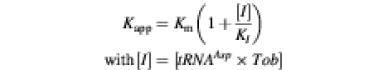Abstract
Aminoglycosides inhibit translation in bacteria by binding to the A site in the ribosome. Here, it is shown that, in yeast, aminoglycosides can also interfere with other processes of translation in vitro. Steady-state aminoacylation kinetics of unmodified yeast tRNAAsp transcript indicate that the complex between tRNAAsp and tobramycin is a competitive inhibitor of the aspartylation reaction with an inhibition constant (KI) of 36 nM. Addition of an excess of heterologous tRNAs did not reverse the charging of tRNAAsp, indicating a specific inhibition of the aspartylation reaction. Although magnesium ions compete with the inhibitory effect, the formation of the aspartate adenylate in the ATP–PPi exchange reaction by aspartyl-tRNA synthetase in the absence of the tRNA is not inhibited. Ultraviolet absorbance melting experiments indicate that tobramycin interacts with and destabilizes the native L-shaped tertiary structure of tRNAAsp. Fluorescence anisotropy using fluorescein-labelled tobramycin reveals a stoichiometry of one molecule bound to tRNAAsp with a KD of 267 nM. The results indicate that aminoglycosides are biologically effective when their binding induces a shift in a conformational equilibrium of the RNA.
Keywords: aminoacyl-tRNA synthetase/antibiotics/fluorescence spectrometry/pharmacology/translation
Introduction
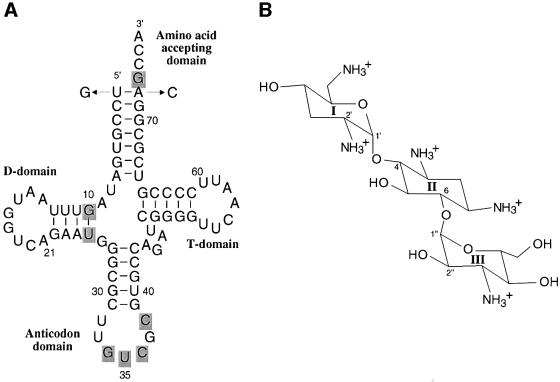
Aminoacylation of transfer RNA (tRNA) is essential for cell replication and growth. It is therefore an attractive target for therapeutic intervention. The following strategies have been explored so far: (i) amino acid analogues (Loftfield, 1973; Vasquez, 1974), (ii) oligonucleotides mimicking tRNA features (Loftfield, 1973), (iii) aminoacyl-adenylate analogues (Sassanfar et al., 1996) and (iv) blocking the formation of initiator tRNAfMet (Loftfield, 1973). Up to now, most of the known natural products targeted against specific aminoacyl-tRNA synthetases (aaRSs), or apparent amino acid specificities of the aaRSs could not be developed into an antibiotic due mainly to their lack of systemic bioavailability (Schimmel et al., 1998). The correct aminoacylation of tRNAs by their cognate synthetase is crucial for the accurate transmission of genetic information. It is determined by specific structural features of the tRNAs (Giegé et al., 1998). Although the global architecture of tRNAs is highly conserved, subtle protein–tRNA interfacial contacts guarantee specific aminoacylation of each tRNA by a specific synthetase. The aminoacylation reaction of yeast tRNAAsp (Figure 1A) by its cognate aspartyl-tRNA synthetase (AspRS) is biochemically (Romby et al., 1985; Pütz et al., 1991; Frugier et al., 1994) and structurally well described (Westhof et al., 1985; Cavarelli et al., 1993).
Fig. 1. Yeast tRNAAsp and the aminoglycoside tobramycin. (A) Sequence of yeast tRNAAsp transcript (Gangloff et al., 1971) showing the change of the first base pair (U1–A72→G1–C72); nucleotides are numbered according to Sprinzl et al. (1998). Identity nucleotides of the aspartylation reaction are shadowed (Pütz et al., 1991; Frugier et al., 1994). The G1–C72 wild-type transcript shows equivalent aspartylation parameters to those of fully modified tRNAAsp and U1–A72 transcripts (Pütz et al., 1991). (B) Structure of the aminoglycoside antibiotic tobramycin, a member of the 2′deoxystreptamine group. The antibiotics of the aminoglycoside family result from modifications of neamine, a two-ring system made of 2-deoxystreptamine (called ring B or II) glycosylated at the 4-position by a 6-membered amino-sugar (called ring A or I) of the glycopyranoside series. Further modifications with various amino-sugars at the 6-position lead to the kanamycin family.
Aminoglycosides are known to interact with ribosomal RNAs inhibiting translation at the ribosome level (Blanchard et al., 1998). They further interfere with translational control (Tok et al., 1999), viral transcription of HIV (Zapp et al., 1993; Mei et al., 1995) and ribozyme activity (reviewed in Schroeder et al., 2000). Here, the interaction of tobramycin, a common aminoglycoside of the 2′deoxystreptamine group (Figure 1B), with the yeast tRNAAsp–AspRS complex was investigated. It is demonstrated that the aminoacylation of tRNAAsp, a primary step in protein synthesis, can be specifically inhibited by tobramycin with binding affinities in the nanomolar range. Tobramycin is not the first compound shown to inhibit aminoacylation. Purpuromycin had already been shown to inhibit the aminoacylation reaction at the level of tRNA charging, but with several molecules of purpuromycin binding with micromolar affinity to all tRNAs (Kirillov et al., 1997). Similarly, tobramycin is not the first compound shown to interfere with an RNA–protein enzymic system. Recently, it has been shown that synthetic benzimidazole derivatives inhibit the processing of the precursor-tRNAs to mature tRNAs by the RNA subunit of Escherichia coli RNase P (M1 RNA), but with I50 values between 5 and 20 µM (Hori et al., 2001).
