Abstract
In the protozoan malaria parasite, Plasmodium falciparum, the telomere-associated sequences (TASs) of the 14 linear chromosomes display a similar higher order organization and form clusters of four to seven telomeres localized at the nuclear periphery. Experimental evidence has shown that the physical tethering of chromosome ends enhances the ectopic recombination between gene families involved in antigenic variation and parasite sequestration. Using FISH analysis, we observed that chromosome ends lacking the subtelomeric region are usually delocalized from telomere clusters, but still remain at the nuclear periphery. This indicates that subtelomeric DNA is necessary for cluster formation but is not essential for peripheral positioning. Intriguingly, these truncated chromosomes have unusually long telomeric tracts (up to three times longer than average length), showing that TASs play a role in telomere length regulation. On these chromosomes, the newly formed telomere frequently extends from truncated genes leading, in some cases, to the transcription of telomeric DNA. The implications of both subtelomeric gene expression and nuclear architecture in the virulence of this serious human pathogen are discussed.
Keywords: malaria/nuclear architecture/telomere-associated sequences/telomere clustering/telomere length
Introduction
Telomeres, the complex of repetitive DNA and associated proteins at chromosome ends, are essential for chromosome stability. They prevent chromosome ends from fusing and from being recognized as damaged DNA. The telomeres from several organisms have been shown to be located at the nuclear periphery. Transcriptional repression and heterochromatin formation are other processes intimately associated with telomeres and assigned to functional subdomains within the nucleus (reviewed in Greider, 1996; Cockell and Gasser, 1999). Telomeric DNA generally consists of tandemly repeated, short G-rich sequences and ends with a 3′ overhang, formed by the degradation of the ultimate primer used for synthesizing the lagging strand during DNA replication (reviewed in de Lange, 1995). It was recently observed that telomeres in mammalian cells, ciliates and trypanosomes end with large ‘T loops’ (for telomere loops), presumably formed by invasion of the 3′ telomeric overhang into the duplex telomeric repeat array. These structures are thought to protect chromosomal termini from degradation and recognition as broken ends (Griffith et al., 1999; Murti and Prescott, 1999; Munoz-Jordan et al., 2001).
Telomere length regulation involves a tight balance of competing forces: telomere shortening and telomere elongation (reviewed in McEachern et al., 2000). In most organisms telomere length appears to fluctuate around a mean value, which is species specific. However, in certain cell types or organisms, this equilibrium can be unbalanced. For example, in human somatic cells continuous telomere shortening has been observed, whereas in African trypanosomes telomeres display an irregular pattern of growth and shortening (Bernards et al., 1983; Pays et al., 1983; Harley et al., 1990; Hastie et al., 1990). Telomere length is controlled by specific telomere-binding proteins. Several have been described: Rap1p, Cdc13p and Rif proteins in Saccharomyces cerevisiae (Berman et al., 1986; Shore and Nasmyth, 1987; Nugent et al., 1996; Virta-Pearlman et al., 1996); Rap1p in Kluyveromyces lactis (Larson et al., 1994); Taz1p and Pot1p in Schizosaccharomyces pombe (Cooper et al., 1997; Baumann and Cech, 2001); TRF1 and TRF2 in humans (Chong et al., 1995; Bilaud et al., 1996; Broccoli et al., 1997); and a TBP in the ciliate Oxytricha nova (Froelich-Ammon et al., 1998). Mutations in these telomere-binding proteins alter the telomere length of each chromosome end.
The telomere adjacent regions are generally composed of a mosaic of non-coding repetitive elements, which seem to have evolved in a species-specific manner (reviewed in Pryde and Louis, 1997) and are considered to serve as molecular buffers of non-functional ‘junk DNA’ next to the telomere repeats (Wilkie et al., 1991). Although their biological role remains ambiguous, specialized functions such as protecting subtelomeric genes from transcriptional silencing due to the telomere-position effect (TPE) may have evolved in eukaryotes (Fourel et al., 1999; Pryde and Louis, 1999).
The protozoan parasite, Plasmodium falciparum, is responsible for the most fatal form of human malaria. The severity of malaria is correlated with the expression of virulence factor genes localized predominantly at chromosome extremities (reviewed in Craig and Scherf, 2001). The haploid nuclear genome of P.falciparum is extremely AT rich (82%) and consists of 14 linear chromosomes varying from 0.7 to 3.4 Mb. At each chromosome end, telomere repeats [GGGTT(T/C)A] are followed by a non-coding subtelomeric region of ∼15–30 kb. This region is composed of six different telomere-associated repetitive elements (TAREs 1–6) and has a highly conserved organization (Figueiredo et al., 2000). These elements are flanked by members of gene families coding for virulence factors, including var genes (Rubio et al., 1996; Hernandez-Rivas et al., 1997), which are responsible for antigenic variation and cytoadhesion. When cultivated in vitro, P.falciparum chromosomes spontaneously undergo breakage, occasionally leading to large terminal deletions. Broken chromosomes are frequently healed by the addition of new telomere repeats, most likely by a plasmodial telomerase (Bottius et al., 1998). Recently, P.falciparum chromosome ends were shown to form clusters of four to seven telomeres at the nuclear periphery, facilitating ectopic recombination among heterologous subtelomeric chromosome regions, including genes coding for virulence factors (Freitas-Junior et al., 2000).
In the work reported here, we studied P.falciparum mutant strains in which spontaneous terminal chromosome truncations occurred. We observed that the loss of the subtelomeric region changed the location of chromosome ends in the nucleus. Furthermore, we present evidence, for the first time, that telomere length on individual chromosome ends can vary drastically and is modulated in cis by the DNA sequences at the telomere junction.
Results
Telomere length varies strikingly among malaria species
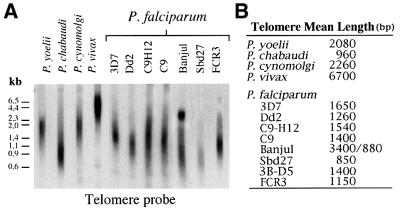
The mean telomere length of five different species of Plasmodium was analysed using the ‘telomere restriction fragment’ (TRF) method (de Lange et al., 1990). This technique takes advantage of the lack of restriction sites in the G-rich repetitive telomeric DNA as a means to liberate intact telomeres. Genomic DNA was digested with four frequently cutting restriction enzymes (AluI, DdeI, MboII, RsaI) and analysed by Southern blot using a P.falciparum telomere-specific probe (Figure 1A). The mean telomere length was determined using a PhosphorImager quantitative analyser (Figure 1B). Figure 1A shows the striking difference observed in the telomere length between the human malaria species P.falciparum and Plasmodium vivax, the rodent malarial species Plasmodium yoelii and Plasmodium chabaudi and the simian parasite Plasmodium cynomolgi. P.vivax exhibits the longest mean telomere length of ∼6700 bp. P.yoelii and P.cynomolgi telomeres have similar mean lengths of 2000 and 2300 bp, respectively, while P.chabaudi has the shortest telomeric DNA tracts averaging 960 bp. Within P.falciparum strains, the mean telomere length variation is less striking, ranging from 850 to 1600 bp. The ‘Banjul’ strain displays an unusual hybridization pattern in which two different-sized peaks are observed: (i) a broad smear represents telomeric DNA similar to the mean length of the other strains and (ii) a sharper peak, corresponding to higher molecular weight telomeric molecules of ∼3400 bp. Hence a large difference exists in the average telomere length among the different malarial species.
