Abstract
There are 15 subtypes of influenza A virus (H1–H15), all of which are found in avian species. Three caused pandemics in the last century: H1 in 1918 (and 1977), H2 in 1957 and H3 in 1968. In 1997, an H5 avian virus and in 1999 an H9 virus caused outbreaks of respiratory disease in Hong Kong. We have determined the three-dimensional structures of the haemagglutinins (HAs) from H5 avian and H9 swine viruses closely related to the viruses isolated from humans in Hong Kong. We have compared them with known structures of the H3 HA from the virus that caused the 1968 H3 pandemic and of the HA–esterase–fusion (HEF) glycoprotein from an influenza C virus. Structure and sequence comparisons suggest that HA subtypes may have originated by diversification of properties that affected the metastability of HAs required for their membrane fusion activities in viral infection.
Keywords: avian influenza/influenza A virus/sialic acid/swine influenza/virus evolution
Introduction
The haemagglutinin (HA) and neuraminidase (NA) membrane glycoproteins of influenza A viruses, which function respectively as the receptor-binding and membrane fusion glycoprotein in cell entry (HA) and as the receptor-destroying enzyme in virus release (NA), are divided into subtypes on the basis of differences in their antigenicities and amino acid sequences. Fifteen subtypes of HA (H1–H15) that share between 40 and 60% sequence identity and nine of NA (N1–N9, 40–60% identity) have been distinguished (World Health Organization, 1980; Rohm et al., 1996). Viruses containing all 15 HAs have been isolated from avian species and more limited numbers from equine, H3 and H7; seals, H3, H4 and H7; whales, H1 and H13; and swine, H1, H3 and H9. Viruses containing three different HA subtypes, H1, H2 and H3, caused pandemics in the last century: in 1918, H1; in 1957, H2; in 1968, H3; and H1 again in 1977. Recent outbreaks of influenza in humans in Hong Kong and Southern China have resulted from infections with H5 and H9 viruses (Claas et al., 1998; Lin et al., 2000).
Influenza viruses that infect humans or the genes for their HAs initially derive from avian viruses either directly by cross-species infection or by gene reassortment during mixed infections. Direct infection is proposed to have occurred in 1918 (Reid et al., 1999), in 1997 (Claas et al., 1998) and in 1999 (Lin et al., 2000) and, on the last two occasions, viruses from local birds were found to have infected humans. Reassortant viruses appear to have caused the pandemics of 1957 and 1968; the 1957 H2 virus differed by three genes, those for HA, NA and the RNA polymerase subunit PB1, from the H1 virus that infected humans between 1918 and 1957; the 1968 H3 virus differed by two genes, those for HA and PB1, from the H2 virus that infected humans between 1957 and 1968 (Kawaoka et al., 1989). In both cases, the genes for the H2 and H3 HAs are proposed to have been contributed by avian viruses, during infection of an unknown host that was infected simultaneously by the prevalent human virus.
In the intrapandemic periods that followed the introduction of the H1 subtype in 1918, the H2 subtype in 1957, the H3 subtype in 1968 and the H1 subtype again in 1977, antigenic drift resulting in ∼15% sequence difference was detected in the HAs as a consequence of variant selection under immune pressure (e.g. Bean et al., 1992).
Influenza type B viruses that, like influenza A viruses, contain both HA and NA surface glycoproteins have only been isolated from humans (Francis, 1940) and seals (Osterhaus et al., 2000) and are not divided into subtypes. Influenza type C viruses have been isolated from humans (Taylor, 1949) and from swine (Yuanji and Desselberger, 1984) but, in contrast to both type A and type B viruses, they contain only one type of glycoprotein, HA–esterase–fusion (HEF), which combines all three functions of: receptor binding, H; membrane fusion, F; and receptor destroying-esterase activity, E (Herrler et al., 1988).
Structural information is currently available for the influenza A HA of the H3 subtype (Wilson et al., 1981) and for an influenza C HEF (Rosenthal et al., 1998). Comparison of these two structures showed their close similarity, and suggested a common subdomain structure and its possible evolutionary significance. Here, we report the structures of HAs of two additional subtypes, H5 and H9, which are closely related to the HAs of the H5 and H9 viruses isolated in 1997 and 1999 from humans in Hong Kong. We find that the membrane-distal domains in the H5 and H9 HAs are rotated about the molecular 3-fold axis of symmetry by 20° when compared with the H3 subtype HA and by 30° when compared with HEF. Furthermore, the subdomains defined in the H3 HA–HEF comparison appear displaced independently as rigid bodies, supporting the suggestion of their structural identity and the possibility that HAs and HEF evolved from three functionally distinct ancestral proteins.
We have described recently (Ha et al., 2001) structures of complexes of the H5 avian and H9 swine HAs with sialylated pentasaccharide analogues of avian and human cell surface receptors, to examine the HA’s receptor-binding function and particularly to explore the basis for the evolution of receptor-binding specificity from a preference of avian viruses for sialic acid receptors in α2,3 linkage to a preference of human viruses for α2,6 linkages. Here we provide evidence that all 15 HA subtypes fit into four major structural classes by correlating differences in the relative positions of subdomains in the H3, H5 and H9 HAs with specific sequence motifs; we suggest generalizations about the mechanism of low pH activation of membrane fusion, the molecule’s second function in virus infection; and we present the possibility that HA subtypes originated in the diversification of HA properties resulting from a required balance between stability and the potential for extensive structural reorganization that occurs during membrane fusion.