Results
Aspartylation activity of tRNAAsp in the presence of tobramycin
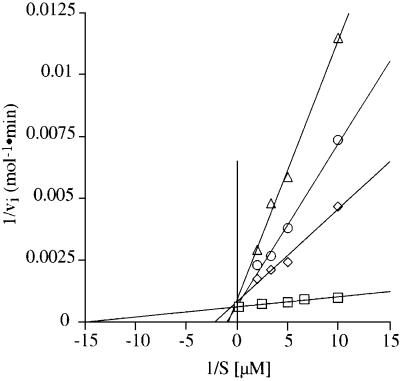
Aminoacylation experiments with yeast AspRS (Figure 1) reported here were carried out on molecules obtained in vitro by transcription with T7 RNA polymerase. Figure 2 compares the aspartylation kinetics of wild-type tRNAAsp transcript in the absence and presence of the aminoglycoside tobramycin. The aspartylation of tRNAAsp decreases with increased concentrations of tobramycin (1–3 mM). The lines in the absence and presence of the antibiotic intercept the y-axis at one point, indicating that tobramycin acts as a competitive inhibitor with respect to tRNAAsp (Figure 2). The Michaelis–Menten parameters kcat and Km in the presence and the absence of tobramycin and/or total tRNA from yeast are summarized in Table I together with the relative kinetic specificity constants (kcat/Km)rel = (kcat/Km)tobramycin/(kcat/Km)–tobramycin. A more intuitive number is also included, namely the loss in aminoacylation efficiency caused by the antibiotic. The presence of tobramycin affects mainly the Km by factors up to 30-fold (for 3 mM tobramycin at an ATP:Mg2+ ratio of 5:15 mM), while the kcat is only decreased 2-fold. As a consequence the relative specificity constants are decreased and the loss in aminoacylation efficiency increases up to 60-fold. Further, aminoacylation experiments using fully modified yeast tRNAAsp reveal that tobramycin inhibits charging to the same extent, showing equivalent kinetic parameters for aminoacylation compared with those of the corresponding in vitro transcripts (data not shown). The addition of tobramycin at various times or directly before the start of the reaction by AspRS does not show a change in the aminoacylation kinetics (data not shown). Tobramycin does not exhibit a time dependence of the inhibition of the aminoacylation reaction of tRNAAsp. Thus, the kinetics measurements indicate that binding of tobramycin leads to spatial conformational changes of the tRNA, resulting in a reduced affinity of tRNAAsp for AspRS or a loss in transition state stabilization of the tRNAAsp–AspRS complex.
Fig. 2. Inhibition of the aspartylation reaction of tRNAAsp transcripts by tobramycin. The double reciprocal plot (Lineweaver–Burk) shows the initial velocity of the aspartylation reaction as a function of tRNAAsp concentration in the absence of tobramycin (circles) and in the presence of tobramycin at 1 (squares), 2 (diamonds) and 3 mM (triangles).
Table I. Kinetic parameters for aspartylation of yeast tRNAAsp transcripts with yeast AspRS in the absence or presence of tobramycin.
| ATP/MgCl2 (mM) | Inhibitortobramycin (mM) | Competitor tRNAtotal [mM] | Km (nM) | kcat (s–1) | kcat/Km (relative) | Loss of specificity (x-fold) |
|---|---|---|---|---|---|---|
| 5/15a | 0 | – | 44 | 0.66 | 1 | 1 |
| 5/15a | 1 | – | 466 | 0.33 | 0.047 | 21 |
| 5/15a | 2 | – | 945 | 0.38 | 0.027 | 37 |
| 5/15a | 3 | – | 1190 | 0.30 | 0.017 | 60 |
| 5/15a | 3 | 10 | 151 | 0.27 | 0.12 | 8 |
| 5/15a | 3 | 30 | 115 | 0.21 | 0.12 | 8 |
| 5/15a | 3 | 90 | 95 | 0.18 | 0.13 | 8 |
| 2/10b | 0 | – | 732 | 0.31 | 0.028 | 35 |
| 2/10b | 3 | – | 1285 | 0.58 | 0.03 | 33 |
aATP:Mg2+ ratio of 1:3.
bATP:Mg2+ ratio of 1:5.
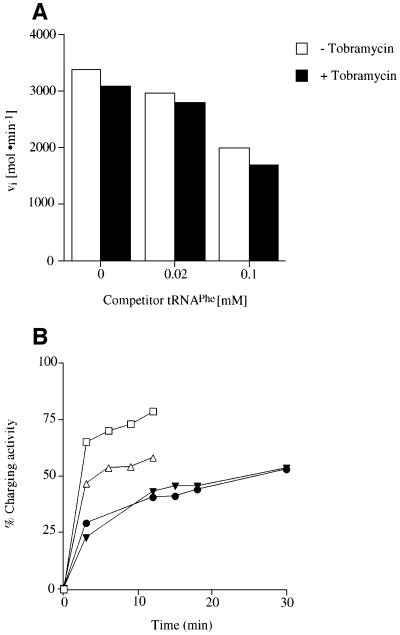
To determine whether the inhibition is specific for the tRNAAsp–AspRS interaction, the effect of tobramycin on tRNAAsp was determined in the presence of an increasing excess of competitor tRNA. For this purpose tRNAAsp is incubated together with up to 2-fold excess of tRNAPhe over tobramycin. No recovery of the aminoacylation activity is observed after the addition of native tRNAPhe (Figure 3A). In the high concentration range of competitor tRNA an inhibition effect of the aminoacylation reaction is noticeable, probably due to the addition of high concentrations of charges and salt. Moreover, assays in the presence of high levels of total tRNA (3- to 50-fold excess over tobramycin) also resulted in no recovery of aspartylation activity (Table I). Further analysis of the kinetic parameters shows only small effects on the aspartylation reaction. A loss of specificity by a factor of 8 is observed, but remains essentially unchanged with increasing concentrations of total tRNA.
Fig. 3. Kinetic measurements of the competition of tobramycin binding to tRNAAsp by other tRNAs. (A) Influence of an excess of competitor tRNA, e.g. tRNAPhe at 0.02 and 0.1 mM (2-fold excess compared with tobramycin), on the aspartylation reaction of tRNAAsp in the absence (unfilled bars) and in the presence of 0.05 mM tobramycin (filled bars). (B) Competition of tobramycin upon binding to tRNAAsp and tRNAPhe within a native mixture of all yeast tRNAs. The level of aminoacylation is expressed as the charging activity for the aspartylation by yeast AspRS in the absence (squares) and presence of 0.3 mM tobramycin (triangles) and as a control for the phenylalanylation by yeast PheRS in the absence (filled circles) and presence of 0.3 mM tobramycin (filled triangles).
