Abstract
The OLE pathway of yeast regulates the abundance of the ER-bound enzyme Δ-9 fatty acid desaturase OLE1, thereby controlling unsaturated fatty acid pools and membrane fluidity. Previously, we showed that this pathway is exquisitely regulated by the ubiquitin/proteasome system. Activation of the pathway involves proteasomal processing of a membrane-bound transcription factor and the subsequent mobilization of the cleaved, ubiquitylated transcription factor from its partner molecule by CDC48UFD1/NPL4, a ubiquitin-selective chaperone-like enzyme. Here we report that the OLE1 protein itself is naturally short-lived and is degraded by ubiquitin/proteasome-dependent ER-associated degradation (ERAD). We found that CDC48UFD1/NPL4 plays a second role in the OLE pathway by mediating ERAD of OLE1. Intriguingly, other ERAD substrates also require CDC48UFD1/NPL4 for degradation, indicating that this enzyme is a novel, constitutive component of the ERAD machinery. We propose that CDC48UFD1/NPL4 functions as a segregase that liberates ubiquitylated proteins from non-modified partners.
Keywords: CDC48/chaperone/OLE pathway/segregase/ubiquitin
Introduction
Membrane fluidity is of central importance for the function and integrity of the membrane system of the cell. It is essential for the mobility of embedded proteins and for forming membrane curvatures, which in turn are required for the formation of organelles, the vesicular system and the nuclear envelope. A crucial parameter that determines membrane fluidity is the balance between saturated and unsaturated fatty acids. Unsaturated fatty acids (UFAs) are formed at the surface of the endoplasmic reticulum (ER) by fatty acid desaturases, which convert coenzyme A (CoA)-bound saturated fatty acids to unsaturated fatty acids by introducing double bonds in the carbon chains (Mitchell and Martin, 1995). The yeast enzyme, OLE1 (Δ-9 fatty acid desaturase), specifically converts palmitoyl (16:0) and stearoyl (18:0) CoA to palmitoleic (16:1) and oleic (18:1) acids by the introduction of a single double bond between carbon 9 and carbon 10 of the respective carbon chains (Mitchell and Martin, 1995). Yeast cells are not able to synthesize polyunsaturated fatty acids, but they can take up these compounds from the environment and incorporate them into their membrane systems (Bossie and Martin, 1989).
UFA levels in yeast are largely controlled by a regulon, coined the OLE pathway, that regulates the abundance of the OLE1 protein in the ER membrane. Previously, we showed that this pathway is uniquely and stringently controlled by the ubiquitin/proteasome system (Hoppe et al., 2000; Rape et al., 2001; for review see Hoppe et al., 2001). The master switches for the OLE pathway are two NF-κB-related transcription factors, SPT23 and MGA2, which are initially synthesized as inactive, ER-bound precursors (p120) (Zhang et al., 1997; 1999; Hoppe et al., 2000). They become activated by two ubiquitin-dependent events. In the first step, SPT23 forms a homodimer at the membrane in which one molecule (processing substrate) is cleaved off the membrane by regulated ubiquitin/proteasome-dependent processing (RUP) (Hoppe et al., 2000; Rape et al., 2001). SPT23 retains its ubiquitin modification after processing and initially remains tethered to its unprocessed, membrane-bound partner (processing template). In the second step, the processed transcription factor is liberated from its partner for nuclear targeting by the activity of the chaperone-like complex CDC48UFD1/NPL4 (Rape et al., 2001). Intriguingly, this enzyme preferentially binds ubiquitylated substrates, and is thereby capable of segregating the ubiquitylated, processed molecule from its non-modified partner molecule (Rape et al., 2001).
Here we describe that yet another key constituent of the OLE pathway, the OLE1 protein itself, is also under the exquisite control of the ubiquitin/proteasome system. We show that OLE1 is a naturally short-lived protein and that its turnover is mediated by ubiquitin/proteasome-dependent ER-associated degradation (ERAD). This finding significantly substantiates the view that ERAD plays an important physiological role besides its well-established function in protein quality control. Intriguingly, we discovered that ERAD of OLE1 and other substrates requires the activity of the ubiquitin-selective CDC48UFD1/NPL4 chaperone-like enzyme. Thus CDC48UFD1/NPL4 has a much broader function than was previously assumed, and indicates that this enzyme is a constitutive component of the ubiquitin/proteasome system.
