Abstract
The Staphylococcus aureus multidrug-binding protein QacR represses transcription of the qacA multidrug transporter gene and is induced by multiple structurally dissimilar drugs. QacR is a member of the TetR/CamR family of transcriptional regulators, which share highly homologous N-terminal DNA-binding domains connected to seemingly non-homologous ligand-binding domains. Unlike other TetR members, which bind ∼15 bp operators, QacR recognizes an unusually long 28 bp operator, IR1, which it appears to bind cooperatively. To elucidate the DNA-binding mechanism of QacR, we determined the 2.90 Å resolution crystal structure of a QacR–IR1 complex. Strikingly, our data reveal that the DNA recognition mode of QacR is distinct from TetR and involves the binding of a pair of QacR dimers. In this unique binding mode, recognition at each IR1 half-site is mediated by a complement of DNA contacts made by two helix–turn–helix motifs. The inferred cooperativity does not arise from cross-dimer protein–protein contacts, but from the global undertwisting and major groove widening elicited by the binding of two QacR dimers.
Keywords: DNA binding cooperativity/multidrug-binding protein/protein–DNA complex/QacR/repressor
Introduction
The emergence of multidrug resistance (MDR) represents a serious health problem. A major component of MDR is the large number of membrane transport systems that efflux drugs. For example, in human cancer cells, resistance to antitumor chemotherapeutic agents is partially mediated by the P-glycoprotein efflux pump (Gottesman et al., 1996; Ambudkar et al., 1999). Currently, >100 possible multidrug transporters have been described and can be divided into five major families: the ATP-binding cassette (ABC), which includes P-glycoprotein; the major-facilitator superfamily (MFS); the multidrug and toxic compound extrusion transporters (MATE); the drug/metabolite transporters (DMT); and the resistance/nodulation/division transporters (RND) (Levy, 1992; Putman et al., 2000; Saier and Paulsen, 2001). Following the discovery of the mammalian P-glycoprotein, the phenomenon of MDR by drug efflux was also described in many bacterial systems, including Staphylococcus aureus (Tennent et al., 1989; Paulsen et al., 1996; Mitchell et al., 1999; Brown and Skurray, 2001; Mayer et al., 2001). Indeed, S.aureus has become a serious clinical problem because of the emergence of strains that are unresponsive to the traditional arsenal of antimicrobial compounds.
Resistance to antiseptics and disinfectants in S.aureus strains is associated typically with the presence of QacA or Smr (QacC), which are plasmid-encoded determinants found in clinical isolates. These determinants vary in their ability to confer resistance to a range of toxic organic cations such as quaternary ammonium compounds (QACs) (Behr et al., 1994; Mayer et al., 2001). QacA is a member of the MFS family of multidrug transporters, which utilize the proton motive force in drug efflux (Brown and Skurray, 2001). The unregulated production of QacA would probably cause the dissipation of the membrane electrochemical gradient and thus would be toxic to the cell. Such toxicity is demonstrated by overexpression of TetA, the MFS transporter responsible for the efflux of tetracycline (Eckert and Beck, 1989). Therefore, the expression of these genes must be under stringent control. The repressor protein QacR was recently demonstrated to regulate the transcription of qacA by binding to an operator site, IR1, which overlaps the transcription start site of the gene (Grkovic et al., 1998).
QacR is a dimeric, 188 residue (23 kDa) gene regulator that is induced off its operator DNA site by binding to one of many structurally dissimilar cationic lipophilic compounds, which are also substrates of the QacA transporter (Grkovic et al., 1998, 2001). Thus, QacR is also a multidrug-binding protein and its structure when bound to six different drugs has been determined recently (Schumacher et al., 2001). QacR is a member of the TetR/CamR family of transcriptional regulators (Aramaki et al., 1995), which all share a highly homologous N-terminal DNA-binding domain of ∼45 residues. Structures of TetR have revealed that this region forms a three-helix bundle that contains a helix–turn–helix (HTH) DNA-binding motif (Hinrichs et al., 1994; Kisker et al., 1995; Orth et al., 2000). Within TetR members, the conserved DNA-binding domains are connected to sequentially diverse ligand-binding domains (Aramaki et al., 1995). Unlike other characterized TetR family members, which bind ∼15 bp operator sites, the DNA site footprinted by QacR, IR1, is unusually long, consisting of a 28 bp inverted repeat that is separated by 6 bp (Grkovic et al., 1998; Orth et al., 2000; Folcher et al., 2001). Because this operator overlaps the qacA transcription start site, it has been suggested that QacR binding prevents the transition of the RNA polymerase–promoter complex into a productively transcribing state rather than blocking the binding of RNA polymerase (Grkovic et al., 1998). Also distinct from other TetR family members, QacR does not autoregulate its own expression. Recently, we demonstrated a more significant difference between QacR and TetR: QacR binds its DNA site with a stoichiometry of two QacR dimers per operator, whereas TetR binds as a single dimer (Grkovic et al., 2001). However, given that QacR probably contains a DNA-binding domain homologous to TetR, the structural basis for its DNA-binding mode is unclear, especially considering that the IR1 site contains a single inverted repeat rather than a pair of inverted repeats, which would be expected for a site recognized by a pair of dimers. Therefore, to determine the mechanism by which QacR recognizes its extended IR1 operator site, we determined the crystal structure of a QacR–IR1 complex to 2.90 Å resolution.
