Abstract
A naturally arising point mutation in the env gene of HIV-1 activates the aberrant inclusion of the cryptic exon 6D into most viral messages, leading to inefficient viral replication. We set out to understand how a single nucleotide substitution could cause such a dramatic change in splicing. We have determined that the exon 6D mutation promotes binding of the SR protein SC35 to the exon. Mutant exon 6D sequences function as a splicing enhancer when inserted into an enhancer-dependent splicing construct. hnRNP H family proteins bind to the enhancer as well; their binding is dependent on the sequence GGGA located just downstream of the point mutation and depletion– reconstitution studies show that hnRNP H is essential for enhancer activity. A polypurine sequence located further downstream in exon 6D binds SR proteins but acts as an exonic splicing silencer. hnRNP H is required for interaction of U1 snRNP with the enhancer, independent of the point mutation. We propose that SC35 binding to the point mutation region may convert the hnRNP H–U1 snRNP complex into a splicing enhancer.
Keywords: HIV-1/hnRNP H/pre-mRNA splicing/SR proteins
Introduction
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus that is dependent on alternative splicing to produce messenger RNAs encoding the various viral proteins. During viral infection, >40 messages are derived by alternative splicing from a single pre-mRNA (Schwartz et al., 1990; Purcell and Martin, 1993). In addition, approximately half of the RNA transcripts remain unspliced and are used as viral genomes and as mRNAs for the gag and pol genes. Alteration in the complex splicing pattern generating the viral mRNAs can dramatically affect HIV-1 infectivity and pathogenesis (Gottlinger et al., 1992; Purcell and Martin, 1993; Wentz et al., 1997).
Some HIV-1 isolates have the ability to include the cryptic exon 6D, located in the env coding region, in spliced messages (Benko et al., 1990; Salfeld et al., 1990). Comparisons of the sequences of HIV variants reveal that exon 6D is conserved in the IIIB family, while other isolates lack one or both 6D splice sites (Gottlinger et al., 1992). Inclusion of exon 6D in the viral mRNA results in the production of the chimeric protein Tev (Tat–Env–Rev fusion protein) and p186Drev (Rev-related protein). Tev has wild-type Tat transactivation activity and low but detectable Rev activity, while p186Drev has no known functional activity (Benko et al., 1990; Salfeld et al., 1990). Exon 6D 5′ and 3′ splice site sequences may also play a role in partially spliced message stability through interactions with the rev-dependent mRNA transport mechanism (Lu et al., 1990; Hammarskjold et al., 1991).
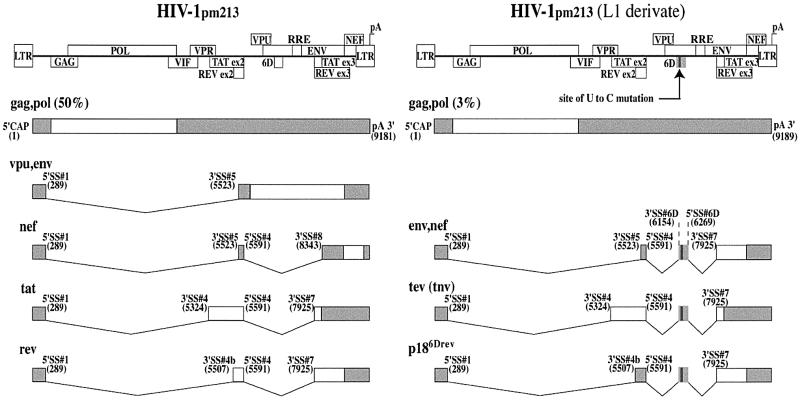
In a previous study, Wentz et al. (1997) identified a mutant HIV-1 isolate (HIV-1pm213 clone L1) related to the HXB2 strain. HIV-1pm213 clone L1 is characterized by an abnormal predominance of mRNAs containing exon 6D. The amount of unspliced mRNAs is dramatically reduced and so are the spliced mRNAs coding for Tat, Rev, Env, Vpu and Nef. This abnormal splicing pattern and altered gene expression results in a dramatic decrease in viral replication. A point mutation converting a U residue to a C residue (U-to-C) within exon 6D is responsible for the loss of balanced splicing and the predominance of exon 6D-containing mRNAs (summarized in Figure 1). This point mutation activates exon inclusion in vivo when mutant but not wild-type exon 6D is inserted between exons 4 and 6 of a cardiac troponin T (cTNT)-derived minigene. The point mutation was proposed to disrupt an exonic splicing silencer (ESS) within the 6D sequence (Wentz et al., 1997). An additional sequence in exon 6D, downstream of the putative ESS, resembles a purine-rich exonic splicing enhancer (ESE). An experimentally induced point mutation in this polypurine element interfered with exon 6D inclusion in the three exon heterologous minigene (Wentz et al., 1997). However, this same mutation of the polypurine sequence in a proviral construct increased the ratio of spliced to unspliced viral messages, indicating that the polypurine run may be acting as an ESS in the viral context (Wentz et al., 1997). The juxtaposition of these two regulatory sequences is similar to bipartite splicing regulatory elements observed in other alternatively spliced genes (Caputi et al., 1994).
Fig. 1. Schematic representation of the splicing patterns of wild-type and U-to-C mutant HIV-1pm213 pre-mRNAs. The structure of the HIV-1 genome is indicated, along with the location of the Rev response element (RRE). Coding exons are represented by open boxes. The main splice products for the wild-type (HIV-1pm213) and mutant (HIV-1pm213, L1 derivate) pre-mRNAs are indicated, together with the 5′ and 3′ splice sites and their positions in the viral transcript. The relative percentage of total HIV-1 messages representing the unspliced gag–pol messages are indicated for both viral clones (Wentz et al., 1997). Shaded boxes represent untranslated regions present in the final messages. Figure based on Wentz et al. (1997).
We set out to learn how a single base substitution in a very rarely used exon could lead to such dramatic changes in pre-mRNA splicing. To do this, we searched for protein factors from HeLa cell nuclear extract that can assemble onto exon 6D using RNA affinity chromatography. HeLa cells represent a relevant system for the study of HIV-1 splicing regulation since the splicing pattern of the viral pre-mRNA in HeLa and human T cells is almost identical (Purcell and Martin, 1993). We have found that the point mutation of exon 6D increases the functional interaction of SR protein splicing factors, predominantly SC35, with the viral substrate. This point mutation also increases the binding of hnRNP H family members to the substrate. In a heterologous splicing enhancer-dependent construct derived from the Drosophila double-sex gene, the point mutation in exon 6D serves as a splicing enhancer in in vitro splicing assays. Mutation of three consecutive G residues downstream of the point mutation interferes with hnRNP H family binding to the substrate RNA and we demonstrate that hnRNP H protein is essential for the splicing enhancer activity of the mutant exon. Our data indicate that the point mutation in exon 6D activates a splicing enhancer whose function is dependent on both SR proteins and hnRNP H activity.
