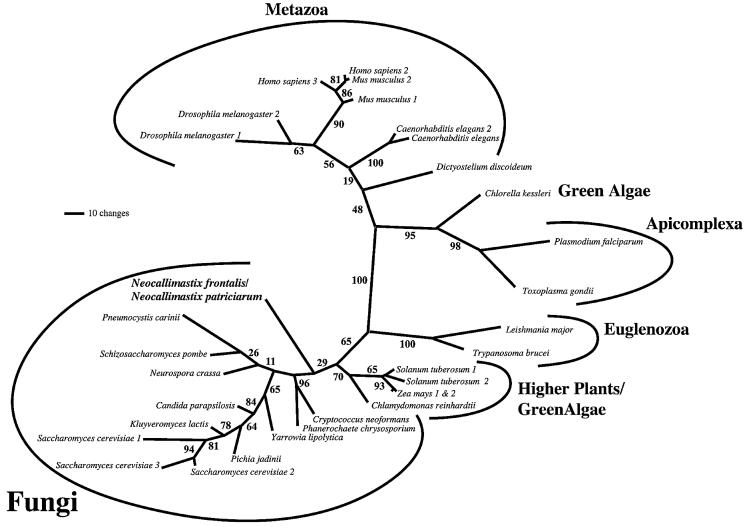
Maximum likelihood trees, estimated under complex models of sequence evolution, incorporating corrections for the presence of invariant sites, site-rate heterogeneity and differential sequence composition, recovered the Neocallimastix sequences as the weakly supported basal branch of a group composed entirely of fungal AAC sequences (Figure 2). This placement is consistent with phylogenies derived from SSU ribosomal RNA (Van der Auwera and De Wachter, 1996), which suggest that chytrids like Neocallimastix are the earliest diverging fungal lineage. In accordance with recent phylogenetic analyses of AAC sequences (Loytynoja and Milinkovitch, 2001), our data suggest that the history of the AAC gene family is complex and features multiple gene duplications and losses, both at the species level and at deeper nodes within the tree. The polyphyly of higher plants and green algae (an accepted clade at the organismal level) may represent an example of gene duplication and differential retention among lineages, as might the branch uniting higher plant and fungal AACs.
Fig. 2. Phylogenetic analysis of AAC family protein sequences. Unrooted maximum likelihood phylogenetic tree of 32 AAC protein sequences. The Neocallimastix sequences are recovered as part of a monophyletic group otherwise defined by mitochondrial AACs of aerobic fungi. The polyphyly of green algal sequences is probably due to gene duplications early in the history of the AAC family, coupled with differential retention of paralogues among lineages (see text). Numbers at the nodes represent bootstrap values.
Localization and functional studies of the hydrogenosomal ADP/ATP carrier
To establish the localization of the ADP/ATP carrier, cellular fractions from N.patriciarum were obtained by mechanical disruption and differential centrifugation (Marvin-Sikkema et al., 1993b). Immunoblotting of fractions showed the presence of a protein that cross-reacted with Neurospora crassa polyclonal anti-AAC serum in the cell-free extract and the hydrogenosomal fraction, but not in the cytosolic fraction (Figure 3A). The apparent molecular mass of the single band (31 kDa) is slightly smaller than the calculated molecular mass of the predicted gene product (33.7 kDa).
Fig. 3. Cellular fractionation and adenine nucleotide exchange in hydrogenosomal membranes. Western blot of N.patriciarum cellular fractions (20 µg) probed with an antiserum raised against N.crassa AAC (A) and the hydrogenosomal marker β-succinyl-CoA synthetase (Brondijk et al., 1996; Benchimol et al., 1997) (B). Lane 1: cell-free extract; lane 2: cytosolic fraction; and lane 3: hydrogenosomal fraction. (C) Uptake experiments with isolated hydrogenosomal membranes fused with liposomes. Fused membranes were loaded with 5 mM ADP and the exchange reaction was started by dilution of the vesicles in buffer containing a final concentration of 1.54 µM [8-14C] ATP (closed circles). Nucleotide exchange was inhibited by the addition of bongkrekic acid and carboxy-atractyloside at a final concentration of 20 and 25 µM, respectively (open circles).
ADP/ATP exchange activity was measured in hydrogenosomal membranes fused with liposomes. The results show that adenine nucleotide exchange takes place across hydrogenosomal membranes and that the activity can be inhibited by the addition of carboxy-atractyloside and bongkrekic acid (Figure 3C).