Results and discussion
Overall QacR–DNA structure
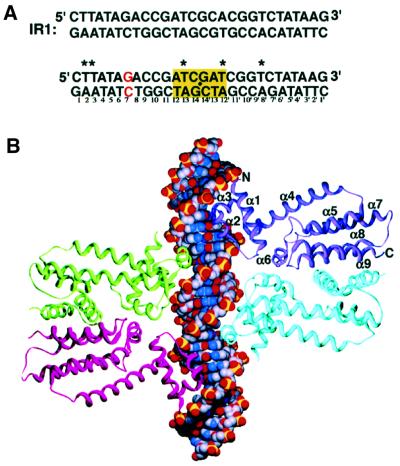
The crystal structure of QacR bound to a symmetrized version of the 28 bp in vivo IR1 operator site (Figure 1A) was solved to 2.90 Å resolution by multiple isomorphous replacement (MIR) and refined to an Rwork and Rfree of 22.2 and 25.8%, respectively (Table I) (Brünger et al., 1998). There are four monomers of QacR (two dimers) and a full IR1-binding site in the crystallographic asymmetric unit (Figure 1). Each of the monomers is essentially identical, with root mean square deviations (r.m.s.ds) of 0.58, 0.63 and 0.64 Å in a pair-wise comparison of residues 4–185 of corresponding Cα atoms. Super imposition of Cα atoms of these residues of each dimer (comparing one dimer with the other) results in an r.m.s.d. of 0.68 Å.
Fig. 1. Structure of the QacR–DNA complex. (A) The extended qacA operator. The top duplex shows the 28 bp IR1 sequence, the qacA regulatory element, bound by QacR. The bottom duplex, which was used for crystallization, differs only in two base pairs of the 6 bp spacer (yellow box). One ‘half-site’ is numbered 1–14 and the other 1′–14′. The qacA transcription start site is shown in red. Asterisks indicate sites of 5-iodouracil replacement used for phase determination. A black dot indicates the location of the two-fold axis. (B) Overall structure of the QacR–DNA complex. The subunits of one dimer are shown as purple and cyan ribbons, and the other as red and green. The DNA is shown as CPK with phosphates, oxygen, carbon and nitrogen colored yellow, orange, gray and blue, respectively. The secondary structural elements of one subunit are labeled. This figure (and Figures 2 and 6) was made with Swiss-PdbViewer (Guex and Peitsch, 1997) and rendered with POVRAY (Persistence of Vision Raytracer version 3.1; http://www.povray.org).
Table I. QacR–DNA structure determination and refinement.
| Crystal | Native 2 | Native 1 | io13 | io12′/8′ | io2 | io3 | Dy/io2 | Uranyl |
|---|---|---|---|---|---|---|---|---|
| Space group | P65 | P65 | P65 | P65 | P65 | P65 | P65 | P65 |
| Cell constants (Å) | ||||||||
| a | 174.0 | 174.7 | 175.0 | 174.5 | 174.0 | 174.6 | 175.3 | 174.7 |
| b | 174.7 | 174.0 | 174.7 | 175.0 | 174.5 | 174.0 | 174.6 | 175.3 |
| c | 152.0 | 152.1 | 151.0 | 152.9 | 151.6 | 151.1 | 151.5 | 153.2 |
| Resolution (Å) | 2.90 | 3.35 | 3.70 | 4.60 | 3.70 | 4.70 | 3.70 | 4.80 |
| Overall Rsym (%)a | 5.4 | 9.9 | 9.7 | 10.9 | 5.0 | 11.4 | 10.7 | 11.5 |
| Overall I/σ(I) | 11.4 | 10.5 | 12.2 | 10.3 | 13.5 | 9.8 | 11.5 | 14.6 |
| High resolution (Å) | 3.03–2.90 | 3.5–3.35 | 4.1–3.7 | 5.4–3.6 | 4.1–3.7 | 5.4–4.7 | 4.1–3.7 | 5.5–4.8 |
| Rsym (%) | 40.1 | 24.9 | 21.0 | 35.5 | 12.3 | 15.0 | 25.2 | 25.0 |
| I/σ(I) | 1.8 | 1.3 | 2.7 | 2.0 | 2.3 | 2.8 | 1.9 | 2.3 |
| Total reflections (no.) | 302 270 | 85 618 | 58 289 | 96 855 | 55 213 | 71 643 | 35 842 | 64 543 |
| Unique reflections (no.) | 59 396 | 39 618 | 26 650 | 22 516 | 27 646 | 23 102 | 26 450 | 21 610 |
| Phasing powerb | 0.81 | 0.95 | 1.19 | 0.84 | 1.02 | 1.00 | ||
| Rcullisc | 0.629 | 0.704 | 0.718 | 0.697 | 0.690 | 0.636 | ||
| Riso (%)d | 8.5 | 10.6 | 9.1 | 9.5 | 10.5 | 17.7 | ||
| Heavy atom sites (no.) | 2 | 4 | 2 | 2 | 4 | 4 | ||
| Overall figure of merite |
|
|
|
|
|
|
|
0.42 |
| Refinement statistics: native 2 | ||||||||
| Completeness (%) | 98.7 | |||||||
| Resolution (Å) | 75.70–2.90 | |||||||
| Rwork/Rfree (%)f | 22.2/25.8 | |||||||
| R.m.s.d. | ||||||||
| bond angles (°) | 1.36 | |||||||
| bond lengths (Å) | 0.009 | |||||||
| B-values (Å2) | 2.5 | |||||||
| Solvent (no., water/sulfate) | 45/5 | |||||||
| Ramachandran analysis | ||||||||
| most favored (%/no.) | 86.5/610 | |||||||
| additional allowed (%/no.) | 12.8/90 | |||||||
| generously allowed (%/no.) | 0.1/1 | |||||||
| disallowed (%/no.) | 0.6/4 | |||||||
aRsym = ΣΣ|Ihkl – Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average value of intensity.
bPhasing power = r.m.s. (|Fh|/E), where |Fh| is the heavy atom structure factor amplitude and E is the residual lack of closure error.
cRCullis = Σ||Fh(obs)| – |Fh(calc)||/Σ|Fh(obs)| for centric reflections, where |Fh(obs)| is the observed heavy atom structure factor amplitudes and |Fh(calc)| is the calculated heavy atom structure factor amplitude.
dRiso = Σ||Fph| – |Fp||/Σ|Fp|, where |Fp| is the protein structure factor amplitude and |Fph| is the heavy atom derivative structure factor amplitude.
eFigure of merit = <|ΣP(α)eiα/ΣP(α)|>, where α is the phase and P(α) is the phase probability distribution.
fRwork = Σ||Fobs| – |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where all reflections belong to a test set of 10% data randomly selected in CNS.