Results
The OLE1 protein is short-lived
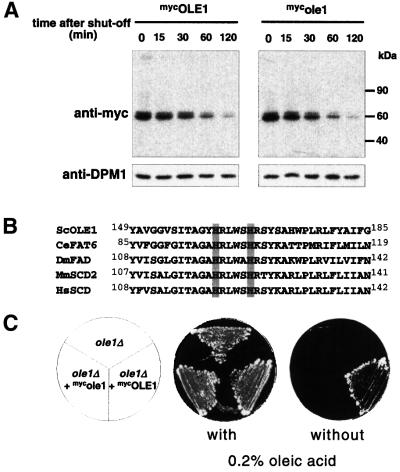
OLE1 is a polytopic integral membrane protein, which spans the ER membrane four times (Stukey et al., 1989). The vast majority of the protein is exposed to the cytosol, including its N- and C-terminus. The yeast enzyme comprises a desaturase enzymatic domain fused to a cytochrome b5 domain, whereas these two activities are found in two separate proteins in mammals (Mitchell and Martin, 1995). Because over- and underexpression of OLE1 are toxic for yeast cells, the level of OLE1 is tightly regulated. Indeed, all known mechanisms that control UFA levels regulate OLE1 protein levels (McDonough et al., 1992; Choi et al., 1996; Gonzales and Martin, 1996). From these observations we hypothesized that OLE1 is in all likelihood a short-lived protein. To investigate this possibility we replaced the fatty-acid controlled promoter of OLE1 with the GAL promoter (this procedure also removed the 5′ UTR responsible for fatty acid-regulated mRNA decay) (Gonzalez and Martin, 1996) and followed the protein level of epitope-tagged OLE1 (mycOLE1) by an expression shut-off experiment. We repressed the GAL promoter by adding glucose to the medium and blocked translation by the addition of cycloheximide. As shown in Figure 1A (left panel), OLE1 is indeed short-lived with a half-life of about 30 min. A caveat for this type of experiment is that the synthetic expression of OLE1 is expected to cause altered UFA levels, which in turn may influence the half-life and/or the physiology of the cell. To circumvent this problem we mutated the OLE1 open reading frame at positions that encode two conserved essential histidine residues of OLE1’s desaturase domain (Shanklin et al., 1994; Figure 1B). Inactivation of OLE1 was confirmed by a genetic experiment, which showed that the mutant myc-tagged protein (mycole1) could not complement the ole1 deletion strain (Figure 1C). We repeated the expression shut-off experiment with this strain and found that the inactive mutant protein has about the same short half-life as wild-type OLE1 (Figure 1A, right panel). Thus, OLE1 is indeed a short-lived protein in yeast cells under normal growth conditions, and we imagine that this property is elementary for the previously described regulatory mechanisms of the OLE pathway that control OLE protein levels. For the reasons stated above, we used the inactive OLE1 enzyme, mycole1, in the following studies.
Fig. 1. OLE1 is short-lived in vivo. (A) Expression shut-off experiments with WT cells expressing mycOLE1 and the mutant variant mycole1 under the control of the GAL1-10 promoter. Cells were grown in YPGal to an OD600 of 0.5 at 23°C and shifted for another 2 h to 37°C. The experiment was started by adding glucose and cycloheximide to the medium. At each time point indicated, the cellular level of both epitope-tagged OLE1 variants was analyzed by anti-myc immunoblots (upper panel). As a control, the blots were reprobed with an antibody against the stable ER membrane protein dolichol phosphate mannose synthase, DPM1 (lower panel). (B) Sequence comparison of Δ-9 fatty acid desaturases from different organisms (Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Mm, Mus musculus; Hs, Homo sapiens). The two conserved His residues, which were replaced by Ala residues in the mutant ole1 variant, are shaded in gray. (C) Growth of the ole1Δ deletion strain expressing either none, mycOLE1 or mutant mycole1 in the presence and absence of oleic acid, respectively. The lethal phenotype of ole1Δ in the absence of unsaturated fatty acids in the growth medium can only be suppressed by expressing the functional desaturase.
Turnover of OLE1 proceeds via ERAD
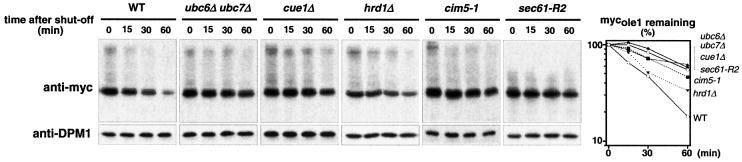
The prevalent pathway for ER-membrane protein turnover is ER-associated degradation (ERAD) (for review see Bonifacino and Weissman, 1998). ERAD is mediated by the cytosolic ubiquitin/proteasome pathway and is believed to require the SEC61 translocon channel for retrograde translocation of ERAD substrates to the cytosol (Wiertz et al., 1996; Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999). To investigate whether OLE1 turnover is mediated by ERAD we used an assortment of mutants deficient in ERAD. As shown in Figure 2, mycole1 was substantially stabilized in a ubc6 ubc7 double deletion mutant, as well as in a cue1 deletion strain. UBC6 is an integral membrane protein of the ER, which together with UBC7 constitute the chief E2 ubiquitin-conjugating enzymes involved in ERAD, whereas CUE1 is an ER-targeting factor for UBC7 (Sommer and Jentsch, 1993; Jungmann et al., 1993; Biederer et al., 1997). Thus, downregulation of OLE1 is indeed ubiquitin-dependent and involves prototypical ERAD E2 enzymes. OLE1 was only weakly stabilized in cells deleted for HRD1 (Figure 2), a gene encoding an ERAD-specific E3 ubiquitin ligase (Bays et al., 2001). We also tested deletion mutants of other ubiquitin ligases (DOA10 and RSP5; Huibregtse et al., 1995; Swanson et al., 2001), but no stabilization of mycole1 was observed in these strains (data not shown). Thus, we assumed that OLE1 degradation may involve additional ligases to those identified previously for ERAD. Degradation of OLE1 is indeed mediated by proteasomes, as mycole1 was significantly stabilized in a cim5-1 mutant (Ghislain et al., 1993), a temperature-sensitive mutant of a gene encoding an essential subunit of the 19S cap of the proteasome (Figure 2). Strong stabilization of mycole1 was also observed in sec61-R2, a strain carrying an allele of SEC61 that is known to exhibit strong defects in retrograde translocation of ERAD substrates but which is proficient in translocation into the ER (Zhou and Schekman, 1999). In conclusion, these data confirmed that downregulation of OLE1 proceeds, at least to a large part, by ERAD, involving the ubiquitin/proteasome system and the SEC61 translocon.
