Abstract
ZipA and FtsA are essential division proteins in Escherichia coli that are recruited to the division site by interaction with FtsZ. Utilizing a newly isolated temperature-sensitive mutation in zipA we have more fully characterized the role of ZipA. We confirmed that ZipA is not required for Z ring formation; however, we found that ZipA, like FtsA, is required for recruitment of FtsK and therefore all downstream division proteins. In the absence of FtsA or ZipA Z rings formed; however, in the absence of both, new Z rings were unable to form and preformed Z rings were destabilized. Consistent with this, we found that an FtsZ mutant unable to interact with both ZipA and FtsA was unable to assemble into Z rings. These results demonstrate that ZipA and FtsA are both required for recruitment of additional division proteins to the Z ring, but either one is capable of supporting formation and stabilization of Z rings.
Keywords: cell division/cytokinesis/FtsZ/Z ring
Introduction
Cytokinesis in Escherichia coli involves the circumferential invagination of the cell envelope at the division site. This involves the coordinated invagination of the cytoplasmic membrane along with the synthesis of septal peptidoglycan. At least nine genes, ftsZ, ftsA, ftsQ, ftsI, ftsL, ftsN, ftsW, ftsK and zipA, are specifically required and their products must be localized to the division site in order for cell division to occur (Lutkenhaus and Addinall, 1997; Rothfield et al., 1999; Margolin, 2000).
The analysis of bacterial cell division has advanced rapidly with the application of fluorescence microscopy (Maddock and Shapiro, 1993; Pogliano et al., 1995), which has allowed the order of addition of proteins to the division site to be determined. The first protein known to localize to the division site is FtsZ, which assembles into the Z ring, a circumferential ring of FtsZ abutting the inner membrane (Bi and Lutkenhaus, 1991; Addinall et al., 1996). It was suggested that the Z ring is a cytoskeletal element that functions as a scaffold to recruit the other division proteins (Bi and Lutkenhaus, 1991; Lutkenhaus, 1993). This is consistent with the ability of FtsZ to polymerize in vitro into protofilaments and sheets of two to three protofilaments (Mukherjee and Lutkenhaus, 1994; Erickson et al., 1996). How these protofilaments are arranged in the Z ring is unknown. Importantly, the assembly of all other known division proteins depends upon the Z ring.
Two division proteins, ZipA and FtsA, interact directly with FtsZ (Hale and de Boer, 1997; Wang et al., 1997). Both of these proteins localize independently to the division site but require FtsZ to localize (Addinall and Lutkenhaus, 1996; Ma et al., 1996; Hale and de Boer, 1999; Liu et al., 1999). ZipA consists of three domains: an N-terminal membrane-spanning domain, a C-terminal domain that binds FtsZ, and a long central region that could serve as a flexible linker (Hale and de Boer, 1997; Liu et al., 1999). Interestingly, the C-terminal region of ZipA can bundle FtsZ protofilaments in vitro (RayChaudhuri, 1999; Hale et al., 2000). Although ZipA is an excellent candidate for linking FtsZ polymers to the membrane, apparently normal Z rings can form in filamentous cells in which the ZipA level is reduced by 90% (Hale and de Boer, 1999; Liu et al., 1999).
FtsA is a member of the actin/DnaK/sugar kinase family of proteins (Bork et al., 1992). It has no membrane spanning domain, although in cell fractionation studies it has been found in both the membrane and the cytosolic fractions (Sanchez et al., 1994), and a FtsA–GFP (green fluorescent protein) shows preference for the membrane (Ma et al., 1996). Although these studies suggest that FtsA is also a candidate for linking FtsZ protofilaments to the membrane, apparently normal Z rings are formed in cells depleted of FtsA or in filaments formed as a result of a temperature-sensitive ftsA mutation (Addinall et al., 1996; Hale and de Boer, 1999). Thus, Z ring formation does not appear to require FtsA or ZipA.
Binding of ZipA or FtsA to FtsZ requires a conserved sequence near the C-terminus of FtsZ (Wang et al., 1997; Liu et al., 1999; Ma and Margolin, 1999a). This region of FtsZ is on the surface of FtsZ protofilaments and is not required for polymerization (Wang et al., 1997). ZipA has been crystallized with a peptide corresponding to the conserved C-terminus of FtsZ (Mosyak et al., 2000). The binding of FtsA to FtsZ has not been studied in such detail, although it is known that the C-terminus of FtsZ is required.
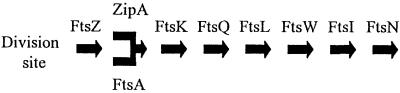
The remaining division proteins are all integral membrane proteins and they assemble to the division site in a defined order. In addition to FtsZ these proteins also require FtsA for localization. Following FtsA the order is FtsK, FtsQ, FtsL, FtsW, FtsI and FtsN (Addinall et al., 1997a; Wang and Lutkenhaus, 1998; Wang et al., 1998; Yu et al., 1998; Chen et al., 1999; Ghigo et al., 1999; Weiss et al., 1999; Chen and Beckwith, 2001). The linear order of addition of these proteins suggests that they may form a complex that carries out cell division. Although the requirement of FtsA for these other proteins to localize is well documented, it has not been determined if ZipA is required.
A key unanswered question is how the Z ring is stabilized at the division site and attached to the membrane. While the function of FtsA has been examined by both depletion studies and the use of temperature-sensitive mutations, the role of ZipA has only been examined in depletion studies (Hale and de Boer, 1999; Liu et al., 1999). Cells filament when depleted of ZipA, however the level of Zip A is only reduced by 90%. It is possible that ZipA could have another function, such as nucleating the Z ring, requiring ZipA at only 10% of the physiological level. To investigate this possibility we have generated a temperature-sensitive zipA mutation. Using this mutation we confirm that ZipA is not required for Z ring formation. However, ZipA is required for recruitment of FtsK, implying that ZipA is also required for recruitment of all downstream division proteins. Furthermore, by combining zipA (Ts) and ftsA (Ts) mutations we were able to examine the effects of inactivating both proteins. The results demonstrate that either protein is capable of supporting Z ring formation and stabilization.