The ‘io’ designator indicates the locations of the 5-iodouracil substitutions numbered in Figure 1A. ‘io12′/8′’ is a double 5-iodouracil-substituted oligodeoxynucleotide. ‘Dy’ and ‘Uranyl’ indicate dysprosium chloride and uranyl acetate, respectively.
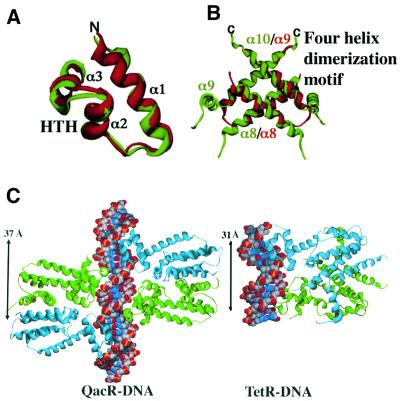
QacR is comprised of nine helices: α1(3–18), α2(25–32), α3(36–42), α4(46–71), α5(75–88), α6(96– 108), α7(110–136), α8(145–162) and α9(168–185). The first three helices form a three-helix bundle DNA-binding domain that contains an HTH motif (α2 and α3), in which α3 is the recognition helix. The DNA-binding domain is structurally similar to that of the DNA-binding domain of TetR (Orth et al., 2000) (r.m.s.d. of 1.6 Å for corresponding Cα residues 2–46 of QacR and 4–48 of TetR), with the most significant difference corresponding to the position of α1. The N-terminus of α1 is oriented differently in QacR to contact the phosphate backbone (Figure 2A). The drug-binding/dimerization domain of QacR is formed by helices 4–9. Although the inducer-binding domains of QacR and TetR display little sequence homology (<20%), they contain a region of significant structural homology that corresponds to two C-terminal antiparallel helices, which form a four-helix dimerization motif in each protein (Figure 2B). In QacR, these helices are α8 and α9 and account for the majority of the 1530 Å2 buried surface area per monomer. The analogous helices in TetR are non-contiguous (α8 and α10). Superimposition of these secondary structures alone results in an r.m.s.d. of 1.4 Å for the 49 corresponding Cα atoms despite a sequence identity of only 11% (Figure 2B).
Fig. 2. Structural homologues with distinct DNA recognition modes. (A) Superimposition of the three-helix bundle DNA-binding domains of QacR (red) and TetR (green) showing the high degree of structural homology within the DNA-binding domains of these proteins (r.m.s.d. = 1.6 Å). (B) Superimposition of the C-terminal antiparallel helices of QacR (red) and TetR (green) that form a four-helix dimerization motif in each protein. In QacR, these helices correspond to contiguous helices α8 and α9, while in TetR the helices are α8 and α10. The region between α8 and α9 is disordered in TetR. The remainder of the helices within the ligand-binding domains of these proteins show little correspondence, and superimpositions of the DNA-binding domains and four-helix dimerization regions must be carried out independently to observe the structural homology. (C) Comparison of two TetR family members bound to DNA: QacR and TetR (Orth et al., 2000). Despite belonging to the same family, QacR and TetR have different modes of recognition; QacR binds as a dimer of dimers and TetR as a single dimer. The bound DNA operator sites of each differ significantly in conformation. Also, note the larger HTH center-to-center distance (denoted by an arrow and measured as the distance between amide nitrogens of QacR residues Gly37 and Gly37′, which corresponds to Pro39 in TetR) of 37 Å in QacR in comparison with 31 Å in TetR.
The QacR–DNA complex structure reveals how two QacR dimers bind the extended 28 bp IR1 operator; each dimer docks on either side of the cognate DNA site such that two HTH motifs, one from each dimer, contact successive DNA major grooves (Figure 1B). This binding mode is consistent with the extended footprint of QacR and our recent results demonstrating that a pair of QacR dimers binds the extended IR1 site (Grkovic et al., 2001).
QacR–DNA contacts
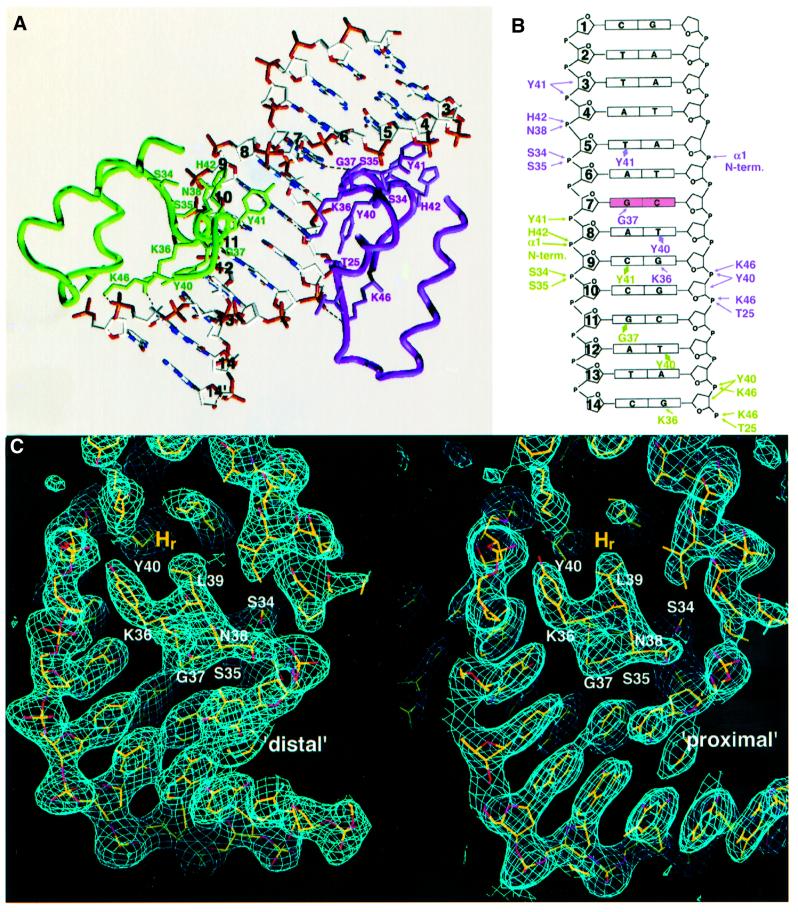
The two QacR dimers make a total of 16 base and 44 phosphate contacts with the IR1 site (Figure 3A and B). Only the DNA major groove is contacted, and the inter actions made by each QacR dimer to each DNA ‘half-site’ (nucleotides 1–14 and 1′–14′), as described below, are essentially identical (Figure 3A and B). Because each QacR monomer within a given dimer docks differently onto the DNA, one distal to the dyad that relates dimers and the other more proximal to the dyad, and recognizes distinct bases, we have designated the DNA contacts as ‘distal’ or ‘proximal’. The distal monomers (purple and red in Figure 1B) make the majority of the base-specific interactions, while the proximal monomers (green and cyan in Figure 1B) make several base and phosphate contacts. This arrangement is consistent with data revealing that the sequence of the outer inverted repeats is essential for specific binding, whereas the central six base pairs appear to be more important for providing the appropriate spacing for QacR binding. Specifically, the studies showed that addition to or removal of two base pairs from the wild-type 6 bp spacer eliminates QacR binding (Grkovic et al., 1998, 2001).
