Abstract
RNA localization is a widespread mechanism for achieving localized protein synthesis. In Saccharo myces cerevisiae, Ash1 is a specific repressor of transcription that localizes asymmetrically to the daughter cell nucleus through the localization of ASH1 mRNA to the distal tip of the daughter cell. This localization depends on the actin cytoskeleton and five She proteins, one of which is a type V myosin motor, Myo4. We show here that a novel RNA-binding protein, Khd1 (KH-domain protein 1), is required for efficient localization of ASH1 mRNA to the distal tip of the daughter cell. Visualization of ASH1 mRNA in vivo using GFP-tagged RNA demonstrated that Khd1 associates with the N element, a cis-acting localization sequence within the ASH1 mRNA. Co-immunoprecipitation studies also indicated that Khd1 associates with ASH1 mRNA through the N element. A khd1Δ mutation exacerbates the phenotype of a weak myo4 mutation, whereas overexpression of KHD1 decreases the concentration of Ash1 protein and restores HO expression to she mutants. These results suggest that Khd1 may function in the linkage between ASH1 mRNA localization and its translation.
Keywords: ASH1/KH domain/mRNA localization/RNA-binding protein/translational control
Introduction
The asymmetric distribution of proteins is vital to cellular function and cell fate determination. One mechanism for achieving asymmetric distribution of a protein is by localizing its mRNA to a distinct site within the cell. Localization of mRNAs is specified by sequences generally found in the 3′ untranslated region (3′ UTR) of the mRNA, and is mediated by cytoskeletal filaments that are required for transport and subsequent anchoring of the mRNA at its final destination (Wilhelm and Vale, 1993; St Johnston, 1995; Nasmyth and Jansen, 1997; Oleynikov and Singer, 1998). The transport, anchoring and translational regulation of localized transcripts are governed by proteins that form large ribonucleoprotein complexes with the mRNAs (Wilhelm and Vale, 1993; Hazelrigg, 1998).
The asymmetric distribution of Ash1 in the budding yeast Saccharomyces cerevisiae provides an excellent opportunity to study the asymmetric segregation of cell fate determinants resulting from mRNA localization. Ash1 is a cell-type specific transcriptional repressor that determines proper mating-type switching by differentially regulating expression of the HO endonuclease (Bobola et al., 1996; Sil and Herskowitz, 1996). Ash1 is found in the nucleus of daughter cells, where it represses HO transcription and ultimately prevents mating-type switching in these cells (Bobola et al., 1996; Sil and Herskowitz, 1996). This transcriptional regulation of HO expression restricts mating-type switching to mother cells (Nasmyth, 1983; Herskowitz, 1988). The asymmetric distribution of Ash1 to daughter cell nuclei is a result of the localization of ASH1 mRNA to the distal tips of daughter cells (Long et al., 1997; Takizawa et al., 1997).
Five genes have been identified that are required for ASH1 mRNA localization; SHE1–SHE5 (Jansen et al., 1996; Long et al., 1997; Takizawa et al., 1997). SHE1 encodes a type V myosin motor, Myo4, which co-localizes with ASH1 mRNA at the tip of daughter cells (Haarer et al., 1994; Bertrand et al., 1998; Munchow et al., 1999; Takizawa and Vale, 2000). Using a live-cell assay, particles containing Myo4 and ASH1 mRNA were observed to move rapidly from mother cells to daughter cells, suggesting that Myo4 plays a direct role in transporting ASH1 mRNA to the bud tip (Bertrand et al., 1998; Beach et al., 1999; Takizawa and Vale, 2000). Immunoprecipitation experiments have revealed that Myo4 associates with ASH1 mRNA and that this association is dependent on SHE2 and SHE3 (Munchow et al., 1999; Takizawa and Vale, 2000). SHE2 encodes an RNA-binding protein that directly binds to ASH1 mRNA (Bohl et al., 2000; Long et al., 2000). The C-terminus of She3 interacts with She2, while its N-terminus interacts with Myo4 (Bohl et al., 2000; Long et al., 2000). Thus, She3 has the properties of an adapter that links Myo4 to the She2–ASH1 mRNA complex. SHE5 is identical to BNI1, which was shown to encode a protein involved in regulating the actin cytoskeleton (Jansen et al., 1996; Kohno et al., 1996; Evangelista et al., 1997). She4 is also hypothesized to be required for proper organization of the actin cytoskeleton (Jansen et al., 1996; Wendland et al., 1996). Taken together, these results suggest that ASH1 mRNA is localized to the bud tip by actomyosin-based transport. Loc1, a nuclear RNA-binding protein, is also involved in ASH1 mRNA localization (Long et al., 2001).
Based on these studies, the following model for ASH1 mRNA localization has been proposed (Bohl et al., 2000; Long et al., 2000, 2001; Takizawa and Vale, 2000; Kwon and Schnapp, 2001). First, the ASH1 mRNA is identified by Loc1 in the nucleus. Secondly, the ASH1 mRNA is transported through the nuclear pores to the cytoplasm, where it binds to the cytoplasmic RNA-binding protein She2. Thirdly, the She2–ASH1 mRNA complex associates with Myo4 via the She3 adapter protein. Finally, the ASH1 mRNA–She2–She3–Myo4 complex is transported to the distal tips of daughter cells along polarized actin filaments.
In cases where protein localization is determined by mRNA localization, it can be expected that translation of the mRNA would be blocked until its proper localization at the distant site. Thus, mRNA localization is likely to be tightly coupled to its translational control (Curtis et al., 1995; St Johnston, 1995; Preiss and Hentze, 1999). Indeed, several examples are known in which translational control is directly linked to protein localization. For example, in Drosophila, translation of maternal oskar mRNA is silenced during transport to the posterior pole of the oocyte and later activated when Oskar protein is required (Macdonald and Smibert, 1996). It has been shown that the protein Bruno binds to the 3′ UTR of oskar mRNA and prevents premature translation (Kim-Ha et al., 1995; Gunkel et al., 1998). It is therefore likely that additional components, such as RNA-binding proteins, contribute to efficient localization of ASH1 mRNA through regulation of its translation.
During our studies on the identification and characterization of RNA-binding proteins required for ASH1 mRNA localization, we identified a previously uncharacterized yeast protein, Khd1. In this study we show that Khd1 is required for the tight anchoring of ASH1 mRNA to the distal tip of the daughter cell. Khd1 both co-localizes and physically associates with ASH1 mRNA. Over expression of Khd1 causes decreased Ash1 protein concentrations. These results suggest that Khd1 functions in the linkage between ASH1 mRNA localization and its translation.
Results
A putative RNA-binding protein involved in proper localization of ASH1 mRNA
To identify proteins required for ASH1 mRNA localization, we carried out a systematic survey of the different candidate RNA-binding proteins and their effects on ASH1 mRNA localization. The yeast genome contains five genes, PUF1/JSN1, PUF2, PUF3, PUF4/YGL014w and PUF5/MPT5, that code for homologs of the Puf family of RNA-binding proteins (Zhang et al., 1997; Olivas and Parker, 2000; Tadauchi et al., 2001) and five genes, MER1, MSL5, PBP2, SCP160 and YBL032w, which code for proteins that contain the KH RNA-binding motif (Engebrecht and Roeder, 1990; Van Dyck et al., 1994; Abovich and Rosbash, 1997; Weber et al., 1997; Mangus et al., 1998). We constructed mutants of each of these nine genes by PCR-mediated gene disruption, except for MSL5, which is an essential gene (Abovich and Rosbash, 1997) (see Materials and methods). Disruptants of each of the nine genes were viable, although mpt5Δ and scp160Δ mutants exhibited temperature-sensitive growth at 37°C. We examined ASH1 mRNA localization in these deletions by in situ hybridization (Table I). ASH1 mRNA was partially delocalized in mpt5Δ, scp160Δ and ybl032wΔ mutants, whereas ASH1 mRNA was properly localized in the other six mutants. As YBL032w had not been characterized previously, we designated it KHD1 (KH-domain protein 1).