Results
Isolation and characterization of a temperature-sensitive zipA mutation
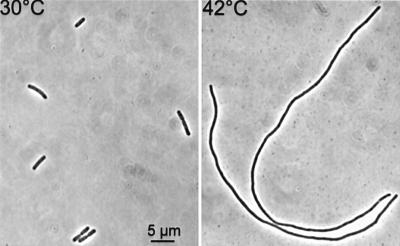
Temperature-sensitive alleles of zipA were obtained by in vitro mutagenesis of a plasmid containing zipA cloned downstream of the arabinose promoter (see Materials and methods). Mutagenized plasmid DNA was transformed into W3110ΩPlac::zipA (Table I). This strain contains the lac promoter upstream of the zipA gene and therefore requires IPTG for growth. Transformants were selected at 30°C on plates containing arabinose but no IPTG and screened at 42°C on the same plates. Five temperature-sensitive alleles were obtained. One of these was crossed onto the chromosome of W3110 as described in Materials and methods to give PS223 [zipA1 (Ts)]. The resultant strain was slightly elongated at 30°C and filamented immediately upon a shift to 42°C (Figure 1).
Table I. Strains and plasmids used in this study.
| Description | Reference | |
|---|---|---|
| Strain | ||
| MCI23 | MC4100 ftsI23 leu::Tn10 | Addinall et al. (1996) |
| MCA12 | MC4100 ftsA12 leu::Tn10 | Addinall et al. (1996) |
| W3110 | wild type | laboratory collection |
| PS223 | W3110 zipA1 | this study |
| PS236 | W3110 ftsA12 leu::Tn10 | this study |
| PS234 | W3110 zipA1 ftsA12 leu::Tn10 | this study |
| PS413 | W3110 ftsI23 leu::Tn10 | this study |
| PS414 | W3110 zipA1 ftsI23 leu::Tn10 | this study |
| W3110ΩPlac-zipA (pAM1) | W3110 zipA::kan-ter-Plac-zipA / pAM1 KanR SpecR | Liu et al. (1999) |
| W3110ΩPBAD-zipA | W3110 zipA::kan-ter-PBAD-zipA | laboratory collection |
| JKD7-2/pKD3c | ftsZ::kan recA56 srl::Tn10/pKD3c carry a copy of ftsZ on a temperature-sensitive replicon, CmR | Dai and Lutkenhaus (1991) |
| PS165 | JKD7-2/pKD3c/pSEB135 | this study |
| PS166 | JKD7-2/pKD3c/pSEB135FL | this study |
| Plasmid | ||
| pBAD-zipA | zipA under the control of PBAD regulated by arabinose, AmpR | laboratory collection |
| pFC13 | temperature-sensitive replicon, CmR | Cornet et al. (1994) |
| pEL3 | temperature-sensitive replicon, AmpR | Armstrong et al. (1984) |
| pJC104 | code for a FtsZ-GFP fusion protein under the control of PBAD promoter | Mukherjee et al. (2001) |
| pLZ1 | pBAD18 carrying cysZ-kan-ter-PBAD-zipA cassette. AmpR KanR | laboratory collection |
| pSEB138 | pFC13 carrying the chromosomal fragment cysZ-zipA-lig, CmR | this study |
| pSEB139A and pSEB139B | pSEB138 in which WT zipA has been replaced by zipA1 or zipA2, CmR | this study |
| pSEB135 and pSEB135FL | ftsZ or ftsZFL under the control of PBAD regulated by arabinose, AmpR | this study |
Fig. 1. The zipA1 (Ts) mutation confers a temperature-sensitive division phenotype. A culture of PS223 [zipA1 (Ts)] growing exponentially at 30°C was shifted to 42°C. Cells were photographed before and 2 h after the shift to 42°C.
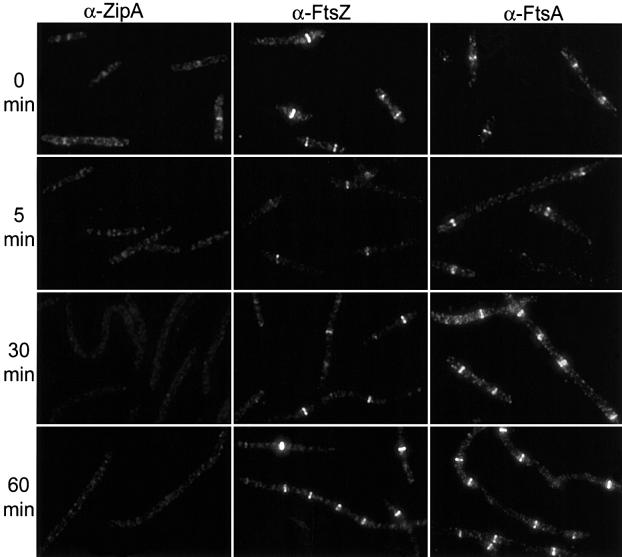
Immunofluoresence microscopy revealed that the mutant was temperature sensitive for ZipA localization. At 30°C, 44% of the cells had ZipA localized to the division site (Figure 2; Table II). This low percentage relative to FtsZ and FtsA (>80%, see below) indicates that this mutant is somewhat impaired for ZipA localization at the permissive temperature. It also suggests that Z rings can form without ZipA. Also, ZipA was sometimes located at quarter positions, but not midcell. This has been observed before with temperature-sensitive mutants (Addinall et al., 1996) and indicates an earlier failed division attempt at midcell. Five minutes after the temperature shift to 42°C the percentage of cells with ZipA localized was reduced to 2.6%, and was further reduced to 0.5% after 30 min (Figure 2; Table II). Immunoblot analysis demonstrated that the ZipA1 protein was stable for at least 60 min at the non-permissive temperature (data not shown). Together, these results demonstrate that the zipA1 (Ts) mutation confers temperature sensitivity to ZipA localization.
Fig. 2. Localization of ZipA, FtsZ and FtsA in zipA1 (Ts) cells. PS223 [zipA1 (Ts)] was grown as in Figure 1. Samples were taken, fixed and processed for fluorescence microscopy as described in Materials and methods. The first, second and third columns are photographs of cells stained with anti-ZipA, anti-FtsZ and anti-FtsA antisera as indicated. Samples were taken at various times after the temperature shift to 42°C as indicated to the left of the figure.