Fig. 2. OLE1 turnover is mediated by ERAD. Expression shut-off experiments with different ERAD mutants expressing the mutant mycole1 variant under the control of the GAL1-10 promoter. Experimental procedures were identical to those described in Figure 1A. Shown are the protein levels for mycole1 (upper panels) and as a control for DPM1 (lower panels). The right panel shows the quantification of the mycole1 decay. The estimated half-lives of mycole1 in WT, ubc6Δ ubc7Δ, cue1Δ, hrd1Δ, cim5-1 and sec61-R2 strains were 25, 80, 80, 35, 55 and 70 min, respectively.
ERAD of OLE1 involves the chaperone-like CDC48UFD1/NPL4 enzyme
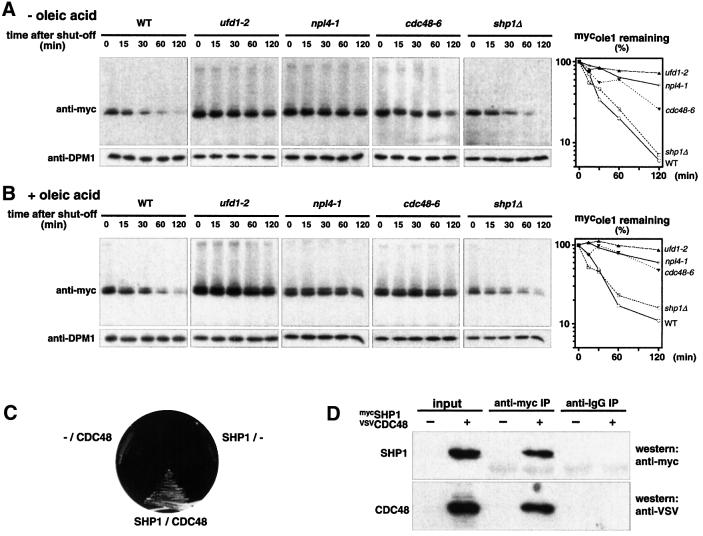
Recently, a chaperone-like complex, designated CDC48UFD1/NPL4, has been described, which appears to be important for several ubiquitin-dependent pathways (Ghislain et al., 1996; Dai et al., 1998; Meyer et al., 2000; Hitchcock et al., 2001; Rape et al., 2001). This complex comprises the catalytic subunit CDC48, an AAA-type ATPase, and two cofactors, UFD1 and NPL4. We showed recently that this complex preferentially binds ubiquitin–protein conjugates and that it is essential for the OLE pathway, as it mediates the mobilization of processed, ubiquitylated SPT23 transcription factor from the membrane (Rape et al., 2001). Since the mechanism of SPT23 activation has some resemblance to ERAD (Hoppe et al., 2000; Rape et al., 2001), we speculated that the CDC48UFD1/NPL4 complex might also be involved in OLE1 degradation. To test this idea, we expressed mycole1 in temperature-sensitive mutants of the essential genes CDC48, NPL4 and UFD1, respectively (the alleles cdc48–6, npl4-1, ufd1-2; Latterich et al., 1995; DeHoratius and Silver, 1996; Hoppe et al., 2000). As shown in Figure 3A, mycole1 was indeed significantly stabilized in all three mutants. But since the CDC48UFD1/NPL4 complex is essential for the production of UFAs (Rape et al., 2001), we considered the possibility that the observed stabilization might be caused by UFA depletion. However, as shown in Figure 3B, addition of oleic acid had essentially no effect on the outcome of the experiment, confirming a direct involvement of the chaperone in ERAD of OLE1.