Results
The point mutation in exon 6D enhances SR protein binding to the exon
In order to study how the point mutation in exon 6D increased HIV-1 splicing, we tested whether the point mutation activated or interfered with the binding of trans-acting splicing factors to the exon. We first examined the binding of the SR protein family of pre-mRNA splicing factors to this region of exon 6D. Purine-rich sequences have been shown previously to serve as binding sites for SR proteins (Lavigueur et al., 1993; Dirksen et al., 1994; Ramchatesingh et al., 1995), and we tested whether the exon 6D polypurine sequence helped to recruit SR proteins to exon 6D. We also tested whether the U-to-C or other mutations at this position such as U-to-G and U-to-A affected splicing factor recruitment.
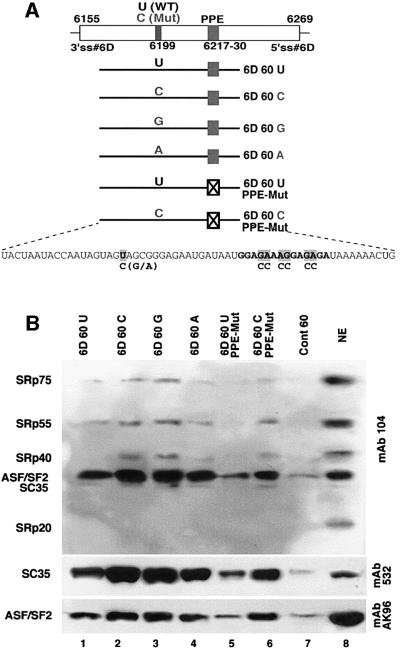
The binding of proteins from HeLa cell nuclear extract to 60 nucleotide RNAs containing the wild-type exon 6D sequence or the U-to-C, U-to-G and U-to-A mutant exon 6D sequences was assayed. A mutation in the polypurine sequence, and a double mutation of both the U-to-C and polypurine sequences were also tested (Figure 2A). A control 60mer substrate RNA containing no known splicing factor binding sites was also tested (Caputi et al., 1999). These RNAs were covalently linked to agarose beads and the RNA-linked beads were incubated in HeLa cell nuclear extract and then washed extensively. Proteins remaining bound to the beads were eluted in SDS-containing buffer and loaded onto SDS–PAGE gels. The gels were transferred to nitrocellulose and probed with mAb104, which recognizes all the members of the SR protein family (Roth et al., 1991), and with monoclonal antibodies specific for SC35 and ASF/SF2. A control lane containing 10 µl of total nuclear extract is also included in these immunoblot experiments. Based on calculations described previously, the amount of nuclear extract loaded in the control lane is roughly a quarter of the nuclear extract from which the eluted proteins were derived in each experiment (Caputi et al., 1999; Caputi and Zahler, 2001).
Fig. 2. SR protein assembly onto exon 6D sequences. (A) Graphic representation of exon 6D and the location of the substrates used in the RNA affinity chromatography assay. The sequence of the wild-type substrate RNA used in the experiment is indicated together with PPE and U-to-C, U-to-A and U-to-G mutations. (B) SR protein binding to the exon 6D substrates. Exon 6D and control RNA substrates were covalently linked to agarose beads and incubated in HeLa cell nuclear extracts. Proteins bound to the substrates were eluted and immunoblotted with antibodies specific for all SR proteins (mAb 104) or antibodies specific for the SR proteins SC35 (mAb 532) and SF2/ASF (mAb AK-96). Lanes 1–7 contain the proteins eluted from beads covalently linked to the substrate RNAs indicated. Lane 8 contains 10 µl of HeLa cell nuclear extract.
The results of the RNA affinity chromatography assay indicate that SR proteins and SC35 in particular assemble on exon 6D. The U-to-A mutation and more dramatically the U-to-C and U-to-G mutations increase binding of SR proteins to the exon (Figure 2B, lanes 1–4). This suggests that the point mutation leads to an increased binding of SR proteins to the exon 6D sequence, consistent with the role of this mutation in enhancing splicing in vivo (Wentz et al., 1997). Mutation of the polypurine sequence decreases SR protein recruitment to the substrate (Figure 2B, lane 5). The substrate RNA containing both the U-to-C mutation and the mutation in the polypurine element binds SR proteins much more strongly than the polypurine mutant alone (Figure 2B, lanes 5 and 6). This result indicates that the U-to-C mutation activates SR protein assembly onto exon 6D, which is more easily seen when the polypurine element binding site for SR proteins is eliminated from the substrate RNA.
Mutant exon 6D sequences act as SR protein-dependent splicing enhancers in an enhancer-dependent splicing substrate
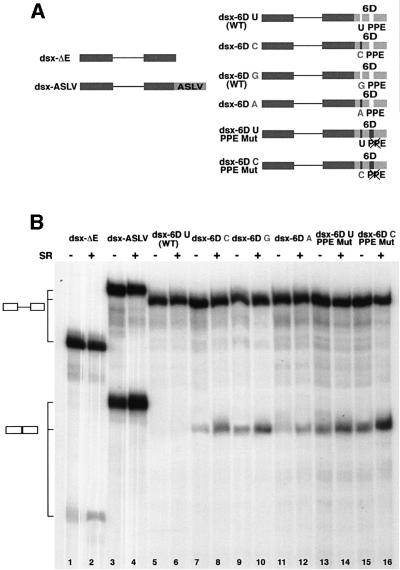
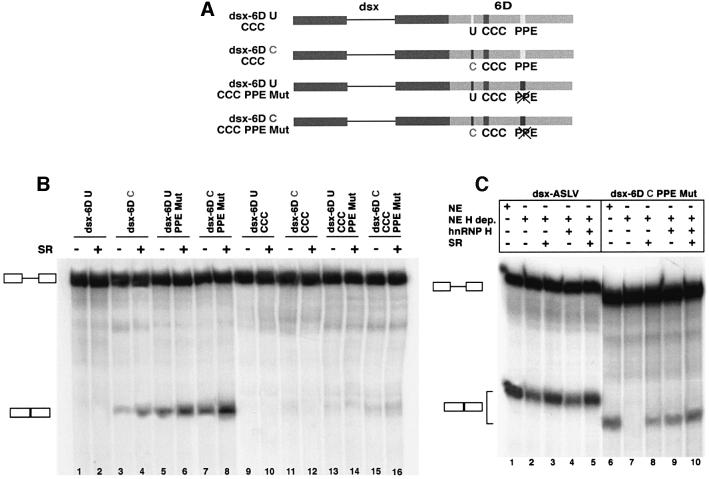
We next sought to determine whether either of these two SR protein interaction sequences in exon 6D functioned as splicing enhancers. SR protein-dependent splicing enhancers have been identified in many alternatively spliced exons (Sun et al., 1993; Ramchatesingh et al., 1995). To test whether the U-to-C mutation functioned as an SR protein-dependent ESE, we inserted the central portion of exon 6D analyzed in Figure 2 into the downstream exon of an enhancer-dependent splicing reporter substrate derived from the Drosophila melanogaster dsx gene (Graveley et al., 1998) (Figure 3A). Variants of the exon 6D sequence containing the U-to-C mutation, mutation of the polypurine element or both mutations were tested. As controls we utilized the dsx-ΔE and the dsx-ASLV constructs (Graveley et al., 1998). The dsx-ΔE construct does not contain splicing enhancer sequences. It is inefficiently spliced in HeLa nuclear extract and additional SR proteins do not dramatically improve this very low level of splicing (Figure 3B, lanes 1 and 2). In dsx-ASLV a strong SR-dependent ESE derived from the avian sarcoma-leukosis virus (ASLV) has been inserted into the downstream exon of the dsx substrate. This substrate splices efficiently in HeLa nuclear extracts and the splicing efficiency is increased by the addition of exogenous SR proteins to the reaction mixture (Figure 3B, lanes and 4).