Table II. Localization of FtsZ, ZipA and FtsA in temperature-sensitive mutants.
| Strain | Protein |
||
|---|---|---|---|
| FtsZ (%) | ZipA (%) | FtsA (%) | |
| PS223 (zipA1) | |||
| 30°C | 85 | 44 | 83 |
| 5 min at 42°C | 31 | 2.6 | 30 |
| 30 min at 42°C | 80 | <0.5 | 87 |
| PS236 (ftsA12) | |||
| 30°C | 95 | 77 | 77 |
| 5 min at 42°C | 64 | 52 | 2.7 |
| 30 min at 42°C | 97 | 90 | 0.4 |
| PS234 (zipA1 ftsA12) | |||
| 30°C | 83 | 35 | 76 |
| 5 min at 42°C | <0.4 | 0.5 | 2.4 |
| 30 min at 42°C | 0.7 | 0.4 | <0.4 |
Although ZipA was not localized at the non-permissive temperature, we found that FtsZ was localized. At the permissive temperature, 85% of the cells had a Z ring (Figure 2; Table II). At 5 min after the temperature shift the percentage was reduced to 31%, indicating that some of the Z rings were destabilized by the shift. However, by 30 min after the temperature shift the percentage was 80%, indicating that the Z rings had re-formed. At later times after the temperature shift the cells had started to filament and the number of Z rings per cell had increased (Figure 2). These results are consistent with the previously published depletion results (Hale and de Boer, 1999; Liu et al., 1999) and support the conclusion that ZipA is not required for Z ring formation. The percentage of cells with localized FtsA paralleled the FtsZ results, indicating that they colocalized (Figure 2; Table II). As previously established, FtsA localizes to the Z ring coincident with its formation and independent of ZipA (Hale and de Boer, 1999; Liu et al., 1999).
FtsK requires ZipA to localize to division sites
The availability of the zipA1 (Ts) mutant allowed us to test whether the localization of other cell division proteins required ZipA. To do this we examined the localization of FtsK in PS223 [zipA1 (Ts)]. Previous results have shown that the localization of FtsK requires FtsZ and FtsA, but does not require FtsQ or FtsI (Wang and Lutkenhaus, 1998; Chen and Beckwith, 2001). This result places FtsK at a key point in the assembly pathway between cytoplasmic components of the division machinery and the membrane components. Immunofluorescence microscopy of FtsK revealed that it was localized at the permissive temperature (Figure 3). However, at 5 and 30 min after the shift to non-permissive temperature, FtsK was not localized. Even though these filaments contain Z rings decorated with FtsA, FtsK is unable to localize, indicating that FtsK requires ZipA, in addition to FtsA, to localize to the Z ring.
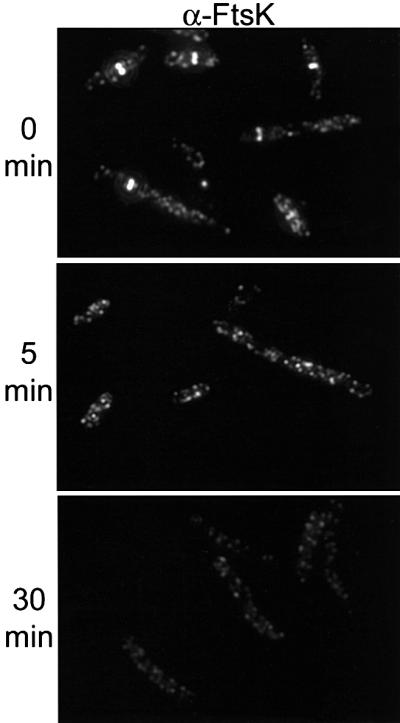
Fig. 3. Localization of FtsK in zipA1 (Ts) cells. PS223 [zipA1 (Ts)] was grown as in Figure 1. Samples were taken at 30°C and after a shift to 42°C, fixed and processed for fluorescence microscopy as described in Materials and methods. The upper panel shows cells before the temperature shift. The middle and lower panels show cells at 5 and 30 min, respectively, after the temperature shift to 42°C.
The fate of Z rings in the zipA1 (Ts) ftsA12 (Ts) double mutant
Both FtsA and ZipA bind to the C-terminus of FtsZ and are candidates for linking FtsZ protofilaments to the membrane. However, Z rings are able to assemble in filamentous cells that are formed due to depletion or inactivation of either of these proteins. This implies that an additional unknown component is required or that either FtsA or ZipA alone can stabilize the Z ring. To test this latter possibility we determined if Z rings could assemble in the absence of both ZipA and FtsA. To do this, a strain containing temperature-sensitive mutations for both genes was constructed. The ftsA12 (Ts) mutation was cotransduced with leu::Tn10 into PS223 [W3110 zipA1 (Ts)] to give PS234 [ftsA12 (Ts) zipA1 (Ts)]. Thirty percent of the tetracycline resistant transductants displayed temperature sensitivity at 37°C, indicative of the presence of the ftsA12 (Ts) mutation; the ftsA12 (Ts) mutation is temperature sensitive at 37°C whereas the zipA1 (Ts) mutation is only temperature sensitive above 40°C. The presence of the ftsA12 (Ts) mutation was confirmed by demonstrating that temperature sensitivity could be cotransduced with tetracycline resistance at a frequency of 26% from this strain to a wild-type strain. The strain carrying both mutations grew slower and was more elongated than strains carrying either mutation alone. The localization of FtsZ, FtsA and ZipA in this double mutant strain was then compared with strains carrying either mutation alone.
The strains used for the immunofluorescence microscopy were isogenic other than the temperature-sensitive mutations. As shown above, both FtsZ and FtsA were localized in the zipA1 (Ts) mutant, but ZipA was not. We also examined the effect of the ftsA12 (Ts) mutation in this strain background, since we had previously used a different strain (Addinall et al., 1996). The results were the same as we observed, that FtsZ and ZipA were localized at the non-permissive temperature but FtsA was not (Table II). At 5 min after the shift the fraction of cells with Z rings decreased from 95 to 64%. By 30 min after the shift the percentage was restored to 97%. These results indicate that inactivation of FtsA leads to a transient drop in the percentage of cells with Z rings, which is rapidly restored.