Fig. 3. Involvement of the CDC48UFD1/NPL4 segregase in OLE1 degradation. (A) Expression shut-off experiments with the ts mutants ufd1-2, npl4-1, cdc48-6 and the deletion strain shp1Δ expressing the mutant mycole1 variant under the control of the GAL1-10 promoter in the absence of oleic acid (at 37°C). Shown are the protein levels for mycole1 (upper panels) and as a control for DPM1 (lower panels). The right panel shows the quantification of the mycole1 decay. The estimated half-lives of mycole1 in WT, ufd1-1, npl4-1, cdc48-6 and shp1Δ strains were 25, 275, 110, 60 and 27 min, respectively. (B) The same experiment as in (A) was performed in the presence of 0.2% oleic acid added 1 h prior to the 37°C shift to the medium. (C) Yeast two-hybrid interaction of CDC48 and SHP1. Open reading frames of SHP1 and CDC48 were cloned into pGAD424 and pGBD, respectively. Transformants were streaked onto SC-Leu-Trp-His plates to test for two-hybrid interaction. Empty vectors are indicated by ‘–’. (D) Co-immunoprecipitation of CDC48 with SHP1. Cytosolic fractions from yeast cells expressing epitope-tagged VSVCDC48 (indicated by ‘+’) and mycSHP1 were subjected to anti-myc and anti-IgG immunoprecipitation, respectively. The panel designated as ‘input’ shows 10% of the amount of protein used for immunoprecipitation.
In mammalian cells, CDC48 (p97) is known to form two alternative complexes. Together with p47 it mediates homotypic membrane fusion, but in combination with mammalian UFD1 and NPL4, the chaperone appears to collaborate with the ubiquitin/proteasome system (Meyer et al., 2000; Hetzer et al., 2001). To investigate whether ERAD of OLE1 specifically requires the CDC48UFD1/NPL4 complex or whether the alternative CDC48/p97-p47 complex may also mediate ERAD, we cloned the presumed p47 homolog from yeast. This protein, termed SHP1, shares 30% identical (50% similar) residues with its mammalian counterpart. By performing two-hybrid assays and co-immunoprecipitation experiments (Figure 3C and D) we found that SHP1 indeed interacts with CDC48. This finding suggests that, similar to mammalian p97, yeast CDC48 can also form two complexes, CDC48UFD1/NPL4 and CDC48SHP1. To examine whether SHP1 is involved in ERAD of OLE1, we generated a shp1 deletion mutant and followed the half-life of mycole1 in this strain. As shown in Figure 3A and B (right panels), mycole1 was as unstable in shp1 mutants as in WT cells. Thus, ERAD of OLE1 does not involve CDC48SHP1 but specifically requires the CDC48UFD1/NPL4 chaperone. Importantly, by cell fractionation studies we detected the stabilized mycole1 protein in the microsomal pellet fraction of ufd1-2 mutant cells. This indicates that the activity of the CDC48UFD1/NPL4 chaperone is required at the membrane prior to proteasomal degradation of the substrate (Figure 4 and Discussion).
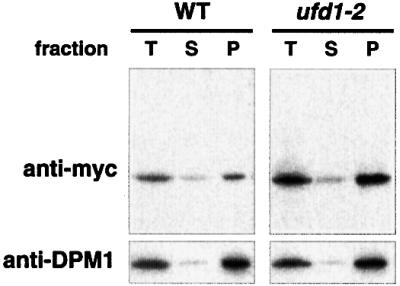
Fig. 4. Accumulation of stabilized OLE1 at the membrane. WT and ufd1-2 cells expressing mutant mycole1 under the control of the GAL1-10 promoter were grown in YPGal to a final OD600 of 1.0. An expression shut-off assay was performed by adding glucose and cycloheximide to the medium. After 30 min, cells corresponding to 50 OD600 (for both yeast strains) were subjected to cell fractionation as described in Materials and methods. Equal amounts of total extract (T), soluble (S) and pellet (P) fraction were analyzed by immunoblotting with anti-myc (upper panel) and the ER membrane protein anti-DPM1 (lower panel) antibody, respectively.
General role of CDC48UFD1/NPL4 in ERAD
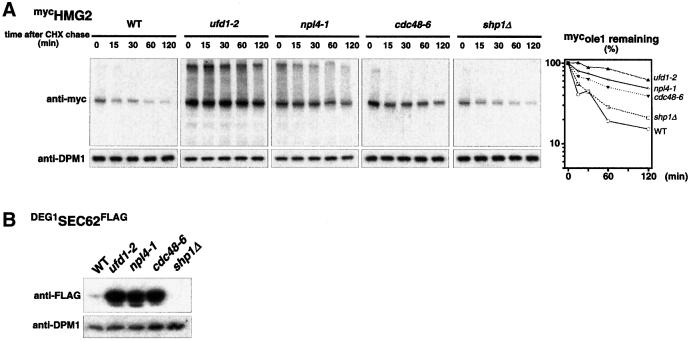
The discovery of the involvement of the CDC48UFD1/NPL4 complex in degradation of OLE1 raised the question of whether this chaperone is uniquely involved in ERAD of this particular substrate, or whether it is generally needed for ERAD. To address this question, we tested two well characterized ERAD substrates: HMG2, a naturally short-lived HMG CoA reductase from yeast (Hampton, 1998) and DEG1SEC62FLAG (Mayer et al., 1998), an engineered short-lived variant of the ER-membrane protein SEC62, bearing the Deg1 degradation signal derived from the short-lived transcription factor MATα2 (Chen et al., 1993). As shown in Figure 5, both proteins are indeed strongly stabilized in cdc48, ufd1 and npl4 mutants at their nonpermissive temperature. Furthermore, these two ERAD substrates were not stabilized in the shp1 mutant (Figure 5A, right panel). From these results we conclude that the ubiquitin-selective CDC48UFD1/NPL4 chaperone is a bona fide component of the ERAD machinery and that its activity is generally required for ERAD.