Fig. 3. In vitro splicing of the dsx–exon 6D substrates. (A) Schematic representation of the dsx–exon 6D splicing substrates. Dsx exonic sequences: dark boxes; ASLV and exon 6D sequences: light boxes. U-to-C, U-to-G, U-to-A and PPE mutations are indicated. (B) In vitro splicing reactions. The radiolabeled pre-RNA substrates indicated were spliced in in vitro splicing reaction mixtures containing HeLa cell nuclear extract. Purified HeLa cell SR proteins (200 ng) were added where indicated. After 2 h of incubation at 30°C, the RNAs were recovered and separated on a 6% polyacrylamide–8 M urea gel. RNA precursors and spliced products are indicated by the schematics on the left.
The substrate containing the wild-type exon 6D sequence gave no detectable splice product (Figure 3B, lanes 5 and 6), indicating that this substrate may contain splicing inhibitory sequences since it is a weaker splicing substrate than the dsx-ΔE parent substrate (Figure 3B, lanes 1 and 2). Splicing is activated in the substrate containing the U-to-C mutation in dsx-6DC and the level of splicing increases upon addition of SR proteins to the splicing mixture (Figure 3B, lanes 7 and 8). The substrate dsx-6DG containing the U-to-G mutation is also spliced efficiently (Figure 3B, lanes 9 and 10) while splicing of the substrate dsx-6DA carrying the U-to-A mutation can be detected only upon addition of exogenous SR proteins to the splicing mixture (Figure 3B, lanes 11 and 12).
Mutation of the polypurine sequence combined with the upstream wild-type U or mutated C increases splicing efficiency of the upstream intron in these substates (Figure 3B, lanes 13–16). This indicates that the polypurine sequence, which has SR protein recruitment activity, is serving as a splicing silencer in this context. The dsx-6DC PPE Mut substrate, which has both the U-to-C mutation and the polypurine sequence, has the strongest level of splicing activity of any of the exon 6D-derived substrates (Figure 3B, lanes 15 and 16). These data indicate that the U-to-C mutation activates an SR protein-dependent splicing enhancer while the polypurine element, although involved in recruiting SR proteins to the same exon, functions as an exonic splicing silencer.
SC35 activates exon 6D splicing
The result of the RNA affinity chromatography assay (Figure 2B) indicates that a 30 kDa species of SR protein is the predominant family member to assemble on the U-to-C mutant exon 6D. SC35 and ASF/SF2 are SR proteins of ∼30 kDa; both are present in roughly equal amounts in HeLa NE and both are recognized by mAb104. Analysis of the single SR proteins shows that binding of both SC35 and SF2 to exon 6D is increased by the U-to-C mutation (Figure 2B). However, a much larger fraction of the total SC35 in nuclear extract assembles onto the exon 6DC PPE Mut substrate RNA than the fraction of total SF2 that assembles onto this same substrate (Figure 2B, lanes 6 and 8). This suggests that SC35 has a higher affinity than SF2 for exon 6D.
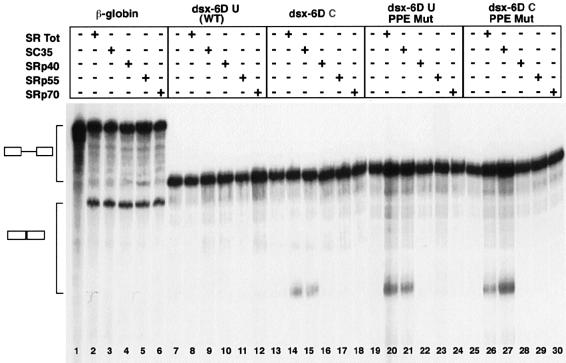
To determine whether individual SR proteins have distinct activity in promoting the splicing of the exon 6D substrates, we complemented splicing-deficient HeLa S100 cytoplasmic extracts with single SR proteins. SR proteins are absent from this splicing-deficient S100 and addition of SR proteins to the extract can complement this deficiency to allow in vitro splicing to occur (Krainer et al., 1991). We purified total SR proteins from HeLa cells and individual SR proteins from calf thymus according to our published protocol (Zahler, 1999). The SC35 purified from calf thymus contains >95% SC35 with only a trace of SF2 detectable.
As a first step, we normalized the amount of the different SR proteins added to the HeLa S100 splicing-deficient extract such that equal amounts of spliced human β-globin first intron substrate were generated with each SR protein added. These levels of SR proteins were picked so that the β-globin substrate splicing was below the maximum achievable in these extracts (Figure 4, lanes 2–6). We next tested these β-globin-normalized levels of SR proteins for activation of splicing of the dsx-6D splicing reporter substrates. Splicing of the dsx-6DU substrate was not activated by complementation of the S100 extracts by total SR proteins or by any of the single SR proteins (Figure 4, lanes 8–12). Only the total HeLa SR protein preparation and purified SC35 were able to efficiently complement the HeLa S100 extract and activate splicing of the dsx-6DC, dsx-6DU PPE Mut and dsx-6DC PPE Mut substrates (Figure 4, lanes 14, 15, 20, 21, 26 and 27). SRp70, SRp55 and SRp40 were unable to stimulate splicing of any of the exon 6D substrate RNAs (Figure 4, lanes 16–18, 22–24 and 28–30) even though they complement the S100 extract to activate splicing of the β-globin substrate with the same efficiency as SC35 (Figure 4, lanes 2–6).
Fig. 4. In vitro splicing of the dsx–exon 6D substrates in HeLa S100 extracts complemented with SR proteins. The radiolabeled pre-RNA substrates indicated were spliced in in vitro splicing reaction mixtures containing HeLa S100 extract. Normalized amounts (∼200 ng) of each SR protein preparation were added to the splicing mixture as indicated. After 2 h of incubation at 30°C, the RNAs were recovered and separated on a 6% polyacrylamide–8 M urea gel. RNA precursors and spliced products are indicated by the schematics on the left.