Fig. 3. QacR–DNA contacts. (A) QacR–DNA contacts in one IR1 ‘half-site’ colored as in Figure 1B. Note that the green and purple subunits are from different dimers (see Figure 1B) and interact with bases in the ‘same’ major groove. Only contacted nucleotides, i.e. 3–14′, are shown. The light blue sphere near the N-terminus of Gly37 (purple subunit) represents a tightly bound solvent molecule between the protomers in each major groove (an identically positioned solvent molecule is found in the other half-site). Selected hydrogen bonds are shown as dashed lines. Essentially identical interactions are observed in the other half-site involving the red and cyan subunits (Figure 1B). (B) Schematic showing QacR–DNA half-site contacts (colored as in Figure 3A). The bases are labeled and shown as rectangles. Hydrogen bonds are indicated by arrows, while van der Waals contacts are shown as diamonds. Phosphate and deoxyribose contacts are indicated by arrows to the appropriate P or sugar ring, the latter of which is numbered. (C) Simulated annealing (SA) composite omit maps shown for a ‘distal’ (left) and a ‘proximal’ (right) recognition helix. The map was calculated following SA at a starting temperature of 2000 K. Carbons, nitrogens, oxygens and phosphates are colored yellow, blue, red and dark yellow, respectively. This figure, in which the recognition helices (Hr) are positioned similarly, highlights the slightly different docking of each HTH motif as well as the pseudo-direct repeat recognized by each motif. This figure and Figure 6 were made with O (Jones et al., 1991).
Identical base contacts are made by the distal monomers (purple and red in Figure 1B) and include hydrophobic interactions between the Cβ methylene carbons of Tyr40 and Tyr41 to the C7 of T8′ and T5, respectively, and a hydrogen bond from the side chain Nε of Lys36 to the O6 of G9′. Residues Thr25, Ser34, Ser35, Asn38, Tyr40, Tyr41, His42 and Lys46 from each helix of the three-helix bundle contribute to 12 distal phosphate contacts. The positive helix dipole of the α1 N-terminus interacts with the phosphate backbone (Figure 3A and B). Contacts from the OH of Tyr40 and Tyr41 to cross-strand phosphates 3 and 9′, respectively, buttress these residues into position for base recognition. Also contributing to specificity is a hydrogen bond between the amide nitrogen of Gly37 and the O6 of G7. The presence of a glycine residue at this position is absolutely essential for tight docking of the recognition helix and leaves no space for a water molecule, suggesting a favorable entropic component for solvent release during binding. Nucleotide G7 is the location of the qacA transcription start site (Grkovic et al., 1998) and its interaction with Gly37 may be an important feature of the repression mechanism (Figures 1B, 3A and B).
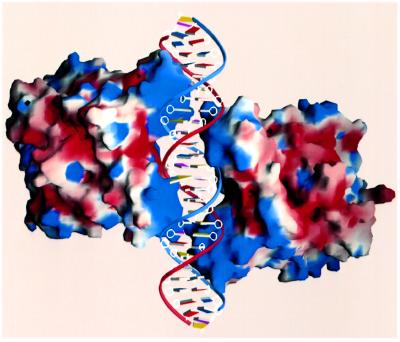
The amide nitrogen of Gly37′ (where the prime indicates the proximal monomer) is 3.8 Å and its Cα is 3.5 Å from the O6 of G11, thereby explaining a preference for guanine over adenine at this position. Moreover, the presence of any side chain on residue 37′ would clash with any base at position 11. The additional proximal base contacts are not specific, consistent with the lack of conservation of the 6 bp spacer, and include those from the Cβ methylene group of Tyr40′ and the Nε amino group of Lys36′ to the C7 of T12′ and the N7 of G14′, respectively (Figure 3A and B). Interestingly, although the distal and proximal DNA sequences differ (Figure 3B), the contacts made by the differently docked distal and proximal HTH motifs to each site are similar, e.g. Gly37 and Gly37′ contact guanines, and Tyr40 and Tyr40′ contact thymines. These sets of similar contacts thus unveil the presence of a pseudo direct repeat in the DNA bound by the distal and proximal HTH motifs. Ten phosphate contacts are supplied by each proximal monomer (five contacts to each strand) and are provided by the same residues that contact the distal region, with the exception of Asn38. Taken together, both the distal and proximal interactions divulge a distinguishing feature of DNA recognition by QacR, which is the high degree of structural complementarity between the surfaces of the binding partners. Formation of this complex results in the burial of a surface area of 4850 Å2 and no solvent is located at the protein–DNA interface. Binding is enhanced further by the complementary electrostatic fields of the highly basic QacR DNA-binding domain and the DNA phosphate backbone (Figure 4).