Fig. 5. General role of the CDC48UFD1/NPL4 segregase in ERAD. (A) Expression shut-off (cycloheximide chase) with WT and ufd1-2, npl4-1, cdc48-6 and the deletion strain shp1Δ expressing mycHMG2. Cells were grown in YPD to an OD600 of 0.5 at 23°C and then shifted for another 2 h to 37°C. After adding cycloheximide to the medium, samples were taken at the time points indicated and protein extracts were prepared. The protein level of epitope-tagged mycHMG2 is shown by performing western blots with an anti-myc antibody (upper panel). As a control, blots were reprobed with an antibody against the stable ER membrane protein DPM1 (lower panel). The right panel shows the quantification of the mycHMG2 decay. The estimated half-lives of mycHMG2 in WT, ufd1-1, npl4-1, cdc48-6 and shp1Δ strains were 20, 175, 110, 70 and 30 minutes, respectively. (B) Steady state level of the unstable DEG1SEC62FLAG variant in WT, ufd1-2, npl4-1, cdc48-6 cells and the deletion strain shp1Δ. Cells were grown in YPGal to an OD600 of 0.5 at 23°C and after another 2 h of growth at 37°C, aliquots were taken and protein samples prepared. The steady state level of the epitope-tagged SEC62 variant was analyzed by using an anti-FLAG antibody (upper panel). As a control, blots were reprobed with an antibody against DPM1 (lower panel).
Discussion
The importance of OLE1
The functional integrity and plasticity of the cellular vesicular system, the organelles and the nuclear envelope are crucial for eukaryotic life. Although homeostasis of fatty acid pools was long thought to be critical for these processes, only fairly recent genetic data have substantiated this view. In particular, genetic screens in yeast produced clear evidence for such ties but also revealed unanticipated links to a variety of additional cellular functions. Stewart and Yaffe (1991), for example, undertook a screen in yeast for mutants with defects in mitochondrial distribution and morphology (mdm mutants). One of the identified gene products, MDM2, turned out to be identical to OLE1, the enzyme investigated in our work (Stewart and Yaffe, 1991). Inactivation of OLE1 results in an ∼2.5-fold decline of UFA (16:1 and 18:1) levels, associated with a fragmentation of the reticular network of mitochondria and an accumulation of defective mitochondrial vesicles in the mother cell during cell division (Stewart and Yaffe, 1991). These phenotypes are a direct consequence of UFA depletion because they can be reverted by UFA supplementation. Deficiency in UFA levels also severely affects nuclear envelope structure whereas vesicular traffic appears to be less sensitive to UFA depletion (for review see Schneiter and Kohlwein, 1997).
Role of ERAD in the OLE pathway
Previous work has revealed that the desaturase activity in yeast is exclusively regulated by adjusting the abundance of the OLE1 enzyme (the OLE pathway). Gene expression of OLE1 was found to be regulated by UFAs both at the level of transcription and mRNA decay (Choi et al., 1996; Gonzalez and Martin, 1996; Hoppe et al., 2000). Based on these findings, we hypothesized that efficient regulation by gene expression requires a short-lived gene product. Indeed, we have shown here that OLE1 is rapidly degraded and that the protein is turned over by ERAD. This finding places OLE1 on the short list of known natural (not abnormal) substrates of ERAD and reinforces the view that ERAD is not solely a protein quality control pathway involved in the elimination of misfolded or misassembled proteins of the ER. Interestingly, another known example of a naturally short-lived protein of the ER in yeast is HMG CoA reductase (HMG2). This enzyme plays a key role in setting the rate for the biogenesis of sterols and its derivatives and is thus a crucial factor for membrane composition along with OLE1. Hampton and colleagues have shown that the turnover of this yeast enzyme is induced by components that act downstream of HMG2 function within the mevalonate pathway, indicating a negative feedback control of HMG2 turnover (for review see Hampton, 1998). In striking resemblance to this finding we recently discovered that OLE1 degradation is also controlled by fatty acid pools (S.Braun and S.Jentsch, in preparation). This finding suggests the intriguing possibility that ERAD of naturally short-lived proteins may be frequently regulated by cues that originate at the ER membrane.