Results
H5 and H9 HA structure determinations
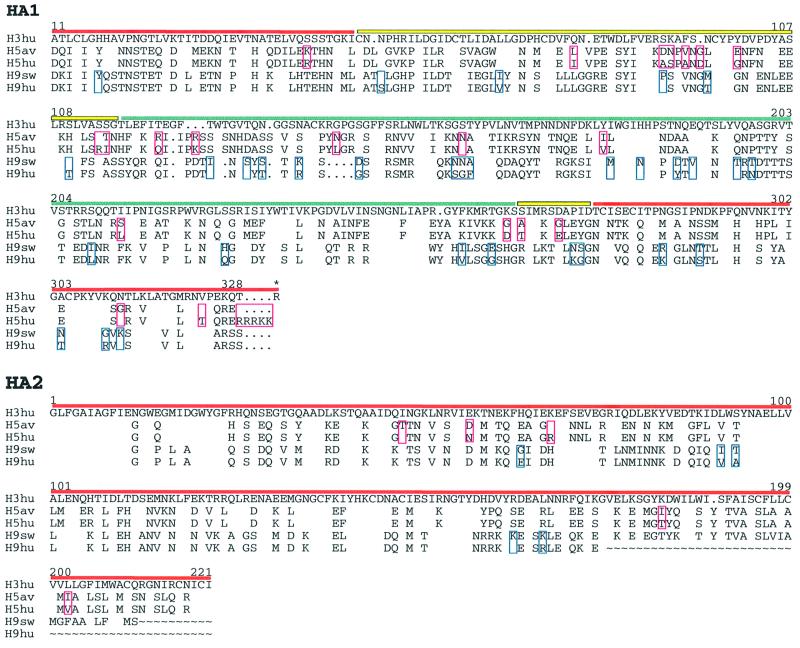
Using X-ray crystallography, we determined the structures of the bromelain-released, soluble ectodomains of the HAs (BHAs) from the H5 subtype influenza A virus A/Duck/Singapore/3/97 and from the H9 subtype influenza A virus A/Swine/Hong Kong/9/98. These HAs have 26 and 39 amino acid substitutions, respectively, relative to the HAs from the 1997 H5 and 1999 H9 human viruses isolated in Hong Kong (Figure 1) [(Claas et al., 1998; Suarez et al., 1998; Subbarao et al., 1998; Bender et al., 1999; Lin et al., 2000; K.Cameron and A.J.Hay, personal communication (H5); D.Markwell and K.F.Shortridge, personal communication (H9)]. The structures were determined to 1.9 (H5) and 1.8 Å (H9) resolution.
Fig. 1. Structure-based sequence alignment of influenza A H3, H5 and H9 HAs. Sequences of human H3hu (A/Aichi/2/68), avian H5av (A/Dk/Sing/97), human H5hu (A/HK/486/97), swine H9sw (A/Sw/9/98) and human H9hu (A/Hong Kong/1073/99). Residues identical to H3 are blank. Numbering is as in the human H3 sequence. Horizontal lines above the sequences indicate subdomains (red, F′ and F; yellow, E′; blue, R). Sequence differences between the H5 human and avian, and H9 swine and human viruses are boxed in red (H5) and blue (H9). An asterisk labels the arginine between HA1 and HA2 that is removed during proteolytic cleavage of HA0. The alignment was generated using GCG (Womble, 2000) and edited manually on the basis of the X-ray structures.
Structure of the H5 and H9 haemagglutinins
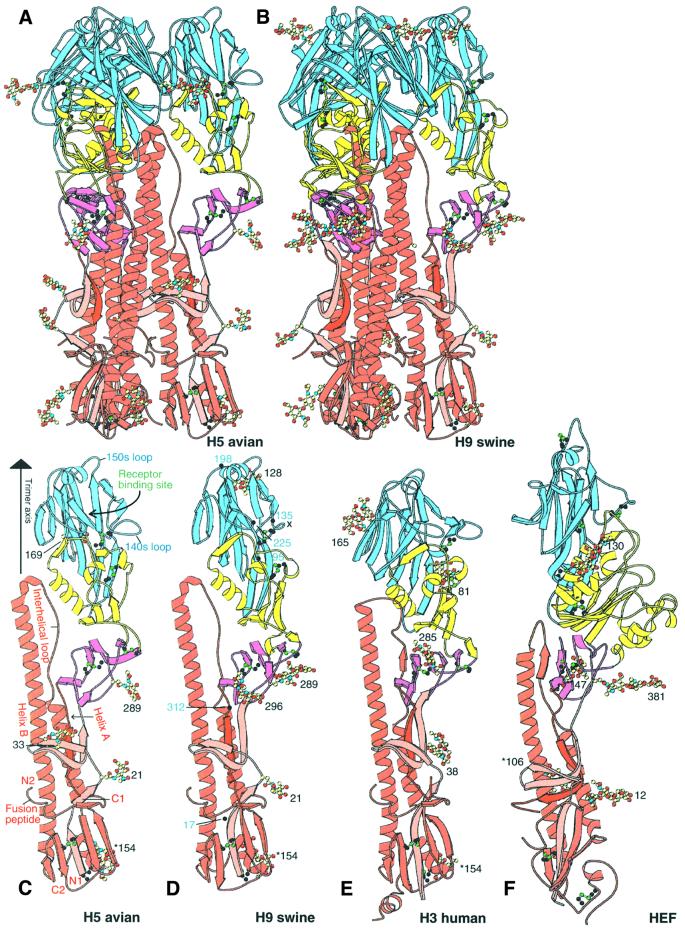
Like the H3 HA and influenza C HEF, H5 and H9 subtype HAs are trimeric molecules in which each monomer contains two disulfide-linked polypeptide chains, HA1 and HA2, generated by proteolytic cleavage of a single chain precursor, HA0 (Figure 2A and B) (Wiley and Skehel, 1987; Skehel and Wiley, 2000). The membrane-proximal stem of the trimer is composed of HA2 (red in Figure 2) and two segments of HA1, residues 1–55 (pink in Figure 2) and 275–329 (purple in Figure 2), centred around a triple-stranded α-helical coiled coil formed by the N-terminal half of the long central α-helix of HA2 (helix B in Figure 2C). On top of the stem, the membrane-distal globular portion of the molecule contains the receptor-binding subdomain (blue in Figure 2) that includes the receptor-binding site (labelled in Figure 2C), and the vestigial fragment of an esterase subdomain (yellow in Figure 2A–E) defined previously by its close structural similarity to a portion (54%) of the 9-O-acetylesterase domain of influenza C HEF (yellow in Figure 2F) (Rosenthal et al., 1998; Zhang et al., 1999).