Fig. 4. The electrostatic surface potential of DNA-bound QacR. Electrostatic surface representation of QacR in the QacR–DNA complex with blue and red regions indicating positive and negative electrostatic regions, respectively. The view is that of Figure 1B rotated around the DNA axis by 180°. Note the highly basic nature of the DNA-binding domain. This figure was made with GRASP (Nicholls et al., 1991).
Structural basis for the inferred cooperative binding of DNA by QacR
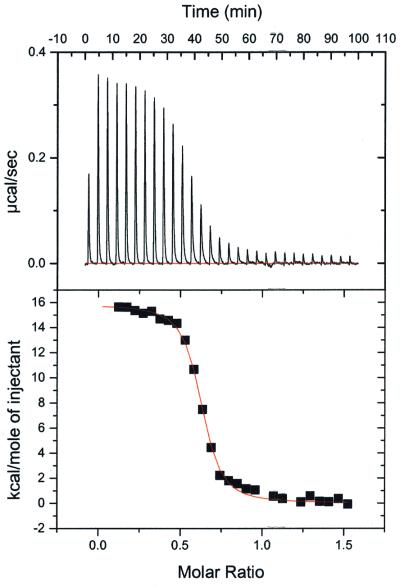
The QacR–DNA structure explains recent data, from gel filtration, chemical cross-linking and dynamic light scattering studies (DLS), which indicated that QacR binds IR1 as a pair of dimers (Grkovic et al., 2001). Notably, DLS and gel filtration experiments carried out in the presence of IR1 revealed molecular weights consonant only with two QacR dimers and a DNA duplex; no single dimer–DNA species were present. These data, combined with the finding that a single band is observed in QacR–IR1 gel shifts, suggest strongly that QacR binds DNA cooperatively as a pair of dimers. In addition, DNA fragments containing a single IR1 half-site are not bound by QacR (Grkovic et al., 1998). To determine the binding affinity of QacR for IR1 and to gain further insight into the DNA-binding mechanism of QacR, an isothermal titration calorimetry (ITC) experiment was carried out (Figure 5). QacR binding to a 36mer containing the IR1 site (see Materials and methods) was found to be endothermic (Figure 5). The apparent Kd was 49 ± 3 nM in the presence of 300 mM NaCl and the stoichiometry was 0.57 DNA duplex to QacR dimer. These data are consistent with a cooperative dimer of dimers binding mechanism.
Fig. 5. ITC data of QacR–IR1 binding. Representative data (raw data on top and curve fit below) from an ITC experiment in which a 36 bp IR1-containing duplex was titrated into the reaction cell containing QacR. Note that the first addition (not used in the fit) was half the volume of the other additions. Thermodynamic values obtained from the curve fit are: ΔS = 86.63 cal/mol/K, ΔH = 1.54 × 104 ± 134.7 kcal/mol, Kd = 49 ± 3 nM, n = 0.570 ± 0.003, where n is the stoichiometry of bound DNA per QacR dimer.
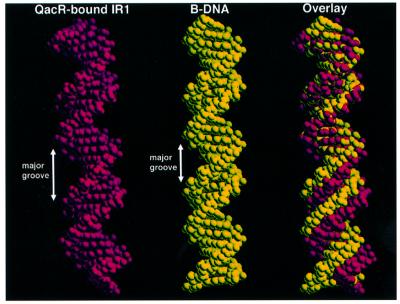
QacR is the only known TetR family member that appears to bind its operator cooperatively. The QacR–DNA structure demonstrates that such cooperative binding cannot be mediated by protein–protein interactions because the closest contacts are >5.0 Å between each of the dimers. Rather, the structure indicates that features of the DNA and protein conformations are key to binding cooperativity. Specifically, although the QacR-bound DNA displays characteristics that are B-DNA-like with respect to shift, slide, tilt, roll and propeller twisting (none) and a global bend of only 3.0° (Ravishanker et al., 1989), the major groove is widened markedly throughout the entire binding site (average major groove width = 12.6 Å, as compared with 11.0 Å for B-DNA) (Figure 6). There are two clustered regions where the DNA major groove is most widened. The first is within the A8C9C10 region (width = 12.9 Å) where the majority of base contacts, mediated from recognition helices of two subunits, occur (Figure 3A and B), and the second is located within the central base pairs of the DNA dyad axis (13.3 Å). Notably, this major groove expansion requires a significant unwinding of the DNA, which is reflected in the observed twist of 32.1° and the 11.2 bp per turn pitch, in contrast to a twist of 34.3° and 10.5 bp per turn observed in canonical B-DNA. The importance of these features in the binding of IR1 by QacR is underscored by the 37 Å center-to-center distances between the recognition helices of each QacR dimer (as measured by the distance between the amide nitrogens of Gly37), which would be only 34 Å for proteins that bind consecutive major grooves of B-DNA. The identical center-to-center distances and the DNA contacts of each QacR dimer suggest that binding of the first dimer, the energetically costly step, invokes an induced fit of the protein and DNA to produce optimal major groove interaction. In turn, a DNA conformation is created that would readily bind the second QacR dimer, thus effectively locking the IR1 site into an unwound conformation.
Fig. 6. Major groove widening of IR1 by QacR binding. Comparison of the QacR-bound IR1 28 bp operator (red) to a B-DNA strand (yellow) of the same sequence (generated using Sybyl6.7). On the right is an overlay in which nucleotides of the first turn were superimposed. This overlay underscores the overall widening of the DNA major groove of the operator site bound by QacR.