Fig. 1. Telomere length varies substantially among Plasmodium species, and to a lesser extent within the same species, as shown by TRF analysis. (A) One microgram of total genomic DNA was digested with four frequently cutting restriction enzymes: AluI, DdeI, MboII, RsaI. TRFs were identified using a P.falciparum telomere-specific probe, as telomere repeats in other Plasmodium species also consist of the same type of degenerate G-rich heptamer (Ponzi et al., 1985; L.M.Figueiredo, unpublished data). The intensity of the signal in each lane was quantified by PhosphorImager analysis and the peak of highest intensity was taken as the mean value for telomere length. Based on the sequences available at the genome database, we calculate that the distance between the telomere and the first restriction site (among the sites of AluI, DdeI, MboII, RsaI) is ∼30 bp in P.falciparum and P.vivax. (B) Summary of the average telomere lengths (in bp).
Large inter- and intra-chromosomal variation of telomere repeat length
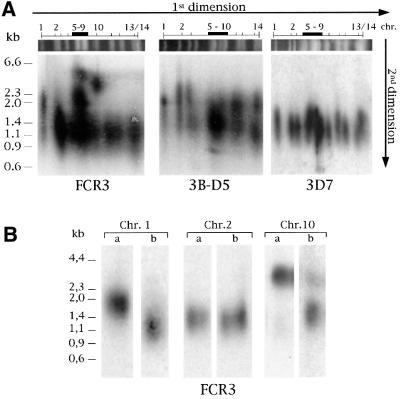
The broad smears observed by TRF analysis in Figure 1 indicate the possibility of multiple size classes of telomeres in malarial parasites. Therefore, we examined the telomere length of individual chromosomes from three genetically different P.falciparum strains (FCR3, 3B-D5 and 3D7) using two-dimensional pulse-field gel electrophoresis (2D-PFGE). This type of analysis consists of separating the 14 P.falciparum chromosomes in a first dimension by PFGE followed by digestion with frequently cutting enzymes to liberate the telomere (as described above for TRF analysis). After separation in a second dimension, chromosome-specific telomere restriction fragments were visualized by Southern blot. A characteristic ‘telomere fingerprint pattern’ (TFP) was observed in the three parasite strains (Figure 2A), revealing in each case that the telomere length is heterogeneous among the 14 chromosomes of the parasite genome. Such fingerprints are stable in laboratory-cultured parasites for at least several months (data not shown). In the FCR3 strain, the majority of the chromosomes have a telomere length of ∼1100 bp (see also Figure 1B), although several chromosomes harbour significantly longer telomeres. Specifically, chromosome 1 and some of the chromosomes in the compression zone (chromosomes 5–9) display an intermediate telomere length of 1900 bp. A few others in the compression zone are estimated to have telomeres of up to 3100 bp. Heterogeneity of telomere length also exists in the 3B-D5 strain, although it is less pronounced, ranging from 1400 to 2500 bp. In contrast, the TFP of the 3D7 strain shows that telomere length is much more homogeneous among the 14 chromosomes, ranging within a tighter interval from 1100 to 1600 bp. Moreover, comparison of the TFPs in Figure 2A reveals that the same chromosome can have telomeres of greater than the average length in one strain and the mean length in another strain.
Fig. 2. (A) TFP is characteristic of each P.falciparum strain. Chromosomes of the genetically different parasite clones FCR3, 3B-D5 and 3D7 were separated by PFGE in a first dimension. Agarose strips containing the 14 chromosomes were next subjected to TRF analysis by digestion with four frequently cutting restriction enzymes (AluI, DdeI, MboII, RsaI), followed by conventional separation on an agarose gel for the second dimension. TRFs were identified using a telomere-specific probe. The observed compression zone is due to the size similarity of chromosomes 5–9/10. (B) Two telomeres from a single chromosome can have a different size, as measured by TRF analysis of both ‘arms’ of three different chromosomes from FCR3. Individual chromosomes (1, 2 and 10) were isolated by PFGE and digested overnight with enzymes that liberate large subtelomeric restriction fragments (SacII for chromosome 1 and BssHII for chromosomes 2 and 10). Restriction fragments were separated by PFGE, excised from the gel and subjected to TRF analysis as described in (A).
The long smears observed for some chromosomes in the TFP indicated that there might be a difference between the lengths of the two telomeres on the same chromosome. To address this question, large subtelomeric restriction fragments from chromosomes 1, 2 and 10 of FCR3 were isolated by PFGE and analysed by TRF analysis (Figure 2B). The mean length of the chromosome 1 telomeres differs: 1800 versus 1150 bp. For chromosome 10, the difference in telomere length is more pronounced: 3600 and 1500 bp. However, in chromosome 2 both telomeres are of identical length (1400 bp). Together, these data show that the number of telomere repeats at a given chromosome end may vary dramatically both inter- and intra-chromosomally, demonstrating that regulation of telomere length can be chromosome end specific.