Complementation studies in the AAC-deficient Saccharomyces cerevisiae strain WB-12
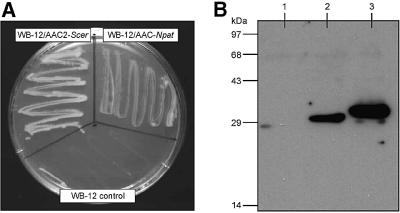
Saccharomyces cerevisiae strain WB-12 is unable to grow on non-fermentable carbon sources, such as glycerol or ethanol, because it lacks functional ADP/ATP carriers (Hatanaka et al., 1999). The growth deficiency can be complemented by introducing a vector expressing yeast AAC2 (AAC2-Scer) behind its own promoter Paac2 (pYES-Paac2-aac2) (Hatanaka et al., 1999) (Figure 4A). A similar expression vector containing the N.patriciarum aac gene behind Paac2 (pYES-Paac2-aacNpat) also complemented the WB-12 strain on YP plates containing glycerol, albeit with smaller colony size (Figure 4A). Growth experiments in YP medium containing either 2% ethanol or 3% glycerol as carbon source showed that the doubling times of the WB-12 strain harbouring pYES-Paac2-aacNpat or pYES-Paac2-aac2 were on average 10.5 and 5.7 h, respectively, while the control strain WB-12 did not grow at all.
Fig. 4. Expression of the hydrogenosomal ADP/ATP carrier of N.patriciarum in S.cerevisiae. (A) The S.cerevisiae strain WB-12 and transformants WB-12/pYES-Paac2-aacNpat (WB-12/AAC-Npat) and WB-12/pYES-Paac2-aac2Scer (WB-12/AAC2-Scer) were inoculated onto YP plates with 3% glycerol, and their growth was examined after 3 days at 30°C. (B) Western blot of isolated mitochondria (20 µg) from WB-12 (lane 1), WB-12/AAC-Npat (lane 2) and WB-12/AAC2-Scer (lane 3) probed with an antiserum raised against N.crassa AAC.
Mitochondria of the three strains were isolated using protoplast formation by lyticase treatment, lysis by mechanical disruption and differential centrifugation (Stuart et al., 2001). Polyclonal antiserum raised against Neurospora crassa AAC recognized epitopes on the AAC from N.patriciarum (AAC-Npat) and AAC2-Scer (Figure 4B). The apparent molecular mass of the expressed AAC-Npat was ∼31 kDa, being in good agreement with the band identified in hydrogenosomes. On an SDS– polyacrylamide gel stained with Coomassie Blue, a faint band at the established molecular weight of AAC-Npat was visible, estimated to be about seven times less protein than in the AAC2-Scer band, which is the most abundant carrier in the inner membranes of yeast mitochondria (data not shown). These results clearly show that AAC-Npat is recognized by the yeast mitochondrial targeting pathways and import machinery. They also establish that the hydrogenosomal carrier is able to functionally complement the yeast mitochondrial ADP/ATP carrier mutant.
Uptake studies on membrane preparations of isolated mitochondria
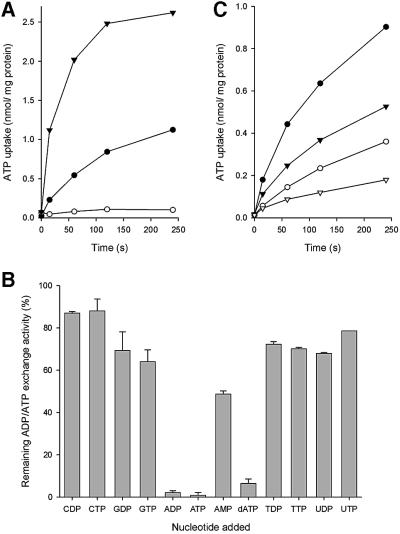
To study the substrate and inhibitor specificity of the hydrogenosomal ADP/ATP carrier, ADP/ATP exchange was measured in mitochondrial membranes fused with liposomes. No significant ADP/ATP exchange was observed in control membranes of the yeast strain WB-12, while high rates were observed in the strain expressing the hydrogenosomal ADP/ATP carrier from N.patriciarum, firmly establishing the ability of the transporter for adenine nucleotide exchange (Figure 5A). The initial uptake rates in the strain expressing AAC2-Scer were approximately five times those of the strain expressing AAC-Npat. These values are in agreement with the observation that AAC2-Scer is expressed at much higher levels than AAC-Npat. The lower transport rates may explain the lower growth rates of the AAC-Npat strain on non-fermentable carbon sources compared with the one expressing AAC2-Scer.
Fig. 5. ADP/ATP exchange in isolated mitochondrial membranes fused with liposomes. ADP/ATP exchange was measured by dilution of fused mitochondrial membranes loaded with 5 mM ADP into a buffer containing a final concentration of 1.54 µM [8-14C] ATP. (A) Uptake in mitochondrial membranes of S.cerevisiae strain WB-12 (open circles), WB-12/pYES-Paac2-aacNpat (closed circles) and WB-12/pYES-Paac2-aac2Scer (closed triangles). (B) The adenine nucleotide exchange activity in mitochondrial membranes of S.cerevisiae strain WB-12/pYES-Paac2-aacNpat in the presence of a 1000-fold excess of unlabelled substrate and expressed as a percentage of activity without added nucleotides. (C) Inhibition of adenine nucleotide exchange (closed circles) by the addition of 20 µM bongkrekic acid (closed triangles), 25 µM carboxy-atractyloside (open circles) and both (open triangles).