Table I. ASH1 mRNA localization in disruptants of genes encoding RNA-binding proteins.
| Genotype | % (n = 100) | |||
|---|---|---|---|---|
| Anchored | Delocalized in the bud | Delocalized in mother and bud | Neck | |
| Wild type | 87 | 12 | 1 | 0 |
| puf1Δ/jsn1Δ | 85 | 14 | 1 | 0 |
| puf2Δ | 69 | 29 | 2 | 0 |
| puf3Δ | 85 | 15 | 0 | 0 |
| puf4Δ | 83 | 16 | 1 | 0 |
| puf5Δ/mpt5Δ | 22 | 61 | 16 | 1 |
| scp160Δ | 23 | 61 | 16 | 1 |
| pbp2Δ | 79 | 20 | 1 | 0 |
| khd1Δ/ybl032wΔ | 53 | 40 | 7 | 0 |
| mer1Δ | 78 | 20 | 2 | 0 |
Anchored: tightly localized ASH1 mRNA at the distal tip; delocalized in the bud: delocalized ASH1 mRNA confined to the bud; delocalized in mother and bud: ASH1 mRNA in both mother cell and bud; neck: ASH1 mRNA at the bud neck.
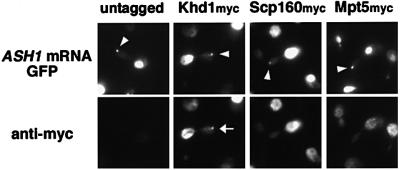
To assess whether Khd1, Scp160 and/or Mpt5 play a direct role in ASH1 mRNA localization, we analyzed whether these proteins co-localized with ASH1 mRNA using a system in which U1A-tagged ASH1 mRNA is marked with green fluorescent protein (GFP) (Takizawa and Vale, 2000). In this experimental system, cells are transformed with two plasmids, U1Ap-GFP and U1Atag-ASH1. U1Ap-GFP expresses a fusion protein of the RNA-binding domain of U1A and a variant of GFP (S65T) in which Ser65 is changed to threonine. U1Atag-ASH1 expresses ASH1 mRNA containing the U1A-binding sequence downstream of the start codon under the control of the GAL1 promoter. Cells expressing U1Ap-GFP and U1Atag-ASH1 display a single large GFP particle localized to the distal tips of daughter cells, and Myo4, She2 and She3 co-localize with this particle (Takizawa and Vale, 2000). We constructed strains harboring myc-tagged versions of Khd1, Scp160 and Mpt5 as described in Materials and methods. These tagged proteins carry 13 repeats of the c-myc peptide at the C-terminus of each protein. These strains displayed normal localization of ASH1 mRNA, indicating that the addition of the myc-tag to the proteins did not impair their function (data not shown). Khd1myc, Scp160myc and Mpt5myc strains were transformed with U1Ap-GFP and U1Atag-ASH1 plasmids and tested for co-localization of the myc-tagged proteins with the GFP signal. Khd1myc co-localized with the GFP-tagged U1Atag-ASH1 RNA particle, whereas Scp160myc and Mpt5myc did not (Figure 1). These results suggest that Khd1 has a direct role in ASH1 mRNA localization. Below, we further characterize the role of Khd1 in the regulation of ASH1 mRNA localization.
Fig. 1. Khd1 co-localizes with the U1Atag-ASH1 RNA particle. Thirteen repeats of the c-myc peptide sequence were inserted at the C-terminus of Khd1, Scp160 and Mpt5 in wild-type cells (10B). All samples expressed both U1Ap-GFP (pPT220) and U1Atag-ASH1 (pPT120). In untagged cells, GFP fluorescence from the U1A-ASH1 RNA particle is visible at the distal tip of the bud (arrowhead), but no staining was detected with the anti-myc antibody. In YKEN203 (Khd1myc) cells, GFP fluorescence from the U1A-ASH1 RNA particle co-localizes with anti-myc immunofluorescence (arrow). In YKEN202 (Scp160myc) and YKEN201 (Mpt5myc) cells, the U1A-ASH1 RNA GFP particle is visible at the distal tip of the bud (arrowhead) but does not co-localize with anti-myc fluorescence.
The N element of ASH1 mRNA is responsible for co-localization with Khd1
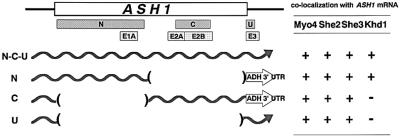
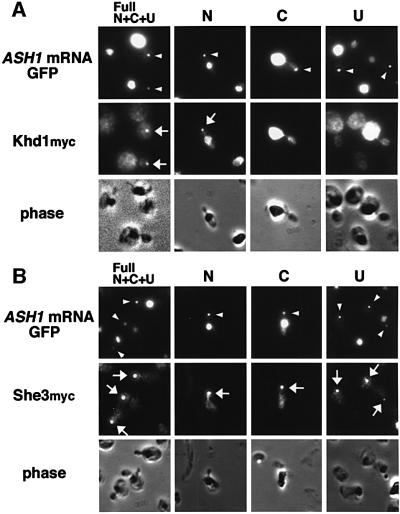
ASH1 mRNA contains three or four cis-acting localization elements: N, C and U (Gonzalez et al., 1999), or E1, E2AB and E3 (Chartrand et al., 1999) (Figure 2). Each of these elements is sufficient for localization of a heterologous reporter mRNA to daughter cells. Two regions (N, C; E1, E2AB) are located in the ASH1 open reading frame (ORF), whereas the U and E3 regions are located in the 3′ UTR. To determine which regions are responsible for the co-localization of ASH1 mRNA with Khd1, we constructed U1A-tagged versions of each element, U1Atag-N, -C and -U, in addition to U1Atag-Full, which contains all of these elements (Figure 2). Each of these constructs produced a bright particle in buds when co-expressed with U1Ap-GFP, indicating that each of the three RNA elements is sufficient to form a particle and localize to buds in the U1Atag constructs that we used (Figure 3). We then tested co-localization of Khd1myc, Myo4myc, She2myc and She3myc to each element (Figures 2 and 3). We found that Myo4myc, She2myc and She3myc co-localized with the GFP signals from all three derivatives of the U1Atag-ASH1 RNA particle (Figures 2 and 3). In contrast, Khd1myc co-localized with U1Atag-N but not with U1Atag-C or U1Atag-U (Figures 2 and 3). These results suggest that Khd1 may have a role different from that of Myo4, She2 and She3, which function in ASH1 mRNA transport (Bertrand et al., 1998; Munchow et al., 1999; Bohl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000).
Fig. 2. Localization elements involved in ASH1 mRNA localization and co-localization with Myo4, She2, She3 and Khd1. ASH1 mRNA contains three or four localization elements [N, C and U in Gonzalez et al. (1999); E1, E2AB and E3 in Chartrand et al. (1999)]. U1Atag-N, U1Atag-C and U1Atag-U are U1A-tagged versions of each element. Right column indicates co-localization of Myo4myc, She2myc, She3myc and Khd1myc to GFP particles of U1A-tagged versions of U1Atag-Full, U1Atag-N, U1Atag-C and U1Atag-U. +, co-localization; –, no co-localization.