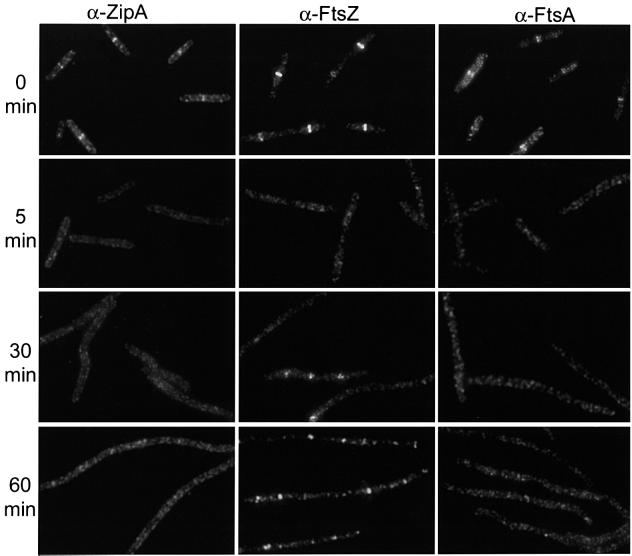
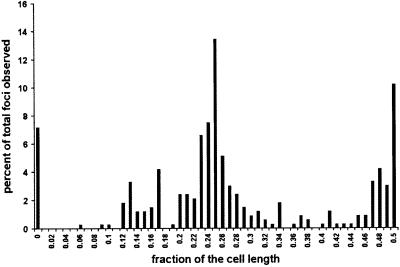
In contrast to the results with the single mutants, the percentage of cells with Z rings in the double mutant strain PS234 decreased from 83 to <1% by 5 min after the temperature shift (Figure 4; Table II). At 30 min and at later times after the shift, spots of fluorescence were observed; however, these spots did not have the sharp appearance of typical Z rings. Higher magnification revealed that the foci of fluorescence were often multiple spots, indicating that FtsZ was present in short arcs that did not extend around the circumference of the cell (Figure 5). Interestingly, measurement of the distribution of these spots indicated that they were present at expected division sites (Figure 6). Most of the foci were present at either the quarter position or midcell, although some were also at the 1/8 position and the poles. These results show that inactivation of FtsA and ZipA leads to a rapid dissolution of preformed Z rings. Also, during continued incubation at the non-permissive temperature FtsZ localizes to division sites; however, Z rings are not formed, instead abnormal structures are formed. These results indicate that the presence of either FtsA or ZipA is required to organize FtsZ filaments into Z rings, but neither is required for FtsZ to initiate localization to the site of division.
Fig. 4. Localization of ZipA, FtsZ and FtsA in ftsA12 (Ts) zipA1 (Ts) cells. A culture of PS234 [zipA1 (Ts) ftsA12 (Ts)] growing exponentially at 30°C was shifted to 42°C. Samples were taken before and at the indicated times after the shift to 42°C. The antibodies used for staining are indicated at the top of the figure.
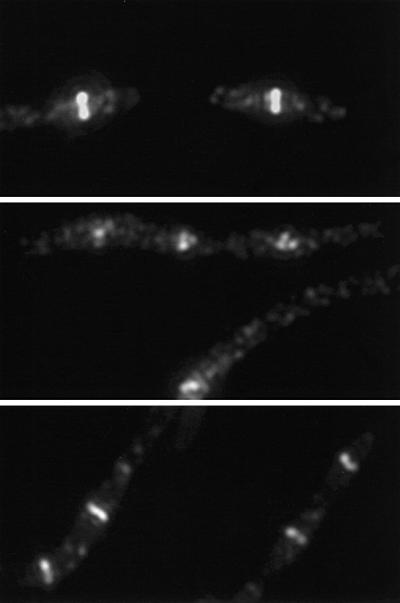
Fig. 5. Localization of FtsZ in ftsA12 (Ts) zipA1 (Ts) cells at 30 and 42°C. Portions of the figures shown in Figures 4 and 7 were magnified ×2. The top two panels show cells of ftsA12 (Ts) zipA1 (Ts) at 30°C (top panel) and 30 min after the temperature shift to 42°C (middle panel). These images illustrate the contrast between the sharp and symmetric appearance of typical Z rings observed at 30°C and the abnormal FtsZ structures observed at 42°C in this strain. The bottom panel shows magnified ftsI23 (Ts) zipA1 (Ts) cells 30 min after a shift to 42°C. These cells display several typical Z rings per cell.
Fig. 6. Distribution of the foci of FtsZ fluorescence in PS234 [zipA1 (Ts) ftsA12 (Ts)] at the non-permissive temperature. Cells from the experiment in Figure 3 were analyzed for the distribution of fluorescent spots. Only the sample taken 30 min after the temperature shift from 30 to 42°C was examined. A total of 156 cells with 334 spots were analyzed. Six cells did not contain any spots. The distance from each spot to the nearest pole was measured and divided by half the total cell length. The graph presented here shows the percentage of spots observed for each fraction of the cell length.
Examination of a zipA (Ts) ftsI23 (Ts) double mutant
As a control for the above experiment we examined a zipA1 (Ts) ftsI23 (Ts) double mutant. In this double mutant strain, ZipA and FtsI are not localized to the division site at the non-permissive temperature; however, because FtsA is still present, Z rings should form. The ftsI23 (Ts) mutation was cotransduced with leu::Tn10 into PS223 [W3110 zipA1 (Ts)] as described in Materials and methods to give PS414 [ftsI23 (Ts) zipA1 (Ts)]. Genetic analysis of the tetracycline resistant colonies revealed that 23% contained both mutations. PS414 grew more slowly than the parent strain, indicating that combining zipA1 (Ts) and ftsI23 (Ts) was deleterious. Consistent with this, cells from the double mutant were more elongated than that observed with either mutation alone. Staining of this double mutant at the permissive temperature revealed a heterogeneous cell length distribution, with some cells containing more than one Z ring, indicating that in some cells division had failed at midcell (Figure 7). Shifting this double mutant to the non-permissive temperature revealed that Z rings were present at this temperature and increased in number with the growth of the filaments. Higher magnification of such filaments revealed typical Z rings in contrast to the spots observed with the zipA1 (Ts) ftsA12 (Ts) double mutant (Figure 5). This result is consistent with the presence of FtsA supporting Z ring formation and stability in the absence of both ZipA and FtsI.
