Abstract
Aberrations in centrosome numbers have long been implicated in aneuploidy and tumorigenesis, but their origins are unknown. Here we have examined how overexpression of Aurora-A kinase causes centrosome amplification in cultured cells. We show that excess Aurora-A does not deregulate centrosome duplication but gives rise to extra centrosomes through defects in cell division and consequent tetraploidization. Over expression of other mitotic kinases (Polo-like kinase 1 and Aurora-B) also causes multinucleation and concomitant increases in centrosome numbers. Absence of a p53 checkpoint exacerbates this phenotype, providing a plausible explanation for the centrosome amplification typical of p53–/– cells. We propose that errors during cell division, combined with the inability to detect the resulting hyperploidy, constitute a major cause for numerical centrosome aberrations in tumors.
Keywords: cancer/cell cycle/chromosomal instability/centrosome anomalies
Introduction
The centrosome is the major microtubule organizing centre (MTOC) of animal cells. It is involved in all microtubule-dependent processes including the formation of the bipolar spindle during mitosis (for review see Kellogg et al., 1994). Although bipolar spindles can form in the absence of centrosomes (Heald et al., 1996; Bobinnec et al., 1998; Fry et al., 1998b; Khodjakov et al., 2000; Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001), these organelles play a dominant role in determining the number of spindle poles in the vast majority of animal cell divisions (Heald et al., 1997). Thus, in order to maintain chromosome stability, it is important that centrosomes duplicate and segregate in synchrony with the nuclear genome (for review see Hinchcliffe and Sluder, 2001; Stearns, 2001).
Aberrations in the centrosome duplication cycle result in the formation of monopolar or multipolar spindles, and it has been long proposed that such aberrations may cause aneuploidy and contribute to cancer development (Boveri, 1914; for recent review see Pihan and Doxsey, 1999; Lingle and Salisbury, 2000; Brinkley, 2001). Interest in this hypothesis has recently been revived by the demonstration that many types of tumors feature a high proportion of cells with centrosomal abnormalities, including excessive numbers of centrosomes and centrosomes with aberrant structures or phosphorylation states (Lingle et al., 1998; Pihan et al., 1998; Lingle and Salisbury, 1999; Sato et al., 1999). Centrosome anomalies have been reported to arise at early stages of tumor formation and to expand concomitant with tumor progression (Pihan et al., 2001; Shono et al., 2001). Furthermore, multipolar spindles can frequently be seen in tumors, and a detailed analysis of colorectal cancer cell lines revealed a strong correlation between centrosome amplification and aneuploidy (Ghadimi et al., 2000). These findings are consistent with the idea that centrosomal anomalies contribute to aneuploidy and cancer development, but in the absence of mechanistic insights into their origins, it remains difficult to decide whether centrosome anomalies constitute a cause or consequence of aneuploidy.
Recent studies have begun to identify proteins involved in the regulation of centrosome duplication. In particular, cyclin-dependent kinase 2 (Cdk2), in association with either cyclin A and/or cyclin E, is required for centrosome duplication, and this most likely constitutes an important aspect of the coordination between centrosome duplication and DNA replication (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999). Furthermore, the murine protein kinase mMps1, as well as the putative chaperone nucleophosmin/B23, have been proposed to act downstream of Cdk2 in this process (Okuda et al., 2000; Fisk and Winey, 2001). Yet another kinase, Zyg-1, was convincingly shown to be required for centrosome duplication in Caenorhabditis elegans (O’Connell et al., 2001).
Centrosome amplification in tumors and cell lines has been linked to numerous genetic aberrations, including the loss of the tumor suppressor protein p53 and the deregulated expression of its regulators (Mdm2) and downstream targets (p21Cip1, Gadd45) (Fukasawa et al., 1996; Carroll et al., 1999; Hollander et al., 1999; Mantel et al., 1999). Other genetic alterations reported to affect centrosome numbers concern proteins involved in the response to DNA damage, including BRCA1, BRCA2 and ATR (Smith et al., 1998; Tutt et al., 1999; Xu et al., 1999) and in ubiquitin-dependent protein degradation, including Skp2 and TSG101 (Xie et al., 1998; Nakayama et al., 2000). However, it is unknown how deregulation of any of these gene products gives rise to extra copies of centrosomes.
Considering the implication of several protein kinases in the centrosome cycle, it is particularly intriguing that centrosome amplification has also been observed in response to overexpression of the Aurora-A kinase (Zhou et al., 1998; for aurora nomenclature, see Nigg, 2001). The precise function of Aurora-A is not known but roles related to centrosome separation, spindle assembly and spindle maintenance have been proposed (for review see Bischoff and Plowman, 1999; Giet and Prigent, 1999; Goepfert and Brinkley, 2000; Nigg, 2001). Interestingly, Aurora-A is considered as a candidate oncogene. It is overexpressed in many tumors and maps to chromosome 20q13, a region frequently amplified in cancers (Sen et al., 1997; Bischoff et al., 1998). Furthermore, overexpression of Aurora-A in NIH 3T3 cells confers a transformed phenotype that is characterized by centrosome amplification and aneuploidy, and Aurora-A transformed cells induce tumors in nude mice (Bischoff et al., 1998; Zhou et al., 1998).
Since overexpression of Aurora-A has been implicated in human pathology, we considered it an attractive model for studying the molecular mechanism(s) underlying centrosome amplification. Specifically, we have asked by what mechanisms overexpression of Aurora-A causes the appearance of extra centrosomes in cultured cells. Our findings lead us to conclude that Aurora-A does not deregulate centrosome duplication during S phase. Instead, we found that multiple centrosomes are almost invariably seen in multinucleated, tetraploid cells, indicating that extra centrosomes arise as a result of aberrations occurring during mitosis and/or cytokinesis. A virtually indistinguishable centrosome amplification phenotype could be observed upon overexpression of other kinases known to regulate cell division, notably Polo-like kinase 1 (Plk1) and Aurora-B, and upon drug-induced inhibition of cytokinesis. We also show that the accumulation of extra copies of centrosomes is strikingly enhanced in p53–/– cells, consistent with the emerging notion that p53 arrests tetraploid cells as part of a G1 checkpoint (Minn et al., 1996; Khan and Wahl, 1998; Lanni and Jacks, 1998; Casenghi et al., 1999; Andreassen et al., 2001). These findings identify errors occurring during cell division as a major source of numerical centrosome aberrations, and they help to explain why cells accumulate extra copies of centrosomes in the absence of a functional p53 pathway. They also have implications for the origin and significance of centrosome anomalies during tumor development.