The next question addressed was whether tobramycin can specifically inhibit tRNAAsp charging within a mixture of various tRNA families. The level of aspartylation and phenylalanylation within a fraction of total tRNAs was monitored in the absence and presence of tobramycin. Figure 3B reveals that tobramycin exhibits the same inhibition potential towards the aspartate system as shown for purified tRNAAsp (see Figure 2). The aspartyl ation reaction is inhibited by ∼20%. However, the phenylalanylation of tRNAPhe within the same pool is not affected (Figure 3B).
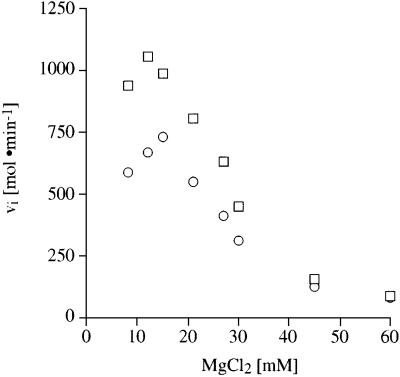
Electrostatic interactions play a dominant role in aminoglycoside–RNA binding (Tor et al., 1998). If the positively charged aminoglycoside is in competition with divalent metal ions, inhibition of the tRNA charging reaction could be overcome by increased concentrations. In one series of experiments, the ATP:MgCl2 ratio was kept constant (1:3), while varying the magnesium ion concentration from 7 to 60 mM. Figure 4 illustrates that the aminoacylation reaction reaches its maximum at 10 mM MgCl2. In the presence of tobramycin this maximum is reduced and shifted to 15 mM MgCl2. Reaction mixtures containing >12 mM MgCl2 reveal a decrease in aminoacylation activity as well as a reduction in the inhibition effect caused by tobramycin. A second set of experiments was carried out under constant ATP conditions (5 mM) with increased MgCl2 concentrations ranging from 1 to 50 mM giving identical results (data not shown).
Fig. 4. Kinetic measurements of the competition of tobramycin binding to tRNAAsp by magnesium ions. Influence of magnesium ions on the aspartylation reaction of tRNAAsp in a buffer solution containing increasing concentrations of MgCl2 up to 60 mM, while keeping the ATP and MgCl2 at a constant ratio of 1:3, in the absence of tobramycin (squares) and in the presence of 0.3 mM tobramycin (circles).
Tobramycin could bind specifically to the catalytic site of yeast AspRS as a structural analogue of ATP. To exclude this possibility, the concentration of ATP in the aspartylation reaction was reduced from 5 to 2 mM ATP (leading to a change in the ATP:MgCl2 ratio from 1:3 to 1:5). The effect of tobramycin on the aspartylation kinetics of tRNAAsp is compared before and after the change of the ATP:MgCl2 ratio (Figure 5A and Table I). A slight change in the initial velocity is observed in the absence of the antibiotic. However, in the presence of tobramycin the inhibitory effect is identical, showing that ATP is not competing with tobramycin for binding to the catalytic site of AspRS.
Fig. 5. Aspartyl-adenylate formation by AspRS in the presence of tobramycin. (A) The Lineweaver–Burk plot shows the kinetics of the aspartylation reaction in the absence of tobramycin in a buffer solution containing 5 mM ATP and 15 mM MgCl2 (1:3) (squares), or 2 mM ATP and 10 mM MgCl2 (1:5) (diamonds) and in the presence of 3 mM tobramycin at an ATP:MgCl2 ratio of (1:3) (circles) or (1:5) (triangles). (B) Influence of tobramycin on the [32P]PPi–ATP exchange reaction catalysed by AspRS.
Aspartyl-adenylate formation by ApsRS is not inhibited by tobramycin
During the first step in the aminoacylation reaction, AspRS forms the activated aspartyl-adenylate in the absence of tRNAAsp. A series of pyrophosphate exchange [PPi] experiments were designed to estimate the degree of inhibition during adenylate formation. AspRS was incubated with radioactively labelled pyrophosphate and aspartic acid in the presence and the absence of the aminoglycoside. The initial rate of the pyrophosphate exchange reaction at all three concentrations of tobramycin (0.3–30 mM) tested is at the same level and no difference at the level of pyrophosphate incorporation either in the absence or presence of the aminoglycoside is visible (Figure 5B). Therefore, tobramycin does not act as a structural analogue of ATP or an amino acid in the aminoacylation reaction and does not interact with the catalytic domain of the AspRS itself during the amino acid activation step.
Ultraviolet absorbance melting experiments reveal a destabilization of the tertiary structure of tRNAAsp upon interaction with tobramycin
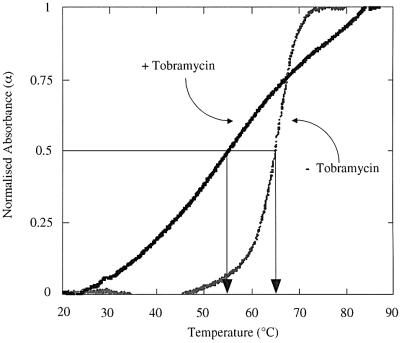
Ultraviolet (UV) absorbance melting curves were performed to assess the effect of the tobramycin–tRNAAsp interaction on the overall structural stability of the tRNAAsp. In the presence of 3 mM MgCl2 the shape of the melting curve of tRNAAsp shows a sharp and symmetrical profile (Figure 6) indicating a highly cooperative melting transition in a quasi all-or-none manner. The calculated melting temperature (Tm) of 65°C and the shape of the transition is in agreement with published data (Puglisi et al., 1993). After the addition of 1 mM tobramycin the melting profile of tRNAAsp changes dramatically with a drop of the Tm of 10°C to 55°C. The Tm and the shape of the melting transition in the presence of the antibiotic resemble that of tRNAAsp in the absence of magnesium ions (Puglisi et al., 1993). This broad, non-cooperative transition indicates several melting domains related to the independent melting behaviour of the helical regions of the tRNA. Therefore, it appears that tobramycin destabilizes the functional tertiary conformation of the tRNAAsp and/or the tRNAAsp–AspRS complex including magnesium ions and the adenylate. Indeed, disruption of tertiary interactions in the core of tRNAAsp results in a decrease of (kcat/Km)rel (Puglisi et al., 1993).