Excess concentrations of competing nucleotides were added to the nucleotide exchange experiments to establish the substrate specificity of the hydrogenosomal ADP/ATP carrier. ATP/ADP exchange was inhibited by >95% by the addition of a 1000-fold excess of adenine nucleotides ADP and ATP, whereas GDP, GTP, UDP, UTP, TDP, TTP, CDP or CTP only marginally affected the exchange rates. AMP and dATP inhibited ATP uptake by 50 and 90%, respectively (Figure 5B). When fused membranes were pre-treated with carboxy-atractyloside or bongkrekic acid, characteristic inhibitors of mitochondrial ADP/ATP carriers, initial nucleotide exchange rates decreased by 69 and 38%, respectively (Figure 5C). When both inhibitors were present, ADP/ATP exchange was inhibited even further (Figure 5C), clearly showing that the inhibitor specificity of the hydrogenosomal carrier is identical to the mitochondrial homologue.
Discussion
Fungal hydrogenosomes were first reported to be bounded by a single membrane, which gave rise to the hypothesis that these organelles were derived from peroxisomes (Marvin-Sikkema et al., 1993a; Hackstein and Vogels, 1997). However, more recent observations by electron microscopy have demonstrated that there are two, tightly apposed membranes surrounding fungal hydrogenosomes (Benchimol et al., 1997; van der Giezen et al., 1997b). Other properties that fungal hydrogenosomes share with mitochondria, but not peroxisomes, include the existence of a transmembrane pH gradient and an alkaline lumen. Free Ca2+ pools and calcium phosphate precipitates have also been detected in fungal hydrogenosomes, suggesting that, like mitochondria, they accumulate this intracellular messenger (Biagini et al., 1997).
A peroxisomal origin for fungal hydrogenosomes was also suggested based upon a cross-reaction on a western blot between a N.frontalis hydrogenosomal protein and an anti-SKL antibody (Marvin-Sikkema et al., 1993a), which recognizes the consensus peroxisomal targeting motif (Rachubinski and Subramani, 1995). However, the gene encoding the cross-reacting protein (hydrogenase) was not isolated and sequenced to confirm the presence of the SKL motif. In contrast, there is strong published data demonstrating that Neocallimastix hydrogenosomal malic enzyme (ME) and β-succinyl-CoA synthetase, which are encoded in the nucleus, are targeted to the hydrogenosome using N-terminal targeting signals, which resemble those of yeast mitochondrial proteins (Brondijk et al., 1996; van der Giezen et al., 1997a). Hydrogenosomal ME is also imported selectively into mitochondria, rather than peroxisomes, in the heterologous host Hansenula polymorpha in an N-terminal pre-sequence-dependent manner (van der Giezen et al., 1998), presumably by the Tom/Tim protein import system (Neupert, 1997; Pfanner, 1998).
The ability to export ATP is central to the function of any energy-generating organelle, including hydrogenosomes. In mitochondria this function is carried out by nuclear encoded proteins called ADP/ATP carriers (AAC), which are sorted to mitochondria by a membrane translocation pathway characteristic for this organelle (Sirrenberg et al., 1996, 1998). The first evidence that fungal hydrogenosomes might also contain AAC-like proteins was provided by inhibitor studies (Marvin-Sikkema et al., 1994). It was observed that bongkrekic acid and carboxy-atractyloside, specific inhibitors of the mitochondrial AAC, interfered with H2 production from malate by Neocallimastix hydrogenosomes. Here we report the discovery of two genes on the nuclear genome of the hydrogenosomal fungi N.frontalis and N.patriciarum, which code for carriers with all the properties of mitochondrial ADP/ATP exchangers.
Phylogenetic analysis of the Neocallimastix AAC protein sequences demonstrates that they share common ancestry with mitochondrial AAC sequences. The position of the Neocallimastix AAC sequences reflects what we know about organismal phylogeny (Van der Auwera and De Wachter, 1996). The simplest explanation of the AAC phylogeny is that the chytrid AAC genes were inherited vertically from a common ancestor they shared with aerobic mitochondria-containing fungi. In contrast, neither our phylogenetic analysis of the entire mitochondrial carrier family (not shown), nor the distribution of indels and residues implicated in function (see Figure 1), provide strong support for inclusion of the recently reported Trichomonas vaginalis hydrogenosomal protein HMP31 (Dyall et al., 2000) within the AAC subfamily. There is already published data to indicate the presence of an AAC in trichomonads (Čerkasov et al., 1978; Steinbüchel and Müller, 1986), but it has not yet been demonstrated that HMP31 actually carries out this function (Dyall et al., 2000).