Analysis of hnRNP binding to the exon 6D regulatory sequences
We next sought to determine whether other RNA binding proteins known to play a role in regulation of alternative splicing interact specifically with the exon 6D regulatory sequences. hnRNP A1 has been shown to regulate alternative splicing of a variety of cellular and viral substrates in a manner opposite to the effects exerted by SR proteins (Mayeda and Krainer, 1992; Mayeda et al., 1993; Caceres et al., 1994; Caputi et al., 1999; del Gatto-Konczak et al., 1999). If the U-to-C mutation disrupts an ESS, one possibility is that it disrupts an hnRNP A1 binding site. A variety of substrate RNAs containing exon 6D sequences with combinations of mutations were immobilized to beads and incubated in HeLa cell nuclear extract. Proteins bound to the beads were identified by immunoblotting (Figure 5). hnRNP A1 bound equally to all six of the different exon 6D substrates tested, indicating that none of these mutations affect the level of hnRNP A1 binding.
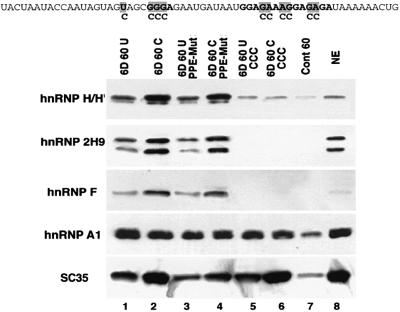
Fig. 5. Analysis of hnRNP H/H′/F/2H9 and A1 assembly onto exon 6D. Sequences of the various substrates used in this RNA affinity chromatography experiment are indicated at the top. Location of the U-to-C, GGGA sequence and PPE mutations are indicated in order from left to right. RNA substrates were covalently linked to agarose beads and incubated in HeLa cell nuclear extracts. Proteins bound to the substrates were eluted and immunoblotted with antibodies specific for the single hnRNPs indicated and SC35. Lane 8 contains 10 µl of HeLa cell nuclear extract.
Members of the hnRNP H group, hnRNP H, hnRNP H′, hnRNP F and hnRNP 2H9, are involved in mRNA processing and exhibit extensive sequence homology (Honore, 2000). They share a common structure of two or three repeats of a similar RNA binding domain named the quasi RNA recognition motif (qRRM) and two glycine-rich auxiliary domains (Matunis et al., 1994; Honore et al., 1995; Mahe et al., 1997). Members of the hnRNP H group have been shown to be involved in splicing regulation as components of both splicing enhancer and splicing silencer complexes (Min et al., 1995; Mahe et al., 1997; Chen et al., 1999; Chou et al., 1999). We have determined that hnRNPs H/H′/F/2H9 have similar binding affinity for specific RNA sequences; the core of the sequence bound by all of these proteins is GGGA (Caputi and Zahler, 2001). A GGGA element is present three nucleotides downstream of the U-to-C mutation in exon 6D and we tested whether hnRNP H proteins interact with this sequence in exon 6D.
Specific binding of hnRNP H family members can be detected in substrates that contain the intact GGGA sequence, while the binding of the hnRNP H family members to the control substrate RNA (Figure 5, lane 7) is equivalent to the binding of these proteins to the two mutant substrates containing disruptions of the GGGA sequence (Figure 5, lanes 5 and 6). This is consistent with the GGGA sequence being the core of the binding sequence for hnRNP H proteins on exon 6D. The binding of hnRNPs H/H′/F/2H9 onto the exon 6D substrate increases with the U-to-C mutation. hnRNPs H/H′/F/2H9 can all be detected as assembling specifically onto the wild-type exon 6D sequences and also onto the U-to-C mutants and the polypurine sequence mutants. However, we can detect a 2- to 3-fold increase in the recruitment of the members of the hnRNP H group to the substrates carrying the U-to-C mutation (Figure 5, lanes 2 and 4) over substrates with the wild-type U at this location (Figure 4, lanes 1 and 3). Mutation of the GGGA sequence leading to loss of hnRNP H family binding has no effect on the efficiency of SR protein binding to exon 6D (Figure 5, lanes 5 and 6 compared with lanes 1 and 2).
HnRNP H is required for exon 6D splicing
We next investigated the role of hnRNPs of the H group in the regulation of exon 6D splicing. To do this, we created dsx–exon 6D splicing substrates in which the GGGA sequence that is required for hnRNP H family binding to exon 6D is mutated (Figure 6A), and tested these substrates in in vitro splicing reactions (Figure 6B). The dsx–exon 6D substrates carrying the U-to-C mutation and the substrates carrying the mutated PPE are capable of being spliced in HeLa cell nuclear extract, while the dsx-6DU substrate containing the wild-type exon 6D sequence does not splice (Figures 3B and 6B, lanes 1–8). When the GGGA sequence was mutated in these substrates, the splicing efficiency dropped dramatically (Figure 6B, lanes 9–16). Addition of SR proteins to the splicing reaction did not detectably improve splicing efficiency (Figure 6B, lanes 10, 12, 14 and 16). These results indicate that the GGGA sequence is an essential part of the complex enhancer regulating exon 6D splicing.
Fig. 6. In vitro splicing of the dsx–exon 6D substrates containing the mutated GGGA sequence. (A) Schematic representation of the dsx–exon 6D splicing substrates with mutations in the GGGA sequence. Dsx exonic sequences are shown as dark boxes and exon 6D sequences as light boxes. The sites of the U-to-C, GGG and PPE mutations are indicated. (B) In vitro splicing reactions. The radiolabeled pre-mRNA substrates indicated were added to splicing reaction mixtures containing HeLa cell nuclear extract and either 0 or 200 ng of total HeLa SR proteins. After 2 h of incubation at 30°C, the RNAs were recovered and separated on a 6% polyacrylamide–8 M urea gel. RNA precursors and spliced products are indicated by the schematics on the left. (C) The radiolabeled pre-mRNA substrates indicated were added to splicing reaction mixtures containing HeLa cell nuclear extract (lanes 1 and 6) or hnRNP H-depleted HeLa cell nuclear extract (lanes 2–5 and 7–10). Reaction mixtures were also complemented with 200 ng total HeLa SR proteins or 200 ng of recombinant hnRNP H as indicated.