Fig. 6. UV absorbance melting experiments of yeast tRNAAsp transcripts in the absence and presence of 1 mM tobramycin. Points are experimental. The calculated Tm is indicated by the arrows.
Fluorescence anisotropy experiments
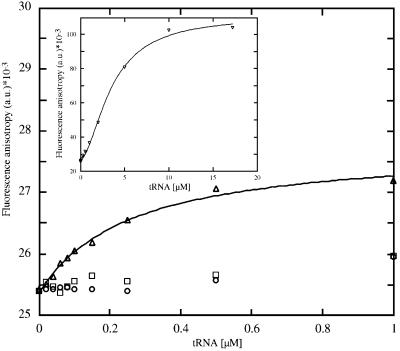
In order to determine quantitatively the specificity and stoichiometry of ligand binding, the fluorescence anisotropy of fluorescein-labelled tobramycin (Tob–Fl) was measured. Figure 8 shows the change in fluorescence anisotropy as a function of tRNA concentration. The anisotropy value r (see Equation 1 in Materials and methods) for Tob–Fl in the absence of tRNA is (25.50 ± 0.17) × 10–3. Addition of tRNAAsp results in a hyperbolic increase in the fluorescence anisotropy to (27.31 ± 0.04) × 10–3 for completely bound Tob–Fl, with 1.2 molecules of Tob–Fl found to bind with high affinity and specificity to tRNAAsp with an R value of 0.99 (see Equation 3 in Materials and methods). Fitting the plot for tRNAAsp with a 1:1 stoichiometry reveals a dissociation constant of 267 nM (see Equation 2 in Materials and methods). An active tRNAAsp anticodon loop variant (Figure 7A) gives similar results (data not shown). An aminoacylation-inactive 3D-folding variant of tRNAAsp (tRNAAsp mutant D, Figure 7B) containing the identity elements for aspartylation, but with a disrupted 3D structure, was also titrated. The initial value of the fluorescence anisotropy of Tob–Fl does not change significantly with the addition of tRNAAsp mutant D. The inset in Figure 8 shows the titration of Tob–Fl with fully modified tRNAPhe. The plot displays a large and slightly sigmoidal increase in the fluorescence anisotropy to ∼(105 ± 0.08) × 10–3 at very high concentrations of tRNAPhe. Upon binding to tRNAPhe the fluorescence anisotropy is reduced dramatically, indicating that the fluorophore is constrained in some manner upon binding to the RNA. The affinity of Tob–Fl for tRNAPhe is much lower, with a KD of 6.0 µM and 1.73 bound molecules of Tob–Fl per tRNAPhe with an R value of 0.99. The KD is increased by a factor of 20 compared with that of the tRNAAsp transcript. Hill plot analysis using the tobramycin–fluorescein binding data with yeast tRNAPhe resulted in a slope value of 1, indicating the absence of a cooperative binding effect (data not shown). The fluorescence anisotropy change measured for tobramycin upon binding to tRNAAsp is rather low (Δr = 2 × 10–3), while in the presence of tRNAPhe a difference in Δr of 80 × 10–3 anisotropy units is observed. A tobramycin binding aptamer interacting with high affinity with Tob–Fl shows a moderate anisotropy change (Δr = 30 × 10–3) (Wang et al., 1996). In this case, tobramycin binds to the RNA deep groove centred about a stem–loop junction site. A portion of the bound tobramycin, ring III and partly II (Figure 1B), is encapsulated between the floor of the deep groove and a looped-out cytosine residue that forms a flap over the binding site in the complex with the aptamer (Jiang et al., 1997). The primary amino group is the site of attachment to the column in the SELEX experiment of the tobramycin aptamer (Wang et al., 1996) and also the site of labelling with the fluorescein dye in our experiments. This amino group on the ring system I reaches slightly out into the solution (Jiang et al., 1997). The moderate change in the anisotropy value in the case of the aptamer can be explained by its degree of freedom. The relatively low anisotropy of Tob–Fl bound to tRNAAsp indicates that the fluorescein is still very mobile. The fluorescein group attached to the tobramycin probably reaches out into the solution, while other amino groups of tobramycin seem to be responsible for the specific binding at the site of the tertiary interaction, possibly between the D and T domains. However, the high anisotropy value found for Tob–Fl bound to tRNAPhe indicates that the reporter dye must be immobilized to a high degree.
Fig. 8. Fluorescence anisotropy measurements of tobramycin–tRNA complex formation. Fluorescence anisotropy (r) of fluorescence-labelled tobramycin (Tob–Fl; 10 nM) as a function of tRNAAsp (triangles), tRNAAsp mutant D (circles) or transcribed E.coli tRNAAsp (squares) concentration. The solid line is calculated by curve fitting to Equation 2. The tRNAAsp mutant D (sequence, Figure 7B) and E.coli tRNAAsp showed only non-specific binding at high concentrations of transcript. The inset displays the fluorescence anisotropy (r) of Tob–Fl solution (10 nM) as a function of native tRNAPhe (inverted triangles) concentration. The solid line is calculated by curve fitting to Equation 2. Please note that the buffer conditions are reduced due a high quenching effect (see Materials and methods).
Fig. 7. Sequences of tRNA variants used in the fluorescence anisotropy measurements. The identity elements for aspartylation are shadowed. Sequence variations from tRNAAsp are highlighted in lower-case letters. (A) The yeast tRNAAsp mutant A is an active anticodon loop variant of tRNAAsp. The anticodon sequence shows a shift of the GUC-identity elements for AspRS, but is active in the aspartylation reaction (J.Pütz and R.Giegé, unpublished data). (B) The 3D structure of the yeast tRNAAsp mutant D is prevented from folding into the native L-shaped structure by the insertion of a series of adenine nucleotides in the D and T domains, which impairs aminoacylation.