Evidence that DNA sequences at the telomere junction are involved in telomere length regulation
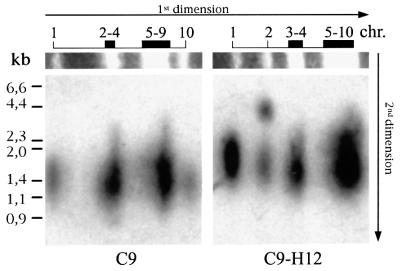
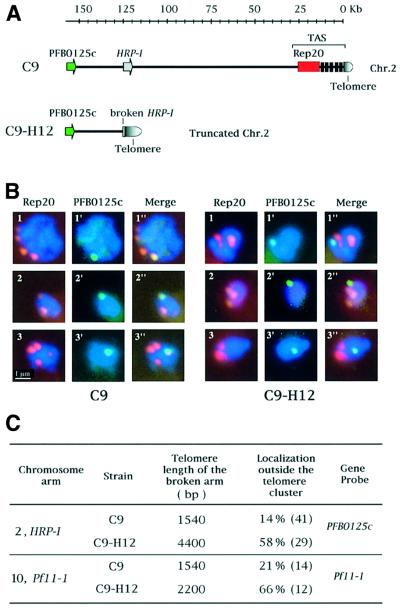
To investigate the molecular basis of the unusually long telomeres on specific chromosome ends, we used two P.falciparum clones with an identical genetic background (C9 and its subclone C9-H12), but which differ in their TFPs (Figure 3). Chromosomes 2 and 10 in C9-H12 migrate further than the corresponding chromosomes in C9, due to the fact that they contain large chromosome terminal deletions. Comparison of the TFPs for C9 and C9-H12 clearly shows that a new size population has arisen in C9-H12, indicating that one or both telomeres of chromosomes 2 and 10 have become much longer (the new mean lengths being 4400 and 2200 bp, respectively), whereas in C9 both telomeres of chromosomes 2 and 10 have the same average length. We cloned and sequenced the chromosome breakpoint on chromosome 2 of C9-H12, revealing that it is localized within the intron of a gene coding for the histidine-rich protein I (HRP-I). The HRP-I gene is normally located ∼120 kb from the telomere, but in C9-H12 the entire 120 kb subtelomeric region has been deleted from chromosome 2 (corresponding to >10% of the chromosome), including all but the promotor and the first exon of HRP-I. This truncation was repaired by the addition of telomere repeats to the breakage site (schematically shown in Figure 4B). We investigated whether the longer telomere observed in chomosome 2 of C9-H12 is located at the truncated chromosome arm. Both terminal regions from BssHII-digested chromosome 2 were isolated using PFGE. Hybridization analysis confirmed that the longer telomere (4400 bp) corresponds to the truncated arm of chromosome 2, while the intact subtelomeric arm of the same chromosome is only 1500 bp, corresponding to the average length for this strain (data not shown). The same type of analysis was performed for chromosome 2 of the C9 strain, which revealed that both intact subtelomeric arms harbour telomeres of 1400 bp, the average length for this strain (data not shown).
Fig. 3. Intact and truncated chromosome ends harbour telomeres of differing length. TFP of C9 and C9-H12 (a subclone of C9) are presented. Ethidium bromide-stained chromosomes 1–10 are shown. Chromosomes 2 and 10 in C9-H12 migrate more quickly than those of the parental strain C9 due to spontaneously generated large chromosomal truncations. The breakage sites in chromosomes 2 and 10 have been localized to HRP-I (this work) and Pf11-1 (Scherf et al., 1992), respectively.
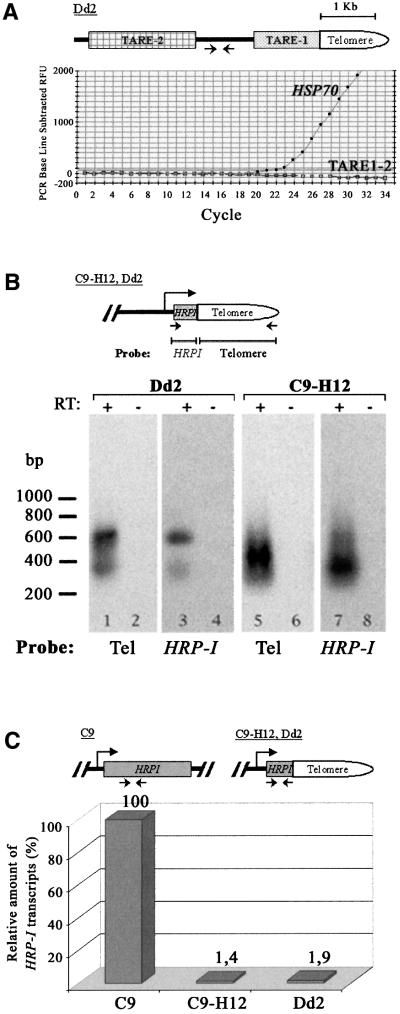
Fig. 4. Telomeric transcription status in intact versus truncated chromosomes. (A) Intact chromosomes ends are not transcribed. Real-time RT–PCR using a pair of oligonucleotides that match the non-repetitive region between TARE1 and TARE2 (see Materials and methods for sequences) of the Dd2 strain shows no amplification on hexamer-synthesized cDNA (open squares). However, a specific product was amplified using HSP70-specific oligonucleotides from the same cDNA (dots). (B) Truncated chromosomes are transcribed. Transcripts from genes that are positioned close to the telomeres can be detected by RT–PCR. Dd2 and C9-H12 cDNA was synthesized from blood-stage RNA using a telomere-specific oligonucleotide as primer for the reverse transcriptase enzyme reaction. Transcription at the breakage site was analysed by PCR using HRP-I gene-specific primers in combination with the telomere primer. Southern blot analysis of the RT–PCR products was performed using DNA probes specific to either HRP-I or the telomere, showing that transcription of truncated genes runs into telomeric repeats. (+) and (–) indicate the presence or absence of reverse transcriptase. (C) There is a decrease in the relative amount of transcripts when HRP-I is telomeric. cDNA from C9, C9-H12 and Dd2 strains was synthesized using an HRP-I-specific oligonucleotide. Quantitative real-time PCR of the HRP-I gene shows that when the gene is truncated (C9-H12 and Dd2), we can detect only 1–2% of the transcripts that exist in C9, where the gene is intact and >100 kb away from the telomere. RFU, reference fluorescence units; RT, reverse transcriptase.
The presence of longer telomeres on truncated chromosomes was confirmed in all additional cases examined (see Table I for summary). A chromosome breakage and healing event had been reported previously for chromosome 10 of C9-H12, resulting in a large subtelomeric deletion of ∼100 kb leaving a truncated Pf11-1 gene adjacent to telomere repeats (Scherf et al., 1992). Southern blot analysis of the chromosome breakpoint using the Pf11-1 gene as a probe revealed that the newly formed telomere is 2200 bp long (data not shown), while the mean telomere length for this strain is 1540 bp. One of the chromosome 1 arms (of the FCR3 strain), which has been described to have undergone chromosome breakage and healing at the RESA gene locus (Cappai et al., 1989), has a longer than average telomere (1800 bp compared with strain mean average length of 1150 bp; see Figure 2B). In the same strain, a truncation in chromosome 10 has also been reported (Scherf et al., 1992) and we again observed that one of the telomeres is longer (3600 bp). The longer telomere of chromosome 2 in the Dd2 strain is at a breakpoint also in the HRP-I gene (analysed in this work, data not shown). Thus, loss of the highly conserved TAS found at the telomere junction appears to lead to a local increase in telomere length. It appears that the sequences found in cis to the telomeric repeats are involved directly in the regulation of telomere length. Alternatively, the proximity of the transcription machinery to the telomeres in truncated chromosome ends might interfere with the telosome, a terminal non-nucleosomal structure described previously (Figueiredo et al., 2000) that is thought to be involved in regulating telomere length.