To test whether hnRNP H is an essential component of the exon 6D splicing enhancer complex, we used RNA affinity chromatography to make an hnRNP H-depleted HeLa cell nuclear extract. This was accomplished by incubating HeLa cell nuclear extract with agarose beads containing a 40 nucleotide RNA whose sequence consists of five eight-base repeats of the hnRNP H family binding site from the rat β-tropomyosin exonic splicing silencer (Chen et al., 1999). After two consecutive rounds of incubation of this extract with the beads, >90% of hnRNP H family proteins were removed from the extract (Figure 7D and data not shown). This extract was then used to test for splicing activity of the dsx-ASLV and dsx-6DC PPE Mut substrates. For dsx-ASLV, the depleted extract showed a reduction in splicing activity (Figure 6C, lanes 1 and 2) that could be restored to full activity by addition of 200 ng of total SR proteins (lanes 3 and 5). In a previous study, we showed that depletion of extracts by RNA affinity chromatography led to a reduction in general splicing activity because of non-specific loss of some SR proteins (Caputi et al., 1999). For the dsx-ASLV construct, addition of 200 ng of recombinant hnRNP H had no effect on splicing activity (lane 4). We complemented with 200 ng of recombinant hnRNP H because we determined that this is the amount of hnRNP H found in 15 µl of our HeLa cell nuclear extract (data not shown). In contrast, the enhancer substrate derived from the 6D exon showed a requirement of at least one hnRNP H family member for splicing to occur. Depletion of hnRNP H from the nuclear extract leads to a loss of splicing activity (lane 7) that can only partially be rescued by addition of SR proteins (lane 8). Addition of recombinant hnRNP H alone shows significant rescue of the splicing enhancer activity (lane 9) and addition of both SR proteins and hnRNP H leads to full restoration of splicing activity (lane 10). The activation of exon 6D splicing at a low level by addition of SR proteins alone (lane 8) may be due to the presence of residual trace amounts of hnRNP H in the depleted extract (data not shown). Altogether, these results indicate that hnRNP H is essential for the exon 6D splicing enhancer activity. We also looked at the level of hnRNP H in HeLa S100 extract given that the enhancer functions in that extract as well (Figure 4). hnRNP H in S100 is found at ∼20% of the level found in an equivalent volume of HeLa nuclear extract (data not shown). Given the high affinity of hnRNP H for the GGGA sequence, it makes sense that splicing of exon 6D substrates can be detected in S100 extracts as well.
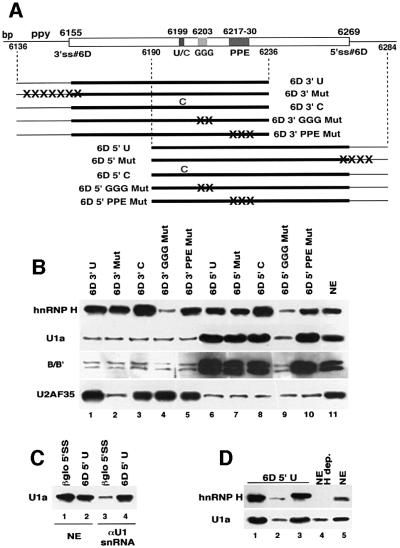
Fig. 7. Assembly of components of the U1 snRNP onto exon 6D requires hnRNP H and is independent of the 5′ end of U1 snRNA. (A) Schematic representation of exon 6D 100mer RNA substrates used in this RNA affinity chromatography experiment. Position of the branch point (bp), polypyrimidine tract (ppy), U-to-C mutation (U/C), GGG sequence, polypurine element (PPE) and 5′ splice site (5′ss) are indicated. The GGG and polypurine mutations are the same as those used previously (see Figure 5A). The 3′ splice site mutation changes the final bases of the intron and first base of the exon from ccactctgtgttagT to agactgtgtaataaA. The 5′ splice site mutation changes the last three bases of the exon and the first three bases of the intron from GAGgta to AAGctt. (B) The exon 6D substrates indicated were covalently linked to agarose beads and incubated in HeLa nuclear extracts. Proteins bound to the substrates after washing were eluted and immunoblotted with antibodies specific for hnRNP H, U1a protein, the Sm epitope and U2AF 35. Lane 11 contains 10 µl of HeLa cell nuclear extract. (C) HeLa cell nuclear extract (lanes 1 and 2) or HeLa cell nuclear extract treated with an antisense oligonucleotide to the 5′ end of U1 snRNA and RNase H (lanes 3 and 4) were incubated with agarose beads containing the 5′ splice junction region of the human β-globin gene first exon (lanes 1 and 3) or the 6D5′U substrate (lanes 2 and 4). Proteins bound to the substrates after washing were eluted and immunoblotted with an antibody specific for U1a protein. (D) The 6D5′U substrate RNA was bound to beads and incubated in 250 µl of either HeLa nuclear extract (lane 1), HeLa nuclear extracted depleted of hnRNP H proteins (lane 2) or hnRNP H-depleted extract complemented with 3 µg of recombinant hnRNP H (lane 3). Proteins bound to the substrates after washing were eluted and immunoblotted with antibodies specific for the hnRNP H or U1a protein. Lane 4 contains 10 µl of hnRNP H-depleted HeLa nuclear extract and lane 5 contains 10 µl of HeLa nuclear extract.
U1 snRNP assembly onto exon 6D is dependent on hnRNP H activity
Spliceosome assembly onto the pre-mRNA proceeds in a stepwise fashion. The first detectable pre-spliceosomal complex is named early or E complex (Reed and Palandjian, 1997). It is characterized by the binding of the U1 snRNP to the 5′ splice site and the U2 auxiliary factor heterodimer (U2AF65/35) to the 3′ splice site sequences. SR proteins are also present in the E complex and are required for its formation (Moore et al., 1993; Graveley, 2000). SR proteins have been shown to interact directly with the 35 kDa subunit of the U2AF heterodimer (U2AF35) and with the U1 snRNP-specific U1-70K protein (Wu and Maniatis, 1993).
We investigated whether enhanced binding of SR proteins in the U-to-C mutants was promoting exon 6D splicing by recruitment of U2AF65/35 and U1 snRNP to the exon 6D-flanking 3′ and 5′ splice sites. To do this, we immobilized to agarose beads RNA substrates of 100 nucleotides, containing the regulatory regions of exon 6D and either the 3′ end of the upstream intron or the 5′ end of the downstream intron (Figure 7A). Beads containing wild-type substrates or substrates with various mutations in the splicing regulatory regions were incubated in HeLa nuclear extract. Proteins bound specifically to the RNA substrates were identified by immunoblotting with antibodies specific for the 35 and 65 kDa (data not shown) U2AF subunits, the U1 snRNP-specific protein U1a, the Sm epitope on snRNP core proteins and hnRNP H (Figure 7B).
Both U2AF subunits assemble onto the substrates containing the 3′ end of the upstream intron (Figure 7B, lanes 1–5) as long as the polypyrimidine tract is intact (lane 2). Mutations to any of the splicing regulatory elements in exon 6D, the U-to-C, the GGG and the polypurine element, have no effect on U2AF recruitment. The U1a component of the U1 snRNP and the Sm B/B′ core snRNP proteins can be detected assembling onto substrates containing the 5′ end of the downstream intron (lanes 6–10). Surprisingly, this assembly occurs on the substrate for which the presumed U1 snRNP-interacting sequence, the 5′ splice site, is mutated (lane 7). Mutations at the U-to-C site or the polypurine element do not affect the level of U1 snRNP protein detected. However, mutation of the GGG sequence, which is the interaction site for hnRNP H, leads to a strong decrease in U1a and B/B′ recruitment. Therefore, there is a strong correlation between the ability to bind hnRNP H and the presence of sequences downstream of the regulatory region (but not the true 5′ splice site) with the ability to recruit U1 snRNP. The U-to-C mutation increases hnRNP H binding to both of the 100mer substrates (lanes 3 and 8).