Functional studies reported here reveal that the substrate specificity of the Neocallimastix hydrogenosomal ADP/ATP carrier is similar to that of typical mitochondrial ADP/ATP carriers (Pfaff and Klingenberg, 1968). Furthermore, adenine nucleotide exchange is inhibited by the mitochondrial AAC inhibitors bongkrekic acid and carboxy-atractyloside, explaining earlier observations that hydrogen production is affected by the addition of these inhibitors (Marvin-Sikkema et al., 1994). Crucially, the gene of N.patriciarum is able to restore growth on non-fermentable carbon sources in an AAC-deficient Saccharomyces strain, demonstrating the similar function of the carrier. Most members of the mitochondrial carrier family are targeted to the mitochondrial inner membrane by cryptic signals within the protein sequence (Sirrenberg et al., 1996, 1998). The observation that the hydrogenosomal AAC of N.patriciarum is targeted correctly to yeast mitochondria suggests that this cryptic signal is also present on this protein.
In recent years, it has become apparent that rather than being exotic exceptions, anaerobic ATP-generating pathways are widespread in eukaryotes and that the biochemistry of mitochondria is more flexible than previously appreciated (van Hellemond et al., 1998; Embley and Martin, 1998). The hydrogenosomes of ciliates and Trichomonas are perhaps the most extreme example of this tendency because published data suggest that they are mitochondria that have converted irreversibly to anaerobic energy generation (Akhmanova et al., 1998; Dyall et al., 2000). This raises the question of whether the ability to modify mitochondria to make hydrogen is a general property of eukaryotes. The origin of fungal hydrogenosomes has a direct bearing on this question because there have been claims that they are different from other hydrogenosomes. In our view, the data presented here and the weight of recently published data now overwhelmingly supports a mitochondrial origin for the fungal organelle as well (Marvin-Sikkema et al., 1994; Benchimol et al., 1997; van der Giezen et al., 1997a,b, 1998). In particular, our demonstration that a hydrogenosomal membrane protein can be correctly imported and function within a bona fide mitochondrion argues strongly that, in fungi, mitochondria and hydrogenosomes are different aspects of the same fundamental organelle.
Materials and methods
Fungal and bacterial strains and growth conditions
Neocallimastix frontalis L2 and N.patriciarum CX were grown anaerobically at 39°C in defined medium supplemented with 20 mM glucose or 20 mM cellobiose (Marvin-Sikkema et al., 1992). Escherichia coli XL1-Blue MRF′ (Stratagene, Amsterdam, The Netherlands) and E.coli DH5α (Bethesda Research Laboratory) were used during cloning procedures. Escherichia coli DH5α was grown at 37°C in Luria–Bertani medium and supplemented with ampicillin (100 µg/ml) when used in plasmid propagation. Escherichia coli XL1-Blue MRF′ was supplemented with MgSO4 (10 mM) and maltose (0.2%) when used in bacteriophage λ experiments.
General DNA techniques and PCR amplification
Standard recombinant DNA techniques were used for nucleic acid preparation and analysis (Sambrook et al., 1989). Fungal DNA was isolated as described previously (Brookman et al., 2000). Two degenerate oligonucleotide primers were designed based on an alignment of AAC sequences from 26 sequences. The primers were: sense, 5′-MGW TAY TTY CCA ACY CAR GC-3′ (Schizosaccharomyces pombe 160–166); and antisense, 5′-CCN SWR GCT ATC ATC AT-3′ (S.pombe 320–325). The numbers in parentheses refer to the amino acid position in the S.pombe AAC (DDBJ/EMBL/GenBank accession No. Q09188). Primers were designed using the most probable codon usage for N.frontalis based on known gene sequences (van der Giezen et al., 1997a). PCR was performed on N.frontalis genomic DNA. The resulting 450 bp fragment was cloned into pGEM-T-Easy (Promega, Southampton, UK) to confirm its sequence, and E.coli DH5α was subsequently transformed for plasmid propagation. Subsequent comparison of nucleotide and derived amino acid sequences was performed using the BLAST algorithm (Altschul et al., 1990). To obtain the genomic sequence, primers outside the coding region of the actual gene were designed which would amplify the complete gene based on the cDNA sequences (all primer sequences are available on request). The genomic and cDNA sequences reported in this work are deposited in DDBJ/EMBL/GenBank under the following accession numbers: N.frontalis gDNA, AY038992; N.patriciarum gDNA, AF384684; N.patriciarum cDNA, AY033433.
Identification and isolation of clones from a cDNA library
The putative N.patriciarum AAC gene was isolated by screening a λZAP II cDNA library, constructed previously (Xue et al., 1992), using the 450-bp probe described above. Positive plaques were isolated, and recombinant pBluescript SK– plasmids were excised according to the manufacturer’s instructions (Stratagene) and subsequently analysed by sequencing.