Fig. 2. H5 avian and H9 swine HA structures compared with H3 HA and HEF. (A) Ribbon diagram of the trimer of H5 (A/Dk/Sing/97) avian HA coloured by subdomains: receptor subdomain R (blue), vestigial enzyme subdomain E′ (yellow), HA2 stem F subdomain (red), F′ subdomain HA1 1–52 (pink), F′ subdomain HA1 275–307 (purple). Oligosaccharides are coloured by atom type. (B) Trimer of H9 swine (A/Sw/9/98) HA. (C) Monomer of H5 (A/Dk/Sing/97) avian HA. Helix A and B, the interhelical loop between them, and the fusion peptide in HA2 are labelled. N1 and C1 indicate the termini of HA1; N2 and C2, those of HA2. The trimer axis, receptor-binding site and two prominent antigenic loops, 140s and 150s, are labelled. Residue numbers mark oligosaccharides, which are coloured by atom type. Carbohydrate attachment sites are at HA1 21, 33, 169 and 289, and at HA2 154. (D) Monomer of the H9 (A/Sw/9/98) swine HA. The x marks the deleted 140s loop. Carbohydrate attachment sites are at HA1 21, 128, 210, 289 and 296, and at HA2 154. (E) Monomer of trimeric H3 (A/Aichi/2/68) human HA (Wilson et al., 1981). Carbohydrate attachment sites are at HA1 8, 22, 38, 81, 165 and 285, and at HA2 154. (F) Monomer of trimeric influenza C virus (C/Johannesburg/1/66) haemagglutinin–esterase–fusion (HEF) protein (Rosenthal et al., 1998; Zhang et al., 1999). These figures and Figures 3B, 4A, B and D were generated using MOLSCRIPT and BOBSCRIPT (Kraulis, 1991; Esnouf, 1997).
The H5 and H9 HAs have 46% sequence identity in HA1 and 62% in HA2. They have 39 and 58%, and 39 and 54% sequence identities, respectively, with the 1968 H3 HA1 and HA2 chains (Figure 2E) and <17% identities with the influenza C virus HEF polypeptides (Figure 2F) (Influenza sequence database at Los Alamos National Laboratory; Macken et al., 2001).
As expected from the level of sequence identity, the overall secondary and tertiary structures of the H5 and H9 HAs are very similar to those of the H3 HA (Figure 2C–E). A prominent loop at HA1 residues 140–147, shown to bind antibodies and undergo antigenic variation in H3 HAs (Wiley et al., 1981), is absent from the H9 HA (x in Figure 2D).The H5 and H9 HAs share three asparagine-linked glycosylation sites (at HA1 21 and 289, and HA2 154) (Figure 2C and D), but have only the HA2 154 site, a site conserved in all known HA sequences of the 15 influenza subtypes, in common with the seven sites of the 1968 H3 HA (Figure 2E), although three pairs of sites in H5 and H3 are near each other in sequence: HA1 33 and 38, 169 and 165, and 289 and 285, respectively. HAs from viruses isolated in the 1997 H5 and 1999 H9 outbreaks in Hong Kong have novel oligosaccharide sites relative to the most closely related avian viruses (H5, HA1 158 and HA2 57; and H9, HA1 95 and 197; Figure 1). A large oligosaccharide at HA1 158 in the prominent antigenic loop at the top of the HA on the edge of the receptor-binding site (labelled 150s loop in Figure 2C) has been shown to decrease receptor binding, possibly altering the host or the cells within a host that the virus could infect (Gambaryan et al., 1998; Lu et al., 1998; Matrosovich et al., 1999; Lin et al., 2000). The novel oligosaccharide sites, in H9 human HA relative to H9 swine HA, at positions 95 and 197, may affect the antigenicity of the human HA.
The H5 human sequences also differ from the H5 avian HA studied here by the insertion of a polybasic furin cleavage site (RRRKK) between HA1 and HA2, which has been correlated in both the H5 and the H7 subtypes with increased pathogenicity (Klenk and Garten, 1994). This insertion is expected only to change the structure of the uncleaved precursor HA0 by lengthening the prominent surface loop at the cleavage site between HA1 and HA2 (between C1 and N2 in Figure 2C and D) (Chen et al., 1998).
Most avian H9 HAs, like the H5 human and avian HAs, have the common avian HA1 residues Gln226 and Gly228 in the receptor-binding site, but the swine, human and a number of avian H9 HAs have HA1 Leu226 and HA1 Gly228, intermediate between the avian sequence and the human HA1 Leu226, HA1 Ser228 pair (Lin et al., 2000; Ha et al., 2001). Two other receptor-binding site residues that are unusual in the H9 swine HA, HA1 Asn183 and Val190, are His183 and Glu190 in human and quail H9 HAs as they are in H3 human HAs (Lin et al., 2000; Ha et al., 2001). The human H9 isolates also contain HA1 His17, like H3 viruses, rather than HA1 Tyr17 as seen in the H9 swine HA studied here. Whether this substitution at HA1 17, which may influence the pH and the mechanism of fusion activation, favours infection in humans is unknown. The remaining residue differences between H5 avian and human viruses, and between H9 swine and human viruses, occur at positions on the surface of the HA, which readily accept mutations (reviewed in Skehel and Wiley, 2000) and where many differences are observed that alter antigenicity within subtypes and probably between subtypes (Nobusawa et al., 1991; Gorman et al., 1992; Webster et al., 1992).