Fig. 3. The N element of ASH1 mRNA is responsible for co-localization of Khd1. (A) Khd1myc co-localizes with U1Atag-N but not with U1Atag-C or U1Atag-U. (B) She3myc co-localizes with U1Atag-N, U1Atag-C and U1Atag-U. Arrowhead, GFP fluorescence from the U1A-ASH1 RNA particle visible at the distal tip of the bud; arrow, anti-myc immunofluorescence that co-localizes with the GFP fluorescence from the U1A-ASH1 RNA particle. Strains used: YKEN203 (Khd1myc), 134 (She3myc).
Khd1 associates with ASH1 mRNA in vivo
Co-localization of ASH1 mRNA and Khd1 suggested that Khd1 is associated with ASH1 mRNA in vivo. To test this possibility, we investigated whether ASH1 mRNA co-immunoprecipitated with Khd1myc using immunoprecipitation and RT–PCR. We used myc-tagged She3 as a positive control, as Munchow et al. (1999) have shown that ASH1 mRNA co-immunoprecipitates with She3myc. Khd1myc and She3myc strains were transformed with a control plasmid and YEpASH1. Cell lysates were prepared from these strains and used for immunoprecipitation with anti-myc monoclonal antibody. The anti-myc antibody efficiently precipitated Khd1myc and She3myc proteins from yeast extracts (Figure 4A). By RT–PCR analysis of the immunoprecipitates, we detected endogenous ASH1 mRNA in immunoprecipitates from Khd1myc and She3myc strains. In contrast, we did not detect ASH1 mRNA in immunoprecipitates from the untagged strain, even when ASH1 was overexpressed (Figure 4A). The PCR product was not seen when reverse transcriptase was omitted, indicating that formation of this band is dependent on RNA (data not shown). These data indicate that Khd1 associates with ASH1 mRNA in vivo.
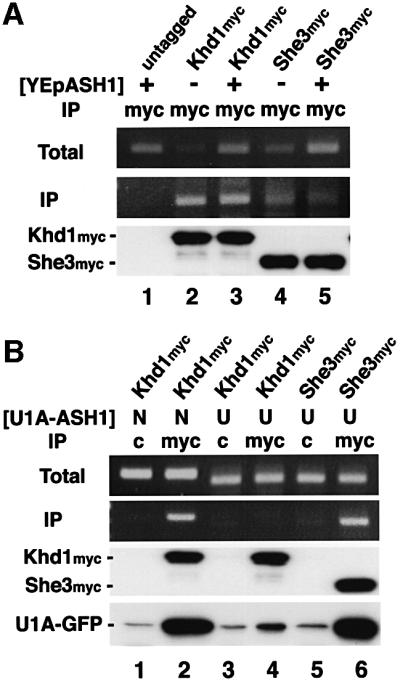
Fig. 4. Khd1 associates with ASH1 mRNA. Khd1 and She3 tagged with the myc epitope were immunoprecipitated using anti-myc antibody 9E10 (myc) or control IgG (c) as described in Materials and methods. Each immunopellet was separated on a 10% SDS–PAGE gel, blotted and probed with anti-myc antibody or anti-GFP antibody for the presence of epitope-tagged proteins (Khd1myc, She3myc or U1Ap-GFP). RNA was extracted from cell extracts (Total) and immuno precipitates (IP) and used as template for RT–PCR. (A) A 360 bp product was amplified using ASH1-specific primers. (B) PCR products of 420 and 380 bp were amplified using specific primers for U1Atag-N (N) and U1Atag-U (U), respectively. One-fifth of each reaction was separated on a 2% agarose gel and stained with ethidium bromide. (A) Lane 1, untagged (YEpASH1); lane 2, Khd1myc (YEplac181); lane 3, Khd1myc (YEpASH1); lane 4, She3myc (YEplac181); lane 5, She3myc (YEpASH1). (B) Lanes 1 and 2, Khd1myc (U1Atag-N + U1Ap-GFP); lanes 3 and 4, Khd1myc (U1Atag-U + U1Ap-GFP); lanes 5 and 6, She3myc (U1Atag-U + U1Ap-GFP). Total amounts of U1Ap-GFP were the same in each cell extract (data not shown). Strains used: 10B (untagged), YKEN203 (Khd1myc), 134 (She3myc).
To examine whether the association of Khd1myc with ASH1 mRNA is mediated by the N element, Khd1myc proteins were immunoprecipitated from the Khd1myc strain co-expressing U1Atag-N or U1Atag-U, and U1Ap-GFP. By RT–PCR analysis of the immunoprecipitates with the anti-myc antibody, the U1Atag-N mRNA was detected in the immunoprecipitates (Figure 4B, lane 2). U1Ap-GFP also co-immunoprecipitated with Khd1myc, suggesting that Khd1myc makes a complex with U1Ap-GFP through the U1Atag-N mRNA. In contrast, the U1Atag-U mRNA and U1Ap-GFP did not co-immunoprecipitate with Khd1myc in the Khd1myc strains co-expressing U1Atag-U and U1Ap-GFP (Figure 4B, lane 4). As a control, when She3myc was co-expressed with U1Atag-U and U1Ap-GFP, the U1Atag-U mRNA and U1Ap-GFP were detected in the She3myc immunoprecipitates (Figure 4B, lane 6). These results support the possibility that the association of Khd1myc with ASH1 mRNA is mediated by the N element.
Genetic interaction between the KHD1 and SHE genes
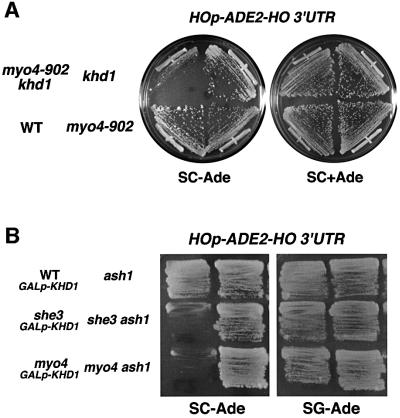
Asymmetric expression of HO is ultimately determined by the localization of ASH1 mRNA (Bobola et al., 1996; Sil and Herskowitz, 1996; Long et al., 1997; Takizawa et al., 1997). Delocalization of ASH1 mRNA in she mutants causes a reduction in HO expression (Jansen et al., 1996; Long et al., 1997; Takizawa et al., 1997). Since the khd1Δ mutation partially affected ASH1 mRNA localization (Table I), we examined the effect of khd1Δ on HO expression using an HOp-ADE2 reporter gene to monitor expression of HO. HOp-ADE2 was constructed by replacing the ho ORF with the ADE2 ORF at the ho locus. Expression of the reporter can thus be assayed in an ade2Δ background by growth on medium lacking adenine (SC-Ade). myo4Δ and she3Δ mutants containing the HOp-ADE2 reporter failed to grow on SC-Ade plates (Figure 5B), demonstrating that inactivation of MYO4 or SHE3 leads to delocalization of ASH1 mRNA, resulting in repression of the HOp-ADE2 reporter. In contrast, the khd1Δ mutation had little effect on HO expression (Figure 5A). The frequency of mating-type switching in the khd1Δ mutant was the same as that in the wild-type strain (data not shown). We then examined whether the khd1Δ mutation affected the phenotype associated with a weak myo4-910 mutation, which by itself had little effect on HO expression. The khd1Δ myo4-910 double mutant showed greatly reduced growth on the SC-Ade plate, indicating a reduced level of HO expression in these cells (Figure 5A). This reduced growth of the khd1Δ myo4-910 double mutant on the SC-Ade plate was dependent on the ASH1 gene, because disruption of the ASH1 gene suppressed the growth defect (data not shown). Thus, the khd1Δ deletion enhanced the effect of the myo4 mutation on HO expression, indicating that the KHD1 gene genetically interacts with MYO4.