Table I. Length and transcription of telomeres at truncated chromosomes.
| Chromosome | Locus of breakage | Strain | Telomere length of broken arm | Mean telomere length for strain | Length ratio | Telomere transcription |
|---|---|---|---|---|---|---|
| 1 | RESA | FCR3 | 1800 (–152) | 1150 | 1.4 | – |
| 2 | HRP-I | Dd2 | 2400 (–289) | 1260 | 1.7 | + |
| 2 | HRP-I | C9-H12 | 4400 (–127) | 1540 | 2.8 | + |
| 10 | Pf11-1 | C9-H12 | 2200 (–86) | 1540 | 1.4 | – |
| 10 | ND | FCR3 | 3600 (ND) | 1150 | 3.1 | ND |
The values obtained from TRF analysis correspond to the size of the telomere repeat tract and a short fragment between the first telomere repeat and the restriction site. In intact chromosome ends, this non-telomeric fragment is very small (∼30 bp). In broken chromosome ends, the size of this fragment (shown in parentheses) was subtracted from telomere length obtained by TRF analysis in order to calculate the ratio between the telomere length of broken chromosomes and the average for the strain. Transcription through the telomeric tract next to the breakage site was detected by RT–PCR: + indicates presence of transcripts, – indicates absence of transcripts. ND, not determined.
Transcription of telomere repeats at a broken chromosome
To verify whether the proximity of genes close to the telomeres in truncated chromosomes has an effect on telomere length regulation, we first sought to demonstrate that intact chromosome ends are not transcribed and then examined the expression of truncated genes. In intact chromosome ends, a non-coding polymorphic region composed of several repetitive elements (TAREs 1–6) separates the telomeric repeats from the first genes (Figueiredo et al., 2000). We designed a pair of oligonucleotides to amplify a 200 bp unique sequence generally found adjacent to P.falciparum telomeres (500–2000 bp). Using real-time RT–PCR, no products were amplified from cDNA, while a control gene generated a specific product (Figure 4A). We were also unable to detect transcripts using specific PCR primers of the TARE2 or TARE1 and the telomere region. Taken together, these results strongly suggest that intact chromosome ends are not transcribed.
To analyse the expression of genes that have been placed close to the telomere by chromosome deletion, we used RT–PCR to synthesize cDNA from asexual ring stage (the early form of the 48 h blood-stage cycle) RNA from C9-H12 and Dd2 clones using a telomere oligonucleotide as the primer for the reverse transcriptase reaction. A specific oligonucleotide matching the HRP-I exon 1 (110 bp from the breakpoint in C9-H12 clone, 290 bp from the breakpoint in Dd2) was used for the PCR. PCR products were transferred to a nylon membrane and hybridized with a HRP-I-specific or a telomere-specific probe (Figure 4B). In both strains, amplification products were detected with a probe for both telomeres and HRP-I (lanes 1, 3, 5 and 7). Cloning and DNA sequence analysis of these products confirmed that they contained the truncated portion of the HRP-I gene and the adjacent telomeric repeats. No products were detected with any of the probes when RT–PCR was performed in the absence of reverse transcriptase (lanes 2, 4, 6, 8 and 10). Two other truncated chromosome ends were analysed: in chromosome 1 of the FCR3 clone, the breakage has removed the promoter of the RESA gene (Cappai et al., 1989); in chromosome 10, of the C9-H12 clone, the truncated gene, Pf11-1, is not normally transcribed in the blood stages (Scherf et al., 1992). In both cases, no transcription is expected to occur and indeed we were not able to amplify any transcripts from these truncated genes using RT–PCR (data not shown, but summarized in Table I).
To compare the relative expression of a gene when in its normal position in the chromosome and when it is telomeric, we quantified the relative transcription of HRP-I in C9, C9-H12 and Dd2 strains using real-time RT–PCR. When HRP-I is truncated and adjacent to the telomere (C9-H12 and Dd2), we detected only 1–2% of the transcripts compared with that of the same gene in the non-truncated chromosome (C9) (Figure 4C). In summary, no transcription was detected in intact chromosome ends, but telomeres can be transcribed if located next to an active promotor.
Distinct localization of truncated chromosomes at the nuclear periphery
Chromosome ends of P.falciparum form clusters of four to seven telomeres at the nuclear periphery (Freitas-Junior et al., 2000). We addressed the question of whether the telomere repeats are sufficient to anchor the chromosomes at the periphery and to form clusters. We compared the nuclear positioning of chromosomes that were either intact at both ends or truncated at one end. Chromosomes 2 and 10 of parasites derived from the same genetic background (C9 and C9-H12) were used for the analysis. Both chromosomes are intact in C9. In C9-H12 however, chromosome 2 is truncated within the HRP-I locus and chromosome 10 is truncated within the Pf11-1 locus. HRP-I is normally transcribed in asexual blood-stage parasites, while Pf11-1 is not (sexual-stage-specific gene) (Scherf et al., 1992). An example for chromosome 2 is shown in Figure 5. Two-colour fluorescence in situ hybridization (FISH) was performed on asexual blood-stage parasites of C9 and C9-H12 strains. Rep20 (or TARE6), a large telomere adjacent repetitive element, was used as a marker for the positioning of the clusters in the nucleus (Freitas-Junior et al., 2000). Hybridization with PFB0125c, a chromosome 2-specific marker located next to the HRP-I locus (Gardner et al., 1998) showed one fluorescent spot located at the periphery of the nucleus in both C9 and C9-H12 strains (Figure 5B, panels 1′–3′). Merging Rep20 and PFB0125c signals (panels 1′′–3′′), we observed that, while in C9 the two signals colocalize in most cells analysed (14% do not colocalize), in C9-H12 the two markers do not colocalize in 58% of the cells (Figure 5C). This result demonstrates that despite the presence of a telomere, localization outside telomere clusters is approximately four times higher for the truncated extremity of chromosome 2 (of C9-H12) than for its intact counterpart (of C9). The same tendency was observed in chromosome 10 using Pf11-1 as a specific subtelomeric chromosome marker: the truncated chromosome end (C9-H12) localizes outside telomere clusters 66% of the time, i.e. three times more frequently than in the intact chromosome (C9) (21%). Thus, lack of a conventional subtelomeric region results in a chromosome end that is found more frequently outside telomere clusters. The question arises of whether the truncated chromosomes are still located at the nuclear periphery.
Fig. 5. Truncated chromosome ends localize preferentially outside telomere clusters. (A) Diagram showing the location of the FISH probes in intact (C9) and truncated (C9-H12) chromosome 2. (B) FISH analysis in C9 and C9-H12 asexual stages. FISH was performed as described in Materials and methods, using Rep20 as marker for chromosome end clusters (in red, panels 1–3) and PFB0125c as a single copy gene marker present in both intact and truncated forms of chromosome 2 (in green, panels 1′–3′). DNA is counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (in blue). (C) Summary of the data obtained by TRF analysis and FISH for chromosomes 2 and 10 of C9 and C9-H12 strains. The number of nuclei analysed is given in parentheses.