The yeast AspRS enzyme is known to aminoacylate both the yeast tRNAAsp and the tRNAAsp from E.coli. In contrast, the bacterial enzyme of AspRS binds both tRNAs with similar affinity but cannot charge yeast tRNAAsp. A crystal structure of an inactive complex between yeast tRNAAsp and E.coli AspRS has recently been published confirming the crucial role of the native and properly adapted tRNA three-dimensional structure (Moulinier et al., 2001). The sequence of yeast tRNAAsp differs from E.coli tRNAAsp in 35 positions, four of which are located in the D domain and six in the T domain with two positions located in the T loop directly. The fluorescence anisotropy binding assay is used to test for binding of Tob–Fl to E.coli tRNAAsp. In contrast to yeast tRNAAsp, the addition of E.coli tRNAAsp does not induce a significant increase in the initial value of the fluorescence anisotropy of Tob–Fl (Figure 8). This corresponds to the behaviour of the aminoacylation-inactive tRNAAsp mutant D (Figure 8) containing the identity elements for aspartylation, but with a disrupted 3D structure. Since Tob–Fl does not bind to E.coli tRNAAsp, inhibition of aminoacylation of yeast AspRS by tobramycin is not expected. This experiment demonstrates that the conformational change necessary for tobramycin binding to a tRNA depends on those parts of the tRNA sequence underlying the tertiary structure.
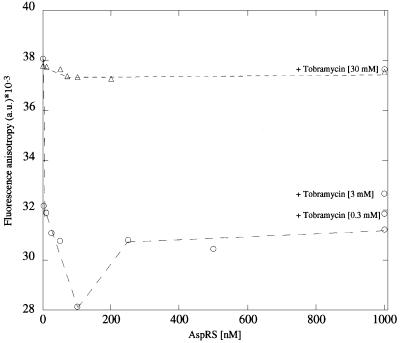
To investigate further the effect induced by tobramycin binding to the tRNAAsp on its interaction with AspRS, tRNAAsp was labelled at the free 5′ terminus with the fluorescent dye erythrosin (tRNAAsp-Ery). The fluorescence anisotropy value of tRNAAsp-Ery is very high (38 ± 0.2 × 10–3) in the absence of AspRS (Figure 9). The fluorescent dye might be restricted in its rotational freedom due to some interaction with the single-stranded 3′ CCA terminus. Upon addition of AspRS the fluorescence anisotropy value dropped down to a minimum of (28 ± 0.16) × 10–3 at 100 nM AspRS. Upon complex formation between tRNAAsp and AspRS a conformational rearrangement occurs that allows the fluorescent dye more rotational freedom. At the end of the titration with AspRS the inhibitor molecule was added in three steps to the tRNAAsp–AspRS complex. The fluorescence anisotropy value increased back to its original value with increasing concentrations of tobramycin. If tobramycin is bound to tRNAAsp, AspRS can no longer bind to the tRNA and the fluorescent dye can fold back at its original position. When tobramycin was allowed to form a complex with tRNAAsp before titration with AspRS, no significant change in the fluorescence anisotropy value was observed during the whole titrational range. Thus, the tobramycin–tRNAAsp complex functions as a competitive inhibitor in the aminoacylation reaction by preventing the synthetase from binding to its substrate.
Fig. 9. Fluorescence anisotropy measurements of tRNAAsp-Ery–AspRS complex formation. Fluorescence anisotropy r of fluorescence-labelled tRNAAsp (tRNAAsp-Ery; 1 nM) as a function of AspRS (circles) and after preincubation of tRNAAsp with 30 mM tobramycin (triangles). In the first case increasing amounts of tobramycin are added at the final AspRS concentration to monitor the effect of the inhibitor on the tRNAAsp-Ery–AspRS complex.
The set of fluorescence anisotropy measurements shows that the inhibitor acts by binding to the tRNA substrate of the enzymic reaction thereby forming an inhibitory complex.
Aminoacylation kinetics were performed under oversaturating inhibitor concentrations. Under these conditions and a measured KD in the lower nanomolar range one can assume that the concentration of the inhibitory complex is equal to the concentration of the substrate, e.g. tRNAAsp.
A detailed analysis shows that the inhibition constant KI using the standard relationship (Equation 4):
is in the lower nanomolar range at 36 nM. If one assumes that tobramycin acts as an inhibitor directly on the enzyme, the calculated KI would increase into the upper micromolar range (125 µM), a value much higher than the measured affinity binding constant.
Discussion
Our steady-state kinetics of the aminoacylation reaction demonstrate that tobramycin specifically inhibits by a competitive mechanism the charging of tRNAAsp in vitro (Figure 2) even in the presence of non-cognate tRNAs (either a mixture of all native tRNAs or in the presence of tRNAPhe) (Figure 3). Tobramycin is in competition with magnesium ions (Figure 4), but not with ATP (Figure 5A). Tobramycin does not act at the level of the formation of activated aspartyl-adenylate by AspRS (Figure 5B). The dissociation constant measured by fluorescence anisotropy is in the nanomolar range (Figure 8). UV absorption melting curves indicate that tobramycin disrupts the native conformation of tRNAAsp (Figure 6). Tobramycin does not bind either to a mutated tRNAAsp, which does not display a tertiary structure, or to E.coli tRNAAsp with sequence variations in both the D and T domains (Figure 8). The non-native tobramycin–tRNAAsp complex acts as a competitive inhibitor of the aspartylation reaction (Figure 9). Chemical and enzymic footprinting studies confirm that the binding of tobramycin alters specifically the native 3D structure of yeast tRNAAsp (to be published elsewhere).
Binding of tobramycin to a tRNA does not lead automatically to a conformational change interfering with the recognition of the tRNA identity elements by the cognate aaRS. Indeed, fluorescence anisotropy measurements show micromolar binding of tobramycin to yeast tRNAPhe (Figure 8), but the phenylalanylation of yeast tRNAPhe by PheRS is not inhibited by tobramycin (Figure 3B). Among aminoglycosides, neomycin B has been demonstrated to inhibit the phenylalanylation of E.coli tRNAPhe with an I50 (e.g. concentration at 50% inhibition of the aminoacylation reaction) in the upper micromolar range (Mikkelsen et al., 2001). Using a crude extract of the tRNA fraction higher concentrations of the aminoglycoside are needed for the inhibition of the aminoacylation reaction. In the yeast tRNAPhe– aminoglycoside crystal complex only one neomycin molecule is observed. Neomycin B is bound to the upper part of the anticodon stem. The same authors tested the inhibition of lead (Pb2+) cleavage of yeast tRNAPhe by various aminoglycosides. They concluded that neomycin B binds probably with two molecules to tRNAPhe with an I50 of 300 µM. Previously, several aminoglycosides had been shown to bind to tRNAPhe at various sites, stabilizing the structure of tRNAPhe in the same manner as polyamines (Kirk and Tor, 1999). The strongest stabilization effect was observed by UV absorbance melting experiments with a ΔTm of +14°C upon addition of neomycin B (10 µM).