Results
Aurora-A does not regulate centrosome duplication
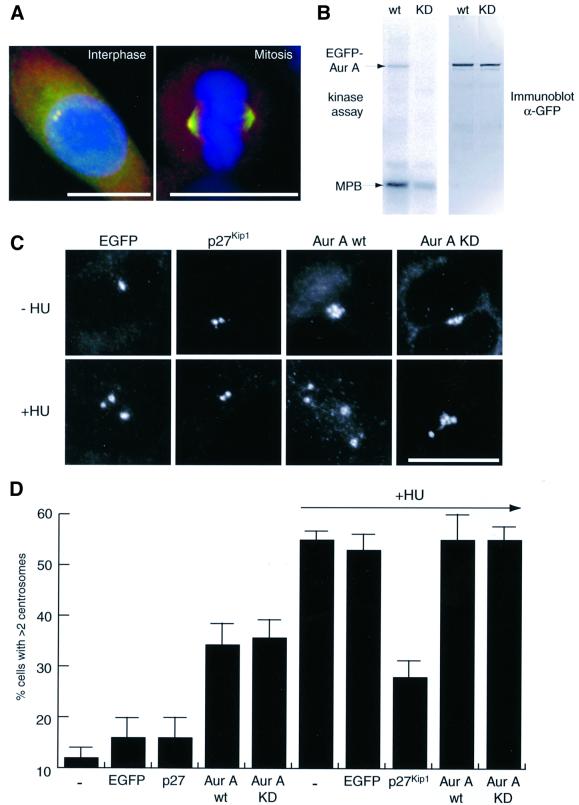
To determine whether Aurora-A plays a direct role in centrosome duplication, GFP-tagged wild-type (wt) and catalytically-inactive (KD) mutant Aurora-A kinases were constructed. Upon transient transfection, both kinases localized to the centrosome during interphase and to the spindle during mitosis (Figure 1A and data not shown), in agreement with the localization of the endogenous protein (Kimura et al., 1997; Bischoff et al., 1998). When immunoprecipitated from transfected cells, the wt Aurora-A phosphorylated both itself and the exogenous substrate myelin basic protein (MBP), whereas the KD mutant was virtually inactive, as expected (Figure 1B). These Aurora-A constructs were then tested for their ability to enhance or inhibit centrosome duplication in a previously developed assay (Meraldi et al., 1999). This assay is based on the observation that hydroxyurea (HU) treatment of Chinese hamster ovary (CHO) cells blocks DNA replication but allows multiple rounds of centrosome duplication to occur, so that by 40 h of treatment >50% of cells contain more than two centrosomes (Balczon et al., 1995; Matsumoto et al., 1999; Meraldi et al., 1999). As shown by immunofluorescent staining with anti-γ-tubulin antibodies (Figure 1C and D), transfection of cells with GFP alone did not significantly affect centrosome numbers in either control or HU-treated cells, whereas overexpression of the Cdk2-inhibitor p27Kip1 strongly inhibited centrosome reduplication in HU-treated cells, in line with our earlier results (Meraldi et al., 1999). Overexpression of wt Aurora-A induced the formation of extra copies of centrosomes even in the absence of HU, confirming previous findings (Zhou et al., 1998). However, Aurora-A did not increase the percentage of cells with extra copies of centrosomes (nor centrosome numbers) in HU-treated cells (Figure 1C and D). Unexpectedly, overexpression of catalytically inactive Aurora-A produced virtually identical results. This KD mutant also caused apparent centrosome amplification in untreated cells, and it did not exert a dominant-negative effect on centrosome reduplication in HU-treated cells (Figure 1C and D). These results are difficult to reconcile with any model invoking a requirement for Aurora-A kinase activity in centrosome duplication.
Fig. 1. Centrosome amplification by Aurora-A does not require kinase activity. (A) Localization of EGFP–Aurora-A mimics distribution of endogenous protein. HeLa cells were transfected for 24 h with EGFP–Aurora-A wt (green) and stained with anti-γ-tubulin antibodies for centrosomes (red) and DAPI for DNA (blue). (B) EGFP–Aurora-A is an active kinase. Wt and KD mutant EGFP–Aurora-A were immunoprecipitated from transfected U2OS cells, using an anti-GFP antibody. In vitro kinase assays were performed in the presence of [γ-32P]ATP and myelin basic protein (MBP) as an exogenous substrate (left hand panel), and equal recovery was confirmed by immunoblotting with anti-GFP antibodies (right hand panel). (C and D) Aurora-A activity is not required for centrosome amplification. CHO cells were transfected for 40 h with the indicated constructs and cultured in the presence or absence of hydroxyurea (HU). (C) Transfected cells were detected by GFP fluorescence and centrosomes visualized with anti-γ-tubulin antibodies. Cells were counted as having normal numbers of centrosomes (one or two visible γ-tubulin dots) or excessive numbers of centrosomes (>2 γ-tubulin dots). (D) Histogram shows results from three independent experiments (400–600 cells each) and bars indicate standard deviations. Scale bars: 10 µm.
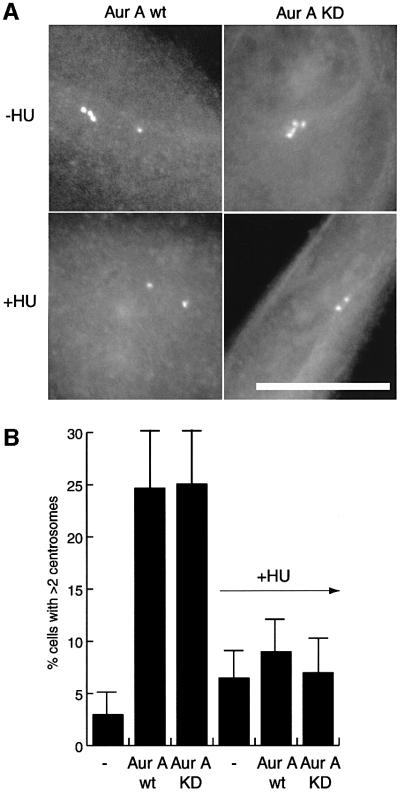
We considered the possibility that overexpression of Aurora-A protein might cause centrosome reduplication in CHO cells by imposing an S phase arrest, thereby de facto mimicking the effects of HU treatment. We thus turned to HeLa cells, which do not reduplicate centrosomes under HU arrest, indicating that they are not proficient for centrosome reduplication under S phase arrest conditions (Figure 2B). Overexpression during 48 h of both wt and catalytically inactive Aurora-A still caused the appearance of extra centrosomes in a substantial fraction of HeLa cells, similar to the results obtained in CHO cells (Figure 2). Most revealingly, however, this increase in centrosome numbers was completely suppressed by addition of HU (Figure 2), demonstrating that Aurora-A could not induce centrosome amplification during S phase arrest. Instead, these results suggested that the generation of extra centrosomes by Aurora-A overexpression required passage of cells through mitosis.
Fig. 2. Aurora-A does not cause centrosome amplification in S phase. HeLa cells were transfected for 48 h with wt or KD mutant EGFP–Aurora-A and cultured in the presence or absence of hydroxyurea (HU). Transfected cells were identified by fluorescence microscopy (A) and the number of centrosomes quantified using the GFP fluorescence of Aurora-A (or anti-C-Nap1 staining; not shown) (B). Histogram shows results from three independent experiments (400–600 cells each) and bars indicate standard deviations. Scale bar: 10 µm.