Fig. 5. Genetic interactions between KHD1 and SHE. (A) Yeast strains YKEN251 (WT HOp-ADE2-HO 3′ UTR), YKEN252 (myo4-910 HOp-ADE2-HO 3′ UTR), YKEN254 (myo4-910 khd1Δ HOp-ADE2-HO 3′ UTR) and YKEN253 (khd1Δ HOp-ADE2-HO 3′ UTR) were streaked on SC-Ade or SC plates and incubated for 3 days at 30°C. (B) Yeast strains YKEN301 (WT GAL1p-KHD1 HOp-ADE2-HO 3′ UTR), YKEN302 (she3Δ GAL1p-KHD1 HOp-ADE2-HO 3′ UTR), YKEN303 (myo4Δ GAL1p-KHD1 HOp-ADE2-HO 3′ UTR), YKEN304 (ash1Δ HOp-ADE2-HO 3′ UTR), YKEN305 (she3Δ ash1Δ HOp-ADE2-HO 3′ UTR) and YKEN306 (myo4Δ ash1Δ HOp-ADE2-HO 3′ UTR) were streaked on SC-Ade or SG-Ade plates and incubated for 3 days at 30°C.
To analyze further the genetic interactions between KHD1 and SHE genes, we examined the effect of KHD1 overexpression on HO expression in myo4Δ and she3Δ mutants. Overexpression of KHD1 from the GAL1 promoter prevented the reduction in HO expression in myo4Δ and she3Δ mutants (Figure 5B). These results suggest a possible genetic interaction between the KHD1 and SHE genes, and imply that Khd1 affects ASH1 mRNA localization at a step different from that of the She proteins.
Overexpression of KHD1 inhibits translation of ASH1 mRNA
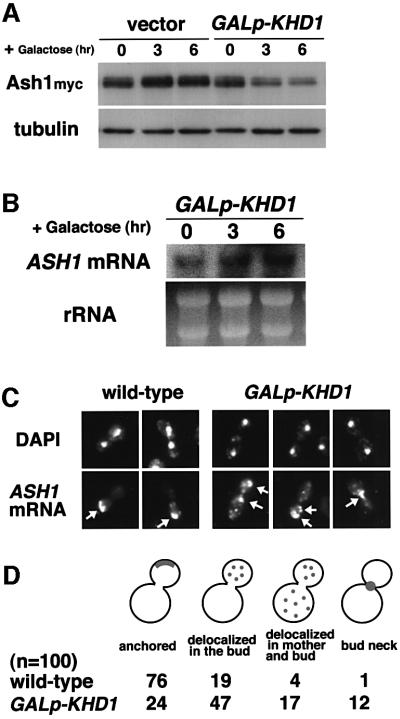
How does KHD1 overexpression suppress the effect of she mutations on HO expression? Since ASH1 negatively regulates the HOp-ADE2 reporter, disruption of the ASH1 gene can suppress a defect in HO expression in she mutants (Figure 5B). This observation raises the possibility that overexpression of KHD1 suppresses the decreased expression of HO observed in she mutations by decreasing Ash1 protein concentrations. To test this possibility, we measured the amounts of myc-tagged Ash1 protein after induction of KHD1 expression from the GAL1 promoter. Western blotting analysis revealed that KHD1 overexpression reduced the concentration of Ash1myc 3.6-fold (Figure 6A). This reduction did not result from toxicity induced by KHD1 overexpression, as the concentration of the unrelated Tub1 protein was not changed (Figure 6A). Overexpression of KHD1 did not affect the concentration of ASH1 mRNA (Figure 6B). These results suggest that KHD1 may be involved in translational control of ASH1 mRNA.
Fig. 6. Overexpression of KHD1 inhibits translation of ASH1 mRNA. (A) Effect of KHD1 overexpression on Ash1myc protein concentration. Yeast cells were cultured in 2% raffinose medium at 30°C and treated with galactose (2%) to induce KHD1 expression from GAL1p-KHD1. At the times indicated, cells were harvested and western blot analysis was performed to assay the concentration of Ash1myc protein. The concentration of tubulin protein was measured as a quantity control. Strain used: K5552 (Ash1myc) transformed with pK736 (GAL1p-KHD1). (B) Effect of KHD1 overexpression on ASH1 mRNA concentration. ASH1 transcripts were quantitated by northern blotting as described in Materials and methods. rRNA was included as a quantity control. (C) Effect of KHD1 overexpression on ASH1 mRNA localization. Yeast cells were cultured in 2% raffinose medium at 30°C and treated with galactose (2%) for 3 h to induce KHD1 expression from GAL1p-KHD1. ASH1 mRNA was stained by digoxigenin-labeled ASH1 antisense probe (ASH1 mRNA; arrow), and DNA was stained by 4,6-diamino-2-phenylindole (DAPI). Strains used: K5552 (ASH1myc; wild type), YKEN307 (ASH1myc; GAL1p-KHD1). (D) The percentages of cells showing different patterns of ASH1 mRNA localization. Localization was determined by RNA in situ hybridization and classified as follows: anchored: tightly localized ASH1 mRNA at the distal tip; delocalized in the bud: delocalized ASH1 mRNA confined to the bud; delocalized in mother and bud: ASH1 mRNA in both mother cell and bud; neck: ASH1 mRNA at the bud neck.
We next examined the effect of KHD1 overexpression on ASH1 mRNA localization. ASH1 mRNA was found to be delocalized in the strain overexpressing KHD1 (Figure 6C and D). In the wild-type strain, 76% of ASH1 mRNA was localized at the distal cortex of the bud. When KHD1 was overexpressed, ASH1 mRNA was localized diffusely within the bud (47%), or mother and bud (17%). These results suggest that the inhibition of ASH1 mRNA translation by KHD1 overexpression might result in a decrease in anchored ASH1 mRNA.
Translation of ASH1 mRNA affects its proper localization
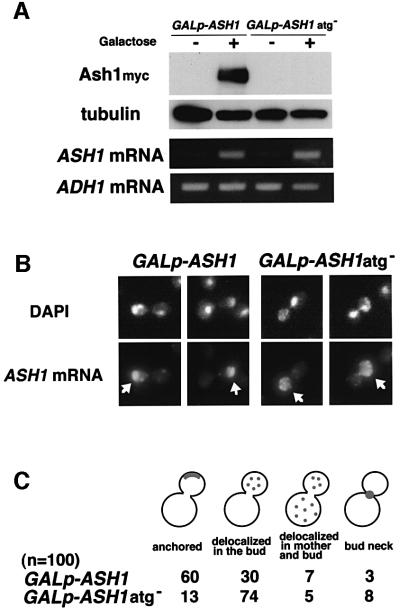
The observation that KHD1 may regulate the localization of ASH1 mRNA via regulation of ASH1 translation raised the possibility that ASH1 mRNA translation could in turn affect ASH1 mRNA localization. ASH1 mRNA is thought to be translated at the distal tips of daughter buds, with Ash1 protein then transported to the proximal, daughter nuclei. To address whether translation of ASH1 mRNA affects its own localization, we compared localization of wild-type ASH1 mRNA with that of an ASH1 mRNA lacking its initiator ATG codon. Both versions of the ASH1 transcript were placed under the control of the GAL1 promoter to create the constructs GAL1p-ASH1 and GAL1p-ASH1atg–. Western blot analysis confirmed that the mRNA derived from GAL1p-ASH1atg– failed to produce Ash1 protein. RT–PCR analysis showed that this transcript was present at the same concentration as wild-type ASH1 mRNA (Figure 7A). However, in comparison to wild-type ASH1 mRNA, ASH1atg– mRNA was found to be somewhat delocalized in the bud (Figure 7B and C). Whereas 60% of wild-type ASH1 mRNA localized at the distal cortex of the bud, 74% of ASH1atg– mRNA localized diffusely within the bud. These results suggest that translation of ASH1 mRNA has a role in anchoring ASH1 mRNA at the distal cortex of daughter cells.