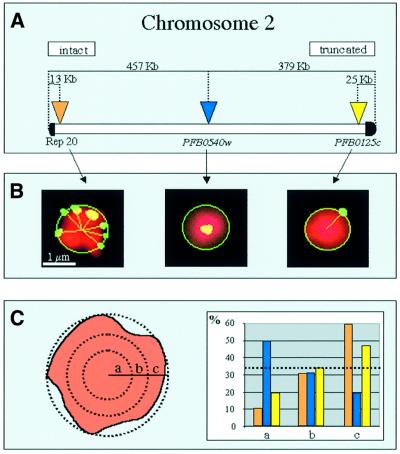
To address this question, we used a computer program to quantify the distance of telomeric probes from the centre of the nucleus and compared these results with the distance measured using a chromosome internal probe. Figure 6A shows the probes used for the FISH analysis, which are derived from P.falciparum chromosome 2, whose complete sequence has been determined recently (Gardner et al., 1998). The results presented in Figure 6B and C show that the distribution of these probes in the nucleus is non-random. The truncated end of the chromosome, represented by the probe PFB0125c, is located in the periphery of the nucleus and has a similar hybridization pattern to the subtelomeric probe, Rep20. In contrast, the chromosome internal probe (PFB0540w) was located in the most central zone of the nucleus. A χ2 analysis performed on the distributions proved that they are significantly different (p <0.01) from each other.
Fig. 6. Intact and truncated chromosome ends are preferentially located at the nuclear periphery. (A) The three probes, Rep 20, PFB0540w and PFB0125c, used to analyse P.falciparum chromosome 2 from the C9 H12 strain, are marked by arrows and the distance from each chromosome extremity is indicated. The truncated and intact extremities are also indicated. (B) FISH analysis was performed using each of the probes indicated in (A) individually. A computer graphic representation of the 2D FISH image is shown. A computer method was used to measure the distance of these signals from the centre of the nucleus. (C) The distance from the periphery of the nucleus was measured by dividing the nucleus into three zones, a = internal, b = intermediate and c = peripheral, as described in Materials and methods. The histogram on the right represents the percentage of signals found in each zone. The dashed line represents the theoretical values of a signal randomly distributed in the three zones mentioned above. The subtelomeric probe Rep20 is represented in orange, the internal probe PFB0540w is represented in blue and the subtelomeric probe from the truncated end of the chromosome is represented in yellow.
Discussion
Chromosome-specific, cis-acting DNA elements can overrule regulation of telomere length
In this work we show that there are large inter- and intra-chromosomal length differences within the 28 telomeres of P.falciparum. Moreover, the same chromosome end that harbours a telomere of average size in one strain can be up to 3-fold longer in another. Our investigations pointed to spontaneously broken chromosomes, which have been healed by the addition of telomere repeats, as a primary event leading to longer telomeres. Once acquired, longer telomeres are stable within the population, resulting in a strain-specific TFP. However, each of the de novo formed telomeres displays a characteristic size. In the C9-H12 strain, for instance, the truncated arm of chromosome 2 has a 4400 bp telomere, while the chromosome 10 truncated arm harbours a smaller telomere of 2200 bp. We postulate that the sequence heterogeneity found in the DNA adjacent to the telomere accounts for the differences seen in telomere length at truncated chromosome ends (see Table I). This could also explain the telomere heterogeneity that has been described in human and murine cells (Zijlmans et al., 1997; Martens et al., 1998), as well as in the human protozoan pathogen Trypanosoma cruzi (Freitas-Junior et al., 1999). How the genomic environment adjacent to telomeres is recognized by the machinery that regulates the telomere length remains unknown. One possibility is that specific proteins bind to the TAS and strengthen a telomere fold-back structure, in which the telomere end is protected. When the TAS is deleted, an unusual environment adjacent to the telomere might disturb the folding back and interfere with the machinery that controls telomere length.
Our findings indicate that the presence of conserved TASs on P.falciparum chromosomes helps to establish a specific and homogeneous telomere length. Consequently, the variation in the mean length observed between different Plasmodium species (960–6700 bp; see Figure 1) may result from the different TAS composition presented by each Plasmodium species. This idea is supported by studies in yeast, which have shown that the telomere–non-telomere junction is important for the regulation of telomere length (Craven and Petes, 1999; Ray and Runge, 1999).
Transcription through telomere repeats at long repeat tracts
It was not possible to amplify specific transcripts from intact chromosome ends using pairs of oligonucleotides derived from the telomeres and adjacent sequences using either classical or real-time RT–PCR, demonstrating that intact chromosome ends are not transcribed. We tested the implications of the proximity of actively transcribed genes to a telomere by analysing the chromosome 2 HRP-I truncation in two genetically distinct strains of P.falciparum: Dd2 and C9-H12. Using RT–PCR, we were able to detect transcripts corresponding to the truncated HRP-I gene during blood stages in the two different strains. These transcripts also contained telomeric repeats, demonstrating that transcription continues into the telomere. Transcription of such truncated chromosomes might explain the signal obtained by Rudenko and Van der Ploeg (1989) when it was shown by nuclear run-on that telomere transcription occurs in P.falciparum and that it is α-amanitin sensitive. It has been shown in yeast that telomeric transcription results in a shortening of the telomere (Sandell et al., 1994); however, in this study we observed that all de novo synthesized telomeres of truncated chromosomes display longer telomeres than intact chromosome ends, whether or not the truncated gene is transcribed. However, a modest increase in length exists for those truncated chromosomes that are transcribed.
Quantitative real-time PCR experiments revealed ∼70 times less HRP-I transcripts when the gene is truncated and next to the telomere (in C9-H12 and Dd2) than when it is intact and internal (in C9). Such a difference may reflect the instability of the HRP-I transcript in C9-H12 and Dd2 due to the lack of a 3′ untranslated region in the mRNA, but it is possible that a telomere repression effect also accounts for this low transcript level. Telomere silencing has been reported in yeast (Gottschling et al., 1990) and human cells (Baur et al., 2001), for genes that are artificially positioned upstream of telomeres. Disrupting the tethering of telomeres in yeast derepresses telomeric silencing (Cockell and Gasser, 1999), indicating that the compartmentalization of telomeres at the nuclear periphery plays a role in gene regulation. It is conceivable that P.falciparum telomeres can also exist in a state that allows transcription and in a state that leads to repression of gene activity. Furthermore, it has been shown in another malaria species, Plasmodium berghei, that the stable insertion of a selectable drug marker adjacent to telomere sequences can be transcribed from the proximity of telomeres (van Dijk et al., 1996; Pace et al., 2000).