Aurora-A overexpression causes multinucleation concomitant with centrosome amplification
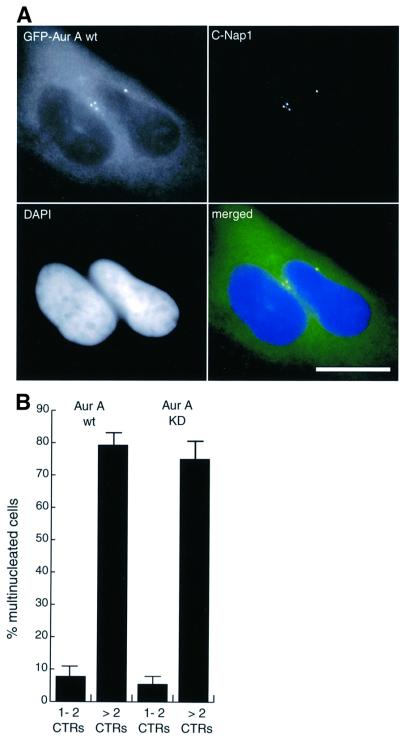
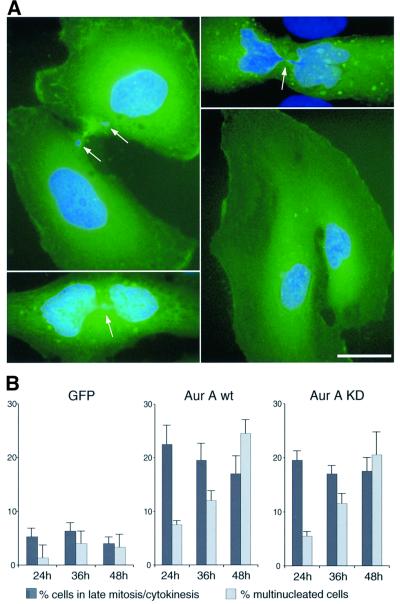
Upon close inspection of cells overexpressing Aurora-A we discovered that most cells harboring increased numbers of centrosome were multinucleated, suggesting that extra centrosomes might have arisen as a consequence of aborted cell divisions (Figure 3A). A detailed quantitative analysis of cells transfected with both wt and catalytically inactive Aurora-A revealed that 75% of cells with multiple centrosomes were indeed multinucleated (Figure 3B). Conversely, <10% of the transfected cells with normal number of centrosomes were multinucleated (Figure 3B). This strong correlation suggested that extra centrosomes arose as a consequence of defects in mitotic progression and cell division, giving rise to tetraploidization. To corroborate this interpretation, the phenotype of cells overexpressing Aurora-A was analyzed in more detail. As shown in Figure 4A, many of the dividing cells overexpressing Aurora-A showed highly aberrant structures, including broad cytoplasmic connections, lagging chromosomes and DNA strands between dividing nuclei. Consistent with a delay in mitotic exit, ∼20% of cells overexpressing Aurora-A were in late mitotic stages or cytokinesis already at 24 h after transfection, whereas only 5% of cells expressing GFP were at comparable stages (Figure 4B). Subsequently, the proportion of multinucleated cells increased progressively, indicating that they arose through cytokinesis failure (Figure 4B).
Fig. 3. Aurora-A overexpression causes multinucleation concomitant with centrosome amplification. (A) HeLa cells were transfected for 48 h with wt or KD mutant EGFP–Aurora-A and analyzed by immunofluorescence microscopy. Transfected cells were identified by GFP-fluorescence (green), centrosomes were stained with anti-C-Nap1 antibodies (red) and DNA with DAPI (blue). Scale bar: 10 µm. (B) Transfected cells were classified according to whether they had normal numbers of centrosomes (one or two fluorescent dots) or more than two centrosomes, and whether they were mononucleated or multinucleated. Histogram shows results from three independent experiments (400–600 cells each) and bars indicate standard deviations.
Fig. 4. Cytokinesis failure in cells overexpressing Aurora-A. (A) HeLa cells were transfected with wt or KD mutant EGFP–Aurora A and analyzed by immunofluorescence microscopy. Transfected cells were identified by GFP fluorescence (green) and DNA was stained with DAPI (blue). Arrows indicate lagging chromosomes and inter connecting DNA bridges. Scale bar: 10 µm. (B) Histogram illustrating the effects of Aurora-A overexpression on the proportion of cells in late mitosis/cytokinesis (dark gray bars) and the frequency of multinucleated cells (light gray bars). Transfected cells were classified at the time points indicated. Results were averaged from three independent experiments (300–600 cells each) and bars indicate standard deviations.
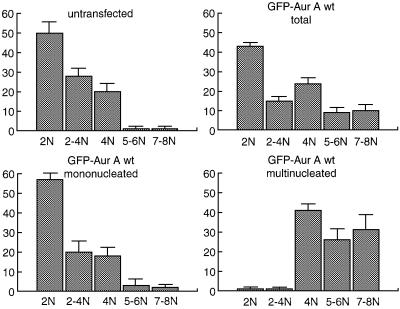
To directly demonstrate that multinucleated cells with extra copies of centrosomes were tetraploid, we quantified their DNA content 48 h after transfection. Compared with the untransfected controls, cells overexpressing Aurora-A showed a substantial increase in the G2 (4N) and >4N (5–8N) populations (Figure 5, upper panels). Most interestingly, a differential analysis of mono- and multinucleated cells showed that this increase in hyperploidy could be attributed almost exclusively to the multinucleated (mostly binucleated) population (Figure 5, lower panels). This demonstrates that the binucleated cells had undergone a duplication of their DNA content and that they continued in the cell cycle. Virtually identical results were obtained upon transfection of the catalytically inactive Aurora-A mutant (data not shown). Taken together, these results demonstrate that mitotic defects triggered by overexpression of wt or catalytically inactive Aurora-A resulted in aberrant cytokinesis and the formation of tetraploid cells. The precise nature of the primary mitotic defect(s) is not known, but the salient proposition emerging from the above results is that any cellular defect leading to abortive cytokinesis and tetraploidization may give rise to extra copies of centrosomes.
Fig. 5. Tetraploidization in Aurora-A overexpressing cells. HeLa cells were transfected with wt EGFP–Aurora-A, stained with propidium iodide and their DNA content measured by quantitative immuno fluorescence microscopy. Cells were classified as having a 2N (G1), 2–4N (S), 4N (G2), 5 or 6N, or 7 or 8N DNA content. Histograms show results from three independent experiments (300–600 cells each) and bars indicate standard deviations.