Aminoglycosides share common structural features with polyamines and one can expect that they can bind non-specifically to all RNAs (Robinson and Wang, 1996). Sequence independent binding in the deep groove of duplex RNA for both of types of cationic ligands is within the millimolar range and sensitive to both pH and salt concentrations (Jin et al., 2000). Tobramycin binding to an RNA duplex enhances thermal stability of a poly(rI)–poly(rC) RNA duplex, decreasing with increasing sodium ion concentrations and pH. Viscometric measurements are consistent with a tobramycin-induced non-specific and non-intercalative binding into the deep groove. Such considerations and the fact that aminoglycosides interact with a great variety of RNA molecules (for recent reviews see Walter et al., 1999; Schroeder et al., 2000) have led to the notion that their binding is not very specific.
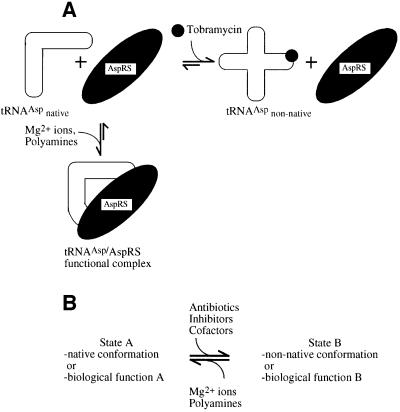
Here, we show that an aminoglycoside antibiotic influences specifically a protein–RNA interface leading to inhibition of function only if the binding is correlated with a conformational change. This observation, together with recent results on the 30S particle of the ribosome (Ogle et al., 2001) and a complex of the A site of the 16S rRNA with aminoglycosides (Vicens and Westhof, 2001), lead us to propose a model for the differences in efficiency of inhibition induced by aminoglycosides in various systems. We have shown that the antibiotic binds to the tRNA substrate of the catalytic reaction and interferes with enzymic function because it induces a conformational change in the substrate (Figure 10A). Small molecules, like magnesium ions, polyamines and related aminoglycosides, may bind non-specifically and stabilize the native folding of RNA molecules, leading to proper biological function. In contrast, when the binding of a small molecule or antibiotic leads to the stabilization of a non-native RNA conformation or to a shift in a conformational equilibrium, biological errors or enzyme inhibition can be promoted (Figure 10B).
Fig. 10. Scheme of inhibition of function by antibiotics. (A) Scheme of inhibition of aspartylation of yeast tRNAAsp by tobramycin. The L-shaped structure of yeast tRNAAsp is stabilized by magnesium ions and cationic polyamines. AspRS is able to recognize the spatial arrangement of the identity elements on the tRNA structure and binds to the native conformation of tRNAAsp. If the functional complex is formed, the tRNAAsp becomes aspartylated by AspRS. Tobramycin also binds with high affinity to tRNAAsp. Binding of tobramycin disrupts and destabilizes the native structure of tRNAAsp, and AspRS is unable to bind to the unfolded tRNAAsp conformation. Thus, the antibiotic, by interacting with the native tertiary structure of tRNAAsp, can interfere with the subsequent productive interaction of tRNAAsp with AspRS. (B) Conformational states of RNA molecules and interaction with ligands. In the native conformation (A) an RNA molecule can perform its natural function. Upon a conformational change, the RNA adopts either a non-native conformation or another conformation related to alternative function (B). Polyamines and specific divalent metal ions (e.g. magnesium ions) usually stabilize the native state of an RNA molecule. In contrast, inhibitors, antibiotics or appropriate cofactors may shift the equilibrium to either a non-native state of the RNA or an alternative conformation, thereby interfering with its function.
Aminoglycosides or novel small molecular RNA binders that are specific for various RNA folds may constitute lead compounds for developing new and highly specific RNA drug targets. Despite the use of high through-put screening (HTS) methods (reviewed in Hermann and Westhof, 2000), adequate filters are necessary for choosing the RNA targets.
Materials and methods
Aminoacylation experiments
Yeast AspRS was purified as described elsewhere (Lorber et al., 1983). Aminoacylation tests were performed in 0.1 M HEPES–KOH pH 7.5, 30 mM KCl, 15 mM MgCl2, 5 mM adenosine triphosphate, 52 µl [l-3H]aspartic acid in the absence and presence of tobramycin at various concentrations in a 100 µl volume. These reactions were performed under steady-state conditions of enzyme (2.5 nM AspRS) and tRNA substrate concentrations (0.05–0.8 µM) and subsaturating concentrations of aspartic acid at 30°C as described (Perret et al., 1990; Pütz et al., 1991). Apparent Km and kcat values obtained at subsaturating concentrations of amino acid, were derived from a Lineweaver–Burk plot. kcat/Km values for replicate experiments varied at most by 15%.
Adenylate formation by AspRS
[32P]PPi–ATP exchange reactions (200 µl) were performed in 100 mM HEPES–KOH pH 7.4, 10 mM MgCl2, 2 mM ATP, 5 mM l-aspartate, 2 mM [32P]PPi (∼2 c.p.m./pmol) and 0.25 µg/ml AspRS. ATP synthesis was determined as described (Kern and Giegé, 1979). Values for replicate experiments varied at most by 5%.
Ultraviolet absorbance melting curves
Absorbance was monitored on a 941 Uvikon UV spectrophotometer equipped with an external waterbath at 258 nm and temperature was increased continuously at 0.5°C/min. Incubation conditions were 10 mM Na-cacodylate pH 6.5, 50 mM NaCl, 3 mM MgCl2 in the absence and presence of tobramycin (1 mM). The data are normalized for differences in tRNA concentration and given as α (fraction of molecules in a dissociated state) (Marky and Breslauer, 1987).