Rotation of the membrane-distal subdomains of H5 and H9 HAs relative to those of the H3 HA
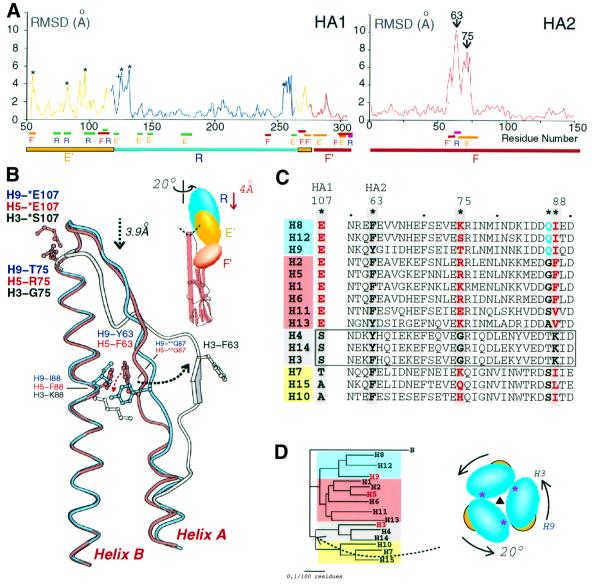
When the central triple-stranded α-helical stem of HA2 of the H5, H9 and H3 HAs are superimposed (helix B in Figure 2C–E), the membrane-distal globular domains of H5 and H9 are rotated about the trimer molecular axis through ∼20° relative to H3 and displaced upwards by ∼4 Å (Figure 3B and D). H5 differs from H9 by only ∼5°. The three subdomains R, E′ and F′ (a two-layer β-sheet subdomain in the stem region, residues HA1 275–307) are each relocated independently by different translations (from 5 to 8 Å) and rotations (from 16 to 19°) around slightly different axes (Figure 3B). Each subdomain has apparently migrated as a rigid body because structural differences within each subdomain among the three HA subtypes are small (r.m.s.d. ∼1 Å). The independent repositioning of the R and E′ subdomains is evidence of their separate identity and stability, and provides further support for the suggestion that HEF and HA derived evolutionarily from distinct ancestral protein domains with independent functions (Rosenthal et al., 1998; Zhang et al., 1999).
Fig. 3. Differences in subdomain locations in H3, H5 and H9 HAs and schematic diagram of key residues determining HA subtype structures. (A) R.m.s.ds between the α-carbon coordinates of the H5 and H3 HAs are plotted against HA sequence number. HA1 blue line = R subdomain superposition, yellow = E subdomain superposition, red = F′ subdomain superposition. An asterisk marks insertions in H5 relative to H3 (HA1 residues 53, 81, 95, 125, 133 and 255) and + marks a deletion at HA1 124. The underlines designate interface residues (F′–E′, orange; R–E′, green; F–E′, red; F–R, red) and the subdomain locations: E′, yellow; R, blue; and F and F′, red. (B) Superposition of the long (B) α-helices of HA2 showing differences in the intrahelical loop and long (B) α-helix N-terminal conformations: H9, blue; H5, red; H3, grey. Insert: schematic diagram indicating that the H5 and H9 globular domains are rotated 20° and displaced distally by 4 Å relative to the H3 HA. Within individual subdomains, the r.m.s.d. overlaps are 1.1, 1.0 and 1.1 Å for R, E′ and F′, respectively. The interhelical loop of HA2 contacts the R, E′ and F′ subdomains of HA1 (dashed line). (C) Sequence alignment of 15 HA subtypes at HA1 107 and HA2 60–90. Residues (*) suggested to determine the conformational differences between subtypes are shaded grey. The groups of subtypes (clades) predicted to have similar structures are shaded in four colours (grey, H3/H4/H14, G-75, K-88; yellow, H7/H10/H15, non-G-75, non-K-88, non-E-107; red, H1/H2/H5/H6/H11/H13, non-G-75, non-K-88, E-107; blue, H8/H9/H12, non-G-75, non-K-88, E-107, Q-87). The strains used to represent each subtype were: H1 (A/USSR/90/77), H2 (A/RI/5+/57), H3 (A/Aichi/2/68), H4 (A/Dk/Czechoslovakia/56), H5 (A/Dk/Singapore/3/97), H6 (A/Sh/Australia/1/72), H7 (A/Eq/Prague/1/56), H8 (A/Tk/Ontario/6118/68), H9 (A/Sw/HK/9/98), H10 (A/Ck/Germany/N/49), H11 (A/Dk/England/56), H12 (A/Dk/Alberta/60/76), H13 (A/Gl/Maryland/704/77), H14 (A/Ma/Gurjev/263/82) and H15 (A/Sh/West Australia/2576/79). (D) Phylogenetic relationships between HA subtypes and influenza B (B/Lee/40) HA based on HA amino acid sequences. The displacement of the HA1 subdomains (insert: HA viewed down the trimer axis from the top; the asterisk marks the interface where H3/H9 differ from H5 near residue 216) probably occurred when the H3, H4, H14 group diverged from the H7, H10 and H15 subtypes. The shaded colours for groups of subtypes correspond to the colours and groups in (C).
The membrane-distal globular domain of HEF is rotated a further 30° relative to the H5 and H9 HAs, or ∼50° relative to H3. The different angular position of the globular membrane-distal domain of HEF, relative to the HAs, apparently results from a completely different interface between this domain and the central stem of the molecule: HEF has an extra 100 residues in the esterase subdomain, E, which are located at the interface with the stem; the central long α-helices of HEF2 are two turns shorter (Figure 2F) than the homologous α-helices in HA; and these three long α-helices diverge at their N-termini, rather than being packed into a coiled coil as in the HAs.