Polyploidization may commonly accompany centrosome amplification
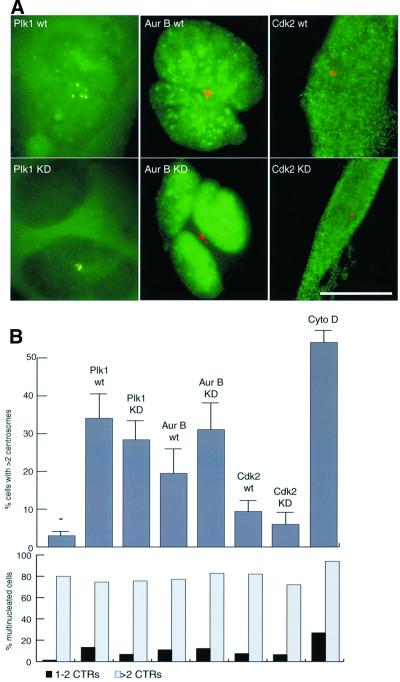
As a first test of the above proposition, we asked whether overexpression of other mitotic kinases, described to induce multinucleation but not previously reported to cause centrosome amplification, would also lead to the appearance of extra copies of centrosomes. HeLa cells were transfected with either Plk1 or Aurora-B, both of which were previously shown to induce multinucleation upon overexpression (Mundt et al., 1997; Terada et al., 1998). Cdk2, which is required for centrosome duplication during S phase (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999) but has never been described to cause multinucleation, was expressed for comparison. As shown in Figure 6, overexpression of both Plk1 and Aurora-B generated extra copies of centrosomes, and as shown above for Aurora-A, this phenotype did not require kinase activity. In contrast, overexpression of Cdk2 did not cause a significant increase in centrosome numbers (Figure 6). Most importantly, the vast majority of all cells harboring extra centrosomes were also multinucleated. This was true for cells overexpressing Plk1 or Aurora-B, as well as for the few cells with extra centrosomes amongst Cdk2-transfected and untransfected populations (Figure 6B). Furthermore, consistent with the finding that catalytically inactive Aurora-B is particularly effective in causing multinucleation (Terada et al., 1998), the KD mutant produced more extensive centrosome amplification than wt Aurora-B (Figure 6B). Taken together, these results strengthened the conclusion that cytokinesis failure represents a widespread cause for the appearance of extra copies of centrosomes. This view was corroborated further by treatment of cells with a cytokinesis-inhibitory drug. Exposure of cells to cytochalasin D for 30 h very effectively reproduced an apparent centrosome amplification phenotype indistinguishable from that seen in response to kinase overexpression (Figure 6B).
Fig. 6. Multinucleation, induced by Plk1, Aurora-B or cytochalasin D, correlates with centrosome amplification. HeLa cells were transfected for 48 h with the indicated constructs or treated for 30 h with cytochalasin D (0.6 µg/ml). (A) Transfected cells were identified by GFP-fluorescence or staining with anti-HA antibodies (green), and centrosome were visualized with anti-C-Nap1 antibodies (red). Scale bar: 10 µm. (B) Cells were classified according to whether they had normal numbers of centrosomes (one or two fluorescent dots) or more than two centrosomes, and whether they were mononucleated or multinucleated. Histogram shows results from three independent experiments (400–600 cells each), and bars indicate standard deviations.
The absence of p53 favors the accumulation of cells with extra centrosomes
One of the most prominent proteins implicated in the generation of extra copies of centrosomes is the tumor suppressor p53 (Fukasawa et al., 1996). Although p53 function has been extensively studied in the context of checkpoint responses to DNA damage (for review see Levine, 1997; Vogelstein et al., 2000), the mechanism(s) underlying centrosome amplification in p53–/– cells has not previously been understood. In view of the results described above, we reasoned that the accumulation of extra centrosomes in p53–/– cells might relate to the occasional failure of such cells to undergo proper cell division, and a need for p53 to arrest (or eliminate) the resulting tetraploid cells (Minn et al., 1996; Khan and Wahl, 1998; Lanni and Jacks, 1998; Casenghi et al., 1999; Andreassen et al., 2001). To examine this hypothesis, we first asked how frequently individual p53–/– cells displayed both centrosome amplification and multinucleation. Examination of p53–/– mouse embryo fibroblasts (MEFs) revealed a 66% correlation between the two phenotypes (data not shown), consistent with the above model. Furthermore, many of the mononucleated cells displayed enormous nuclei, strongly suggesting that they were polyploid (data not shown). We also examined the ability of multinucleated cells bearing extra copies of centrosomes to proceed through the cell cycle, using bromodeoxyuridine incorporation as a measure of S phase entry. Multinucleated wt MEFs were severely compromised in their ability to proceed, while p53–/– MEFs readily re-entered S phase regardless of their ploidy and centrosome content (data not shown, but see Khan and Wahl, 1998; Lanni and Jacks, 1998; Casenghi et al., 1999; Andreassen et al., 2001).
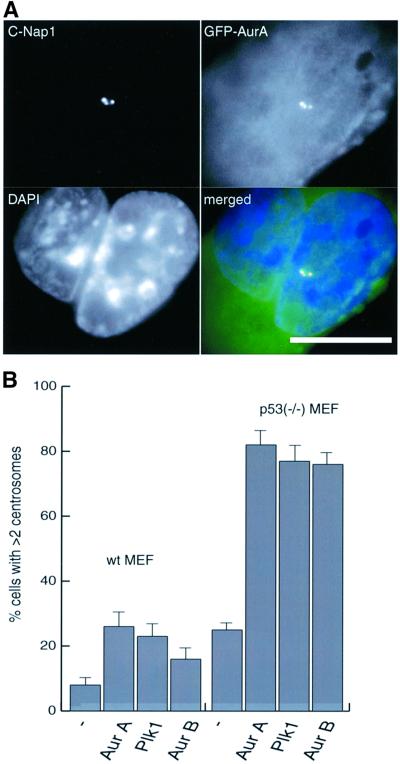
As a more rigorous test of our hypothesis, we next asked whether the centrosomal phenotypes induced by kinase overexpression would be exacerbated in a p53–/– background. Thus, p53–/– MEFs and wt MEFs were transfected for 48 h with Aurora-A, Aurora-B or Plk1. As shown in Figure 7, up to 80% of the transfected p53–/– MEFs had more than two centrosomes, whereas such a phenotype was seen in only 25% of the transfected wt MEFs (Figure 7B). In all cases a strong correlation existed between the presence of extra centrosomes and multiple nuclei (Figure 7A, and data not shown). Taken together, these results demonstrate that the centrosome amplification phenotype seen in cells lacking a functional p53 checkpoint arises through a combination of defective mitosis/cytokinesis and the inability to prevent the resulting polyploid cells from further cell cycle progression.
Fig. 7. The absence of p53 favors the generation of extra copies of centrosomes. Wt MEFs or p53–/– MEFs were transfected for 48 h with wt EGFP–Aurora-A, EGFP–Plk1 or EGFP–Aurora-B and processed for immunofluorescence microscopy. (A) Transfected cells were identified by GFP fluorescence (green), centrosomes were stained with anti-C-Nap1 antibodies (red) and DNA with DAPI (blue). Scale bar: 10 µm. (B) Centrosome numbers were counted as described in the legend to Figure 1. Histogram shows results from three independent experiments (400–600 cells each), and bars indicate standard deviations.