Role of CDC48UFD1/NPL4 in ERAD
Inspired by our recent identification of the ubiquitin-selective chaperone-like enzyme CDC48UFD1/NPL4, we discovered that this enzyme complex is also needed for ERAD of OLE1. Most importantly, our studies revealed that CDC48UFD1/NPL4 is a general, novel component of the ERAD machinery and that its function is not restricted to OLE1 turnover. Interestingly, CPY*, an aberrantly folded, soluble ERAD substrate, is also stabilized in mutants deficient in the CDC48UFD1/NPL4 complex (our unpublished data). Because CPY* is thought to be recognized as an ERAD substrate either in the ER lumen or within the Golgi (Vashist et al., 2001), the cyto/nucleoplasmic CDC48UFD1/NPL4 enzyme is unlikely to play a role in ERAD-substrate recognition. A significant fraction of CDC48UFD1/NPL4 localizes to the cytosolic face of ER membranes or to the nuclear envelope (Rape et al., 2001), suggesting that its activity is particularly important at this location. Consistent with this finding, we found that stabilized ERAD substrates accumulate at the ER membrane in mutants deficient in CDC48UFD1/NPL4 activity (Figure 4). This result also indicates that the activity of the enzyme is needed prior to proteasomal degradation.
But which step in the ERAD pathway might require a chaperone-like activity? An important clue comes from our recent work on the SPT23 transcription factor (Rape et al., 2001), showing that CDC48UFD1/NPL4 exhibits a pronounced specificity towards ubiquitylated proteins. Contrary to a recent report (Hitchcock et al., 2001), CDC48UFD1/NPL4 is not required for SPT23 processing but for the segregation of ubiquitylated, processed SPT23 transcription factor from its unprocessed SPT23 partner molecule (Rape et al., 2001). We therefore assume that CDC48UFD1/NPL4 in general has the property to segregate ubiquitylated proteins from non-modified partners. Because the conventional terms ‘chaperone’ or ‘unfoldase’ may be misleading, as they do not describe the segregating activity of CDC48UFD1/NPL4 precisely, we suggest the term ‘segregase’ for this or similar activities (e.g. also for the activity of NSF in untethering SNARE complexes). Notably, CDC48 is assembled into a homohexameric ring, which, upon ATP hydrolysis, undergoes a dramatic rotational outward movement similar to the diaphragm in a camera lens (Zhang et al., 2000). It seems highly likely that bound protein complexes are untethered by this movement. Because of its ubiquitin specificity we assume that CDC48UFD1/NPL4 acts downstream of the ubiquitylation enzymes in ERAD pathways. As shown previously (Biederer et al., 1997), ubiquitylation of ERAD substrates may prevent the substrates from slipping back to the ER. A possible function of CDC48UFD1/NPL4 may therefore be to segregate the ubiquitylated ERAD substrates from the proteins of the translocon channel. Importantly, membrane extraction of ERAD substrates also requires the activity of the proteasome (Mayer et al., 1998), suggesting that ubiquitylation, CDC48UFD1/NPL4 segregase activity and proteasomal degradation are tightly coupled, both physically and mechanistically. Specifically, we propose that ubiquitylated ERAD substrates might be directly translocated from the SEC61 channel into the proteasome by the joint forces of two types of hexameric AAA-ATPases, the CDC48 chaperone and the six RPT subunits of the 19S cap of the proteasome. An interesting speculation is that proteasomes may directly attach to the segregase through RPT–CDC48 interaction.
Segregase or proteasome—alternative fates for ubiquitin–protein conjugates?
This and our previous work (Rape et al., 2001) have revealed two important physiological roles of the ubiquitin-selective CDC48UFD1/NPL4 segregase within the ubiquitin pathway. Importantly, as shown for the processed SPT23 transcription factor, segregation of a substrate by CDC48UFD1/NPL4 does not always result in immediate proteasomal degradation (Rape et al., 2001). We therefore propose that ubiquitin–protein conjugates (possibly dependent on the type of ubiquitylation; e.g. mono versus multi) may be targeted either to the proteasome for degradation or to the segregase for protein complex disassembly. However, in cases where proteolysis requires the segregation of a proteolytic substrate from other proteins, e.g. in ERAD or in selective degradation of subunits of oligomeric protein complexes, the segregase and the proteasome may cooperate in degradation. Intriguingly, vertebrate CDC48UFD1/NPL4 was recently shown to also be required for nuclear envelope assembly (Hetzer et al., 2001). As this process might involve SNARE proteins, CDC48UFD1/NPL4 could play a similar role to the CDC48-related segregase NSF, which untethers v/t-SNARE complexes after the vesicles have fused with the target membrane. It remains to be seen whether ubiquitylation of SNAREs is an important event in nuclear envelope assembly.
Materials and methods
Cloning and yeast techniques
Standard yeast protocols were used (Guthrie and Fink, 1991). Media and plates were supplemented with 0.2% oleic acid dissolved in Nonidet P40 (0.2% final) when indicated. Strains were derivatives of DF5 (Hoppe et al., 2000). ufd1-2 was constructed by a PCR-based strategy. The shp1Δ deletion strain was constructed by a PCR method described by Knop et al. (1999); npl4-1 was a gift of P.Silver, cdc48-6 of K.U.Fröhlich, cue1Δ and hrd1Δ of T.Sommer, cim5-1 of C.Mann and sec61-R2 of R.Schekman. Mutations were introduced into the DF5 background by repetitive mating and tetrad dissection except for cim5-1 and sec61-R2.