We next chose to investigate further the finding that components of the U1 snRNP could be detected interacting with the exon 6D splicing enhancer in the absence of the wild-type 5′ splice site (Figure 7B, lane 7). Interaction of U1 snRNP with a 5′ splice site involves base pairing between U1 snRNA and the 5′ splice site region. To test whether this base pairing was required for the interaction of U1 snRNP with the substrate RNA, we cleaved off the 5′ end of U1 snRNA by treating HeLa cell nuclear extract with a DNA oligonucleotide complementary to the 5′ end of U1 snRNA and RNase H (Bruzik and Steitz, 1990). We tested this U1 snRNP-inactivated extract for the ability to assemble the U1 snRNP onto the 5′ splice site of the first intron of the human β-globin gene and the 5′ splice site of the intron downstream of exon 6D in an RNA affinity chromatography experiment (Figure 7C). In untreated HeLa cell nuclear extract, U1a protein can be detected on both of the β-globin and exon 6D 5′ splice site substrates (Figure 7C, lanes 1 and 2). In the U1 snRNP-inactivated HeLa nuclear extract, the U1a protein can be detected at only background levels on the β-globin 5′ splice site (lane 3) consistent with its inability to interact by base pairing with the 5′ splice site. In contrast, the exon 6D 5′ splice site substrate assembles the same level of U1a in both the U1 snRNP-inactivated and untreated HeLa cell extracts (lanes 4 and 2). This result suggests that assembly of the U1 snRNP onto the exon 6D 5′ splice site region does not require interactions between the 5′ end of U1 snRNA and the 5′ splice site.
The results in Figure 7B point to a requirement for the hnRNP H binding site in the recruitment of U1 snRNP components to the exon 6D substrates containing the 5′ end of the downstream intron (Figure 7B, lane 9). To test whether hnRNP H was required for the observed binding of U1a protein to these substrates, we compared binding of U1a and hnRNP H to the exon 6D 100mer substrate containing the 5′ end of the downstream intron using HeLa nuclear extract, hnRNP H-depleted HeLa nuclear extract and hnRNP H-depleted HeLa nuclear extract complemented with recombinant hnRNP H to the level found in HeLa nuclear extracts (Figure 7D, lanes 1–3, respectively). U1a binding decreases dramatically in the hnRNP H-depleted extract (lane 2) and returns to the level of the untreated extract (lane 1) when the hnRNP H-depleted extract is complemented with recombinant hnRNP H (lane 3). The slightly retarded mobility of the recombinant hnRNP H in lane 3 is due to the presence of a His6 tag on the protein. These results indicate that U1 snRNP assembly onto exon 6D substrates is dependent on hnRNP H protein activity.
Discussion
Properly balanced splicing of the single HIV-1 pre-mRNA is essential for the viral life cycle and is dependent on the host cell splicing machinery. A single nucleotide change (U-to-C) in an HIV-1 isolate has been shown to induce an abnormal splicing pattern in which the majority of mRNAs contain the tev-specific exon 6D (Wentz et al., 1997). The abnormal splicing pattern leads to a dramatic decrease in viral production and infectivity. The results presented here demonstrate that a complex regulatory region promotes exon 6D inclusion in HIV-1 messages. This region consists of three distinct elements: the single nucleotide U-to-C mutation, the GGGA sequence and a polypurine element.
We determined that these elements are conserved in other HIV-1 isolates. This is important because the HIV-1 viral isolate NL4-3, unlike the HXB2 strain, which we have been referring to as the wild-type exon 6D strain in this manuscript, does not contain a functional exon 6D 5′ splice site and does not use exon 6D. When the U-to-C or the PPE mutation was inserted into the NL4-3 exon 6D, an overall increase in viral singly and multiply spliced messages was observed (Wentz et al., 1997). This suggests that the exon 6D splicing regulatory region may control the overall splicing efficiency of the viral pre-mRNA, not just the inclusion of exon 6D in the viral messages. The exon 6D regulatory region is located in the highly variable V1 loop of the env gene, which would make it seem unlikely that there would be strong sequence conservation of the regulatory elements (Myers et al., 1996). However, analysis of this variable V1 loop region indicates that the three elements that we have characterized in exon 6D are highly conserved among different HIV-1 isolates of the B subtype (Figure 8). This is consistent with a model in which the exon 6D regulatory region is important for the global regulation of HIV-1 splicing.
Fig. 8. Exon 6D sequence alignments. The exon 6D splicing regulatory region has been aligned in divergent HIV-1 isolates of the B subtype (Myers et al., 1996). Lower-case bases indicate aligned nucleotides, upper-case bases indicate misaligned nucleotides, dots indicate gaps. Shaded areas indicate the elements of the exon 6D splicing regulatory region (U-to-C mutation, GGGA sequence and polypurine element, respectively, from left to right). The region coding for the variable V1 loop is also indicated. The HXB2 isolate, from which the exon 6D sequence used in this study was derived, is underlined.
Previous studies showed that when exon 6D was inserted as the central exon of a cardiac troponin T (cTNT) three exon, two intron minigene, the mutation of the polypurine element decreased splicing efficiency, thus defining a splicing enhancer. However, when the polypurine element was mutated in the viral genome an increase in multiply spliced messages was detected in transfected cells, indicating that the polypurine element is a splicing silencer in the viral context (Wentz et al., 1997). Polypurine elements similar to the one present in exon 6D are able to bind SR proteins and enhance splicing in several systems (Humphrey et al., 1995; Ramchatesingh et al., 1995). Here, we have shown that although the exon 6D PPE promotes SR protein binding to the substrate, it serves as a splicing silencer when the central portion of exon 6D is inserted into the second exon of the dsx enhancer–reporter substrate. This is in agreement with the results obtained when the PPE was mutated in the viral context (Wentz et al., 1997). Therefore, the splicing context in which this polypurine element is found plays an important role in determining whether it functions as an ESE or an ESS.
It has been proposed previously that the U-to-C mutation may interrupt an ESS element leading to relief from splicing inhibition (Wentz et al., 1997). Our results indicate that the U-to-C mutation creates an ESE. The U-to-C mutation has the ability to promote assembly of several SR proteins, and it has the highest affinity for SC35. Functional binding sequences for several SR proteins have been identified by selection affinity experiments (Tacke and Manley, 1995; Liu et al., 1998; Cavaloc et al., 1999). The U-to-C mutation changes the exon 6D AGUAG sequence to AGCAG, which matches the SC35 binding consensus sequence AGC(G)AG (Tacke and Manley, 1995; Cavaloc et al., 1999). The mutation of this same U to a G also matches the SC35 consensus sequence and also efficiently enhances splicing (Figure 3B, lanes 9 and 10). Changing this same U to an A stimulates splicing only weakly (Figure 3B, lanes 11 and 12). These data are all consistent with a model in which the U-to-C mutation in exon 6D creates an SC35 binding site, which in turn stimulates splicing of this exon.