Phylogenetic analysis
Reference AAC sequences were recovered from databases and unfinished genome projects by similarity searches. Preliminary Cryptococcus neoformans sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org. Preliminary Pneumocystis carinii data was obtained from http://www.uky.edu/Projects/Pneumocystis/ and Phanerochaete chrysosporium from http://www.jgi.doe.gov/programs/whiterot.htm. Sequences were aligned using CLUSTAL_W (Thompson et al., 1994) and alignments refined manually. Taxon sampling was chosen to represent eukaryotic taxonomic diversity, with all available fungal and protist sequences, but only a representative selection of plant and metazoan sequences retained. Regions of ambiguous alignment and residues with gaps were excluded from the analysis, leaving a final dataset of 32 taxa and 252 amino acid positions. Baysian searches of treespace were performed with the program MrBayes (Huelsenbeck, 2000) using the JTT-f amino acid substitution matrix with one invariable and four variable gamma rate categories. Two hundred thousand Monte Carlo Markov Chain generations were performed with trees sampled every 100 generations. For compilation of the Baysian consensus topologies, a ‘burn-in’ of 201 trees was used. For the maximum likelihood (ML) topology shown, branch lengths were calculated under the same model in TREE-PUZZLE 4 (Strimmer and von Haeseler, 1996). One hundred ML bootstrap replicates were performed using the custom software MrBoot (Peter Foster, NHM), which automates MrBayes analyses of resampled datasets generated by the custom software p4 (Peter Foster, NHM). For bootstrap replicate analyses, 20 001 search generations were sampled every 50 generations and a burn-in of 100 trees was used.
Cloning of the yeast expression vectors
The yeast aac2 gene was amplified by PCR using primers to introduce a SacI site and an in-frame NcoI site at the 5′ end of the gene, as well as a NheI and XhoI site after the stop codon at the 3′ end of the gene. The PCR was carried out with the Expand polymerase-mix (Roche, Basel, Switzerland) according to the instructions of the manufacturer using 30 cycles consisting of 1 min denaturation at 93°C, 1 min annealing at 55°C and 1 min extension at 73°C. The PCR product was cloned into pYES3-CT (Invitrogen, Groningen, The Netherlands) using the SacI and XhoI cloning sites. The vector, designated pYES-Pgal-aac2, was isolated by standard genetic techniques and the construction of the plasmid was confirmed by restriction analysis and sequencing (Sambrook et al., 1989). In a second cloning step, the Pgal promoter region of the pYES-Pgal-aac2 vector was removed by restriction with SpeI and NcoI, and replaced by the 600 bp constitutive promoter region upstream of the yeast aac2, which was also amplified by PCR to introduce these two restriction sites. The aac gene of N.patriciarum was amplified by PCR to introduce a NcoI site at the start of the gene and a NheI site after the stop codon. The fragment was restricted and ligated into the pYES-Paac2-aac2 vector, previously restricted with these two enzymes, resulting in the expression vector pYES-Paac2-aacNpat.
All cloned vectors were isolated by miniprep (Qiagen, Crawley, UK) and confirmed by PCR, restriction analysis and sequencing (Cambridge Bioscience, Cambridge, UK). The restriction enzymes and ligase were purchased from New England Biolabs (Hitchin, UK).
Transformation of yeast WB-12 and growth experiments
Saccharomyces cerevisiae strain WB-12 (MATα ade2-1 trp1-1 ura3-1 can1-100 aac1::LEU2 aac2::HIS3), lacking functional yeast AAC1 and AAC2 ADP/ATP carriers, was used for all complementation experiments (Hatanaka et al., 1999). The AAC2 expression vector pYES-Paac2-aac2 and the AAC-Npat expression vector pYES-Paac2-aacNpat were transformed into strain WB-12 using standard techniques, and transformants were selected on SC medium minus Trp plates (Invitrogen). The strains were plated on YP (1% yeast extract and 2% bactopeptone) agar plates containing 3% glycerol. To determine growth rates on non-fermentable carbon sources, strains were grown overnight in selective medium (SC medium minus Trp), washed twice in YP and diluted to an A600 of 0.1 in YP medium containing 3% glycerol or 2% ethanol. Growth at 30°C was monitored for 3 days by measuring the optical density at A600.
Isolation of membranes and preparation of fused liposomes
Yeast mitochondria and Neocallimastix hydrogenosomes were isolated according to the procedures described by Stuart et al. (2001) and Marvin-Sikkema et al. (1993b), respectively. Liposomes were prepared by mixing E.coli total lipid extract and egg yolk phosphatidylcholine in a 3:1 ratio (w/w) in 50 mM potassium phosphate pH 7 containing 100 mM potassium chloride (buffer A) at a final lipid concentration of 20 mg/ml. To prepare membrane vesicles fused to liposomes and loaded with ADP, membranes were mixed with liposomes in a ratio of 1:5 protein to lipid (w/w) in 3 ml of buffer A supplemented with 5 mM ADP. The mixture was frozen in liquid nitrogen, slowly thawed at room temperature, sonicated in an ultrasonicator (Misonix) with a 3.2 mm tip at 15% output (20 cycles of 3 s with 10 s pauses) and stored in liquid nitrogen. The mixture of fused membranes was slowly thawed at room temperature, extruded through 400 nm polycarbonate filters and centrifuged in a Beckmann MLA130 rotor for 20 min at 55 000 r.p.m. and 4°C. The pellet was resuspended in 200 µl of buffer A containing 5 mM ADP, 1 µM valinomycin and 1 µM nigericin. To remove external ADP the suspension was applied to a 3.5 ml bed volume Sephadex G-75 gelfiltration column, previously equilibrated with buffer A containing 1 µM valinomycin and 1 µM nigericin at 4°C. The fused membranes were applied to the column, eluted in 800 µl of buffer A, and used for transport assays. For inhibitor studies, 20 µM bongkrekic acid and/or 25 µM carboxy-atractyloside were present during the entire procedure.