Fluorescent labelling of in vitro transcribed tRNAAsp molecules
tRNAAsp transcripts were prepared by in vitro transcription (Pütz et al., 1991) in the presence of [γ-32P]ATP. Transcription was started by the addition of α-sulfur-substituted guanosine monophosphate (GMP-αS) at 40°C. Additions of GMP-αS were repeated every 20 min for 2 h. Full-length tRNAAsp transcripts were purified using PAGE. tRNAAsp transcripts (20 pmol) were labelled with a 1 mM solution of the appropriate dye (erythrosin-5-iodoacetamide, Molecular Probes Europe) in 30% dimethylsulfoxide (DMSO), 50 µM dithiothreitol (DTT), 20 mM EDTA, 50 mM Tris buffer pH 8.3 at 40°C for 2 h in the dark. Labelled tRNAAsp transcripts (tRNAAsp-Ery) were purified using PAGE.
Fluorescence anisotropy measurements
5-carboxyfluorescein-labelled tobramycin was synthesized and verified as described elsewhere with some modifications (Wang et al., 1996). Tobramycin (50 mg, ∼107 µmol) was dissolved in 500 µl H2O and 500 µl DMSO. Five hundred microlitres of 5-carboxyfluorescein succimidyl ester DMSO solution (10 mg, ∼21 µmol) were added and stirred for 1 h at 5°C. The reaction was stopped by adding 10 ml of H2O. To purify the labelled product the solution was passed twice through a weakly cationic exchange column (Amberlite CG50; Sigma) equilibrated with H2O at pH 5.5, washed with 500 ml of H2O, 500 ml of 0.025 M ammonium hydroxide and eluted with 0.25 M ammonium hydroxide. The product (Tob–Fl) was further purified on a C18-reversed phase column (PR-ODS; Beckman) using a linear gradient of triethylamine acid (TEAAC), 0.1 M, pH 6.0 and acetonitrile from 5 to 100%. The product was lyophilized and verified by NMR and mass spectroscopy [FABMS: 826 (M+H); Wang et al., 1996]. The 6′amino group of tobramycin is preferentially acetylated, since it is the only unhindered primary amino group (Tangy et al., 1983; Wang et al., 1996). Steady-state fluorescence anisotropy experiments were performed using an Aminco SLM 8100 fluorescence spectrophotometer with two photomultiplier tubes (T set-up).
The anisotropy (r) is calculated according to Equation 1:
with
The fluorescence intensity I is measured using vertical excitation and emission polarizers, and vertical and horizontal emission polarizers. G is the correction factor and the subscripts v and h refer to fluorescence with vertical and horizontal polarizers, respectively, in the order excitation, emission. Anisotropy values were integrated over 10 s and an average of 10 measurements was used, or until a reproducible value was measured (error <0.3%). The anisotropy of fluorescein attached to tobramycin is 25.53 × 10–3 compared with 12 × 10–3 for free dye in solution. Binding studies were carried out by titrating a solution of Tob–Fl with aliquots of a tRNA stock solution. Binding of Tob–Fl leads to a decrease in the rotation of the fluorescein upon formation of the RNA–inhibitor complex measured as anisotropy. The dissociation constant was determined from anisotropy measurements using a 10 nM Tob–Fl solution in 20 mM HEPES–KOH pH 7.4, 1 mM MgCl2, 5 mM KCl at 20°C.
The dissociation constant (KD) is calculated by assuming a 1:1 complex by fitting the data according to Equation 2:
where A0, A and ΔA stand for the fluorescence anisotropy in the absence and presence of RNA and the difference in anisotropy between the anisotropy at 1 nM fluorescently labelled compound at an infinite concentration of RNA minus the anisotropy in the absence of RNA, respectively. [RNA]0 and [Inh]0 are the initial concentrations of RNA and inhibitor, respectively.
The binding stoichiometries of the ligand are calculated by fitting the data to a simple two-state model, where the change in anisotropy is induced by the binding of n inhibitor molecules with an apparent association constant KA. The proportion α of inhibitor molecules bound to the RNA will be given by Equation 3:
Acknowledgments
Acknowledgements
We would like to express our gratitude to F.Candau and G.Paddon-Jones, Institut Charles Sadron (CNRS-UPR 22, Strasbourg, France) for help in synthesis and purification of Tob–Fl; G.Keith for providing native yeast tRNAAsp, tRNAPhe and total tRNA; D.Kern, G.Eriani and H.Roy for providing help in the aminoacyl-adenylate reaction IBMC (CNRS-UPR 9002, Strasbourg, France); G.Duportail, Laboratoire de Pharmacologie et Physico-Chimie des Interactions Cellulaires et Moléculaires (CNRS-UMR 7034, Illkirch, France) for the use of the SLM fluorescence spectrophotometer; and Nükhet Cavusoglu from the Laboratoire de Spectrométrie de Masse Bio-organique (CNRS-UMR 7509, CNRS, Strasbourg, France) for the mass spectrometry experiments. We thank the Fondation pour la Recherche Médicale for a grant (FRM 20(00106/6-E) for the purchase of a UV spectrophotometer Uvikon 943 (Bio-Tek Instruments). F.W. is supported by a European Community Research Network Contract (FMRX-CT97-0154).