Structural basis of the differences in orientation of the membrane-distal domains
While the structural basis for the difference in orientation of the membrane-distal domains of HEF relative to HA may be the 100 extra residues in the HEF1 E subdomain (yellow in Figure 2F) and the tertiary structure differences in HEF2, structural differences that could account for the differences in the H3 and the H5 and H9 HA orientations (Figure 3B and D) are very few. In the two inter-subdomain interfaces, R–E′ and E′–F′ (Figure 3B, insert), of all three HA subtypes, the tertiary structures are completely conserved. Details at the homologous interfaces differ because of the packing of different side chains, but generally a residue at a specific position in one subdomain contacts residues in the same sequence positions across an interface in all three HA subtypes. This repacking results in small, local deformations in the interfaces while preserving the approximate location of the main chains and many side chains on both sides of interfaces. Such a preservation of local structure even in interfaces is similar to the repacking seen in interiors of immunoglobulin domains and globin subunits where many similar structures have been observed and analysed from highly divergent sequences (Lesk and Chothia, 1980, 1982).
The impression gained by visually comparing the HA structures is supported by a quantitative comparison of the tertiary structures of the three HAs (Figure 3A). When the R subdomains of the H5 and H3 HAs are superimposed (Figure 3A), the r.m.s.d. between the main chain α-carbon positions in the whole R subdomain and in interface regions of R (residues underlined in Figure 3A) are uniformly low, averaging 1.6 Å. A similar result is found for superpositions of the E′ subdomains (residues HA1 52–116, yellow in Figure 3A) and of the F′ subdomains (residues HA1 275–307, red in Figure 3A). The few high peaks in these plots (asterisks in Figure 3A) are all at points of sequence insertion or deletion between the different subtypes. These data suggest that there are no dramatic differences in secondary or tertiary structure or main chain repackings in the intersubdomain interfaces within HA1 that account for the 20° difference in azimuthal orientation of the membrane-distal globular domains.
Comparisons of the HA2 structures, however, reveal a significant difference in both the secondary and tertiary structures of the H5 and H9 HAs relative to the H3 HA (Figure 3A and B). These differences are confined to HA2 residues 60–75 (r.m.s.d. values of 7–10 Å, Figure 3A), which form a loop between the short and long α-helices, helix A and helix B, respectively, in Figure 3B. This loop of HA2 forms the interface with both the membrane-distal globular subdomains R and E′ and the more lateral subdomain F′ (insert in Figure 3B, underlines in Figure 3A). The remainder of the HA2 chains from all three subtypes superimpose very well with an average r.m.s.d. in α-carbon positions of 0.9 Å (Figure 3A).
The largest differences between the H3 and the H5 and H9 HAs are seen at two sites in the interhelical loop: at the top of the loop near HA2 residue 75 and at the middle of the loop near HA2 residue 63 (Figure 3B). Since the R and E′ subdomains of HA1 only contact the triple-helical stem of HA2 near the C-terminus of the interhelical loop and the F′ subdomain of HA1 contacts the middle section of the HA2 loop (Figure 3B), the different orientations of the R, E′ and F′ subdomains of HA1 may result from the structural differences at these two positions.
At the C-terminus of the loop, residue HA2 75 is glycine in all H3 HAs, which permits a sharp turn into the long α-helix because glycine can adopt φ, ψ dihedral angles (91°, –130°) not permitted for other residues. Neither H5 nor H9 HAs have glycine residues at this position and both have 3.9 Å ‘taller’ turns before the long α-helix, helix B, begins (Figure 3B). These turns in H5 and H9 HAs appear to be stabilized by hydrogen bonds between conserved HA1 Glu107 in a neighbouring E′ subdomain and the backbone amide groups of HA2 residues 75 and 76 (Figure 3B). The effects of the ‘taller’ turns appear to propagate through the subdomain interface to alter the positions of E′ and R.
The difference in the paths of the loops near HA2 residue 63 (Figure 3B) similarly affects the location of the F′ subdomain. The nature of HA2 residue 88 (Figure 3C) in the long α-helix appears to determine how HA2 residue 63 in the middle section of the interhelical loop packs in the trimer. In the H3 subtype, HA2 Lys88 repels HA2 Phe63, creating a bulging interhelical loop (white atoms, dashed black arrow in Figure 3B). In H9, HA2 Ile88 packs against HA2 Tyr63, holding the loop against the long α-helix (blue in Figure 3B). The loop in H5 is similar to that in H9, with HA2 Phe88 packed against HA2 Phe63 (red in Figure 3B) but, in H5, HA2 Phe63 is permitted to approach another long helix of the trimer by the presence of a small residue, HA2 Gly87, while HA2 Gln87 of H9 prevents HA2 Tyr63 from occupying the equivalent position (H9-**Q87, H5-**G87 and dotted red arrow in Figure 3B).
An additional structural difference between H9 and H5 HAs is observed at the R subdomain trimer interface (Figure 3D), where a four-strand β-sheet (HA1 residues 162–173, 240–248, 200–206 and 208–215) is shifted ∼3 Å in H5 relative to H9 and H3 HAs. A glutamate at HA1 residue 216, which forms a hydrogen bond with the backbone nitrogen of HA1 residue 212 across the interface in the H5 HA, may be diagnostic of an H5-like interface. The observation that this R subdomain trimer interface is similar in H3 and H9 but different in H5 suggests that H5 diverged from one of them after they diverged from each other. This order is consistent with proposed phylogenetic trees based on sequence analysis, which suggest that H5 diverged from an H5/H9 ancestor after that ancestor diverged from an H3/H9/H5 ancestor (Figure 3D) (Nobusawa et al., 1991; Webster et al., 1992).