Discussion
The present study was aimed at elucidating the origin of the centrosome amplification phenotype that is frequently observed in a variety of genetically altered cells and tumors. As a tractable model system, we have studied the consequences of overexpressing the Aurora-A kinase and other mitotic kinases in tissue culture cells of both p53+/+ and p53–/– background. Our results lead us to conclude that abortive cell division and concomitant tetraploidization constitute the prevalent mechanism leading to centrosome amplification. In a p53–/– background, this cellular phenotype is viable, potentially setting the stage for chromosomal instability and the propagation of aneuploid cells.
How does overexpression of Aurora-A give rise to extra copies of centrosomes?
The fact that overexpression of Aurora-A leads to extra copies of centrosomes (Zhou et al., 1998) has generally been interpreted to reflect excessive centrosome duplication. However, our present results indicate that Aurora-A is not involved in centrosome duplication. In fact, arresting HeLa cells during S phase, which is the time when centrosomes duplicate, prevented Aurora-A from causing an increase in centrosome numbers. Instead, we discovered that Aurora-A expressing cells bearing extra copies of centrosomes were almost invariably multinucleated, identifying a failure during cell division and concomitant tetraploidization as a primary cause for numerical centrosome aberrations. That cells overexpressing Aurora-A experienced problems with cell division could be deduced from their accumulation in cytokinesis and the high frequency of interconnecting DNA bridges and other aberrations. The exact function of Aurora-A remains to be uncovered, but a role in mitotic progression is consistent with its spindle-association and the peak of its expression during M phase (Giet and Prigent, 1999).
With regard to the purported role of Aurora-A in tumorigenesis, it seems unlikely that the centrosome phenotype alone could explain the ability of this kinase to transform fibroblasts. The ability of Aurora-A to transform NIH 3T3 cells was reported to require kinase activity (Bischoff et al., 1998), whereas extra copies of centrosomes arise in response to overexpression of both wt and catalytically inactive Aurora-A (this study). Taken at face value, this suggests that additional functions of Aurora-A are required for transformation.
Is transient tetraploidization a major cause for numerical centrosome aberrations in tumor tissues?
The presence of multiple centrosomes in p53–/– cells has been proposed to reflect multiple rounds of centrosome duplication within a single cell cycle (Fukasawa et al., 1996). Similarly, numerical centrosome aberrations in tumors and cell lines are commonly referred to as being due to disturbed centrosome replication, centrosome hyperamplification or centrosome overduplication (Carroll et al., 1999; Mantel et al., 1999; Nakayama et al., 2000; Sato et al., 2000). Contrary to the perception implicit in this terminology, we propose that many, and perhaps most, numerical centrosome anomalies arise through failures in cell division and subsequent propagation of polyploid cells.
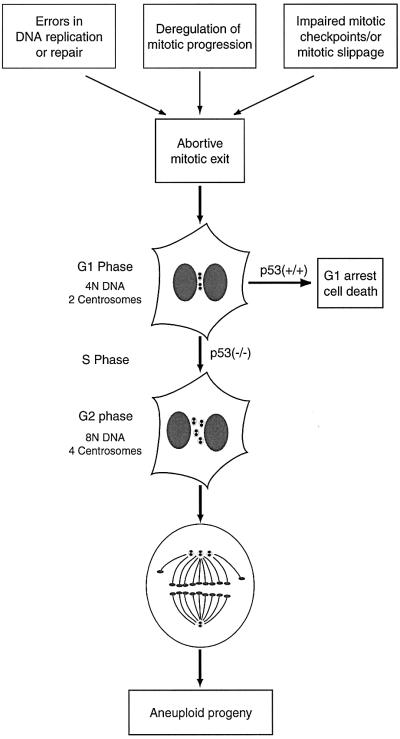
Apparent centrosome amplification has been attributed to deregulation of proteins implicated in the p53 pathway [p53, p21Cip1, Mdm2, Gadd45 (Fukasawa et al., 1996; Carroll et al., 1999; Hollander et al., 1999; Mantel et al., 1999)], DNA repair [ATR, BRCA1, BRCA2 (Smith et al., 1998; Tutt et al., 1999; Xu et al., 1999)], protein degradation [Skp2, TSG101 (Xie et al., 1998; Nakayama et al., 2000)] and mitosis [Aurora-A, survivin (Zhou et al., 1998; Li et al., 1999)]. This raises the question of whether the absence or malfunction of these different gene products produces centrosome anomalies by distinct mechanisms, or, alternatively, whether distinct primary defects converge onto a common secondary defect, which then gives rise to an increase in centrosome numbers. Our results lead us to favor the latter model. A common mechanism, centered on mitotic failure leading to tetraploidization, would readily explain why multiple genes, although acting in pathways ostensibly unrelated to centrosome duplication, all lead to a similar centrosome amplification phenotype (Figure 8). In support of this model, we have shown that overexpression not only of Aurora-A, but also of Plk1 and Aurora-B, two mitotic kinases previously known to induce multinucleation (Mundt et al., 1997; Terada et al., 1998) but never described to induce centrosome amplification, also generate extra copies of centrosomes. Emphasizing the generality of the postulated mechanism, straight inhibition of cytokinesis by cytochalasin D, a drug interfering with contractile ring assembly, produced a centrosome amplification phenotype that was virtually indistinguishable from that seen upon overexpression of mitotic kinases.
Fig. 8. Defects in cell division as a major route to numerical centrosome aberration and aneuploidy. According to this model, tetraploidization (rather than enhanced centrosome duplication) is a prevalent cause for numerical centrosome aberrations. Specifically, we propose that a number of distinct primary cell cycle errors lead to abortive mitotic exit, thereby generating tetraploid cells with 4N DNA and two centrosomes. In a p53+/+ background, such cells will be arrested in G1 and/or eliminated. This occurs most likely as a consequence of DNA damage checkpoint activation, although a mechanism monitoring centrosome status cannot presently be excluded. In the absence of a functional p53 checkpoint, however, such cells will progress to an extra round of DNA replication and centrosome duplication, giving rise to polyploid progeny with four centrosomes. In subsequent mitoses, these cells will frequently form multipolar spindles, thus causing chromosomal instability and aneuploidy.
Our findings also provide a plausible explanation for why p53–/– cells display increased centrosome numbers. We show that p53–/– MEFs bearing extra copies of centrosomes were very often multinucleated, suggesting that the centrosome anomalies seen in these cells also arose through a mechanism involving aborted mitoses. Furthermore, the absence of p53 strongly enhanced the phenotype produced by overexpression of mitotic kinases. These observations fall in line with the demonstration that p53 function is required to impose a G1 arrest in response to tetraploidization (Minn et al., 1996; Khan and Wahl, 1998; Lanni and Jacks, 1998; Casenghi et al., 1999; Andreassen et al., 2001). A survey of the literature further shows that polyploid cells have also been associated with mutations in BRCA1, BRCA2, ATR, Skp2 or TSG101 (Smith et al., 1998; Xie et al., 1998; Tutt et al., 1999; Xu et al., 1999; Nakayama et al., 2000), again suggesting that a defect in mitotic exit may have caused the appearance of extra centrosomes in most, if not all, of the corresponding cells and tissues. Finally, we emphasize that most cultured cell lines contain subpopulations of cells with multiple centrosomes, and when examined carefully, these subpopulations very often show a striking multinucleation/polyploidy phenotype (this study, and data not shown). We thus believe that extra copies of centrosomes frequently arise through failure of cell division and concomitant tetraploidization. To what extent deregulation of centrosome duplication per se constitutes an alternative mechanism for centrosome amplification remains to be determined.