Implication of telomere-associated regions in cluster organization of chromosome ends
Clustering of P.falciparum telomeres at the periphery of the nucleus has been demonstrated recently in sexual and asexual stages of the parasitic life cycle. The conserved organization of the subtelomeric region is thought to be important for the physical alignment of heterologous chromosome ends (Freitas-Junior et al., 2000). Telomere clusters probably account for the high level of genetic diversity observed in these chromosome compartments and generate a boundary for recombination between var genes found at telomeric and internal locations (Rubio et al., 1996; Freitas-Junior et al., 2000). Here, we have presented evidence that TASs are indeed important for the nuclear architecture by forming the genetic elements that mediate the clustering of chromosome ends. Analysis of two different chromosomes that have lost their entire TAS revealed that the truncated chromosome ends are frequently dissociated from a cluster (58 and 66%, respectively, for chromosomes 2 and 10). However, the same chromosome from the parental strain in which the subtelomeric elements are intact is found within clusters (only 14 and 21%, respectively, are not). No telomeric transcription is detected in the truncated chromosome 10, indicating that delocalization from clusters is due to the lack of the subtelomeric region rather than a result of telomeric transcription. A recent study has shown that a plasmid containing TARE6 (or Rep20), the major repetitive element on the subtelomeric region, colocalizes with telomere clusters in transfected P.falciparum (R.A.O’Donnell and B.Crabb, personal communication). Together, these results allow us to conclude that TAS is necessary and sufficient for telomere clustering.
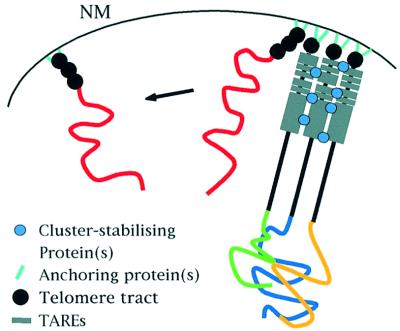
Both intact and truncated chromosome ends localize at the perinuclear space, suggesting that the subtelomeric region is not required for anchoring chromosome ends to the nuclear periphery. We propose a model for the spatial organization of P.falciparum chromosome ends in which the telomeres and the conserved TASs have a distinct role in the chromosome architecture (Figure 7). The model predicts that the subtelomeric regions are involved in the formation and stability of telomere clusters. This could be mediated by specific TAS-binding proteins, ‘cluster-stabilizing proteins’, that would cross-link the different ends. Telomere repeat-specific proteins may mediate localization at the periphery, as has been shown in yeast, where telomeres are tethered to the nuclear membrane by interactions with a number of proteins (Laroche et al., 1998; Galy et al., 2000). At this point, we cannot rule out the possibility that the organization of the cluster contains inherent properties necessary for telomere length regulation. Consequently, the dissociation of a telomere from a cluster might also cause the observed increase in telomere length.
Fig. 7. Model for the spatial organization of chromosome ends in the nucleus of P.falciparum is shown. The model predicts a distinct role for telomeres and TASs in the nuclear architecture. We hypothesize that perinuclear localization of chromosome ends is mediated by putative ‘anchoring proteins’, which bind to the telomere and anchor it to nuclear membrane structural elements. As lack of TASs frequently delocalizes truncated chromosomes from clusters, we propose that TASs play an important role in the physical clustering of chromosome ends perhaps through interactions with specific cluster-stabilizing proteins, which could stabilize the association of chromosome ends. NM, nuclear membrane.
In P.falciparum blood-stage parasites, the resolution of FISH is ∼10 kb, meaning that two individual signals are visualized for probes with a 10 kb distance from each other (data not shown). It is therefore surprising that a probe localized ∼100 kb from one of the extremities of chromosome 2 colocalizes with telomere clusters. Nevertheless, similar results have been observed earlier in FISH studies of three different chromosomes ends (10, 11 and 13; Freitas-Junior et al., 2000) using chromosome-specific probes ∼100 kb from the closest Rep20 element. In contrast, a combination of two internal probes from chromosome 2 separated by 150 kb gave an almost negligible colocalization index (8.7%, background presumably due to random colocalization). To explain such results, we hypothesize that P.falciparum subtelomeric chromosome regions are more compact than chromosome internal regions. This could be explained by the telomere folding back over the subtelomeric regions, as has been observed in yeast (de Bruin et al., 2001). A large number of homologues to yeast proteins involved in nuclear architecture of the telomere, such as Sir proteins, the Ku and the Mlp complexes, have been found in the P.falciparum genome database (Scherf et al., 2001), suggesting that yeast can serve as a model to study the telomere biology of this parasite.
In conclusion, our observations provide an original concept giving new insights into the complex processes of control of telomere length and nuclear architecture. Moreover, this work opens new avenues into the understanding of the recombination processes that occur between heterologous chromosomes, as well as the epigenetic processes that control the transcription of subtelomeric gene families involved in antigenic variation in malaria.
Materials and methods
Strains and culture conditions
Plasmodium falciparum intra-erythrocytic stages were cultivated as described (Trager and Jensen, 1976). Genetically distinct strains from different geographical areas used in this study were: 3D7 (Walliker et al., 1987), FCR3 (Scherf et al., 1988), Dd2 and 3B-D5 (Wellems et al., 1990), C9 and C9-H12 (Scherf et al., 1992), Banjul and Sbd27 (Kahane et al., 1987). C9-H12 is a subclone from the C9 strain generated by selection for a knob– phenotype (generally associated with a spontaneous HRP-I deletion in chromosome 2). Genomic DNA from P.cynomolgi and P.vivax was kindly provided by P.H.David and P.yoelii and P.chabaudi by D.Mattei.
TRF analysis
Genomic DNA (500 ng) was digested overnight with 5 U of four frequently cutting enzymes (AluI, DdeI, MboII and RsaI), separated on a 1% agarose gel and subjected to Southern blot analysis. The membrane was hybridized with an α-32P-labelled DNA probe specific for Plasmodium telomeres (Bottius et al., 1998) and signals were quantified by PhosphorImager (Molecular Dynamics). Peaks were used to calculate the mean telomere length.
Telomere fingerprint pattern
A TFP was obtained by 2D-PFGE analysis. Individual chromosomes were fractionated in the first dimension using a Pulsaphor/Gene Navigator PFGE apparatus (Pharmacia), subsequently digested with AluI, DdeI, MboII, RsaI and separated in the second dimension as described (Hernandez-Rivas and Scherf, 1997). Transfer and hybridization with a telomere-specific probe were as described below.
Southern blotting
DNA products resolved on agarose gels were transferred to Hybond N+ membrane (Amersham) as recommended by the manufacturer. Probes were labelled with [α-32P]dATP by random priming. HRP-I and Pf11-1 probes were as described previously (Scherf and Mattei, 1992; Scherf et al., 1992). The telomere-specific probe consisted of 20 P.falciparum telomere repeats (Bottius et al., 1998). The Rep20 probe was provided by G.Langsley (Patarapotikul and Langsley, 1988). The probe for the PFB0125c gene (Gardner et al., 1998) was obtained by PCR amplification (5′-GGAGAAAAATGCAGATGTGG-3′ and 5′-GCCTCATATAAGGTTTCATATC-3′) and cloning into pCR2.1 vector. Blots were washed twice for 20 min in 2× SSC 0.1% SDS, and once for 30 min in 0.2× SSC 0.1% SDS, at 65°C.