Discussion
A possible structural classification of HA subtypes
The 15 influenza HA subtypes were defined initially by immunodiffusion reactions of detergent-disrupted viruses with anti-HA hyperimmune sera (World Health Organization, 1980) and the validity of the classification was confirmed subsequently by sequencing virus genes for HA (reviewed in Nobusawa et al., 1991; Webster et al., 1992). The observation of similar R, E′ and F′ subdomain orientations in H5 and H9 HAs that differ from those in H3 HAs (Figure 3B) suggests that the 15 HA subtypes may fall into a smaller number of structural classes. Analysis of the sequences of HAs from all 15 subtypes in fact indicates that their structures can be divided into four classes based on predicted three-dimensional structure differences at the N-terminus of the long α-helix, helix B, and in the interhelical loop of HA2 based on differences in residues HA1 107, and HA2 63, 75, 87 and 88. We have examined >1000 HA sequences and show one representative from each subtype in Figure 3C.
The amino acids determining the structure at the N-terminus of the long α-helix of HA2, residue 75 in HA2 and 107 in HA1 (Figure 3B), are Gly75 and Ser107 in the H3, H4 and H14 subtypes (clade shaded grey Figure 3C and D) and, therefore, HAs of these three subtypes are likely to contain an H3-like sharp turn between the long α-helix and the interhelical loop (Figure 3B). The H1, H2, H5, H6, H8, H9, H11, H12 and H13 subtypes do not have glycine at HA2 residue 75 and cannot make an H3-like sharp turn. They can all form the ‘taller turn’ (Figure 3B), which in each subtype can also be stabilized by hydrogen bonds from Glu107 of HA1 (Figure 3B; blue and red shaded clades in Figure 3C and D). HA2 residue 87 is glutamine in H8 and H12 as well as H9 subtype HAs, suggesting an H9-like structural class (H8, H9, H12) based on this interhelical loop conformational difference between H5 and H9 (clade shaded blue in Figure 3C and D). The H7, H10 and H15 sequences also lack a glycine residue at HA2 75, but in addition lack the stabilizing Glu107 of HA1 (green-shaded clade in Figure 3C and D), suggesting that HAs of these subtypes probably resemble H5 and H9 HAs in subdomain position due to a ‘taller turn’, despite lacking the capping residue HA1 Glu107 and despite their evolutionary closeness to HAs of the H3, H4 and H14 subtypes (Nobusawa et al., 1991; Webster et al., 1992).
Overall, therefore, the five residues we have identified share two properties: they are located at positions where the three known HA structures are most different, and unique combinations of them form four classes that correspond to the four clades in the phylogenetic tree of HA sequences. There are no other sets of residues that meet these two criteria. Other regions of the HA, such as the receptor-binding subdomain and the surface antigenic sites, are under selective pressure by variation in the receptors of potential hosts and by host immune responses, respectively (Wiley and Skehel, 1987; Connor et al., 1994). These sites have been defined by genetic, biochemical and structural methods (reviewed in Skehel and Wiley, 2000), and evolution of both receptor-binding specificity and antigenicity are readily apparent among the strains of the individual subtypes that have infected humans (Matrosovich et al., 2000). If variation of these properties occurred from subtype to subtype, sets of residues might also be identified in these sites that correlated with the major clades. Some variations in antigenic regions like the loss of the HA1 140 loop in the H9 subtype (Figure 2D) and in the locations of oligosaccharide side chains are apparent, but we have not found any other set of residues that varies on a subtype basis.
Ionizable residues and fusion pH refolding in different HA subtypes
Mutant selection and mutagenesis studies have identified ionizable residues that affect the pH at which the structural changes in HA required for membrane fusion occur in H3 HA (Daniels et al., 1985, 1987; Doms et al., 1986; Steinhauer et al., 1996). The wide distribution of these residues in the interfaces between all the structural elements that rearrange at low pH (Bullough et al., 1994) suggested that any destabilization of these interfaces could affect the pH of refolding and that the trigger for refolding may be distributed rather than localized. Most of the residues that influence the pH of refolding, however, were found subsequently to be in the same chemical environment in the uncleaved precursor HA0, which does not undergo the low pH-induced, irreversible refolding required for fusion (Chen et al., 1998). Only three ionizable residues, His17 of HA1 and Asp109 and Asp112 of HA2, changed from solvent-accessible to completely buried environments between the uncleaved and cleaved H3 subtype structures, raising the possibility that they may have a special role in triggering refolding (Chen et al., 1998).
We have discussed before the roles of the two acidic residues HA2 Asp109 and Asp112, which are buried by the fusion peptide on cleavage of the precursor HA0 (Chen et al., 1998). Neither H5 nor H9 HAs analysed here contain a histidine at the third buried position, HA1 residue 17; in both cases, HA1 17 is tyrosine (Table I). Although this is not strictly a subtype-specific property, it is notable that among the vast majority of HA sequences, six subtypes comprising two clades have histidine at HA1 17 (H3, H4, H7, H10, H14 and H15); the other nine subtypes comprising the other two clades have HA1 Tyr17.
Table I. Ionizable residues that might affect the pH stability of haemagglutinins in different clades.