Implications for the role of centrosomes in aneuploidy and tumorigenesis
Multipolar spindles were observed in tumors more than a hundred years ago, giving rise to the hypothesis that centrosome anomalies foster chromosome mis-segregation, aneuploidy and tumor progression (Boveri, 1914; Pihan and Doxsey, 1999; Lingle and Salisbury, 2000; Brinkley, 2001). Consistent with this hypothesis, but by no means proving it, a strong correlation has been established between centrosome anomalies and chromosomal instability in tumor cells (Lingle et al., 1998; Pihan et al., 1998; Carroll et al., 1999). Of particular relevance to our present findings, aneuploidy in solid tumors is frequently preceded by a transient state of tetraploidy (Shackney et al., 1989; Levine et al., 1991; Li et al., 1999). A priori, one could argue that centrosome anomalies prompt errors during cytokinesis, thereby generating a transient tetraploid state. To us it seems more likely, though, that a tetraploid state and extra copies of centrosomes arise concomitantly, as a consequence of defects in mitotic progression and aborted cytokinesis. According to this interpretation, centrosome anomalies are a by-product, rather than the cause, of tetraploidization. This implies that centrosome numbers may constitute convenient surrogate markers for polyploidy in tumor tissues. Furthermore, centrosome anomalies are expected to favor chromosome mis-segregation during subsequent cell divisions, and this may explain why the tetraploid state in tumor tissues is not stable but generally progresses to an aneuploid state (Figure 8). Thus, although deregulation of centrosome duplication may not constitute a major primary cause for centrosome amplification in tumors, the hypothesis that centrosome anomalies contribute to tumor development remains attractive and worthy of further investigation.
Materials and methods
Preparation of expression plasmids
To generate an EGFP–Aurora-A expression plasmid, the Aurora-A cDNA was amplified by PCR from pBluescript II SK-Aurora-A (Meraldi and Nigg, 2001), thereby introducing the unique restriction sites XhoI and BamHI at the 5′ and 3′ ends, respectively. Similarly, the Aurora-B cDNA was amplified by PCR from an EST (Image; DDBJ/EMBL/GenBank accession No. H57656), introducing the unique restriction sites EcoRI and BamHI at each end. The PCR products were checked by sequencing (Bischoff et al., 1998; Shindo et al., 1998) and inserted into the EGFP-C1 vector (Clonetech). A catalytically inactive EGFP–Aurora-A was generated by replacing a HindIII–EcoRI fragment with a corresponding fragment from pBluescript II SK-Aurora-A K162R (Meraldi and Nigg, 2001), and catalytically inactive Aurora-B was generated by site-directed mutagenesis, altering lysine 106 to arginine.
Cell culture
HeLa, U2OS, wt MEF and p53–/– MEF cells were grown at 37°C in a 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% heat-inactivated fetal calf serum (FCS), penicillin and streptomycin (100 IU/ml and 100 µg/ml, respectively) (all Gibco-BRL). CHO cells were grown under the same conditions except that DMEM was replaced by Ham’s F-12 medium. To block cytokinesis, HeLa cells were treated with cytochalasin D (0.6 µg/ml) for 30 h.
Transfection experiments and analysis of centrosome numbers
For transfection experiments, cells were seeded onto HCl-treated coverslips at a density of 1 × 105 cells per 35 mm dish. HeLa and CHO cells were transfected with 5 µg of plasmid DNA using calcium phosphate precipitates. For this purpose, DNA was incubated for 5 min in 100 µl of 0.25 M CaCl2, then mixed with 100 µl HBS solution (50 mM HEPES, 1.5 mM Na2HPO4, 0.28 M NaCl) and added dropwise to the cells. After 16 h cells were washed three times with phosphate-buffered saline (PBS) before addition of fresh medium. Alternatively CHO cells were treated after 6 h with 10% glycerol in PBS and then washed three times with PBS. Wt and p53–/– MEFs were transfected with 2 µg of DNA using the Fugene transfection reagent (Roche Diagnostics). Cells were fixed at the indicated times and analyzed by indirect immunofluorescence microscopy. Transfected cells were identified through the enhanced green fluorescent protein (EGFP) tag in the case of Aurora-A, -B, Plk1 and p27Kip1 or through staining with the anti-hemagglutinin (HA) tag monoclonal antibody 12CA5 in the case of Cdk2. As judged by fluorescence intensity, all constructs were expressed at comparable levels. To visualize centrosomes, cells were counter-stained using antibodies against C-Nap1 (Fry et al., 1998a) or γ-tubulin (Fry et al., 1998b). For all quantitative analyses described in this study, at least three independent experiments were performed for each transfection or drug treatment, counting 400–600 cells in each case.
Immunofluorescence microscopy
Immunofluorescence microscopy was performed using a Zeiss Axioplan II microscope and an APOCHROMAT 63× 1.4 oil immersion objective. Pictures were taken using a Micromax CCD camera (Princeton, Instruments) and Metaview software (Universal Imaging Corp.), and images were processed using Adobe Photoshop (Adobe Systems, Mountain View, CA). For antibody staining cells were grown on acid-treated coverslips and fixed with methanol at –20°C for 6 min. Then, coverslips were washed three times in PBS, blocked with 3% bovine serum albumin (BSA) in PBS for 10 min, and again washed three times with PBS. All subsequent antibody incubations were carried out in PBS containing 3% BSA. Antibody reagents were anti-C-Nap1 antibodies [affinity-purified; 1 µg/ml (Fry et al., 1998a)], anti-γ-tubulin antibodies [purified IgG 5 µg/ml (Fry et al., 1998b)] and 12CA5 anti-HA (1:50 tissue culture supernatant). Incubations with primary antibodies were for 1 h at room temperature, followed by three washes with PBS. Secondary reagents were biotinylated donkey anti-rabbit (1:200, Amersham) followed by Texas Red-conjugated streptavidin (1:100, Amersham) and Alexa Fluor 488 conjugated goat-anti-mouse IgG (1:1000, Molecular Probes). DAPI (1 µg/ml) was included in the last incubation to stain DNA. Following three final washes with PBS, coverslips were mounted in 80% glycerol, 3% DABCO (in PBS) mounting medium.