Transport assay
Transport was initiated by diluting the membrane vesicles 2-fold in buffer A containing 1 µM valinomycin and 1 µM nigericin, and 3.1 µM [8-14C]adenosine 5′-triphosphate (Amersham Pharmacia Biotech, Little Chalfont, UK). The experiments were performed at 30°C with constant stirring in total volumes of 300 µl. The uptake was quenched by adding 4 ml of ice-cold phosphate-buffered saline (PBS), followed by immediate filtration over cellulose nitrate filters (0.2 µm pore size). The filters were washed once with 2 ml ice-cold PBS and transferred to a scintillation vial, then 2 ml of Ultima Gold AB scintillation liquid was added and levels of radioactivity were determined by a Packard TriCarb 2100 TR–liquid scintillation analyser. In the competition studies the exchange activity was determined by the accumulation of radiolabelled ATP after 15 s in the presence or absence of a 1000-fold higher concentration of non-labelled nucleotides.
Protein electrophoresis and western blotting
Neocallimastix cell fractionation was performed as described by Marvin-Sikkema et al. (1993b). SDS–PAGE was performed as described previously (Laemmli, 1970). Western blotting was carried out using a semi-dry electroblotter (Hoefer, Little Chalfont, UK) according to methods described previously (Bjerrum and Heegaard, 1988). The primary antisera against Neurospora crassa AAC and N.frontalis β-succinyl-CoA synthetase were added at a titre of 1:50 000 and the anti-rabbit secondary antibodies coupled to horseradish peroxidase (Sigma, Poole, UK) at a dilution of 1:20 000. An electrochemiluminescence kit (Pharmacia) was used for the detection of secondary antibodies.
Acknowledgments
Acknowledgements
We would like to thank Dr H.Terada for providing S.cerevisiae strain WB-12. Neocallimastix frontalis L2 and N.patriciarum CX were kindly provided by Drs A.J.Richardson and C.S.Stewart. The antiserum raised against the N.crassa AAC was kindly provided by Dr L.Palmieri. We would like to thank Dr J.E.Walker for critical reading of the manuscript. Preliminary C.neoformans sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of C.neoformans was accomplished with support from the National Institute of Allergy and Infectious Diseases (grant No. 1 U01 AI48594-01). Preliminary P.carinii data was obtained from http://www.uky.edu/Projects/Pneumocystis/. The P.chrysosporium genome project was performed under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, by the University of California, Lawrence Livermore National Laboratory under contract No. W-7405-ENG-48, the Lawrence Berkeley National Laboratory under contract No. DE-AC03-76SF00098, and Los Alamos National Laboratory under contract No. W-7405-ENG-36. This project was supported by a Leverhulme Trust grant (F/696A) to T.M.E. and by an EMBO long-term fellowship (ALTF 520-1997) to M.v.d.G. D.S.H. is funded by the Natural History Museum (London, UK). D.J.S. was supported by an EC Marie Curie long-term fellowship (MCFI-2100-01607), and E.R.S.K. and M.H. were supported by the Medical Research Council.
References
- Akhmanova A., Voncken,F., van Alen,A.T., van Hoek,H.A., Boxma,B., Vogels,G., Veenhuis,M. and Hackstein,J.H. (1998) A hydrogenosome with a genome. Nature, 396, 527–528. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Benchimol M., Durand,R. and Almeida,J.C.A. (1997) A double membrane surrounds the hydrogenosomes of the anaerobic fungus Neocallimastix frontalis. FEMS Microbiol. Lett., 154, 277–282. [DOI] [PubMed] [Google Scholar]
- Biagini G.A., van der Giezen,M., Hill,B., Winters,C. and Lloyd,D. (1997) Ca2+ accumulation in the hydrogenosomes of Neocallimastix frontalis L2: a mitochondrial-like physiological role. FEMS Microbiol. Lett., 149, 227–232. [Google Scholar]
- Bjerrum O.J. and Heegaard,N.H.H. (1988) CRC Handbook of Immunoblotting of Proteins. CRC Press, Boca Raton, FL.