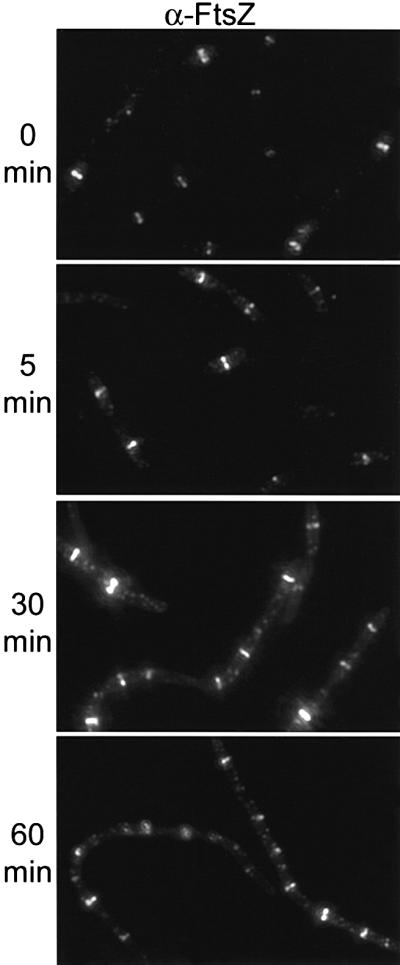
Fig. 7. Localization of FtsZ in ftsI23(Ts) zipA1 (Ts) cells. A culture of PS414 [zipA1 (Ts) ftsI23 (Ts)] was analyzed as described in the legend to Figure 4.
Behavior of an FtsZ mutant unable to interact with ZipA and FtsA
We showed that FtsZ is unable to form rings in a ftsA12 (Ts) zipA1 (Ts) double mutant, so we were curious as to the fate of an FtsZ mutant protein that is unable to interact with both FtsA and ZipA. The prediction is that such a mutant would not assemble into rings. Based upon numerous studies which have shown that the extreme, conserved C-terminal region of FtsZ is essential for interaction with FtsA and ZipA (Liu et al., 1999; Ma and Margolin, 1999b; Hale et al., 2000; Yan et al., 2000; Haney et al., 2001), we used site-directed mutagenesis to replace residues F377 and L378 with alanine to give FtsZFL. Using the yeast two-hybrid system, we determined that this mutant interacts with SulA and FtsZBs as well as wild-type FtsZ, but did not interact with FtsA or ZipA (data not shown). This is in accordance with the study by Haney et al. (2001) who found that each of these mutations greatly reduced the interaction of FtsZ with FtsA and abolished its interaction with ZipA.
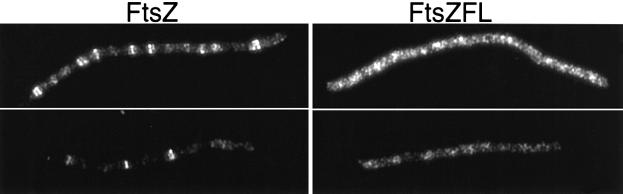
In order to check the ability of this FtsZFL mutant to form rings, we expressed it in cells following depletion of FtsZ. For that purpose we used the strain JKD7-2/pKD3c, which has its chromosomal copy of ftsZ interrupted by a kanamycin resistance gene and FtsZ provided by a copy of the gene on the thermosensitive replicon pKD3c. This strain is able to grow normally at 30°C but is unable to replicate the pKD3c plasmid at 42°C, resulting in FtsZ depletion and filamentation. After 2 h at 42°C, no FtsZ could be detected by western blot or immunofluorescence microscopy (data not shown). After 90 min at 42°C arabinose was added to induce expression of ftsZ or the ftsZFL allele encoded by the plasmids pSEB135 or pSEB135FL, respectively. Wild-type FtsZ was able to assemble into typical rings within the filaments, visible as sharp fluorescent bands (Figure 8). Also, some spirals were present, an indication of excess ftsZ expression. In contrast, the FtsZFL protein did not assemble into rings, even though western blot analysis demonstrated that it was induced to the same level as the control. Occasionally with FtsZFL abnormal structures or spots could be observed; however, typical Z rings were not present. This result indicates that a mutant FtsZ that is unable to interact with FtsA and ZipA is unable to assemble into rings.
Fig. 8. Localization of FtsZ and mutant FtsZFL. Cultures of PS165 (pSEB135-PBAD::ftsZ) or PS166 (pSEB135FL-PBAD::ftsZFL) were placed at 42°C to deplete the endogenous FtsZ. After 90 min 0.02% arabinose was added, and samples were taken 90 min later and stained for FtsZ as described in Materials and methods. Since the level of induction by arabinose is variable from cell to cell in the same culture (Siegele and Hu, 1997), each column shows two cells from each culture, illustrating two different levels of expression of FtsZ (left column) or FtsZFL (right column).
Discussion
The first clearly documented step in bacterial cell division is the assembly of the Z ring at the division site. Coincident with formation of the Z ring is the recruitment of FtsA and ZipA, which bind directly and independently to FtsZ. This is followed by the recruitment of additional cell division proteins to form a fully competent septal ring capable of carrying out cell division. In this study we have further investigated the role of ZipA in assembly of the septal ring utilizing a newly isolated temperature-sensitive mutation and have come to two important conclusions. First, we find that ZipA, like FtsA, is required for the recruitment of FtsK to the division site. Along with previous results this suggests that both FtsA and ZipA are required for the assembly of all additional known cell division proteins. Secondly, we find that either ZipA or FtsA is required for Z ring assembly and stability, suggesting overlapping roles for these proteins in maintaining Z ring integrity.
Since it was known that Z rings form in the absence of either FtsA or ZipA it was suggested that an additional factor was required (Addinall and Lutkenhaus, 1996; Hale and de Boer, 1999; Liu et al., 1999). It was therefore surprising to find that no Z rings formed when both proteins were inactivated. Simultaneous inactivation of both ZipA and FtsA had two effects. First, preformed Z rings rapidly disappeared, suggesting that FtsA and ZipA have the ability to stabilize Z rings. Secondly, new Z rings did not form even though cells continued to increase in cell length. FtsZ was, however, able to localize in spots and arcs at regular intervals, suggesting it was localizing to division sites. These results argue that FtsA and ZipA are not involved in nucleating Z ring assembly, but at least one of them is required for FtsZ filaments to assemble into a Z ring.