Fig. 7. Translation of ASH1 mRNA is required for proper localization of ASH1 mRNA. (A) Expression of Ash1myc protein and ASH1 mRNA. Yeast cells were cultured in 2% raffinose medium at 30°C and treated with galactose (2%) to induce ASH1 expression from GAL1p-ASH1myc and GAL1p-ASH1atg–myc. At the times indicated, cells were harvested and western blot analysis was performed to assay the concentration of Ash1myc protein. Tubulin protein was included as a quantity control. RNAs were also extracted from cell extracts and used as templates for RT–PCR. A 360 bp product was amplified using ASH1-specific primers. One-fifteenth of each reaction was separated on a 2% agarose gel and stained with ethidium bromide. ADH1 mRNA was included as a quantity control. (B) Comparison of ASH1 mRNA localization in TTC356 (GAL1p-ASH1myc; wild-type) and in TTC360 (GAL1p-ASH1atg–myc). Wild-type ASH1 mRNA was localized at the distal cortex of the bud; ASH1atg– mRNA was localized diffusely within the bud. ASH1 mRNA was stained by digoxigenin-labeled ASH1 antisense probe (ASH1 mRNA; arrow), and DNA was stained by DAPI. (C) The percentages of cells showing different patterns of ASH1 mRNA localization. Localization was determined by RNA in situ hybridization and classified as in Figure 6.
Discussion
Translation of ASH1 mRNA is required for proper localization of ASH1 mRNA
ASH1 mRNA is transported as part of a complex con sisting of ASH1 mRNA–She2–She3–Myo4 to the distal tips of daughter cells using polarized actin filaments (Bohl et al., 2000; Long et al., 2000, 2001; Takizawa and Vale, 2000; Kwon and Schnapp, 2001). ASH1 mRNA is thought to be translated at the distal tips of daughter buds and then transported to the proximal daughter nuclei. It remains to be determined how ASH1 mRNA is anchored at the bud tip and how translation of ASH1 mRNA is regulated. In this study, we found that the tight anchoring of ASH1 mRNA to the distal tip of the daughter cell requires translation of ASH1 mRNA. Compared with wild-type ASH1 mRNA, ASH1atg– mRNA, which lacks the initiator ATG codon, was somewhat delocalized in the bud (Figure 7B and C). Overexpression of KHD1 inhibits translation of ASH1 mRNA and impairs the proper localization of ASH1 mRNA (Figure 6C and D). Gonzalez et al. (1999) also demonstrated that the tight anchoring of ASH1 mRNA to the distal cortex depends on the translation of ASH1 mRNA, especially of the C-terminal sequences. They suggested that Ash1 protein might have a role in the tight anchoring of ASH1 mRNA. Another possibility is that the translational machinery, including ribosomes co-localized with ASH1 mRNA at the distal tip, might be involved in the tight anchoring of ASH1 mRNA.
A novel RNA-binding protein, Khd1, is required for efficient localization of ASH1 mRNA
To identify RNA-binding proteins involved in translation, anchoring or transport of ASH1 mRNA, we carried out a systematic survey of the different candidate RNA-binding proteins and their effect on ASH1 mRNA localization (Table I). ASH1 mRNA was found to be partially delocalized in mpt5, scp160 and khd1 deletion mutants. Khd1 co-localized with the GFP signal from the U1Atag-ASH1 RNA particle, whereas Scp160 and Mpt5 did not (Figure 1). In addition, co-immunoprecipitation studies indicated that Khd1 is associated with ASH1 mRNA through the N element (Figure 4). These results suggest that Khd1 has a direct role in some aspect of ASH1 mRNA localization.
ASH1 mRNA contains three or four localization elements, each of which is sufficient for localization of a reporter mRNA to daughter cells (Figure 2; Chartrand et al., 1999; Gonzalez et al., 1999). However, each element is not sufficient for the tight anchoring of ASH1 mRNA at the distal tip (Gonzalez et al., 1999). When we tested each element independently (Figures 2 and 3), we found that Myo4, She2 and She3 co-localized with the GFP signals from all three elements of the ASH1 RNA particle, whereas Khd1 co-localized with the N element (U1Atag-N) but not the C or U elements (U1Atag-C or U1Atag-U). In the khd1Δ mutant, Myo4 still co-localized with the GFP signals from U1Atag-N in the bud (data not shown), suggesting that KHD1 is not required for localization of the N element in the bud and its co-localization with Myo4. Furthermore, localization of ASH1 mRNA in the khd1Δ myo4-910 double mutant was similar to that in the myo4-910 single mutant (data not shown), suggesting that the khd1Δ mutation does not exacerbate partial defect in ASH1 mRNA localization of a weak myo4-910 mutation. Taken together, these results suggest that Khd1 might have a different role from Myo4, She2 and She3, which are thought to function in ASH1 mRNA transport.
Khd1 contains three KH RNA-binding motifs. The KH domain was first identified in the human heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Siomi et al., 1993; Krecic and Swanson, 1999). Overexpression of KHD1 resulted in a decrease in Ash1p expression, suggesting that KHD1 may be involved in the translational control of ASH1 mRNA via interaction with its N element. This proposed site of Khd1 action contrasts with the fact that hnRNP-K1 governs translation by binding to the 3′ UTR of their target mRNAs (Ostareck et al., 1997; Shyu and Wilkinson, 2000). Interestingly, chicken zipcode-binding protein (ZBP-1) and its Xenopus ortholog, Vera/Vg RBP, which have four KH domains in addition to one RNA recognition motif (RRM), are required for mRNA localizations of β-actin and Vg1 mRNAs (Ross et al., 1997; Deshler et al., 1998; Havin et al., 1998). Our finding that Khd1 is also involved in mRNA localization and its translation in yeast may provide a means to address functions of KH domain proteins in mRNA localization and their possible relationship to translation.
Khd1 may function in the linkage between ASH1 mRNA localization and its translation
We imagine that Khd1 inhibits the translation of ASH1 mRNA during the time it is being transported. As Khd1 seems to localize around the nuclear membrane, possibly on the endoplasmic reticulum, Khd1 may associate with ASH1 mRNA soon after its export from the nucleus, and this Khd1–ASH1 mRNA complex is then transported by the She machinery. Overexpression of KHD1 from the GAL1 promoter resulted in decreased concentrations of Ash1p, perhaps due to increased inhibition in ASH1 mRNA translation. Interestingly, strong overexpression of KHD1 by the TDH3 promoter is toxic to cell growth (data not shown), whereas deletion of ASH1 is not. This suggests that Khd1 may be involved in translating other mRNAs in addition to ASH1. A recent report has identified other mRNAs, in addition to ASH1, that are transported by the She machinery (Takizawa et al., 2000). It would be interesting to test whether Khd1 also acts on the translation and/or localization of these mRNAs.