The OLE1 ORF was amplified from a cDNA library and cloned into YIplac211 containing three N-terminal myc tags. Expression was driven by the GAL1-10 promoter. The physiologically inactive ole1 variant was constructed by site-directed mutagenesis replacing two conserved, essential histidine residues into alanine (H161A and H166A; see Figure 1B). Both constructs were inserted into the URA locus of ura3-52 strains by linearizing the vectors with EcoRV. VSVCDC48 (in YEplac112) was expressed under the control of its own promoter, mycSHP1 (in p425) under the control of the 361 bp MET25 promoter fragment. The Deg1SEC62FLAG variant (in Ycplac22) has been described (Mayer et al., 1998). Cloning details will be provided upon request. The mycHMG2 construct (pRH244) was a gift of R.Hampton.
Expression shut-off experiments
Strains expressing mycole1 under the control of the GAL1-10 promoter were grown in YPGal to an OD600 of 0.5 at 23°C and shifted to 37°C for 2–3 h. Transcription and translation were arrested by adding glucose and cycloheximide to a final concentration of 2% and 0.5 mg/ml, respectively. For each time point of the expression shut-off experiment, samples of 1 OD600 of yeast cells were harvested and protein extracts were performed by NaOH lysis and TCA precipitation as described (Knop et al., 1999). Detection of mycole1 and DPM1 was performed by immunoblots with polyclonal anti-myc (Santa Cruz) and monoclonal anti-DPM1 (Molecular Probes) antibodies. Quantification of chemiluminescence signals was performed by using a CCD camera (LAS 1000, Fujifilm) and the software programs Image Reader LAS 1000 V1.1 and Image Gauge V3.01 (Fujifilm).
Strains expressing mycHMG2 under the control of the ADH promoter were grown in YPD under the conditions described above. Translational arrest was performed by adding cycloheximide to the medium to a final concentration of 0.5 mg/ml. Cell lysis and sample preparation were done as described above.
Membrane fractionation
Fractionation of mycole1 was performed as described (Rape et al., 2001). In brief, total cell extracts were prepared by glass bead disruption. After removal of cell debris by centrifugation at 700 g, crude membrane and cytosolic fractions were fractionated by high spin centrifugation at 20 000 g. Soluble and pellet fractions were probed by western blotting with polyclonal anti-myc antibodies (Santa Cruz Biotechnology) or with monoclonal anti-DPM1 antibodies (Molecular Probes).
Immunoprecipitation of VSVCDC48
WT strains expressing mycSHP1 (from pMET25) and VSVCDC48 were grown in YPD to an OD600 of 0.5 at 23°C and then shifted for 1 h to methionine-free SC medium. Lysis of 25 OD600 of yeast cells were done by glass bead disruption in 150 µl lysis buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 mM PMSF). After diluting the extract to a final volume of 500 µl, cytosolic fractions were prepared as described above. These extracts were incubated with 2 µg of polyclonal anti-myc (Santa Cruz) antibodies in the presence of 0.05% Triton X-100 for 90 min at 4°C. Protein G-agarose beads (Roche) were added for another 60 min at 4°C. Subsequently, the samples were washed five times with lysis buffer containing 0.05% Triton X-100, then with lysis buffer without Triton, and finally incubated in HU buffer (Knop et al., 1999) for 15 min at 65°C. Proteins were separated on 12% polyacrylamide gels and analyzed by immunoblots with monoclonal anti-myc (Santa Cruz) and anti-VSV (Roche) antibodies, respectively.
Two-hybrid interaction between VSVCDC48 and mycSHP1
SHP1 and CDC48 were cloned into pGAD424 (Clontech) and pGBD (James et al., 1996), respectively. PJ69-4A cells (James et al., 1996) were transformed with various combinations of bait and prey plasmids according to standard protocols and plated on SC-Leu-Trp plates. Transformants were streaked onto SC-Leu-Trp-His plates to test for two-hybrid interaction.
Acknowledgments
Acknowledgements
We thank K.U.Fröhlich, R.Hampton, R.Schekman, P.Silver and T.Sommer for mutants and plasmids. We thank T.Braun for early contributions to this work, A.Buchberger for help and discussions, and other members of the department for help and advice. Our work was supported by the Max Planck Society and grants to S.J. from the Deutsche Forschungsgemeinschaft, the European TMR Ubiquitin Network and Fonds der Chemischen Industrie.
Note added in proof
After submission of our work, several papers describing related findings appeared: Ye et al. (2001) Nature, 414, 652–656; Bays et al. (2001) Mol. Biol. Cell, 12, 4114–4128; Rabinovich et al. (2002) Mol. Cell. Biol., 22, 626–634; Jarosch et al. (2002) Nature Cell Biol., in press.