References
- Blanchard S.C., Fourmy,D., Eason,R.G. and Puglisi,J.D. (1998) rRNA chemical groups required for aminoglycoside binding. Biochemistry, 37, 7716–7724. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.C. and Moras,D. (1993) Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Frugier M., Söll,D., Giegé,R. and Florentz,C. (1994) Identity switches between tRNAs aminoacylated by class I glutaminyl- and class II aspartyl-tRNA synthetases. Biochemistry, 33, 9912–9921. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Keith,G., Ebel,J.P. and Dirheimer,G. (1971) Structure of aspartate-tRNA from brewer’s yeast. Nature New Biol., 230, 125–126. [DOI] [PubMed] [Google Scholar]
- Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T. and Westhof,E. (2000) Rational drug design and high-throughput techniques for RNA targets. Comb. Chem. High Throughput Screen., 3, 219–234. [DOI] [PubMed] [Google Scholar]
- Hori Y. et al. (2001) Synthetic inhibitors of the processing of pretransfer RNA by the ribonuclease P ribozyme: enzyme inhibitors which act by binding to substrate. Biochemistry, 40, 603–608. [DOI] [PubMed] [Google Scholar]
- Hyun Ryu D. and Rando,R.R. (2001) Aminoglycoside binding to human and bacterial A-site rRNA decoding region constructs. Bioorg. Med. Chem., 9, 2601–2608. [DOI] [PubMed] [Google Scholar]
- Jiang L., Suri,A.K., Fiala,R. and Patel,D.J. (1997) Saccharide–RNA recognition in an aminoglycoside antibiotic–RNA aptamer complex. Chem. Biol., 4, 35–50. [DOI] [PubMed] [Google Scholar]
- Jin E., Katritch,V., Olson,W.K., Kharatisvili,M., Abagyan,R. and Pilch,D.S. (2000) Aminoglycoside binding in the major groove of duplex RNA: the thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol., 298, 95–110. [DOI] [PubMed] [Google Scholar]
- Kern D. and Giegé,R. (1979) The 32PPi–ATP isotope-exchange reaction catalyzed by the yeast valyl-tRNA synthetase: order of substrate binding and effect of tRNA. FEBS Lett., 103, 274–281. [DOI] [PubMed] [Google Scholar]
- Kirillov S., Vitali,L.A., Goldstein,B.P., Monti,F., Semenkov,Y., Makhno,V., Ripa,S., Pon,C.L. and Gualerzi,C.O. (1997) Purpuromycin: an antibiotic inhibiting tRNA aminoacylation. RNA, 3, 905–913. [PMC free article] [PubMed] [Google Scholar]
- Kirk S.R. and Tor,Y. (1999) tRNA(Phe) binds aminoglycoside antibiotics. Bioorg. Med. Chem., 7, 1979–1991. [DOI] [PubMed] [Google Scholar]
- Loftfield R.B. (1973) Inhibitors of amino acid activation. In Hochster,R.M., Kates,K. and Quastel,J.H. (eds), Metabolic Inhibitors. Academic Press, New York, pp. 107–130.
- Lorber B., Kern,D., Dietrich,A., Gangloff,J., Ebel,J.P. and Giegé,R. (1983) Large scale purification and structural properties of yeast aspartyl-tRNA synthetase. Biochem. Biophys. Res. Commun., 117, 259–267. [DOI] [PubMed] [Google Scholar]
- Marky L.A. and Breslauer,K.J. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- Mei H.-Y., Galan,A.A., Halim,N.S., Mack,D.P., Moreland,D.W., Sanders,K.B., Truong,H.N. and Czarnik,A.W. (1995) Inhibition of an HIV-1 Tat-derived peptide binding to TAR RNA by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett., 5, 2755–2760. [Google Scholar]
- Mikkelsen N.E., Johansson,K., Virtanen,A. and Kirsebom,L.A. (2001) Aminoglycoside binding displaces a divalent metal ion in a tRNA–neomycin B complex. Nature Struct. Biol., 8, 510–514. [DOI] [PubMed] [Google Scholar]
- Moulinier L., Eiler,S., Eriani,G., Gangloff,J., Thierry,J.C., Gabriel,K., McClain,W.H. and Moras,D. (2001) The structure of an AspRS–tRNA(Asp) complex reveals a tRNA-dependent control mechanism. EMBO J., 20, 5290–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen,D.E., Clemons,W.M.,Jr, Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia,A., Grosjean,H., Ebel,J.P., Florentz,C. and Giegé,R. (1990) Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature, 344, 787–789. [DOI] [PubMed] [Google Scholar]
- Puglisi J.D., Pütz,J., Florentz,C. and Giegé,R. (1993) Influence of tRNA tertiary structure and stability on aminoacylation by yeast aspartyl-tRNA synthetase. Nucleic Acids Res., 21, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Puglisi,J.D., Florentz,C. and Giegé,R. (1991) Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science, 252, 1696–1699. [DOI] [PubMed] [Google Scholar]
- Robinson H. and Wang,A.H. (1996) Neomycin, spermine and hexaaminecobalt (III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res., 24, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P., Moras,D., Bergdoll,M., Dumas,P., Vlassov,V.V., Westhof,E., Ebel,J.P. and Giegé,R. (1985) Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J. Mol. Biol., 184, 455–471. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Kranz,J.E., Gallant,P., Schimmel,P. and Shiba,K. (1996) A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug. Biochemistry, 35, 9995–10003. [DOI] [PubMed] [Google Scholar]
- Schimmel P., Tao,J. and Hill,J. (1998) Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J., 12, 1599–1609. [PubMed] [Google Scholar]
- Schroeder R., Waldsich,C. and Wank,H. (2000) Modulation of RNA function by aminoglycoside antibiotics. EMBO J., 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangy F., Capmau,M.L. and Le Goffic,F. (1983) Photo-induced labelling of Escherichia coli ribosomes by a tobramycin analog. Eur. J. Biochem., 131, 581–587. [DOI] [PubMed] [Google Scholar]
- Tok J.B., Cho,J. and Rando,R.R. (1999) Aminoglycoside antibiotics are able to specifically bind the 5′-untranslated region of thymidylate synthase messenger RNA. Biochemistry, 38, 199–206. [DOI] [PubMed] [Google Scholar]
- Tor Y., Hermann,T. and Westhof,E. (1998) Deciphering RNA recognition: aminoglycoside binding to the hammerhead ribozyme. Chem. Biol., 5, R277–R283. [DOI] [PubMed] [Google Scholar]
- Vasquez D. (1974) Inhibitors of protein synthesis. FEBS Lett., 40, S63–S84. [DOI] [PubMed] [Google Scholar]
- Vicens Q. and Westhof,E. (2001) Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure, 9, 647–658. [DOI] [PubMed] [Google Scholar]
- Walter F., Vicens,Q. and Westhof,E. (1999) Aminoglycoside–RNA interactions. Curr. Opin. Chem. Biol., 3, 694–704. [DOI] [PubMed] [Google Scholar]
- Wang Y., Killian,J., Hamasaki,K. and Rando,R.R. (1996) RNA molecules that specifically and stoichiometrically bind aminoglycoside antibiotics with high affinities. Biochemistry, 35, 12338–12346. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas,P. and Moras,D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol., 184, 119–145. [DOI] [PubMed] [Google Scholar]
- Zapp M.L., Stern,S. and Green,M.R. (1993) Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell, 74, 969–978. [DOI] [PubMed] [Google Scholar]