Inactivation of Khd1 in wild-type strains causes only partial delocalization of ASH1 mRNA and has little effect on HO expression. This is in contrast to the much more severe phenotypes caused by the she mutations (Table I). Similarly, the frequency of mating-type switching is not affected in a khd1Δ strain, whereas it is greatly reduced in she mutants (Jansen et al., 1996). A requirement for Khd1 can be seen, however, in a strain containing a weak Myo4 mutation: combining a khd1Δ mutation with the weak myo4-910 allele was found to cause reduced HO expression (Figure 5A). Thus, it appears that when the She machinery is intact and ASH1 mRNA is rapidly transported (Bertrand et al., 1998), Khd1 function is not crucial for asymmetric localization of the ASH1 mRNA. However, when there is some other defect in ASH1 mRNA transport, the role of Khd1 in HO expression is manifested.
The weak phenotype of the khd1Δ mutant may also suggest the existence of a protein that functions redundantly with Khd1. The PBP2 gene encodes a KH protein that has the greatest structural similarity to Khd1. We inactivated PBP2 and examined the phenotype in a khd1Δ background. However, we found that the pbp2Δ mutation had no additive effect on ASH1 mRNA localization or HO expression (data not shown). Furthermore, Pbp2 did not co-localize with the GFP signal from the U1Atag-ASH1 RNA particle (data not shown). These results indicate that Pbp2 is not redundant with Khd1 for ASH1 mRNA localization. We have observed that another KH domain protein, Scp160, is required for optimal ASH1 mRNA localization and HO expression (Table I). The Scp160 protein, however, did not co-localize with the GFP signal from the U1Atag-ASH1 RNA particle (Figure 1), and thus its role in ASH1 localization is probably indirect. It has recently been reported that Scp160 localizes to membrane-bound polysomes and that its deletion causes pleiotropic defects, including temperature-sensitive growth and increased ploidy (Weber et al., 1997; Lang and Fridovich-Keil, 2000; Frey et al., 2001). Scp160 might be a component of the general translational machinery involved in the translation of various mRNA including ASH1. In conclusion, our studies on localization of ASH1 mRNA have begun to reveal the ways in which KH-domain proteins modulate mRNA localization and possibly translation.
Materials and methods
Strains and general methods
Escherichia coil DH5α was used for DNA manipulations. The yeast strains used in this study are described in Table II. Standard procedures were followed for yeast manipulations (Kaiser et al., 1994). The media used in this study included rich medium, synthetic complete medium with glucose (SC), synthetic minimal medium with glucose (SD) and sporulation medium (Kaiser et al., 1994). SC lacking amino acids or other nutrients (e.g. SC-Leu lacks leucine and SC-Ade lacks adenine) was used to select transformants and to score ADE2 reporter activity. SG and SR are identical to SC except that they contain 2% galactose and raffinose, respectively, instead of 2% glucose. Recombinant DNA procedures were carried out as described previously (Sambrook et al., 1989).
Table II. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| W303 | MATa ade2 trp1 can1 leu2 his3 ura3 GAL psi+ | Sil and Herskowitz (1996) |
| K1107 | MATa HOp-LacZ-HO 3′ UTR | Nasmyth (1987) |
| K5552 | MATα ASH1-myc | Jansen et al. (1996) |
| 10B | MATα HOp-ADE2-HO 3′ UTR | Tadauchi et al. (2001) |
| TTC356 | MATa GALp-ASH1-myc | this study |
| TTC360 | MATa GALp-ASH1-myc (atg–) | this study |
| YKEN111 | MATα HOp-ADE2-HO 3′ UTR puf1Δ/jsn1Δ::CgHIS3 | this study |
| YKEN113 | MATα HOp-ADE2-HO 3′ UTR puf2Δ::CgHIS3 | this study |
| YKEN112 | MATα HOp-ADE2-HO 3′ UTR puf3Δ::CgHIS3 | this study |
| YKEN110 | MATα HOp-ADE2-HO 3′ UTR puf4Δ::CgHIS3 | this study |
| YKEN109 | MATα HOp-ADE2-HO 3′ UTR puf5Δ/mpt5Δ::CgHIS3 | this study |
| YKEN123 | MATα HOp-ADE2-HO 3′ UTR scp160Δ::CgHIS3 | this study |
| YKEN124 | MATα HOp-ADE2-HO 3′ UTR pbp2Δ::CgHIS3 | this study |
| YKEN125 | MATα HOp-ADE2-HO 3′ UTR khd1Δ::CgHIS3 | this study |
| YKEN126 | MATα HOp-ADE2-HO 3′ UTR mer1Δ::CgHIS3 | this study |
| YKEN201 | MATα HOp-ADE2-HO 3′ UTR PUF5/MPT5myc::kanMX6 | this study |
| YKEN202 | MATα HOp-ADE2-HO 3′ UTR SCP160myc::kanMX6 | this study |
| YKEN203 | MATα HOp-ADE2-HO 3′ UTR KHD1myc::kanMX6 | this study |
| YKEN204 | MATα HOp-ADE2-HO 3′ UTR SHE2myc::kanMX6 | this study |
| 101 | MATα MYO4myc | Jansen (1996) |
| 134 | MATα SHE3myc | Jansen (1996) |
| YKEN251 | MATα HOp-ADE2-HO 3′ UTR | this study |
| YKEN252 | MATα HOp-ADE2-HO 3′ UTR myo4-910 | this study |
| YKEN253 | MATα HOp-ADE2-HO 3′ UTR khd1Δ::CgHIS3 | this study |
| YKEN254 | MATα HOp-ADE2-HO 3′ UTR khd1Δ::CgHIS3 myo4-910 | this study |
| YKEN301 | MATα HOp-ADE2-HO 3′ UTR kanMX6::GAL1p-KHD1 | this study |
| YKEN302 | MATα HOp-ADE2-HO 3′ UTR she3Δ kanMX6::GAL1p-KHD1 | this study |
| YKEN303 | MATα HOp-ADE2-HO 3′ UTR myo4Δ kanMX6::GAL1p-KHD1 | this study |
| YKEN304 | MATα HOp-ADE2-HO 3′ UTR ash1Δ | this study |
| YKEN305 | MATα HOp-ADE2-HO 3′ UTR she3Δ ash1Δ | this study |
| YKEN306 |
MATα HOp-ADE2-HO 3′ UTR myo4Δ ash1Δ |
this study |
| YKEN307 | MATα ASH1-myc kanMX6::GAL1p-KHD1 | this study |
Plasmids
Plasmids used in this study are described in Table III. Plasmid pPT120 expresses U1Atag-ASH1 from the GAL1 promoter (Takizawa and Vale, 2000). Plasmid pPT220 expresses U1A-GFP-GST-NLS from the TDH3 promoter (Takizawa and Vale, 2000). Plasmid pK404 is YEplac195 carrying GAL1p-U1Atag-ASH1 3′ UTR. The fragment containing GAL1p-U1Atag and the fragment containing ASH1 3′ UTR were inserted into YEplac195. Plasmid pK622 is pRS426 carrying GAL1p-U1Atag-ASH1 coding region (1–804)-ADH1 3′ UTR. The fragment containing GAL1p-U1Atag-ASH1 coding region (1–804) and the fragment containing ADH1 3′ UTR were inserted into pRS426. Plasmid pK852 is pRS426 carrying GAL1p-U1Atag-ASH1 coding region (828–1764)-ADH1 3′ UTR. The fragment containing GAL1p-U1Atag-ASH1 coding region (828–1764) and the fragment containing ADH1 3′ UTR were inserted into pRS426. Plasmid pK736 is YEpURA3 plasmid carrying GAL1p-KHD1. Plasmid pCgHIS3 is pUC19 carrying the Candida glabrata HIS3 gene (Sakumoto et al., 1999).