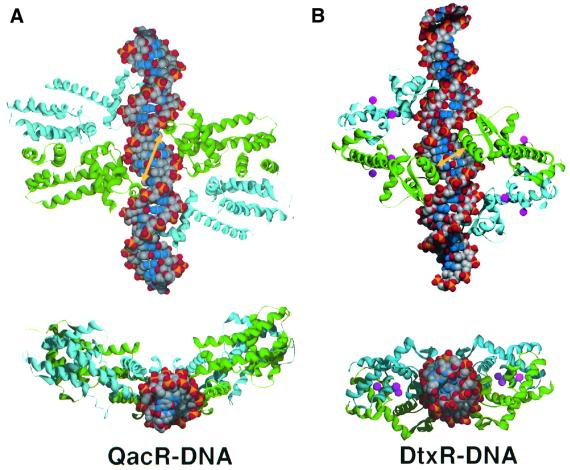
Other proteins that bind DNA as cooperative multimers include the Escherichia coli MetJ (Somers et al., 1992), phage P22 Arc protein (Raumann et al., 1994), E.coli tryptophan repressor, TrpR (Lawson and Carey, 1993), homeodomain protein pax (Wilson et al., 1995) and diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae (White et al., 1998; Pohl et al., 1999; Chen et al., 2000). The DNA-binding mechanism of QacR resembles most closely that utilized by DtxR, which also binds DNA cooperatively as a tetramer (White et al., 1998; Chen et al., 2000). As observed in the QacR–IR1 complex, there are no cross-dimer contacts in DtxR, and the cooperativity is also presumed to arise from alterations in the DNA conformation (White et al., 1998). However, a key distinction between the DNA-binding modes of QacR and DtxR is the location of the proximal recognition helices (Figure 7). In DtxR, these helices are separated by 5 bp, whereas the proximal recognition helices of QacR are separated by 10 bp. Moreover, in QacR, the proximal recognition helix of one dimer is 5 bp from the distal recognition helix of the other dimer (Figures 1B and 7). In contrast, in DtxR, the distal recognition helices are 10 bp from the proximal helices of the other dimer (Figure 7). A further difference is found in the placement of each dimer on the DNA, which can be cast in terms of the locations of the molecular dyads that relate each dimer. Whereas the molecular 2-fold axes relating each DtxR monomer of a dimer lie offset but in the same plane and antiparallel to each other, the corresponding QacR 2-fold axes do not lie in the same plane. The result is the creation of a triangular cavity between each dimer and free access to the opposite side of the IR1 site (Figure 7). QacR provides the first example where two consecutive major grooves are contacted by two HTH motifs from separate dimers.
Fig. 7. Comparison of (A) QacR and (B) DtxR: DNA binding by pairs of dimers. The yellow arrows highlight the center-to-center approaches of the proximal recognition helices of each dimer. Note the 10 bp separation of the QacR proximal helices, which contrasts with the 5 bp separation of the corresponding helices of DtxR (White et al., 1998). Monomers within a dimer are colored cyan and green, and the nickel ions bound by DtxR are colored magenta. The bottom view is shown looking down the DNA helical axis of each. The DNA is shown as CPK with phosphates, oxygen, carbon and nitrogen colored yellow, orange, gray and blue, respectively.
Structurally homologous repressors with distinct DNA-binding mechanisms
As would be anticipated for proteins belonging to the same family, QacR and TetR share structurally homologous DNA-binding motifs that consist of a three-helix bundle with a HTH that inserts into the DNA major groove (Figure 2A and C). Furthermore, both proteins sink their recognition helices deeply into the major groove floor such that no water-mediated base contacts are utilized in base pair binding. Neither QacR nor TetR (Orth et al., 2000) binds the DNA minor groove. An unusual feature of QacR, which is shared by TetR and probably all TetR/CamR family members, is its short recognition helix. The seven residues of this helix are fully engaged in DNA binding, with the only exception of QacR residue Leu39 (Figure 3C), which is part of the hydrophobic interior of the protein. The recognition helix of TetR is engaged similarly but recruits Arg28, from outside the recognition helix, to make a base pair-specific contact (Orth et al., 2000). QacR has taken a more drastic approach to ensure DNA-binding specificity and high affinity by employing two cooperatively binding dimers. Another notable difference between these complexes is the conformation of the bound DNA. TetR kinks its binding site and induces a 17° bend towards the protein, which optimizes the positioning of its HTH motifs for specific base interactions within each DNA half-site. This DNA distortion also includes localized major groove widening. QacR, on the other hand, smoothly widens its entire IR1-binding site major groove and bends its DNA site by only 3°. These distinctions are reflected in the different HTH center-to-center distances observed in QacR and TetR; in QacR, this distance is 37 Å compared with 31 Å for TetR (Figure 2C). The contacts made to each symmetric DNA half-site by each TetR HTH motif are essentially identical, as is usually observed in protein–DNA interactions involving a dimer binding to a palindromic sequence. In contrast, QacR binding represents a departure from this common mode of recognition in that each HTH motif of a given dimer makes a different set of contacts, each with a non-palindromic site. Thus, sequence-specific DNA binding by QacR and TetR displays significant mechanistic differences (Figure 2C).
In conclusion, the crystal structure of the QacR–IR1 complex uncovers a new mode of DNA binding by a TetR family member that involves the cooperative binding of a pair of dimers to an extended site. Moreover, such binding requires each HTH motif to contact different bases within the non-symmetrical IR1 half-site. The inferred cooperativity does not result from direct protein–protein contacts, but rather from global DNA undertwisting and major groove widening caused by the binding of two QacR dimers. A mechanism is suggested in which binding of the first dimer forces the DNA into the ‘widened’ conformation, which in turn favors the binding of the second dimer. Whether other TetR family members bind in such a manner remains to be ascertained. Finally, this study demonstrates that even structurally homologous proteins of the same family, which share a homologous function, such as transcription repression, can utilize significantly different mechanisms of action. Thus, the structure of one family member is clearly insufficient to describe all members.