We have characterized a third element of the exon 6D splicing regulatory region, a GGGA sequence three nucleotides downstream of the U-to-C mutation. The core sequence capable of binding all members of the hnRNP H family is GGGA (Caputi and Zahler, 2001). Mutation of the GGGA sequence in exon 6D leads to loss of hnRNP H group recruitment to exon 6D (Figure 5). This loss of hnRNP H group binding is coupled with complete loss of splicing in the dsx–exon 6D splicing substrates. Depletion–reconstitution experiments indicate that hnRNP H is essential for the function of the exon 6D splicing enhancer (Figure 6). Mutation of the GGGA sequence has no effect on SR protein recruitment to exon 6D, while the U-to-C mutation does increase by 2- to 3-fold the amount of hnRNP H proteins that assemble on exon 6D (Figure 5). One explanation for this result is that the U-to-C point mutation improves the overall consensus of hnRNP H group protein binding to this site characterized by the GGGA sequence at its core. Another possibility is that, since the U-to-C mutation increases recruitment of SR proteins to exon 6D, the SR proteins help to recruit hnRNP H family proteins to exon 6D. While mutation of the GGGA sequence eliminates hnRNP H family binding, it does not decrease SR protein binding to exon 6D, indicating that while SR protein binding to the substrate may help in the binding of hnRNP H proteins, the binding of SR proteins is not dependent on a cooperative interaction with hnRNP H proteins.
SR protein-dependent ESEs are thought to act by recruiting U2AF and U1 snRNP to the flanking splice sites of an alternatively spliced exon with weak consensus splice junctions, thus promoting inclusion of the alternative exon. Our results show that U2AF and the U1 snRNP assemble on the regions flanking exon 6D independent of the U-to-C mutation. U2AF binding to a polypyrimidine tract independent of a splicing enhancer has been characterized previously (Kan and Green, 1999). We demonstrated that hnRNP H interacting with the GGGA element is required for assembly of U1 snRNP onto exon 6D and that this assembly does not require that the 5′ end of U1 snRNA base pairs with the 5′ splice site. That this U1 snRNP interaction does not depend on the 5′ end of U1 snRNA or the presence of a 5′ splice site is consistent with the observation that the U-to-C mutation can stimulate overall viral splicing in a provirus derived from the NL4-3 HIV-1 isolate, which lacks a 5′ splice site downstream of exon 6D (Wentz et al., 1997). We propose that hnRNP H interaction with the conserved GGGA sequence functions to bring U1 snRNP to the pre-mRNA through protein–protein interactions, but that this complex is inactive in functioning as a splicing enhancer, perhaps due to the function of the polypurine ESS. Binding of SC35 to the U-to-C mutation may promote a change in this hnRNP H–U1 snRNP complex that turns it into an active splicing enhancer complex which stimulates the overall splicing of the HIV-1 pre-mRNA.
Materials and methods
Substrate RNA synthesis, immobilization of RNA on agarose beads and RNA affinity chromatography assays
Substrate RNA synthesis from oligonucleotides, covalent linkage to adipic acid dihydrazide–agarose beads and RNA affinity chromatography assays were performed as described previously (Caputi et al., 1999; Caputi and Zahler, 2001). The sequences of the substrates are indicated in Figures 2, 4 and 7 except for the Cont 60 substrate whose sequence has been described previously (Caputi et al., 1999). The β-globin substrate RNA used in the experiment in Figure 7C contains the last 21 nucleotides of the first exon and the first 39 nucleotides of the first intron of the human β-globin gene.
Plasmid constructs
Plasmids dsx-ΔE and dsx-ASLV used as templates for generating in vitro splicing substrates were a generous gift from Dr B.R.Graveley (University of Connecticut) and Dr T.Maniatis (Harvard University). A series of exon 6D-containing splicing substrates were constructed by inserting pairs of kinased complementary annealing DNA oligonucleotides into the BstbI site of the dsx-ΔE plasmid.
Preparation of hnRNP H-depleted HeLa cell nuclear extract
A 40 nucleotide hnRNP H group high-affinity binding substrate RNA containing five repeats of the eight-nucleotide rat β-tropomyosin hnRNP H binding site sequence GUGGGGAC was transcribed by T7 RNA polymerase from an oligonucleotide substrate. HeLa cell nuclear extract (0.25 ml) was treated with two consecutive rounds of 5 nmol of hnRNP H binding site RNA immobilized to agarose beads at 30°C for 15 min. Depleted extract was then used in pre-mRNA splicing and RNA affinity assays. The depletion resulted in removal of >90% of hnRNP H family proteins from the extracts.
snRNA inactivation in HeLa cell nuclear extract
Experiments in which the 5′ end of U1 snRNA was decapitated in HeLa cell nuclear extract with antisense oligodeoxynucleotides were performed as described by Bruzik and Steitz (1990). HeLa nuclear extract was incubated at 30°C for 30 min in the presence of 13 µM oligodeoxynucleotide (anti-U1 5′-TGCCAGGTAAGTAT) and 5 U of RNase H.
In vitro pre-mRNA splicing assays and SR protein preparation
Capped, 32P-labeled run-off transcripts were synthesized by in vitro transcription using T7 RNA polymerase. HeLa cell nuclear and S100 extracts were prepared and splicing reactions were performed in a total volume of 25 µl containing 15 µl of HeLa cell nuclear extract or S100 as described (Mayeda and Krainer, 1999). The reaction mixtures were incubated at 30°C for 2 h. RNAs recovered from the splicing reaction mixtures were separated on an 8 M urea–6% polyacrylamide gel and visualized with a PhosphorImager (Molecular Dynamics). Total SR proteins were prepared from HeLa cells and individual SR proteins were prepared from calf thymus as described previously (Zahler, 1999). Recombinant His6-tagged hnRNP H protein (Chou et al., 1999) was kindly provided by Dr M.Y.Chou and Dr Douglas Black.