DNA quantification
For quantitative analyses of DNA content, cells were grown on acid-treated coverslips and fixed at room temperature by consecutive treatment with 20% ethanol in PBS, 80% ethanol in PBS and pure ethanol, 10 min each. The coverslips were then dried, incubated with 3 µg/ml propidium iodide and 0.2 mg/ml RNase in PBS for 1 h at room temperature and mounted as described above. Immunofluorescence microscopy was performed using a Neofluar 10× 0.3 objective, and the DNA content of each cell was measured with Metaview software by integrating the propidium iodide signal intensity of the nuclei over a fixed area of 30 × 30 µm. For background subtraction, a corresponding area without nuclei was measured using identical settings. Transfected cells were identified by their GFP signal.
Cell extracts, immunoprecipitations, kinase assays and immunoblotting
U2OS cells were seeded at a density of 1 × 105 cells per 35 mm dish and transfected with 5 µg of expression plasmids encoding either wt or catalytically inactive EGFP–Aurora-A. After 8 h, nocodazole (0.5 µg/ml) was added, and 24 h later, cells were collected, washed once with PBS, once with PBS containing 1 mM PMSF and lysed in 150 µl lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1% NP-40, 20 mM β-glycero phosphate, 0.3 mM NaVO4, 1 mM DTT, 1 mM PMSF and aprotinin, leupeptin and pepstatin at 10 µg/ml each) for 30 min at 4°C. Lysates were clarified by centrifugation at 16 000 g for 10 min at 4°C. After preclearance of the lysates with protein G–Sepharose beads, wt and mutant EGFP–Aurora-A were immunoprecipitated using 2 µg of affinity-purified sheep anti-GFP antibody (gift of F.Barr) and protein G–Sepharose beads (Amersham Pharmacia Biotech). Immuno precipitates were washed three times with lysis buffer and twice with kinase buffer (50 mM HEPES pH 7.4, 10 mM MgCl2, 5 mM MnCl2, 5 µm ATP, 1 mM DTT and 5 mM β-glycerophosphate). Samples were then incubated for 30 min at 30°C in 50 µl of kinase buffer together with 2µ Ci of [γ-32P]ATP and myelin basic protein (0.5 mg/ml) as substrate. Kinase reactions were stopped by addition of 0.5 vol. of gel sample buffer and boiling for 5 min.
Proteins were resolved by electrophoresis on a 12% SDS– polyacrylamide gel and transferred onto nitrocellulose membranes, using a semi-dry blotting apparatus (Hoefer). To verify equal loading, proteins were visualized by Ponceau S staining, and membranes were soaked with blocking buffer (5% low-fat dried milk in PBS + 0.1% Tween-20). EGFP–Aurora-A (wt and K162R) was detected using anti-GFP monoclonal antibodies (gift of M.Maniak) and visualized using alkaline-phosphatase-conjugated anti-mouse IgG secondary antibodies (Promega). Phosphorylation was monitored by autoradiography, using a phosphoimager (Fujifilm).
Acknowledgments
Acknowledgements
We thank C.J.Sherr (St Jude Children’s Research Hospital, Memphis, TN) and H.te Riele (The Netherlands Cancer Institute, Amsterdam) for p53–/– and wt MEF, C.Regnard for the GFP-p27Kip1 construct, F.Barr (MPI, Martinsried) and M.Maniak (University of Kassel) for antibodies, and U.Hacker for excellent technical assistance. P.M. was supported by a postdoctoral fellowship from the Swiss National Science Foundation, and R.H. acknowledges receipt of an EMBO longterm fellowship.
References
- Andreassen P.R., Lohez,O.D., Lacroix,F.B. and Margolis,R.L. (2001) Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell., 12, 1315–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R., Bao,L., Zimmer,W.E., Brown,K., Zinkowski,R.P. and Brinkley,B.R. (1995) Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol., 130, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J.R. and Plowman,G.D. (1999) The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol., 9, 454–459. [DOI] [PubMed] [Google Scholar]
- Bischoff J.R. et al. (1998) A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J., 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov,A., Mir,L.M., Rieder,C.L., Edde,B. and Bornens,M. (1998) Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol., 143, 1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. (1914) Zur Frage der Entstehung maligner Tumoren. Fischer Verlag, Jena.
- Brinkley B.R. (2001) Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol., 11, 18–21. [DOI] [PubMed] [Google Scholar]
- Carroll P.E., Okuda,M., Horn,H.F., Biddinger,P., Stambrook,P.J., Gleich,L.L., Li,Y.Q., Tarapore,P. and Fukasawa,K. (1999) Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene, 18, 1935–1944. [DOI] [PubMed] [Google Scholar]
- Casenghi M., Mangiacasale,R., Tuynder,M., Caillet-Fauquet,P., Elhajouji,A., Lavia,P., Mousset,S., Kirsch-Volders,M. and Cundari,E. (1999) p53-independent apoptosis and p53-dependent block of DNA rereplication following mitotic spindle inhibition in human cells. Exp. Cell Res., 250, 339–350. [DOI] [PubMed] [Google Scholar]
- Fisk H.A. and Winey,M. (2001) The mouse mps1p-like kinase regulates centrosome duplication. Cell, 106, 95–104. [DOI] [PubMed] [Google Scholar]
- Fry A.M., Mayor,T., Meraldi,P., Stierhof,Y.D., Tanaka,K. and Nigg,E.A. (1998a) C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2105. J. Cell Biol., 141, 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi,P. and Nigg,E.A. (1998b) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J., 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K., Choi,T., Kuriyama,R., Rulong,S. and Vande Woude,G.F. (1996) Abnormal centrosome amplification in the absence of p53. Science, 271, 1744–1747. [DOI] [PubMed] [Google Scholar]
- Ghadimi B.M., Sackett,D.L., Difilippantonio,M.J., Schrock,E., Neumann,T., Jauho,A., Auer,G. and Ried,T. (2000) Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosom. Cancer, 27, 183–190. [PMC free article] [PubMed] [Google Scholar]
- Giet R. and Prigent,C. (1999) Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci., 112, 3591–3601. [DOI] [PubMed] [Google Scholar]
- Goepfert T.M. and Brinkley,B.R. (2000) The centrosome-associated Aurora/Ipl-like kinase family. Curr. Top. Dev. Biol., 49, 331–342. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize,R., Blank,T., Sandaltzopoulos,R., Becker,P., Hyman,A. and Karsenti,E. (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature, 382, 420–425. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize,R., Habermann,A., Karsenti,E. and Hyman,A. (1997) Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization120. J. Cell Biol., 138, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E.H. and Sluder,G. (2001) ‘It takes two to tango’: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev., 15, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li,C., Thompson,E.A., Maller,J.L. and Sluder,G. (1999) Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science, 283, 851–854. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Miller,F.J., Cham,M., Khodjakov,A. and Sluder,G. (2001) Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science, 291, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Hollander M.C. et al. (1999) Genomic instability in Gadd45a-deficient mice. Nature Genet., 23, 176–184. [DOI] [PubMed] [Google Scholar]