- Boulay F., Lauquin,G.J., Tsugita,A. and Vignais,P.V. (1983) Photolabeling approach to the study of the topography of the atractyloside binding site in mitochondrial adenosine 5′-diphosphate/adenosine 5′-triphosphate carrier protein. Biochemistry, 22, 477–484. [DOI] [PubMed] [Google Scholar]
- Brondijk T.H.C., Durand,R., van der Giezen,M., Gottschal,J.C., Prins,R.A. and Fèvre,M. (1996) scsB, a cDNA encoding the hydrogenosomal protein β-succinyl-CoA synthetase from the anaerobic fungus Neocallimastix frontalis. Mol. Gen. Genet., 253, 315–323. [DOI] [PubMed] [Google Scholar]
- Brookman J.L., Mennim,G., Trinci,A.P., Theodorou,M.K. and Tuckwell,D.S. (2000) Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 185 rRNA. Microbiology, 146, 393–403. [DOI] [PubMed] [Google Scholar]
- Čerkasov J., Čerkasovová,A., Kulda,J. and Vilhelmova,D. (1978) Respiration of hydrogenosomes of Tritrichomonas foetus; I. ADP-dependant oxidation of malate and pyruvate. J. Biol. Chem., 253, 1207–1214. [PubMed] [Google Scholar]
- Dalbon P., Brandolin,G., Boulay,F., Hoppe,J. and Vignais,P.V. (1988) Mapping of the nucleotide-binding sites in the ADP/ATP carrier of beef heart mitochondria by photolabeling with 2-azido[α-32P] adenosine diphosphate. Biochemistry, 27, 5141–5149. [DOI] [PubMed] [Google Scholar]
- Dyall S.D., Koehler,C.M., Delgadillo-Correa,M.G., Bradley,P.J., Plumper,E., Leuenberger,D., Turck,C.W. and Johnson,P.J. (2000) Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol., 20, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T.M. and Hirt,R.P. (1998) Early branching eukaryotes? Curr. Opin. Genet. Dev., 8, 624–629. [DOI] [PubMed] [Google Scholar]
- Embley T.M. and Martin,W. (1998) A hydrogen-producing mitochondrion. Nature, 396, 517–519. [DOI] [PubMed] [Google Scholar]
- Embley T.M., Horner,D.S. and Hirt,R.P. (1997) Anaerobic eukaryote evolution: hydrogenosomes as biochemically modified mitochondria? Trends Ecol. Evol., 12, 437–441. [DOI] [PubMed] [Google Scholar]
- Fenchel T. and Finlay,B.J. (1995) Ecology and Evolution in Anoxic Worlds. Oxford University Press, Oxford, UK.
- Fiore C., Trezeguet,V., Le Saux,A., Roux,P., Schwimmer,C., Dianoux,A.C., Noel,F., Lauquin,G.J., Brandolin,G. et al. (1998) The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie, 80, 137–150. [DOI] [PubMed] [Google Scholar]
- Florin L., Tsokoglou,A. and Happe,T. (2001) A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J. Biol. Chem., 276, 6125–6132. [DOI] [PubMed] [Google Scholar]
- Gawaz M., Douglas,M.G. and Klingenberg,M. (1990) Structure-function studies of adenine nucleotide transport in mitochondria. II. Biochemical analysis of distinct AAC1 and AAC2 proteins in yeast. J. Biol. Chem., 265, 14202–14208. [PubMed] [Google Scholar]
- Hackstein J.H.P. and Vogels,G.D. (1997) Endosymbiotic interactions in anaerobic protozoa. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol., 71, 151–158. [DOI] [PubMed] [Google Scholar]
- Hatanaka T., Hashimoto,M., Majima,E., Shinohara,Y. and Terada,H. (1999) Functional expression of the tandem-repeated homodimer of the mitochondrial ADP/ATP carrier in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 262, 726–730. [DOI] [PubMed] [Google Scholar]
- Horner D., Hirt,R. and Embley,T. (1999) A single eubacterial origin of eukaryotic pyruvate: ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol. Biol. Evol., 16, 1280–1291. [DOI] [PubMed] [Google Scholar]
- Horner D.S., Foster,P.G. and Embley,T.M. (2000) Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. Evol., 17, 1695–1709. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P. (2000) MrBayes: Bayesian inference of phylogeny. Department of Biology, University of Rochester, NY.