RT–PCR
Ring-stage total RNA was isolated from infected erythrocytes using the TRIzol method (as described in Kyes et al. (2000). One microgram of total RNA was used for the first-strand cDNA synthesis. A telomere antisense oligonucleotide [5′-GGCGCG (T G/A AACCC)3-3′] was used as a specific primer to initiate the first strand cDNA synthesis using the M-MLVL Superscript II reverse transcriptase. The reaction was performed as suggested by the supplier (Gibco-BRL). PCR was carried out for 35 cycles as follows: 95°C for 10 s, 50°C for 30 s, 72°C for 1 min, followed by a final extension period of 5 min at 72°C. The primers used were the antisense telomere oligonucleotide and the specific primer 5′-CCGGGATCCATGAAAAGTTTTAAGAACAA-3′ for the HRP-I gene from chromosome 2. RT–PCR products were separated on a 2% agarose gel and analysed by Southern blot. The products were purified from agarose (Qiagen Gel Extraction Kit), cloned in pCR2.1 vector (TOPO TA cloning kit; Invitrogen) and sequenced.
For quantitative real-time PCR amplified products were designed to be <200 bp in size for optimal RT–PCR measurements. For the HRP-I gene, the 5′-TTATTAGAGCACTTCAAAACCC oligonucleotide was used to synthesize the cDNA and then paired with oligonucleotide 5′-ATACTTTGAGGAGAAAGAAGGC to PCR amplify exon 1 of the HRP-I gene. In order to amplify putative transcripts close to the telomeres of intact chromosome ends, cDNA was synthesized from random hexamers and two oligonucleotides that match the non-repetitive region between TARE1 and TARE2 were used. The TARE1–2A primer is 5′-AGAAAACATAACATCATAAGTCC-3′ and the TARE1–2B primer is 5′-TAATAAAGTACTCGGTTG GGC-3′. The P.falciparum calmodulin and HSP70 housekeeping genes were used as standard controls. PCR was performed on an iCycler apparatus (Bio-Rad) using the SYBR green detection system (Perkin-Elmer). In three independent experiments (in which each reaction was carried out in triplicate), the following PCR programme was used: 10 min at 95°C; 35 cycles of 15 s at 95°C/30 s at 55°C/30 s at 60°C. The iCycler apparatus measured the fluorescence of each sample in every cycle during the elongation step.
FISH
FISH analysis of asexual blood-stage parasites (trophozoites) was performed as described (Freitas-Junior et al., 2000). The FISH data presented here are two-dimensional images of three-dimensional parasite nuclei. Thus, accidental overlap of the hybridization signals cannot be ruled out. In these experiments colocalization is defined as total or partial overlap of the hybridization signal.
Image analysis
The spatial localization of telomeres in the nuclei was assessed by a computerized method that automatically counts the number of telomeres (Olivo-Marin, 2002) and measures their distance from the centre of the nucleus, using a representation corresponding to an orthogonal projection of the three-dimensional sphere into a two-dimensional circle. The program computes the normalized distance Δi = di/R of each telomere i to the centre of its corresponding nucleus, where the radius of nucleus R is determined by thresholding the DAPI image and d is the distance between the probe and centre of the nucleus. The values obtained for Δ were accumulated from >100 cells and grouped into three concentric circles of radii 0.487 (internal zone a), 0.72 (intermediate zone b) and 1 (peripheral zone c). Signals found outside the DAPI staining, but in close vicinity (0.2 of radial distance) were considered to be part of the peripheral zone. These radial values define the partition of a unitary sphere into three zones of equal volume (33.3% each). Statistical significance was assessed by a χ2 test, confirming that the distribution for each probe is significantly non-random (p <0.05 for the three probes).
Acknowledgments
Acknowledgements
We thank U.Nehrbass, L.Pirrit and J.Lowell for valuable comments and discussion, R.Tahar for advice on quantititative real-time PCR and Christophe Zimmer for help with imaging analysis. This work was supported by an EC grant (QLK-CT-2000-00109 and CT98-0362) and by the grants from Fundação para a Ciência e a Tecnologia, PRAXIS XXI/BD/16020/98, Portugal (L.M.F.) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (L.H.F.J.). Chromosome 2 was sequenced by a consortium composed of The Institute for Genome Research, along with the Naval Medical Research Center (USA), with support from NIAID/NIH, the Burroughs Welcome Fund and the Department of Defence. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania (USA) and Monash University (Melbourne, Australia), supported by the Burroughs Welcome Fund.
References
- Baumann P. and Cech,T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Baur J.A., Zou,Y., Shay,J.W. and Wright,W.E. (2001) Telomere position effect in human cells. Science, 292, 2075–2077. [DOI] [PubMed] [Google Scholar]
- Berman J., Tachibana,C.Y. and Tye,B.K. (1986) Identification of a telomere-binding activity from yeast. Proc. Natl Acad. Sci. USA, 83, 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A., Michels,P.A., Lincke,C.R. and Borst,P. (1983) Growth of chromosome ends in multiplying trypanosomes. Nature, 303, 592–597. [DOI] [PubMed] [Google Scholar]
- Bilaud T., Koering,C.E., Binet-Brasselet,E., Ancelin,K., Pollice,A., Gasser,S.M. and Gilson,E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottius E., Bakhsis,N. and Scherf,A. (1998) Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol., 18, 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Cappai R., van Schravendijk,M.R., Anders,R.F., Peterson,M.G., Thomas,L.M., Cowman,A.F. and Kemp,D.J. (1989) Expression of the RESA gene in Plasmodium falciparum isolate FCR3 is prevented by a subtelomeric deletion. Mol. Cell. Biol., 9, 3584–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L., van Steensel,B., Broccoli,D., Erdjument-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cockell M. and Gasser,S.M. (1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev., 9, 199–205. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- Craig A. and Scherf,A. (2001) Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol. Biochem. Parasitol., 115, 129–143. [DOI] [PubMed] [Google Scholar]
- Craven R.J. and Petes,T.D. (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics, 152, 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Zaman,Z., Liberatore,R.A. and Ptashne,M. (2001) Telomere looping permits gene activation by a downstream UAS in yeast. Nature, 409, 109–113. [DOI] [PubMed] [Google Scholar]
- de Lange T. (1995) Teleomere dynamics and genome instability in human cancer. In Blackburn,E. and Greider,C. (eds), Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- de Lange T., Shiue,L., Myers,R.M., Cox,D.R., Naylor,S.L., Killery,A.M. and Varmus,H.E. (1990) Structure and variability of human chromosome ends. Mol. Cell. Biol., 10, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo L.M., Pirrit,L.A. and Scherf,A. (2000) Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol., 106, 169–174. [DOI] [PubMed] [Google Scholar]