References
- Bays N.W., Gardner,R.G., Seelig,L.P., Joazeiro,C.A. and Hampton,R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman,A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie M.A. and Martin,C.E. (1989) Nutritional regulation of yeast Δ-9 fatty acid desaturase activity. J. Bacteriol., 171, 6409–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT α2 repressor. Cell, 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Choi J.Y., Stukey,J., Hwang,S.Y. and Martin,C.E. (1996) Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem., 271, 3581–3589. [DOI] [PubMed] [Google Scholar]
- Dai R.M., Chen,E., Longo,D.L., Gorbea,C.M. and Li,C.C. (1998) Involvment of valosin-containing protein, an ATPase co-purified with IκBα and 26S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem., 273, 3562–3573. [DOI] [PubMed] [Google Scholar]
- DeHoratius C. and Silver,P.A. (1996) Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell, 7, 1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Udvardy,A. and Mann,C. (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature, 366, 358–362. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Dohmen,R.J., Levy,F. and Varshavsky,A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J., 15, 4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C.I. and Martin,C.E. (1996) Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. J. Biol. Chem., 271, 25801–25809. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol., 194. [PubMed] [Google Scholar]
- Hampton R.Y. (1998) Genetic analysis of hydroxymethylglutaryl-coenzyme A reductase regulated degradation. Curr. Opin. Lipidol., 9, 93–97. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Meyer,H.H., Walther,T.C., Bilbao-Cortes,D., Warren,G. and Mattaj,I.W. (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nature Cell Biol., 3, 1086–1091. [DOI] [PubMed] [Google Scholar]
- Hitchcock A.L., Krebber,H., Frietze,S., Lin,A., Latterich,M. and Silver,P.A. (2001) The conserved Npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell, 12, 3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski,K., Rape,M., Schlenker,S., Ulrich,H.D. and Jentsch,S. (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell, 102, 577–586. [DOI] [PubMed] [Google Scholar]
- Hoppe T., Rape,M. and Jentsch,S. (2001) Membrane-bound-transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell Biol., 13, 344–348. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M., Scheffner,M., Beaudenon,S. and Howley,P.M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl Acad. Sci. USA, 92, 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J., Reins,H.A., Schobert,C. and Jentsch,S. (1993) Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature, 361, 369–371. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Latterich M., Frohlich,K.U. and Schekman,R. (1995) Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell, 82, 885–893. [DOI] [PubMed] [Google Scholar]
- Mayer T.U., Braun,T. and Jentsch,S. (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J., 17, 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough V.M., Stukey,J.E. and Martin,C.E. (1992) Specificity of unsaturated fatty acid-regulated expression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem., 267, 5931–5936. [PubMed] [Google Scholar]
- Meyer H.H., Shorter,J.G., Seemann,J., Pappin,D. and Warren,G. (2000) A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J., 19, 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.G. and Martin,C.E.(1995) A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae Δ-9 fatty acid desaturase. J. Biol. Chem., 270, 29766–29772. [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman,R. and Romisch,K. (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J., 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R.K., Bohmler,S., Bordallo,J., Sommer,T. and Wolf,D.H. (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature, 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Rape M., Hoppe,T., Gorr,I., Kalocay,M., Richly,H. and Jentsch,S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell, 107, 667–677. [DOI] [PubMed] [Google Scholar]
- Schneiter R. and Kohlwein,S.D. (1997) Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell, 88, 431–434. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Whittle,E. and Fox,B.G. (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, sterolyl-CoA desaturase, and are conserved in alkane hydrolase and xylene monooxygenase. Biochemistry, 33, 12787–13794. [DOI] [PubMed] [Google Scholar]
- Sommer T. and Jentsch,S. (1993) A protein translocation defect linked to ubiquitin-conjugation at the endoplasmic reticulum. Nature, 365, 176–179. [DOI] [PubMed] [Google Scholar]
- Stewart L.C. and Yaffe,M.P. (1991) A role for unsaturated fatty acids in mitochondrial movement and inheritance. J. Cell Biol., 115, 1249–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey J.E., McDonough,V.M. and Martin,C.E. (1989) Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem., 264, 16537–16544. [PubMed] [Google Scholar]
- Swanson R., Locher,M. and Hochstrasser,M. (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and MATα2 repressor degradation. Genes Dev., 15, 2660–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Kim,W., Belden,W.J., Spear,E.D., Barlowe,C., and Ng,D.T. (2001) Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol., 155, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella,D., Bogyo,M., Yu,J., Mothes,W., Jones,T.R., Rapoport,T.A. and Ploegh,H.L. (1996) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature, 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Zhang S., Burkett,T,J, Yamashita,I. and Garfinkel,D.J. (1997) Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 4718–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Skalsky,Y. and Garfinkel,D.J. (1999) MGA2 or SPT23 is required for transcription of the Δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics, 151, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. (2000) Structure of the AAA ATPase p97. Mol. Cell, 6, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Zhou M. and Schekman,R. (1999) The engagement of Sec61p in the ER dislocation process. Mol. Cell, 4, 925–934. [DOI] [PubMed] [Google Scholar]