- Klingenberg M. (1989) Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch. Biochem. Biophys., 270, 1–14. [DOI] [PubMed] [Google Scholar]
- Kuan J. and Saier,M.H. (1993) The mitochondrial carrier family of transport proteins: structural, functional and evolutionary relationships. Crit. Rev. Biochem. Mol. Biol., 28, 209–233. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lawson J.E. and Douglas,M.G. (1988) Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J. Biol. Chem., 263, 14812–14818. [PubMed] [Google Scholar]
- Lawson J.E., Gawaz,M., Klingenberg,M. and Douglas,M.G. (1990) Structure-function studies of adenine nucleotide transport in mitochondria. I. Construction and genetic analysis of yeast mutants encoding the ADP/ATP carrier protein of mitochondria. J. Biol. Chem., 265, 14195–14201. [PubMed] [Google Scholar]
- Loytynoja A. and Milinkovitch,M.C. (2001) Molecular phylogenetic analyses of the mitochondrial ADP-ATP carriers: The Plantae/Fungi/Metazoa trichotomy revisited. Proc. Natl Acad. Sci. USA, 98, 10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin-Sikkema F.D., Lahpor,G.A., Kraak,M.N., Gottschal,J.C. and Prins,R.A. (1992) Characterization of an anaerobic fungus from llama faeces. J. Gen. Microbiol., 138, 2235–2241. [DOI] [PubMed] [Google Scholar]
- Marvin-Sikkema F.D., Kraak,M.N., Veenhuis,M., Gottschal,J.C. and Prins,R.A. (1993a) The hydrogenosomal enzyme hydrogenase from the anaerobic fungus Neocallimastix sp. L2 is recognized by antibodies, directed against the C-terminal microbody protein targetting signal SKL. Eur. J. Cell Biol., 61, 86–91. [PubMed] [Google Scholar]
- Marvin-Sikkema F.D., Pedro Gomes,T.M., Grivet,J., Gottschal,J.C. and Prins,R.A. (1993b) Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2. Arch. Microbiol., 160, 388–396. [DOI] [PubMed] [Google Scholar]
- Marvin-Sikkema F.D., Driessen,A.J.M., Gottschal,J.C. and Prins,R.A. (1994) Metabolic energy generation in hydrogenosomes of the anaerobic fungus Neocallimastix: Evidence for a functional relationship with mitochondria. Mycol. Res., 98, 205–212. [Google Scholar]
- Müller M. (1993) The hydrogenosome. J. Gen. Microbiol., 139, 2879–2889. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Pfaff E. and Klingenberg,M. (1968) Adenine nucleotide translocation of mitochondria. 1. Specificity and control. Eur. J. Biochem., 6, 66–79. [DOI] [PubMed] [Google Scholar]
- Pfanner N. (1998) Mitochondrial import: crossing the aqueous intermembrane space. Curr. Biol., 8, R262–R265. [DOI] [PubMed] [Google Scholar]
- Rachubinski R.A. and Subramani,S. (1995) How proteins penetrate peroxisomes. Cell, 83, 525–528. [DOI] [PubMed] [Google Scholar]
- Rotte C., Stejskal,F., Zhu,G., Keithly,J.S. and Martin,W. (2001) Pyruvate:NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol. Biol. Evol., 18, 710–720. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saraste M. and Walker,J.E. (1982) Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett., 144, 250. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Bauer,M.F., Guiard,B., Neupert,W. and Brunner,M. (1996) Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature, 384, 582–585. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Folsch,H., Stuart,R.A., Neupert,W. and Brunner,M. (1998) Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature, 391, 912–915. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A. and Müller,M. (1986) Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol., 20, 57–65. [DOI] [PubMed] [Google Scholar]
- Strimmer K. and von Haeseler,A. (1996) Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol., 13, 964–969. [Google Scholar]
- Stuart J.A., Harper,J.A., Brindle,K.M., Jekabsons,M.B. and Brand,M.D. (2001) A mitochondrial uncoupling artifact can be caused by expression of uncoupling protein 1 in yeast. Biochem. J., 356, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: Improving the sensitivity of progressive multiple alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. and De Wachter,R. (1996) Large-subunit rRNA sequence of the chytridiomycete Blastocladiella emersonii and implications for the evolution of zoosporic fungi. J. Mol. Evol., 43, 476–483. [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Rechinger,K.B., Svendsen,I., Durand,R., Hirt,R.P., Fèvre,M., Embley,T.M. and Prins,R.A. (1997a) A mitochondrial-like targeting signal on the hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis: evidence for the hypothesis that hydrogenosomes are modified mitochondria. Mol. Microbiol., 23, 11–21. [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Sjollema,K.A., Artz,R.R.E., Alkema,W. and Prins,R.A. (1997b) Hydrogenosomes in the anaerobic fungus Neocallimastix frontalis have a double membrane but lack an associated organelle genome. FEBS Lett., 408, 147–150. [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Kiel,J.A.K.W., Sjollema,K.A. and Prins,R.A. (1998) The hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis is targeted to mitochondria of the methylotrophic yeast Hansenula polymorpha. Curr. Genet., 33, 131–135. [DOI] [PubMed] [Google Scholar]
- van Hellemond J.J., Opperdoes,F.R. and Tielens,A.G.M. (1998) Trypanosomatidae produce acetate via a mitochondrial acetate: succinate CoA-transferase. Proc. Natl Acad. Sci. USA, 95, 3036–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn,J. and Herrmann,R.G. (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem., 180, 535–545. [DOI] [PubMed] [Google Scholar]
- Walker J.E. (1992) The mitochondrial transporter family. Curr. Biol., 2, 519–526. [Google Scholar]
- Whatley J.M., John,P. and Whatley,F.R. (1979) From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc. R. Soc. Lond. B, 204, 165–187. [DOI] [PubMed] [Google Scholar]
- Xue G.P., Orpin,C.G., Gobius,K.S., Aylward,J.H. and Simpson,G.D. (1992) Cloning and expression of multiple cellulase cDNAs from the anaerobic rumen fungus Neocallimastix patriciarum in Escherichia coli. J. Gen. Microbiol., 138, 1413–1420. [DOI] [PubMed] [Google Scholar]