- Fourel G., Revardel,E., Koering,C.E. and Gilson,E. (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J., 18, 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Porto,R.M., Pirrit,L.A., Schenkman,S. and Scherf,A. (1999) Identification of the telomere in Trypanosoma cruzi reveals highly heterogeneous telomere lengths in different parasite strains. Nucleic Acids Res., 27, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Bottius,E., Pirrit,L.A., Deitsch,K.W., Scheidig,C., Guinet,F., Nehrbass,U., Wellems,T.E. and Scherf,A. (2000) Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P.falciparum. Nature, 407, 1018–1022. [DOI] [PubMed] [Google Scholar]
- Froelich-Ammon S.J., Dickinson,B.A., Bevilacqua,J.M., Schultz,S.C. and Cech,T.R. (1998) Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev., 12, 1504–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin,J.C., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Gardner M.J. et al. (1998) Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science, 282, 1126–1132. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Position effect at S.cerevisiae telomeres: reversible repression of Pol II transcription. Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Hastie N.D., Dempster,M., Dunlop,M.G., Thompson,A.M., Green,D.K. and Allshire,R.C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature, 346, 866–868. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rivas R. and Scherf,A. (1997) Separation and mapping of chromosomes of parasitic protozoa. Mem. Inst. Oswaldo Cruz, 92, 815–819. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rivas R., Mattei,D., Sterkers,Y., Peterson,D.S., Wellems,T.E. and Scherf,A. (1997) Expressed var genes are found in Plasmodium falciparum subtelomeric regions. Mol. Cell. Biol., 17, 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahane B., Sibilli,L., Scherf,A., Jaureguiberry,G., Langsley,G., Ozaki,L.S., Guillotte,M., Muller-Hill,B., Pereira da Silva,L. and Mercereau-Puijalon,O. (1987) The polymorphic 11.1 locus of Plasmodium falciparum. Mol. Biochem. Parasitol., 26, 77–85. [DOI] [PubMed] [Google Scholar]
- Kyes S., Pinches,R. and Newbold,C. (2000) A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol., 105, 311–315. [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin,S.G., Gotta,M., Gorham,H.C., Pryde,F.E., Louis,E.J. and Gasser,S.M. (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol., 8, 653–656. [DOI] [PubMed] [Google Scholar]
- Larson G.P., Castanotto,D., Rossi,J.J. and Malafa,M.P. (1994) Isolation and functional analysis of a Kluyveromyces lactis RAP1 homologue. Gene, 150, 35–41. [DOI] [PubMed] [Google Scholar]
- Martens U.M., Zijlmans,J.M., Poon,S.S., Dragowska,W., Yui,J., Chavez,E.A., Ward,R.K. and Lansdorp,P.M. (1998) Short telomeres on human chromosome 17p. Nature Genet., 18, 76–80. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Cross,G.A.M., de Lange,T. and Griffith,J.D. (2001) T-loops at trypanosome telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti K.G. and Prescott,D.M. (1999) Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl Acad. Sci. USA, 96, 14436–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- Olivo-Marin J.-C. (2002) Extraction of spots in biological images using multiscale products. Pattern Recogn., in press. [Google Scholar]
- Pace T., Scotti,R., Janse,C.J., Waters,A.P., Birago,C. and Ponzi,M. (2000) Targeted terminal deletions as a tool for functional genomics studies in Plasmodium. Genome Res., 10, 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarapotikul J. and Langsley,G. (1988) Chromosome size polymorphism in Plasmodium falciparum can involve deletions of the subtelomeric pPFrep20 sequence. Nucleic Acids Res., 16, 4331–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E., Laurent,M., Delinte,K., Van Meirvenne,N. and Steinert,M. (1983) Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res., 11, 8137–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzi M., Pace,T., Dore,E., and Frontali,C. (1985) Identification of a telomeric DNA sequence in Plasmodium berghei. EMBO J., 4, 2991–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F.E. and Louis,E.J. (1997) Saccharomyces cerevisiae telomeres. A review. Biochemistry, 62, 1232–1241. [PubMed] [Google Scholar]
- Pryde F.E. and Louis,E.J. (1999) Limitations of silencing at native yeast telomeres. EMBO J., 18, 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A. and Runge,K.W. (1999) The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol., 19, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio J.P., Thompson,J.K. and Cowman,A.F. (1996) The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J., 15, 4069–4077. [PMC free article] [PubMed] [Google Scholar]
- Rudenko G. and Van der Ploeg,L.H. (1989) Transcription of telomere repeats in protozoa. EMBO J., 8, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L., Gottschling,D.E. and Zakian,V.A. (1994) Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl Acad. Sci. USA, 91, 12061–12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A. and Mattei,D. (1992) Cloning and characterization of chromosome breakpoints of Plasmodium falciparum: breakage and new telomere formation occurs frequently and randomly in subtelomeric genes. Nucleic Acids Res., 20, 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Hilbich,C., Sieg,K., Mattei,D., Mercereau-Puijalon,O. and Muller-Hill,B. (1988) The 11-1 gene of Plasmodium falciparum codes for distinct fast evolving repeats. EMBO J., 7, 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Carter,R., Petersen,C., Alano,P., Nelson,R., Aikawa,M., Mattei,D., Pereira da Silva,L. and Leech,J. (1992) Gene inactivation of Pf11-1 of Plasmodium falciparum by chromosome breakage and healing: identification of a gametocyte-specific protein with a potential role in gametogenesis. EMBO J., 11, 2293–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Figueiredo,L.M. and Freitas-Junior,L.H. (2001) Plasmodium telomeres: a pathogen’s perspective. Curr. Opin. Microbiol., 4, 409–414. [DOI] [PubMed] [Google Scholar]
- Shore D. and Nasmyth,K. (1987) Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell, 51, 721–732. [DOI] [PubMed] [Google Scholar]
- Trager W. and Jensen,J.B. (1976) Human malaria parasites in continuous culture. Science, 193, 673–675. [DOI] [PubMed] [Google Scholar]
- van Dijk M.R., Janse,C.J. and Waters,A.P. (1996) Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science, 271, 662–665. [DOI] [PubMed] [Google Scholar]
- Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- Walliker D., Quakyi,I.A., Wellems,T.E., McCutchan,T.F., Szarfman,A., London,W.T., Corcoran,L.M., Burkot,T.R. and Carter,R. (1987) Genetic analysis of the human malaria parasite Plasmodium falciparum. Science, 236, 1661–1666. [DOI] [PubMed] [Google Scholar]
- Wellems T.E., Panton,L.J., Gluzman,I.Y., do Rosario,V.E., Gwadz, R.W., Walker-Jonah,A. and Krogstad,D.J. (1990) Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature, 345, 253–255. [DOI] [PubMed] [Google Scholar]
- Wilkie A.O., Higgs,D.R., Rack,K.A., Buckle,V.J., Spurr,N.K., Fischel-Ghodsian,N., Ceccherini,I., Brown,W.R. and Harris,P.C. (1991) Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell, 64, 595–606. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens,U.M., Poon,S.S., Raap,A.K., Tanke,H.J., Ward,R.K. and Lansdorp,P.M. (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA, 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]