Table III. Plasmids used in this study.
| Plasmid | Relevant markers | Source |
|---|---|---|
| YEplac181 | LEU2, 2 µm | Gietz and Sugino (1988) |
| YEplac195 | URA3, 2 µm | Gietz and Sugino (1988) |
| pRS426 | URA3, 2 µm | Sikorski and Hieter (1989) |
| pAS191 | LEU2, 2 µm, ASH1 | Sil and Herskowitz (1996) |
| pPT120 (U1Atag-Full) | HIS3, 2 µm, GAL1p-U1Atag-ASH1 (1–1764)-ASH1 3′ UTR | Takizawa and Vale (2000) |
| pPT220 | TRP1, CEN-ARS, TDH3p-U1A-GFP-GST-NLS | Takizawa and Vale (2000) |
| pK114 (U1Atag-Full) | URA3, 2 µm, GAL1p-U1Atag-ASH1 (1–1764)-ASH1 3′ UTR | this study |
| pK404 (U1Atag-U) | URA3, 2 µm, GAL1p-U1Atag-ASH1 3′ UTR | this study |
| pK622 (U1Atag-N) | URA3, 2 µm, GAL1p-U1Atag-ASH1 (1–804)-ADH1 3′ UTR | this study |
| pK852 (U1Atag-C) | URA3, 2 µm, GAL1p-U1Atag-ASH1 (828–1764)-ADH1 3′ UTR | this study |
| pK736 | URA3, 2 µm, GAL1p-KHD1 | this study |
| pFA6a-13Myc-kanMX6 | 13MYC-ADH1 3′ UTR-kanMX6 | Longtine et al. (1998) |
| pFA6a-kanMX6-GAL1p-3HA | kanMX6-GAL1p-3HA | Longtine et al. (1998) |
| pCgHIS3 | C.glabrata HIS3 in pUC19 | Sakumoto et al. (1999) |
Deletion of the genes encoding RNA-binding proteins
The deletions of PUF1/JSN1, PUF2, PUF3, PUF4/YGL014w, and PUF5/MPT5, SCP160, PBP2, YBL032w and MER1 were constructed by the PCR-based gene deletion method (Baudin et al., 1993; Schneider et al., 1996; Sakumoto et al., 1999). Primer sets were designed such that 46 bases at the 5′ end of the primers were complementary to those at the corresponding region of the target gene, and 20 bases at their 3′ end were complementary to the pUC19 sequence outside the polylinker region in plasmid pCgHIS3 containing the C.glabrata HIS3 gene as a selectable marker. Primer sets for PCR were designed to delete the ORF completely. The PCR products were used to transform strain 10B by selection for His+. The disruption was verified by colony-PCR amplification (Huxley et al., 1990) to confirm that replacement had occurred at the expected locus.
Construction of Khd1myc, Scp160myc and Mpt5myc strains
Khd1myc, Scp160myc and Mpt5myc strains were prepared by the method of Longtine et al. (1998) using pFA6a-13Myc-kanMX6.
Construction of GAL1p-KHD1 strains
The GAL1p-KHD1 strain was prepared by the method of Longtine et al. (1998) using pFA6a-kanMX6-GAL1p-3HA.
Localization of ASH1 mRNA
In situ RNA hybridization with digoxigenin-labeled ASH1 antisense probe was performed as described previously (Takizawa et al., 1997).
Induction and imaging of U1Atag-ASH1 RNA particles
Co-localization of myc-tagged proteins with U1Atag-ASH1 RNA particles was examined as described previously (Takizawa and Vale, 2000). Cells containing U1Ap-GFP and U1Atag-ASH1 were grown overnight at 30°C in synthetic media containing 2% raffinose. Overnight cultures were adjusted to an optical density (OD) of 0.5 (600 nm) in synthetic media containing 2% raffinose and incubated for 2 h at 30°C. Galactose was added to 0.2%, and the cultures incubated for 2 h at 30°C. Cells were examined by phase-contrast microscopy using a ×63/NA 1.4 lens. Images were captured with a cooled charged-coupled device and digital images displayed by using Adobe Photoshop. For co-localization experiments, samples were fixed after induction in 3.7% formaldehyde for 1 h. Cells were washed and made into spheroplasts in SP buffer (100 mM phosphate buffer pH 7.0, 1.2 M sorbitol containing 30 mM mercaptoethanol, 40 mg/ml zymolyase 100T) for 30 min at 37°C. Cells were washed and spread on polylysine-coated, multiwell test slides and then incubated with monoclonal anti-myc antibody 9E10 (Santa Cruz, CA) at a 1:1000 dilution in blocking buffer [phosphate-buffered saline (PBS), 1% bovine serum albumen (BSA)] for 1 h. After washing, cells were incubated with rhodamine-conjugated goat anti-mouse IgG (Boehringer Mannheim) in blocking buffer for 1 h. Cells were washed and mounted in mounting buffer (PBS, 90% glycerol, 1 mg/ml ρ-phenylenediamine, 0.1 µg/ml 4′,6-diamidino-2-phenylindole).
Immunoprecipitation and mRNA detection
Exponentially growing cells (3 × 108) were disrupted with glass beads in 200 ml extraction buffer [25 mM HEPES–KOH pH 7.5, 150 mM KCl, 2 mM MgCl2 containing 20 mM vanadyl ribonucleoside complexes (Sigma), 200 U/ml RNasin (Life Technologies, Grand Island, NY), 0.1% NP-40, 1 mM DTT, 0.2 mg/ml heparin, 1 mM PMSF, 10 µg/ml each of aprotinin, leupeptin and pepstatin]. Extracts were cleared by centrifugation (10 min at 4000 g). Monoclonal anti-myc antibody 9E10 was added to the cleared extracts and incubated for 1 h on ice following incubation with protein A agarose for 1 h at 4°C. Beads were washed four times in wash buffer (25 mM HEPES–KOH pH 7.5, 150 mM KCl, 2 mM MgCl2) and were eluted in 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM EDTA, 1% SDS for 10 min at 65°C. Eluted samples were extracted with phenol-chloroform, ethanol precipitated, resuspended in RQ1 DNase buffer and treated with RQ1 DNase (Promega, Madison, WI). The remaining RNA was extracted, precipitated and resuspended in H2O. RT–PCR was performed with 1 µl RNA as template using the ‘Access’-RT–PCR kit (Promega) and the conditions suggested by the manufacturer. The number of amplification cycles was adjusted to avoid reaching a plateau phase during PCR.
Preparation of yeast extracts and western blot analysis
Yeast cells were grown to an OD600 of 0.5–1.0 and treated with 2% galactose to activate the GAL1 promoter. After treatment, yeast cultures were quickly chilled, and cells were collected by rapid centrifugation. The pellet was washed twice and then suspended in breaking buffer (4% SDS, 40 mM Tris–HCl pH 7.0, 8 M urea, 0.1 mM EDTA, 1% 2-mercapto ethanol). Glass beads (0.4–0.6 mm diameter) were added to this suspension, and cells were broken by vigorous vortexing for 5 min at room temperature. Beads and cell debris were removed by centrifugation at 10 000 g at room temperature. Protein concentrations of the cell extracts were measured at OD280. Cell extracts were subjected to SDS–PAGE on 7% acrylamide gels followed by electroblotting onto Hybond N+ membrane (Amersham). Blots were blocked by incubation for 15 min at room temperature in TBS-M buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl with 4% non-fat dry milk). Blots were then incubated with monoclonal anti-myc antibody 9E10 diluted 1:2000 (to detect Ash1myc) or anti-tubulin antibody diluted 1:1000 (to detect tubulin) in TBS-M buffer overnight at 4°C. After three washes with TBS buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl), blots were incubated for 2 h with peroxidase-conjugated secondary antibody (Calbiochem) diluted 1:3000 with TBS-M buffer. After three final washes with TBS buffer, blots were detected using an enhanced chemiluminescence detection kit (Amersham).