Materials and methods
Crystallization and data collection of the QacR–28mer DNA complex
Mutant QacR (C72A/C141S with a His6 tag) was overexpressed and purified using Ni2+-NTA chromatography as described (Grkovic et al., 1998, 2001). This mutation was made to prevent the serious cross-linking and oxidation problems observed in the wild-type protein. The double mutant QacR is fully active and its DNA binding and drug binding are essentially identical in the wild-type protein (Grkovic et al., 2001). Several oligodeoxynucleotides, containing the IR1 site and ranging from 26 to 44 bp, were used in crystallization trials and produced 12 crystal forms that were not of diffraction quality. Diffraction quality crystals were obtained with the palindromic 28 bp operator site. To crystallize the complex, QacR (in 20 mM Tris pH 7.5, 100 mM NaCl), at a concentration of 10–20 mg/ml, was mixed with DNA at a molar ratio of ∼1:1 (QacR dimer:DNA duplex). Several concentration ratios were tested. Molar ratios of 2:1 and 1:1 produced crystals, the latter being optimal. The QacR–DNA solution was mixed 1:1 with a reservoir solution of saturated lithium sulfate, 10 mM magnesium sulfate and 0.1 M HEPES pH 7.5. Crystals grew slowly over a period of several months, with typical dimensions of 0.6 × 0.5 × 0.3 mm, and diffracted weakly (3.3 Å resolution) on in-house sources. Because the crystallization condition is a suitable cryo-solvent, the crystals can be looped and flash-frozen directly from the drop. The crystals take the space group P65 with a = b = 174.7 Å and c = 151.9 Å. X-ray diffraction data for the initial native and all derivatives were collected at 100 K with an RAXIS IV and a Rigaku RU300-HB rotating anode using Yale focusing mirrors. The data were processed with BIOTEX (MSC/Rigaku, The Woodlands, TX). A high-resolution native data set to 2.90 Å resolution was collected later at 100 K at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline BL 9-2 and processed with MOSFLM.
Structure determination and refinement of the QacR–DNA complex
The structure of the QacR–IR1 complex was solved by MIR using four iodine sites from DNA, in which thymines were substituted with 5-iodouracil (Figure 1A), and dysprosium and uranyl heavy atom sites obtained from 2- and 3-day soaks, respectively (Table I). The crystals contain two QacR dimers and a 28 bp duplex in the asymmetric unit. The dysprosium and uranium sites were located between acidic resides of the DNA-binding domain and the DNA phosphate backbone. Heavy atom parameters were refined, MIR phases calculated (Table I), and phase extension to 3.3 Å resolution and solvent leveling were carried out in PHASES (Furey and Swaminathan, 1997). Because the crystals contain 80% solvent, the initial solvent-leveled 3.3 Å resolution MIR map was of excellent quality and revealed the entire phosphate backbone, most of the DNA bases and the majority of the QacR helices. Two-fold averaging further improved the electron density to allow tracing of not only the 28mer duplex, but both QacR dimers as well. CNS (Brünger et al., 1998) was also used in phase refinement and density modification (solvent flipping). The resulting electron density map was used in conjunction with the non-averaged and averaged PHASES maps for final model building using the ‘native 1’ data set to 3.35 Å resolution. This model was subjected to simulated annealing (SA) in CNS (Brünger et al., 1998). This partially refined model was used as the starting model for refinement against the SSRL 2.90 Å resolution native data. The model was first subjected to rigid body refinement in CNS followed by SA. This was followed by multiple rounds of rebuilding in O (Jones et al., 1991) and refinement (combined SA/positional/thermal parameter refinement) in CNS (Brünger et al., 1998). The final Rwork is 22.2% and the Rfree is 25.8% using all data (75.70–2.90 Å) when refining individual isotropic B-factors (highly constrained), and an Rwork of 22.0% and Rfree of 25.5% (using all data) when refining grouped B-factors (side chains, main chain and nucleotides). The final model includes the entire 28 bp DNA duplex, residues 2–188 and the first histidine of the His6 tag for three subunits, and residues 2–186 for the fourth subunit. The model shows excellent stereochemistry, with only four Ramachandran outliers (Laskowski et al., 1993), corresponding to residue Tyr92 in all subunits (Table I). Notably, Tyr92 has been shown to be a key residue in the multidrug induction mechanism of QacR (Schumacher et al., 2001).
ITC
For ITC experiments, purified QacR and the IR1-containing deoxyoligonucleotide (with the sequence 5′-AATCCTTATAGACCGATCGATCGGTCTATAAGGATT-3′, where the IR1 site is in bold) were dialyzed extensively against 50 mM Tris pH 7.5, 300 mM NaCl, 50 mM imidazole and 5% glycerol. Samples were degassed by vacuum aspiration for 5 min prior to loading, and all titrations were performed at 25°C. Calorimetric assays were performed using a VP-ITC from MicroCal Inc. (MicroCal, Northhampton, MA). The reaction cell (∼1.4 ml) was filled with the degassed solutions. The stirring speed was 300 r.p.m. and the thermal power was recorded every 10 s. The protein solution was contained in the cell at a concentration of 0.008 mM and the titrated ligand DNA concentration was 0.05 mM. Thermogram analysis was performed using the Origin 5.0 package supplied with the instrument.
Coordinates
Coordinates and structure factors for the QacR–DNA complex have been deposited with the Protein Data Bank under the accession code 1JT0.
Acknowledgments
Acknowledgements
M.A.S. is a Burroughs Wellcome Career Development Awardee in Biomedical Sciences. S.G. was the recipient of an Australian Postgraduate Award. This work was supported by grants AI48593 from the National Institutes of Health to R.G.B. and Project Grant 153818 from the National Health and Medical Research Council (Australia) to R.A.S. Intensity data collection at the Stanford Synchrotron Radiation Laboratory (SSRL) was carried out under the auspices of the SSRL biotechnology program, which is supported by the National Institutes of Health, National Center for Research Resources Biomedical Technology Program and the Department of Energy, Office of Biological and Environmental Research.