| H9 | H5 | H3 | H7 | |
|---|---|---|---|---|
| HA1 | ||||
| 17 | Tyr | Tyr | His | His |
| HA2 | ||||
| 51 | Lys | Lys | Lys | Lys |
| 103 | Glu | Glu | Glu | Glu |
| 105 | Gln | Glu | Gln | Gln |
| 106 | Lys | Arg | His | His |
| 110 | Glu | Phe | Val | Leu |
| 111 | His | His | Thr | Thr |
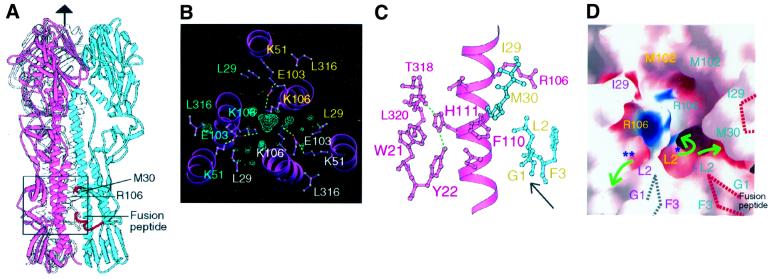
In the H5 HA structure, we observe that His111 of HA2 is the only solvent-inaccessible, basic residue, and that it is near the fusion peptide (arrow in Figure 4C), behind HA2 Phe110 (Figure 4C). In H5, HA2 His111 may become ionized at low pH and contribute to the triggering of refolding. HA2 111 is histidine in the nine subtypes where HA1 17 is tyrosine (Table I). In the H5 and H9 HA structures, HA2 Arg106 and HA2 Lys106, respectively, are also located in this solvent cavity with restricted access via small channels to bulk solvent (Figure 4C, green arrows in Figure 4D). These basic residues at HA2 106 are on the long α-helix, helix B of HA2, pointing toward each other and toward the HA2 trimer axis of symmetry just above the fusion peptide, at the point where the long α-helix unfolds and reverses direction at low pH (Bullough et al., 1994) (Figure 4). In H9 HA, HA2 Lys106 is hydrogen-bonded to HA2 Glu103, which in turn hydrogen-bonds to HA2 Lys51 (dashed lines in Figure 4C). There is a strong peak (>5σ) of electron density with approximately tetrahedral shape buried on the trimer axis above the three HA2 Lys106 residues in the trimer that may be an anion (possibly phosphate, with a refined temperature factor of 29.5 Å2), which could stabilize the HA2 Lys106 positive charges (blue mesh in Figure 4B). In the H5 HA, where the charge on HA2 Arg106 is apparently balanced by the salt bridge hydrogen bond to HA2 Glu105 (glutamine in H9) (but a dipole–dipole repulsion may persist), there are water molecules or monovalent ions near the trimer axis below HA2 Arg106, but there is apparently no large ion above or near the positions of the three Arg106 residues in the trimer. In the H3 HA, HA2 His106 is located in approximately the same position, probably hydrogen-bonded to HA2 Lys51 and HA2 Gln105. The two clades with HA2 His111 that lack HA1 His17 contain arginine or lysine at HA2 position 106, suggesting that if these residues prove to be important in refolding at low pH, they may have evolved as alternatives to HA1 His17 as components of the triggering mechanism.
Fig. 4. H5 and H9 residues implicated in the low pH-induced refolding required for membrane fusion. (A) An H5 HA trimer with monomers coloured blue, pink and white. The box indicates approximately the area detailed in (B–D). Red segments of the fusion peptide and near HA1 Met30 are indicated by red dotted lines in (D). (B) H9 HA difference in electron density (contoured at 4σ) suggests an ion bound between three HA2 Lys106 residues at the centre of the triple-helical stem region in H9. HA2 Glu103 is hydrogen-bonded to both HA2 Lys51 and HA2 Lys106. A number of water molecules are found around the putative counter-ion and below HA2 Lys106 (not shown). (C) H5 HA2 His111 is buried by HA1 Leu320, HA2 Trp21 and Phe110, and is located near the fusion peptide (arrow), HA2 residues G1, L2, F3 coloured blue. (D) In the HA1–HA2 trimer, residue HA2 106 is accessible to solvent only through narrow channels (green arrows) after the fusion peptide (G1, L2 and F3) fits between the helices following HA0 cleavage to HA1 and HA2 (Chen et al., 1998). For clarity, one helix in the front of the cavity and peripheral residues were omitted. This figure was generated using GRASP (Nicholls et al., 1991).
A difference in solvent accessibility in uncleaved and cleaved HAs of the cavity near HA2 positions 111 and 106 is caused by HA2 residues Gly1, Leu2 and Phe3 of the fusion peptide, which occlude most of the channel below HA1 Met30 (Figure 4D) that connects the central cavity to the bulk solvent. It is possible that changes in ionization at low pH may be relieved, in the relatively open cavity of uncleaved HA0 near residues HA2 106 and the putative anion, by local rearrangements. However, following HA0 cleavage, when that cavity is nearly occluded from bulk solvent by the refolded fusion peptide, changes in ionization may contribute to the triggering of the structural changes in HA required for membrane fusion.
Origin of influenza HA subtypes
Within subtypes, HA sequences have ∼80–90% or more sequence identity; between subtypes ∼40–50% (reviewed in Nobusawa et al., 1991). Subtypes may have originated by neutral drift as the result of geographical isolation or by Darwinian selection of fitness for a niche. Phylogenetic evidence indicates that they evolved in avian hosts, where influenza viruses replicate efficiently and infection is usually asymptomatic (reviewed in Gorman et al., 1992). Selective pressures responsible for their diversification in avian species are unknown; antigenic variation within avian subtypes, for example, is not extensive (Gorman et al., 1992).
Our observations that the structural differences between H5 and H9 and H3 HAs may be traceable to differences in a few residues of HA2 in the F subdomain suggest that HA subtypes may have originated as a consequence of residue differences that have the potential to affect the stability of the HA, which is critical for its membrane fusion function during virus entry into cells. HA2 residues 63, 87 and 88 make critical contacts in an intramolecular interface between the long α-helix, helix B, and the interhelical loop. These interactions are broken during induction of the low pH conformation required for membrane fusion. HA2 residue 75 and HA1 residue 107 determine the stability and shape of the N-terminus of the coiled coil in neutral pH HA that is extended by the structural changes induced at low pH by refolding of the interhelical loop into an α-helix and by incorporation of the short α-helix into a new long α-helix (Bullough et al., 1994). Both sets of residues that appear critical for the observed structural differences between HAs of different subtypes, therefore, are at locations that are required to undergo refolding. Diversification of such residues may have resulted in the variation of properties of HAs related to their membrane fusion activity. The extensive changes in HA structure that are required for activation of HA-mediated membrane fusion at low pH or elevated temperatures are irreversible; premature activation results in inactivation of infectivity (reviewed in Skehel and Wiley, 2000). pH and temperature variations in different host species or at different ecological, anatomical or intracellular sites experienced before or during infection may, therefore, have provided the selective pressure for the origin of such a wide variety of influenza HA subtypes.