Acknowledgments
Acknowledgements
We thank M.Ota and K.Shikii for technical assistance, R.P.Jansen for materials and members of the Herskowitz laboratory for helpful discussions. This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture and Science of Japan, and the Senri Life Science Foundation (to K.I.); by special grants for CREST, Advanced Research on Cancer from the Ministry of Education, Culture, and Science of Japan, and the Asahi Glass Foundation (to K.M.); and by a research grant from the United States National Institutes of Health (to I.H.).
References
- Abovich N. and Rosbash,M. (1997) Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell, 89, 403–412. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier,K.O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D.L., Salmon,E.D. and Bloom,K. (1999) Localization and anchoring of mRNA in budding yeast. Curr. Biol., 9, 569–578. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand,P., Schaefer,M., Shenoy,S.M., Singer,R.H. and Long,R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- Bobola N., Jansen,R.P., Shin,T.H. and Nasmyth,K. (1996) Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell, 84, 699–709. [DOI] [PubMed] [Google Scholar]
- Bohl F., Kruse,C., Frank,A., Ferring,D. and Jansen,R.P. (2000) She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J., 19, 5514–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P., Meng,X.H., Singer,R.H. and Long,R.M. (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol., 9, 333–336. [DOI] [PubMed] [Google Scholar]
- Curtis D., Lehmann,R. and Zamore,P.D. (1995) Translational regulation in development. Cell, 81, 171–178. [DOI] [PubMed] [Google Scholar]
- Deshler J.O., Highett,M.I., Abramson,T. and Schnapp,B.J. (1998) A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol., 8, 489–496. [DOI] [PubMed] [Google Scholar]
- Engebrecht J. and Roeder,G.S. (1990) MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol. Cell. Biol., 10, 2379–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell,K., Longtine,M.S., Chow,C.J., Adames,N., Pringle,J.R., Peter,M. and Boone,C. (1997) Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science, 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Frey S., Pool,M. and Seedorf,M. (2001) Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem., 276, 15905–15912. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Buonomo,S.B., Nasmyth,K. and von Ahsen,U. (1999) ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol., 9, 337–340. [DOI] [PubMed] [Google Scholar]
- Gunkel N., Yano,T., Markussen,F.H., Olsen,L.C. and Ephrussi,A. (1998) Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev., 12, 1652–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B.K., Petzold,A., Lillie,S.H. and Brown,S.S. (1994) Identification of MYO4, a second class V myosin gene in yeast. J. Cell Sci., 107, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Havin L., Git,A., Elisha,Z., Oberman,F., Yaniv,K., Schwartz,S.P., Standart,N. and Yisraeli,J.K. (1998) RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev., 12, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T. (1998) The destinies and destinations of RNAs. Cell, 95, 451–460. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. (1988) Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev., 52, 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C., Green,E.D. and Dunham,I. (1990) Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet., 6, 236. [DOI] [PubMed] [Google Scholar]
- Jansen R.P., Dowzer,C., Michaelis,C., Galova,M. and Nasmyth,K. (1996) Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell, 84, 687–697. [DOI] [PubMed] [Google Scholar]
- Kaiser C.A., Adams,A. and Gottschling,D.E. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kim-Ha J., Kerr,K. and Macdonald,P.M. (1995) Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell, 81, 403–412. [DOI] [PubMed] [Google Scholar]
- Kohno H. et al. (1996) Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J., 15, 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Kwon S. and Schnapp,B.J. (2001) RNA localization: SHEdding light on the RNA-motor linkage. Curr. Biol., 11, R166–R168. [DOI] [PubMed] [Google Scholar]
- Lang B.D. and Fridovich-Keil,J.L. (2000) Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res., 28, 1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Singer,R.H., Meng,X., Gonzalez,I., Nasmyth,K. and Jansen,R.P. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science, 277, 383–387. [DOI] [PubMed] [Google Scholar]
- Long R.M., Gu,W., Lorimer,E., Singer,R.H. and Chartrand,P. (2000) She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J., 19, 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Gu,W., Meng,X., Gonsalvez,G., Singer,R.H. and Chartrand,P. (2001) An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol., 153, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.R., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M. and Smibert,C.A. (1996) Translational regulation of maternal mRNAs. Curr. Opin. Genet. Dev., 6, 403–407. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Amrani,N. and Jacobson,A. (1998) Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol., 18, 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchow S., Sauter,C. and Jansen,R.P. (1999) Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci., 112, 1511–1518. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (1983) Molecular analysis of a cell lineage. Nature, 302, 670–676. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. and Jansen,R.P. (1997) The cytoskeleton in mRNA localiz ation and cell differentiation. Curr. Opin. Cell Biol., 9, 396–400. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y. and Singer,R.H. (1998) RNA localization: different zipcodes, same postman? Trends Cell Biol., 8, 381–383. [DOI] [PubMed] [Google Scholar]
- Olivas W. and Parker,R. (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J., 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- Preiss T. and Hentze,M.W. (1999) From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev., 9, 515–521. [DOI] [PubMed] [Google Scholar]
- Ross A.F., Oleynikov,Y., Kislauskis,E.H., Taneja,K.L. and Singer,R.H. (1997) Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol., 17, 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumoto N. et al. (1999) A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast, 15, 1669–1679. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schneider D., Bruton,C.J. and Chater,K.F. (1996) Characterization of spaA, a Streptomyces coelicolor gene homologous to a gene involved in sensing starvation in Escherichia coli. Gene, 177, 243–251. [DOI] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A. and Herskowitz,I. (1996) Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific tran scription of the yeast HO gene. Cell, 84, 711–722. [DOI] [PubMed] [Google Scholar]
- Siomi H., Matunis,M.J., Michael,W.M., and Dreyfuss,G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res., 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. (1995) The intracellular localization of messenger RNAs. Cell, 81, 161–170. [DOI] [PubMed] [Google Scholar]
- Tadauchi T., Matsumoto,K., Herskowitz,I. and Irie,K. (2001) Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J., 20, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A. and Vale,R.D. (2000) The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl Acad. Sci. USA, 97, 5273–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., Sil,A., Swedlow,J.R., Herskowitz,I. and Vale,R.D. (1997) Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature, 389, 90–93. [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi,J.L., Wilhelm,J.E. and Vale,R.D. (2000) Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science, 290, 341–344. [DOI] [PubMed] [Google Scholar]
- Van Dyck L., Jonniaux,J.L., de Melo Barreiros,T., Kleine,K. and Goffeau,A. (1994) Analysis of a 17.4 kb DNA segment of yeast chromosome II encompassing the ribosomal protein L19 as well as proteins with homologies to components of the hnRNP and snRNP complexes and to the human proliferation-associated p120 antigen. Yeast, 10, 1663–1673. [DOI] [PubMed] [Google Scholar]
- Weber V., Wernitznig,A., Hager,G., Harata,M., Frank,P. and Wintersberger,U. (1997) Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem., 249, 309–317. [DOI] [PubMed] [Google Scholar]
- Wendland B., McCaffery,J.M., Xiao,Q. and Emr,S.D. (1996) A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol., 135, 1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J.E. and Vale,R.D. (1993) RNA on the move: the mRNA localization pathway. J. Cell Biol., 123, 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos,M., Puoti,A., Durkin,E., Fields,S., Kimble,J. and Wickens,M.P. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature, 390, 477–484. [DOI] [PubMed] [Google Scholar]