- Kellogg D.R., Moritz,M. and Alberts,B.M. (1994) The centrosome and cellular organization. Annu. Rev. Biochem., 63, 639–674. [DOI] [PubMed] [Google Scholar]
- Khan S.H. and Wahl,G.M. (1998) p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res., 58, 396–401. [PubMed] [Google Scholar]
- Khodjakov A. and Rieder,C.L. (2001) Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol., 153, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole,R.W., Oakley,B.R. and Rieder,C.L. (2000) Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol., 10, 59–67. [DOI] [PubMed] [Google Scholar]
- Kimura M., Kotani,S., Hattori,T., Sumi,N., Yoshioka,T., Todokoro,K. and Okano,Y. (1997) Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J. Biol. Chem., 272, 13766–13771. [DOI] [PubMed] [Google Scholar]
- Lacey K.R., Jackson,P.K. and Stearns,T. (1999) Cyclin-dependent kinase control of centrosome duplication. Proc. Natl Acad. Sci. USA, 96, 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni J.S. and Jacks,T. (1998) Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol., 18, 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Levine D.S., Sanchez,C.A., Rabinovitch,P.S. and Reid,B.J. (1991) Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc. Natl Acad. Sci. USA, 88, 6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ackermann,E.J., Bennett,C.F., Rothermel,A.L., Plescia,J., Tognin,S., Villa,A., Marchisio,P.C. and Altieri,D.C. (1999) Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nature Cell Biol., 1, 461–466. [DOI] [PubMed] [Google Scholar]
- Lingle W.L. and Salisbury,J.L. (1999) Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am. J. Pathol., 155, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W.L. and Salisbury,J.L. (2000) The role of the centrosome in the development of malignant tumors. Curr. Top. Dev. Biol., 49, 313–329. [DOI] [PubMed] [Google Scholar]
- Lingle W.L., Lutz,W.H., Ingle,J.N., Maihle,N.J. and Salisbury,J.L. (1998) Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl Acad. Sci. USA, 95, 2950–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C., Braun,S.E., Reid,S., Henegariu,O., Liu,L., Hangoc,G. and Broxmeyer,H.E. (1999) p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood, 93, 1390–1398. [PubMed] [Google Scholar]
- Matsumoto Y., Hayashi,K. and Nishida,E. (1999) Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol., 9, 429–432. [DOI] [PubMed] [Google Scholar]
- Meraldi P. and Nigg,E.A. (2001) Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci., 114, 3749–3757. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas,J., Fry,A.M., Bartek,J. and Nigg,E.A. (1999) Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nature Cell Biol., 1, 88–93. [DOI] [PubMed] [Google Scholar]
- Minn A.J., Boise,L.H. and Thompson,C.B. (1996) Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev., 10, 2621–2631. [DOI] [PubMed] [Google Scholar]
- Mundt K.E., Golsteyn,R.M., Lane,H.A. and Nigg,E.A. (1997) On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun., 239, 377–385. [DOI] [PubMed] [Google Scholar]
- Nakayama K. et al. (2000) Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J., 19, 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. (2001) Cell division mitotic kinases as regulators of cell division and its checkpoints. Nature Rev. Mol. Cell. Biol., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- O’Connell K.F., Caron,C., Kopish,K.R., Hurd,D.D., Kemphues,K.J., Li,Y. and White,J.G. (2001) The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell, 105, 547–558. [DOI] [PubMed] [Google Scholar]
- Okuda M. et al. (2000) Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell, 103, 127–140. [DOI] [PubMed] [Google Scholar]
- Pihan G.A. and Doxsey,S.J. (1999) The mitotic machinery as a source of genetic instability in cancer. Semin. Cancer Biol., 9, 289–302. [DOI] [PubMed] [Google Scholar]
- Pihan G.A., Purohit,A., Wallace,J., Knecht,H., Woda,B., Quesenberry,P. and Doxsey,S.J. (1998) Centrosome defects and genetic instability in malignant tumors. Cancer Res., 58, 3974–3985. [PubMed] [Google Scholar]
- Pihan G.A., Purohit,A., Wallace,J., Malhotra,R., Liotta,L. and Doxsey,S.J. (2001) Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res., 61, 2212–2219. [PubMed] [Google Scholar]
- Sato N., Mizumoto,K., Nakamura,M., Nakamura,K., Kusumoto,M., Niiyama,H., Ogawa,T. and Tanaka,M. (1999) Centrosome abnormalities in pancreatic ductal carcinoma. Clin. Cancer Res., 5, 963–970. [PubMed] [Google Scholar]
- Sato N., Mizumoto,K., Nakamura,M. and Tanaka,M. (2000) Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp. Cell Res., 255, 321–326. [DOI] [PubMed] [Google Scholar]
- Sen S., Zhou,H. and White,R.A. (1997) A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene, 14, 2195–2200. [DOI] [PubMed] [Google Scholar]
- Shackney S.E., Smith,C.A., Miller,B.W., Burholt,D.R., Murtha,K., Giles,H.R., Ketterer,D.M. and Pollice,A.A. (1989) Model for the genetic evolution of human solid tumors. Cancer Res., 49, 3344–3354. [PubMed] [Google Scholar]
- Shindo M. et al. (1998) cDNA cloning, expression, subcellular localization, and chromosomal assignment of mammalian aurora homologues, aurora-related kinase (ARK) 1 and 2. Biochem. Biophys. Res. Commun., 244, 285–292. [DOI] [PubMed] [Google Scholar]
- Shono M., Sato,N., Mizumoto,K., Maehara,N., Nakamura,M., Nagai,E. and Tanaka,M. (2001) Stepwise progression of centrosome defects associated with local tumor growth and metastatic process of human pancreatic carcinoma cells transplanted orthotopically into nude mice. Lab. Invest., 81, 945–952. [DOI] [PubMed] [Google Scholar]
- Smith L. et al. (1998) Duplication of ATR inhibits MyoD, induces aneuploidy and eliminates radiation-induced G1 arrest. Nature Genet., 19, 39–46. [DOI] [PubMed] [Google Scholar]
- Stearns T. (2001) Centrosome duplication. A centriolar pas de deux. Cell, 105, 417–420. [DOI] [PubMed] [Google Scholar]
- Terada Y., Tatsuka,M., Suzuki,F., Yasuda,Y., Fujita,S. and Otsu,M. (1998) AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J., 17, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A., Gabriel,A., Bertwistle,D., Connor,F., Paterson,H., Peacock,J., Ross,G. and Ashworth,A. (1999) Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol., 9, 1107–1110. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- Xie W., Li,L. and Cohen,S.N. (1998) Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl Acad. Sci. USA, 95, 1595–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Weaver,Z., Linke,S.P., Li,C., Gotay,J., Wang,X.W., Harris,C.C., Ried,T. and Deng,C.X. (1999) Centrosome amplification and a defective G2–M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell., 3, 389–395. [DOI] [PubMed] [Google Scholar]
- Zhou H., Kuang,J., Zhong,L., Kuo,W.L., Gray,J.W., Sahin,A., Brinkley,B.R. and Sen,S. (1998) Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nature Genet., 20, 189–193. [DOI] [PubMed] [Google Scholar]