References
- Ambudkar S.V., Dey,S., Hrycyna,C.A., Ramachanra,M., Pastan,I. and Gottesman,M.M. (1999) Biochemical, cellular and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol., 39, 361–398. [DOI] [PubMed] [Google Scholar]
- Aramaki H., Yagi,N. and Suzuki,M. (1995) Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng., 8, 1259–1266. [DOI] [PubMed] [Google Scholar]
- Behr H., Reverdy,M.E., Mabilat,C., Freney,J. and Fleurette,J. (1994) Relationship between the level of minimal inhibitory concentrations of five antiseptics and the presence of qacA gene in Staphylococcus aureus. Pathol. Biol., 42, 438–444. [PubMed] [Google Scholar]
- Brown M.H. and Skurray,R.A. (2001) Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol., 3, 163–170. [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen C.S., White,A., Love,J., Murphy,J.R. and Ringe,D. (2000) Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry, 39, 10397–10497. [DOI] [PubMed] [Google Scholar]
- Eckert B. and Beck,C.F. (1989) Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol., 171, 3557–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folcher M., Morris,R.P., Dale,G., Salah-Bey-Hocini,K., Viollier,P.H. and Thompson,C.J. (2001) A transcriptional regulator of a pristina mycin resistance gene in Streptomyces coelicolor. J. Biol. Chem., 276, 1479–1485. [DOI] [PubMed] [Google Scholar]
- Furey W.B. and Swaminathan,S. (1997) PHASES-95: a program package for the processing and analysis of diffraction data from macromolecules. Methods Enzymol., 277, 590–620. [DOI] [PubMed] [Google Scholar]
- Gottesman M.M., Pastan,I. and Ambudkar,S.V. (1996) P-glycoprotein and multidrug resistance. Curr. Opin. Genet. Dev., 6, 610–617. [DOI] [PubMed] [Google Scholar]
- Grkovic S., Brown,M.H., Roberts,N.J., Paulsen,I.T. and Skurray,R.A. (1998) QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem., 273, 18665–18673. [DOI] [PubMed] [Google Scholar]
- Grkovic S., Brown,M.H., Schumacher,M.A., Brennan,R.G. and Skurray, R.A. (2001) The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol., 183, 7102–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hinrichs W., Kisker,C., Duvel,M., Muller,A., Tovar,K., Hillen,W. and Saenger,W. (1994) Structure of the Tet repressor–tetracycline complex and regulation of antibiotic resistance. Science, 264, 418–420. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kisker C., Hinrichs,W., Tovar,K., Hillen,W. and Saenger,W. (1995) The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol., 247, 260–280. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lawson C.L. and Carey,J. (1993) Tandem binding in crystals of a trp repressor/operator half-site complex. Nature, 366, 178–182. [DOI] [PubMed] [Google Scholar]
- Levy S. (1992) Active efflux mechanisms for antibiotic resistance. Antimicrob. Agents Chemother., 36, 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S., Boos,M.S., Beyer,A., Fluit,A.C. and Schmitz,F.J. (2001) Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother., 47, 896–897. [DOI] [PubMed] [Google Scholar]
- Mitchell B.A., Paulsen,I.T., Brown,M.H. and Skurray,R.A. (1999) Bioenergetics of the staphylococcal multidrug export protein QacA: identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem., 274, 3541–3548. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B.H. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Orth P., Schnappinger,D., Hillen,W., Saenger,W. and Hinrichs,W. (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nature Struct. Biol., 7, 215–219. [DOI] [PubMed] [Google Scholar]
- Paulsen I.T., Brown,M.H., Littlejohn,T.G., Mitchell,B.A. and Skurray,R.A. (1996) Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl Acad. Sci. USA, 93, 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl E., Holmes,R.K. and Hol,W.G. (1999) Crystal structure of a cobalt-activated diphtheria toxin repressor–DNA complex reveals a metal-binding SH3-like domain. J. Mol. Biol., 292, 653–667. [DOI] [PubMed] [Google Scholar]
- Putman M., van Veen,H.W and Konings,W.N. (2000) Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev., 64, 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raumann B.E., Rould,M.A., Pabo,C.O. and Sauer,R.T. (1994) DNA recognition by β-sheets in the Arc repressor–operator crystal structure. Nature, 367, 754–757. [DOI] [PubMed] [Google Scholar]
- Ravishanker G., Swaminathan,S., Beveridge,D.L., Lavery,R. and Sklenar,H. (1989) Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCCGAATTGCGCG) double helix: ‘curves’, dials and windows. J. Biomol. Struct. Dynam., 6, 669–699. [DOI] [PubMed] [Google Scholar]
- Saier M.H. and Paulsen,I.T. (2001) Phylogeny of multidrug transporters. Semin. Cell Dev. Biol., 12, 205–213. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Miller,M.C., Grkovic,S., Brown,M.H., Skurray,R.A. and Brennan,R.G. (2001) Structural mechanisms of QacR induction and multidrug recognition. Science, 294, 2158–2163. [DOI] [PubMed] [Google Scholar]
- Somers W.S. and Phillips,S.E. (1992) Crystal structure of the met repressor–operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature, 359, 387–393. [DOI] [PubMed] [Google Scholar]
- Tennent J.M., Lyon,B.R., Midgley,M., Jones,I.G., Purewal,A.S. and Skurray,R.A. (1989) Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J. Gen. Microbiol., 135, 1–10. [DOI] [PubMed] [Google Scholar]
- White A., Ding,X., vanderSpek,J.C., Murphy,J.R. and Ringe,D. (1998) Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature, 394, 502–507. [DOI] [PubMed] [Google Scholar]
- Wilson D.S., Guenther,B., Desplan,C. and Kuriyan,J. (1995) High resolution crystal structure of a paired (pax) homeodomain dimer on DNA. Cell, 82, 709–719. [DOI] [PubMed] [Google Scholar]