To reinforce the above conclusions we analyzed an FtsZ mutant impaired in its ability to interact with both ZipA and FtsA. This FtsZ mutant was unable to form rings, which is in accordance with an observation reported by Ma and Margolin (1999b). These authors showed that an FtsZ mutant deleted of the 12 C-terminal residues (FtsZ C1) interacted with itself, but not with FtsA or ZipA, and localized into regularly spaced dots of fluorescence (indicative of abnormal structures). In the same study, these authors also reported that FtsZ mutants that interacted with ZipA but not FtsA were able to form Z rings. These results support our findings that either ZipA or FtsA is sufficient for Z rings to assemble.
How are these two non-homologous proteins able to stabilize Z rings? One possibility is that they accomplish this by anchoring FtsZ filaments to the membrane. This possibility appears more likely for ZipA, which contains an N-terminal membrane-spanning domain. However, it might also be possible for FtsA since it is a cytosolic protein that appears to have affinity for the membrane based upon cell fractionation studies and the localization of a FtsA–GFP fusion (Sanchez et al., 1994; Ma et al., 1996; Hale and de Boer, 1997). This possibility needs to be investigated further.
Another possibility is that ZipA and FtsA support Z ring formation by promoting bundling of filaments through binding to the C-terminus of FtsZ. We have noted that the C-terminal region of FtsZ is not required for polymerization and actually has an anti-bundling effect (Wang et al., 1997). Protofilaments formed by FtsZ320, which is missing the C-terminal 63 residues, bundle more readily and more extensively than those formed by full-length FtsZ. ZipA has been shown to bundle FtsZ filaments under some in vitro conditions (RayChaudhuri, 1999; Hale et al., 2000). Whether FtsA can also promote bundling has not been reported. Perhaps, Z-ring stability results from a combination of bundling and membrane anchoring. It is worth noting that Z rings form and appear stable as long as FtsA or ZipA is present; however, we do not know if these rings are structurally the same. They appear similar by fluorescence microscopy but it is quite possible that they are structurally different.
With either the ftsA12 (Ts) or zipA1 (Ts) mutation we observed a decrease in the percentage of cells with Z rings immediately upon the temperature shift, indicating some instability of the Z rings when these proteins are delocalized. This decrease was only transient as the Z rings were rapidly restored and additional Z rings formed at new sites as the cells filamented. One explanation for this is that the rings that disappear are those in the process of constriction. It has been noted that rings in the process of constriction are vulnerable to a block to division, whereas rings that have not progressed to the constriction stage appear more stable to a block to division (Addinall et al., 1996; Pogliano et al., 1997). This implies that a constricting ring has quite different properties to a ring that has not initiated constriction.
Although FtsA is quite conserved in prokaryotes it is not as conserved as FtsZ. ZipA is even less well conserved and appears to be present in only Gram-negative bacteria (Hale and de Boer, 1997). This suggests that other proteins can substitute to stabilize Z rings in bacteria. In Bacillus subtilis, Z rings switch from a medial position during vegetative growth to a polar location during sporulation (Levin and Losick, 1996). Efficient polar Z ring formation requires the multifunctional SpoIIE protein, which is also found in a polar ring pattern (Levin et al., 1997; Khvorova et al., 1998). The recruitment of SpoIIE to these rings requires FtsZ but is independent of other known cell division genes, including FtsA. Recently, SpoIIE was shown to bind directly to FtsZ and it was suggested that SpoIIE stabilized polar Z rings (Lucet et al., 2000). SpoIIE could be performing this role during sporulation, whereas FtsA could be carrying out this role during vegetative growth.
Previous localization studies have indicated that division proteins are assembled to the division site in a linear order (summarized in Figure 9). In the established pathway, FtsA plays a critical role as it is positioned between the FtsZ ring and membrane components. All downstream division proteins, which are integral membrane proteins, depend upon FtsA for recruitment. The work presented here indicates that ZipA is also required, since FtsK did not localize in its absence and FtsK is required for recruitment of downstream proteins (Wang and Lutkenhaus, 1998; Chen and Beckwith, 2001). It will be interesting to determine if either FtsA or ZipA, or both, interact directly with FtsK.
Fig. 9. Dependency pathway for recruitment of cell division proteins to the division site. The first clearly defined step in cell division is the assembly of FtsZ to form the Z ring. FtsA and ZipA colocalize with FtsZ, and as shown in this study the presence of either one is sufficient for Z ring formation. The other proteins are then recruited and both ZipA and FtsA are required.
Materials and methods
Media
All experiments were done in Luria–Bertani (LB) medium at 30 or 42°C as indicated. Antibiotics were added at the following concentrations: ampicillin (Amp) 100 µg/ml, chloramphenicol (Cm) 20 μg/ml, tetracycline (Tet) 12 µg/ml, and kanamycin (Kan) 25 µg/ml. Glucose (0.2%), IPTG (0.5 mM) and arabinose (as required) were added as specified in the text.
Strain and plasmid constructions
P1 transduction. The ftsA12 (Ts) and ftsI23 (Ts) mutations were introduced into W3110 or W3110 zipA1 (Ts) by P1 cotransduction with leu::Tn10 using phage lysates prepared on MCA12 and MCI23, respectively. Transductants were selected on tetracycline plates at 30°C. Cotransduction of the desired thermosensitive (Ts) mutation was then confirmed by subcloning colonies at 40 or 42°C (for single mutants), and by preparing P1 lysates on the newly obtained transductants (double mutants) and scoring for contransduction into W3110. The percentage cotransduction for leu::Tn10 and ftsA12 (Ts) was 26–30%, and for leu::Tn10 and ftsI23 (Ts) it was 20–23%.
Construction of pBADzipA and W3110ΩPBADzipA. PCR was used to obtain DNA fragments containing cysZ and zipA as described by Liu et al. (1999). The zipA PCR fragment was cloned into pBAD18 between the EcoRI and SalI sites to give pBADzipA with zipA under the control of the PBAD promoter, which is regulated by arabinose. As described in Liu et al. (1999), cysZ was then cloned downstream of araC into pBADzipA, and a fragment containing a kanamycin resistance gene followed by four transcriptional terminators was inserted between cysZ and araC to give pLZ1. The DNA fragment from pLZ1 containing cysZ to zipA was cloned into the temperature-sensitive replicon pEL3 (Armstrong et al., 1984). Replacement of the cysZ–zipA intergenic region on the chromosome with the fragment cysZ-kan-ter-PBAD-zipA was done by insertion and resolution as described by Liu et al. (1999). Kanamycin-resistant colonies were obtained only in the presence of 1% arabinose. An arabinose-dependent, ampicillin-sensitive colony was designated W3110ΩPBADzipA strain.