Protein analysis
Proteins were separated on 12% polyacrylamide–SDS gels and visualized by Coomassie Blue staining or electroblotted onto a nitrocellulose membrane and probed with antibodies. mAb 4B10 against hnRNP A1 (Piñol-Roma et al., 1988) was provided by Dr G.Dreyfuss (University of Pennsylvania). mAb IS-2H9 against hnRNP 2H9 (Mahe et al., 1997) was provided by Dr J.P.Fuchs (INSERM, Strasbourg, France). Rabbit polyclonal anti-hnRNP H/H′ and F antisera (Min et al., 1995; Chou et al., 1999) were provided by Dr D.L.Black (University of California, Los Angeles). Rabbit polyclonal anti-U2AF35 and anti-U2AF65 antibodies were provided by Dr T.Maniatis (Harvard University). Anti-SF2/ASF mAb AK-96 was provided by Dr A.R.Krainer (Cold Spring Harbor Laboratories). Rabbit polyclonal anti-U1a antibody was provided by Dr I.Mattaj (EMBL, Heidelberg). Anti-SC35 monoclonal antibody mAb 532 was provided by Dr J.Stevenin (INSERM, Strasbourg, France). Immunoblots were stained using the appropriate horseradish peroxidase-conjugated secondary antibody and detected using the ECL chemiluminescence kit (Pierce).
Acknowledgments
Acknowledgements
We are grateful to Drs G.Dreyfuss, D.Black, I.Mattaj, A.Krainer, J.Stevenin, T.Maniatis and J.P.Fuchs for their kind gifts of antibodies. We are indebted to Dr D.Black for the gift of recombinant hnRNP H protein. Many thanks to Drs T.Maniatis and B.R.Graveley for the dsx in vitro splicing constructs and to Dr M.L.Hammarskjold for helpful discussions. This work is supported by the University of California Universitywide AIDS Research Program, grant R99-SC-085.
References
- Benko D.M., Schwartz,S., Pavlakis,G.N. and Felber,B.K. (1990) A novel human immunodeficiency virus type 1 protein, tev, shares sequences with tat, env and rev proteins. J. Virol., 64, 2505–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzik J.P. and Steitz,J.A. (1990) Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell, 62, 889–999. [DOI] [PubMed] [Google Scholar]
- Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- Caputi M. and Zahler,A.M. (2001) Determination of the RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem., 276, 43850–43859. [DOI] [PubMed] [Google Scholar]
- Caputi M., Casari,G., Guenzi,S., Tagliabue,R., Sidoli,A., Melo,C.A. and Baralle,F.E. (1994) A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res., 22, 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Mayeda,A., Krainer,A.R. and Zahler,A.M. (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J., 18, 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y., Burgeous,C.F., Kister,L. and Stevenin,J. (1999) The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA, 5, 468–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.D., Kobayashi,R. and Helfman,D.M. (1999) Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev., 13, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.Y., Rooke,N., Turck,C.W. and Black,D.L. (1999) hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol., 19, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Gatto-Konczak F., Olive,M., Gesnel,M.C. and Breathnach,R. (1999) hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen W.P., Hampson,R.K., Sun,Q. and Rottman,F.M. (1994) A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J. Biol. Chem., 269, 6431–6436. [PubMed] [Google Scholar]
- Gottlinger H.G., Dorfman,T., Cohen,E.A. and Haseltine,W.A. (1992) The role of the tnv RNA splicing signals in replication of HIV-1 IIIB isolate. Virology, 189, 618–628. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R., Hertel,K.J. and Maniatis,T. (1998) A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J., 17, 6747–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold M.L., Li,H., Lu,X. and Prasad,S. (1991) Rev regulation of HIV env RNA is dependent on the position of splice sites. In Haseltine,W.A. and Wong-Staal,F. (eds), Genetic Structure and Regulation of HIV. Raven Press, New York, NY.
- Honore B. (2000) The hnRNP 2H9 gene, which is involved in the splicing reaction, is a multiply spliced gene. Biochim. Biophys. Acta, 1492, 108–119. [DOI] [PubMed] [Google Scholar]
- Honore B. et al. (1995) Heterogeneous nuclear ribonucleoproteins H,H′ and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem., 270, 28780–28789. [DOI] [PubMed] [Google Scholar]
- Humphrey M.B., Bryan,J., Cooper,T.A. and Berget,S.M. (1995) A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol. Cell. Biol., 15, 3979–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan J.L. and Green,M.R. (1999) Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev., 13, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A.R., Mayeda,A., Kozak,D. and Binns,G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA binding proteins, U1 70K and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Lavigueur A., La Branche,H., Kornblihtt,A.R. and Chabot,B. (1993) A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev., 7, 2405–2417. [DOI] [PubMed] [Google Scholar]
- Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Heimer,J., Rekosh,D. and Hammarskjold,M.L. (1990) U1 small nuclear RNA plays a direct role in the formation of a rev-regulated immunodeficiency virus env mRNA that remains unspliced. Proc. Natl Acad. Sci. USA, 87, 7598–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe D., Mahl,P., Gattoni,R., Fischer,N., Mattei,M.G., Stevenin,J. and Fuchs,J.P. (1997) Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem., 272, 1827–1836. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Xing,J. and Dreyfuss,G. (1994) The hnRNP F protein: unique primary structure, nucleic acid-binding properties and subcellular localization. Nucleic Acids Res., 22, 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1999) Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol., 118, 309–314. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Helfman,D.M. and Krainer,A.R. (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol., 13, 2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H., Chan,R.C. and Black,D.L. (1995) The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev., 9, 2659–2671. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357.
- Myers G., Foley,B., Mellors,J., Korber,B., Jeang,K. and Wain-Hobson,S. (eds), (1996) Human Retroviruses and AIDS 1996: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory, Los Alamos, NM.
- Piñol-Roma S., Choi,Y.D., Matunis,M.J. and Dreyfuss,G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA binding proteins. Genes Dev., 2, 215–227. [DOI] [PubMed] [Google Scholar]
- Purcell D.F.J. and Martin,M.A. (1993) Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J. Virol., 67, 6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchatesingh J., Zahler,A.M., Neugebauer,K.M., Roth,M.B. and Cooper,T.A. (1995) A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol., 15, 4898–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, Oxford, UK, pp. 103–129.
- Roth M.B., Zahler,A.M. and Stolk,J.A. (1991) A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol., 115, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salfeld J., Gottlinger,H.G., Sia,R.A., Park,R.E., Sodroski,J.G. and Haseltine,W.A. (1990) A tripartite HIV-1 tat–env–rev fusion protein. EMBO J., 9, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Felber,B.K., Benko,D.M., Fenyo,E.M. and Pavlakis,G.N. (1990) Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol., 64, 2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Mayeda,A., Hampson,R.K., Krainer,A.R. and Rottman,F.R. (1993) General splicing factor ASF/SF2 promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev., 7, 2598–2608. [DOI] [PubMed] [Google Scholar]
- Tacke R. and Manley,J.L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J., 14, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz P.M., Moore,B.E., Cloyd,M.V., Berget,S.M. and Donehower,L.A. (1997) A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type I induces dominant aberrant splicing and arrests virus production. J. Virol., 71, 8542–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Y. and Maniatis,T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Zahler A.M. (1999) Purification of SR protein splicing factors. Methods Mol. Biol., 118, 419–432. [DOI] [PubMed] [Google Scholar]