Construction of PS223 [zipA1(Ts)] mutant strain. A library of mutated zipA genes expressed from the PBAD promoter was constructed according to the protocol described in Diaz et al. (1991). Following this protocol, pBADzipA DNA was treated for 2 h with nitrous acid. The zipA gene was amplified by PCR using the oligos described earlier (Liu et al., 1999) and the acid treated pBADzipA DNA as template. These PCR fragments were cloned into pBAD18. The resultant plasmids were transformed into W3110ΩPlac-zipA and selected at 30°C on plates containing Spec, Kan, Amp, arabinose (0.06%) and glucose (0.2%), but no IPTG. Prior testing indicated that pBADzipA complemented under these conditions. Colonies were tested for thermosensitivity at 42°C on the same media. Plasmid DNA extracted from Ts colonies was then sequenced. Five clones (all carrying multiple mutations in zipA) were isolated and grouped into two categories after sequencing. Class 1 (two clones) carried M44V, T211L, Y229C and N281S, and class 2 (three clones) carried T211L, R231C and F244S. Only mutations from class 1 could be recombined into the chromosome using the following procedure. First, pSEB138 containing wild-type zipA was constructed by amplifying a 2.6 kb PCR fragment from the chromosome of W3110 with the oligos 5′CysZ 5′-AACATATGCCGCCACATCGCGTGTTTATC and 3′Lig 5′-TCAAGCTGACGCAGTGAACC (cysZ-zipA-lig fragment), digesting with PvuII and BssHII, and cloning into pFC13 (Cornet et al., 1994) cut with BssHII and FspI. pFC13 is a temperature-sensitive replicon unable to replicate at 42°C. pSEB139A and pSEB139B were obtained by replacing the VspI–AflII fragment carrying zipA from pSEB138 with the VspI–AflII fragment from pBADzipA carrying the mutated zipAs. pSEB138, pSEB139A and pSEB139B were introduced into W3110ΩPBADzipA and transformants were selected on Cm, glucose plates at 30°C with no arabinose. The copy of zipA on the plasmid was able to compensate for the lack of zipA expression from the chromosome. Homologous recombination between the plasmid and the chromosome could replace the PBAD-driven copy of zipA by the zipA on the plasmid under control of its own promoter. Transformants were grown in liquid culture to OD540 = 1 and then plated on Cm, glucose plates at 42°C. Colonies obtained at 42°C were pooled and used to start a liquid culture in LB + glucose at 30°C, and grown for 20 generations. The culture was plated at 30°C on LB containing glucose. Two-hundred and fifty colonies were then tested for thermosensitivity at 42°C, arabinose independence, and sensitivity to Cm and Kan. Seven clones meeting these criteria were isolated. The zipA gene from these clones was sequenced to confirm the presence of the mutations on the chromosome. All four mutations were present.
Construction of pSEB135. The ftsZ gene was amplified by PCR using the oligonucleotide FtsZ2Hybrid 5′-ACCGGATCCCTATGTTTGAACCAATGG for the 5′ end of the gene, and the following oligonucleotides for the 3′ end: FtsZ3′PstI 5′-AGTCTGCAGTTAATCAGCTTGCTTACG to amplify ftsZ, and FtsZ3′Flmut 5′-TCTCTGCAGTTAATCAGCTTGCTTACGAGCGGCTGCTGGGATATCCAG to amplify a fragment coding for the FtsZFL protein. The latter oligonucleotide allowed us to replace the residues F377 and L378 with alanine. The PCR fragments were digested with BamHI and PstI and cloned into pGAD424 (Clontech, Palo Alto, CA) digested with the same enzymes, creating pSEB133 and pSEB133FL. In these plasmids the Gal4act domain is fused to FtsZ or the mutant FtsZFL, respectively. These plasmids allowed us to test the interactions of these two proteins with SulA, ZipA, FtsA and B.subtilis FtsZ in the yeast two-hybrid system using plasmids described previously (Wang et al., 1997). pSEB135 and pSEB135FL were constructed by cloning the DraI–KpnI fragments from pSEB133 or pSEB133FL into pJC104 (Mukherjee et al., 2001) digested with KpnI and PvuII. This step replaced the C-terminal part of FtsZ fused to GFP by the C-terminal part of wild-type ftsZ or the ftsZFL mutant. These plasmids allowed us to express FtsZ or FtsZFL under the control of the PBAD promoter, which is regulated by arabinose. These plasmids were introduced into JKD7.2/pKD3c to produce strains PS165 and PS166. These strains allowed us to express plasmid encoded FtsZ or FtsZFL in conditions where the chromosomal FtsZ is depleted. The stability and the comparable level of expression of these two plasmid-encoded proteins were established by western blot using an anti-FtsZ antibody on samples taken after 3 h of expression at a level sufficient to allow the wild-type protein to complement the FtsZ depletion.
Fluorescence microscopy
An overnight culture grown at 30°C was diluted 100 times in LB and grown at 30°C to OD540 = 0.1. The culture was then shifted to a 42°C water bath. Samples were taken and fixed just before the temperature shift and at 5, 30 and 60 min after the shift. Expression of FtsZFL was done in cells depleted of chromosomally encoded FtsZ. Overnight cultures of PS166 (FtsZFL) and PS165 (FtsZ) were grown at 30°C in LB supplemented with Kan, Amp and 0.2% glucose, and diluted 1/100 in the same medium. After 90 min at 30°C, the cultures were shifted to 42°C for 90 min and cells from 5 ml of each culture were collected by centrifugation. The cells were washed once in LB, resuspended in 20 ml of LB containing Kan, Amp and arabinose, and grown at 37°C for 90 min. Fixation of the cells and preparation for immunofluorescence staining were done as described earlier (Addinall and Lutkenhaus, 1996; Addinall et al., 1997b). Samples were observed and photographed with a Nikon Optiphot fluorescence microscope equipped with an E Plan oil immersion lens (Nikon Instruments Inc., Melville, NY) with a 100× objective and a MagnaFire CCD camera S99802 from OPTRONICS (Goleta, CA). For the Cy3 fluorescence, an Omega Optical XF34 filter block with a 450–490 nm excitation filter and a 520 nm barrier filter was used. Images were imported to Adobe Photoshop software to be assembled. The localization of the various proteins was determined by an indirect immunostaining procedure with rabbit polyclonal antibodies against FtsZ (1/1000), FtsA (1/400), ZipA (1/4000) or FtsK (1/200), and secondary goat anti-rabbit immunoglobulin G antibodies coupled to Cy3 (1/100) from Jackson Immuno Research Laboratories (West Grove, PA). These antisera have been described previously (Liu et al., 1999).
Acknowledgments
Acknowledgements
This work was supported by grant No. GM29764 from the National Institutes of Health.
References
- Addinall S.G. and Lutkenhaus,J. (1996) FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol., 178, 7167–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S.G., Bi,E. and Lutkenhaus,J. (1996) FtsZ ring formation in fts mutants. J. Bacteriol., 178, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S.G., Cao,C. and Lutkenhaus,J. (1997a) FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol., 25, 303–309. [DOI] [PubMed] [Google Scholar]
- Addinall S.G., Cao,C. and Lutkenhaus,J. (1997b) Temperature shift experiments with an ftsZ84 (Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol., 179, 4277–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K.A., Acosta,R., Ledner,E., Machida,Y., Pancotto,M., McCormick,M., Ohtsubo,H. and Ohtsubo,E. (1984) A 37 × 103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J. Mol. Biol., 175, 331–348. [DOI] [PubMed] [Google Scholar]
- Bi E.F. and Lutkenhaus,J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature, 354, 161–164. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander,C. and Valencia,A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.C. and Beckwith,J. (2001) FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol., 42, 395–413. [DOI] [PubMed] [Google Scholar]
- Chen J.C., Weiss,D.S., Ghigo,J.M. and Beckwith,J. (1999) Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol., 181, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet F., Mortier,I., Patte,J. and Louarn,J.M. (1994) Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol., 176, 3188–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K. and Lutkenhaus,J. (1991) ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol., 173, 3500–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J.J., Rhoads,D.D. and Roufa,D.J. (1991) PCR-mediated chemical mutagenesis of cloned duplex DNAs. Biotechniques, 11, 204–206. [PubMed] [Google Scholar]
- Erickson H.P., Taylor,D.W., Taylor,K.A. and Bramhill,D. (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. USA, 93, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo J.M., Weiss,D.S., Chen,J.C., Yarrow,J.C. and Beckwith,J. (1999) Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol., 31, 725–737. [DOI] [PubMed] [Google Scholar]
- Hale C.A. and de Boer,P.A. (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E.coli. Cell, 88, 175–185. [DOI] [PubMed] [Google Scholar]
- Hale C.A. and de Boer,P.A. (1999) Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol., 181, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.A., Rhee,A.C. and de Boer,P.A. (2000) ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol., 182, 5153–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney S.A., Glasfeld,E., Hale,C., Keeney,D., He,Z. and de Boer,P. (2001) Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem., 276, 11980–11987. [DOI] [PubMed] [Google Scholar]
- Khvorova A., Zhang,L., Higgins,M.L. and Piggot,P.J. (1998) The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J. Bacteriol., 180, 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A. and Losick,R. (1996) Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev., 10, 478–488. [DOI] [PubMed] [Google Scholar]
- Levin P.A., Losick,R., Stragier,P. and Arigoni,F. (1997) Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol. Microbiol., 25, 839–846. [DOI] [PubMed] [Google Scholar]
- Liu Z., Mukherjee,A. and Lutkenhaus,J. (1999) Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol., 31, 1853–1861. [DOI] [PubMed] [Google Scholar]
- Lucet I., Feucht,A., Yudkin,M.D. and Errington,J. (2000) Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J., 19, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. (1993) FtsZ ring in bacterial cytokinesis. Mol. Microbiol., 9, 403–409. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. and Addinall,S.G. (1997) Bacterial cell division and the Z ring. Annu. Rev. Biochem., 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Ma X. and Margolin,W. (1999a) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol., 181: 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. and Margolin,W. (1999b) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol., 181, 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ehrhardt,D.W. and Margolin,W. (1996) Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA, 93, 12998–13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock J.R. and Shapiro,L. (1993) Polar location of the chemoreceptor complex in the Escherichia coli cell. Science, 259, 1717–1723. [DOI] [PubMed] [Google Scholar]
- Margolin W. (2000) Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev., 24, 531–548. [DOI] [PubMed] [Google Scholar]
- Mosyak L., Zhang,Y., Glasfeld,E., Haney,S., Stahl,M., Seehra,J. and Somers,W.S. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J., 19, 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus,J. (1994) Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol., 176, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Saez,C. and Lutkenhaus,J. (2001) Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J. Bacteriol., 183, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Pogliano,K., Weiss,D.S., Losick,R., and Beckwith,J. (1997) Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl Acad. Sci. USA, 94, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K., Harry,E. and Losick,R. (1995) Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol., 18, 459–470. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D. (1999) ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J., 18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Justice,S. and Garcia-Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–438. [DOI] [PubMed] [Google Scholar]
- Sanchez M., Valencia,A., Ferrandiz,M.J., Sander,C. and Vicente,M. (1994) Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J., 13, 4919–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D.A. and Hu,J.C. (1997) Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl Acad. Sci. USA, 94, 8168–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. and Lutkenhaus,J. (1998) FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol., 29, 731–740. [DOI] [PubMed] [Google Scholar]
- Wang L., Khattar,M.K., Donachie,W.D. and Lutkenhaus,J. (1998) FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol., 180, 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang,J., Mukherjee,A., Cao,C. and Lutkenhaus,J. (1997) Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol., 179, 5551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D.S., Chen,J.C., Ghigo,J.M. and Beckwith,J. (1999) Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol., 181, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Pearce,K.H. and Payne,D.J. (2000) A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun., 270, 387–392. [DOI] [PubMed] [Google Scholar]
- Yu X.C., Tran,A.H., Sun,Q. and Margolin,W. (1998) Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol., 180, 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]