Materials and methods
Haemagglutinin purification
HAs were prepared by bromelain digestion of purified egg-grown viruses. For A/Duck/Singapore/3/97 (H5N1), digestion was for 2 h at 37°C in Tris 10 mM pH 8.0, 30 mM mercaptoethanol, virus protein:bromelain 20:1. For A/Swine/Hong Kong/9/98 (H9N2), digestion was similar except for 25 mM mercaptoethanol and virus protein:bromelain 5:1. Digestions were terminated using complete protease inhibitor cocktail (Roche Molecular Biochemicals). The BHAs were purified by sucrose gradient centrifugation (5–20% sucrose, 10 mM Tris pH 7.4, 60 h, SW27 Beckman, 20 000 r.p.m., 4°C), dialysed into 10 mM NaCl, 10 mM Tris pH 7.4, absorbed to a Q15 Sartorius anion exchange membrane, eluted in 200 mM NaCl, 10 mM Tris pH 7.4, and concentrated by ultrafiltration using a PM10 Amicon membrane.
Crystal growth and ligand soaking
Hanging drop crystallizations used 1 µl of BHA, 10 mg/ml, 10 mM HEPES buffer pH 7.5 mixed with 1 µl of precipitant solution (H5, 20% MME-PEG 2000, 2 mM NiCl2, 100 mM HEPES pH 7.5; H9, 1.4 M Na citrate, 100 mM HEPES pH 7.5). Crystals were transferred to 200 µl of well solution and then to solutions with increasing amounts of cryoprotectant in 2 h steps (H5, 20% glycerol; H9, 1.5 M xylitol). Crystals were flash-cooled in liquid nitrogen.
Data collection, structural determination and refinement
Both H5 and H9 native diffraction data were collected at beamline 14BMC at the Adanced Photon Source (Argonne National Laboratory), 100K; H9 derivative data were collected at beamline X25C at the National Synchrotron Light Source (Brookhaven National Laboratory), 100K. All data sets were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997).
Because the H9 HA trimer was located on the 3-fold symmetry axis of a P63 lattice, molecular replacement reduced to a single, angular parameter search of 120°. Conventional molecular replacement (MR) and the direct search with AMORE using the X31 H3 subtype HA coordinates (PDB: 1HGE) (H9 HA has 42% sequence identity with the H3 HA) produced a consistent solution. However, the MR-phased electron density map could not be traced in the membrane-distal domain due to domain motions. The MR phases were useful in finding heavy atom sites in derivatives by difference Fourier methods in combination with difference and anomalous Patterson methods. MIRAS phases calculated with MLPHARE (CCP4, 1994) were improved by solvent flattening and histogram matching using DM (CCP4, 1994). Models were built with O (Jones et al., 1991). One saccharide was visible at Asn21 and two at HA1 asparagines 128, 289 and 296, and HA2 154.
The H5 HA structure was determined by MR using H9 HA coordinates (H5 and H3 share 44% sequence identity; H5 and H9 52%). The electron density map calculated from the H9 MR phases was interpretable. Because the difference between H9 and X:31 indicated domain motions, rigid body refinement of each individual domain in the search probe, followed by solvent flipping and histogram matching in CNS (Brünger et al., 1998), was used to improve the electron density map. One saccharide was visible at HA1 Asn21 and two at HA1 asparagines 33, 169 and 289, and HA2 154. C-terminal segments after HA1 324 and HA2 160 are missing in the final model and are presumed to be disordered. One Ni (2+) ion, coordinated by two histidines (HA2 His142), joins two neighbouring molecules at the membrane-proximal end of HA in the H5 crystal lattice.
Both structures were refined with CNS (Brünger et al., 1998). Statistics are summarized in Table II. Coordinates have been deposited in the Protein Data Bank (accession Nos: H5, 1JSM; H9, 1JSD).
Table II. Crystallographic data statistics.
| |
|
H5 |
H9 |
|---|---|---|---|
| Data collection | |||
| Resolution (Å) | 40.0–1.9 | 30.0–1.8 | |
| Unique reflections | 59 121 (3643) | 91 194 (9090) | |
| Completeness (%) | 90.5 (56.4) | 100.0 (100.0) | |
| Redundancy | 6 | 9 | |
| I/<σ> | 17.5 | 12.9 | |
|
Rmerge (%) |
|
7.0 (20.6) |
8.3 (48.2) |
| Refinement | |||
| Rcryst (%) | 24.1 (40.4) | 21.8 (32.5) | |
| Rfree (%) | 27.1 (53.2) | 22.9 (33.2) | |
| R.m.s.d. bond lengths (Å) | 0.009 | 0.007 | |
| R.m.s.d angles (°) | 1.5 | 1.3 | |
| B r.m.s.d. for bonded main chain and side chain atoms | 0.929, 1.632 | 0.540, 0.540 | |
| No. of atoms | |||
| protein | 3738 | 3725 | |
| water | 357 | 282 | |
| carbohydrate |
|
101 | 125 |
| H9 derivatives and phasing |
Pt(NH3)2Cl2 |
K2PtCl4 |
K3IrCl6 |
| Resolution (Å) | 25.0–2.6 | 25.0–2.6 | 25.0–2.7 |
| Unique reflections | 30 731 | 28 769 | 30 645 |
| Completeness (%) | 98.8 | 93.9 | 100.0 |
| Redundancy | 5 | 4 | 5 |
| I/<σ> | 15.5 | 14.2 | 15.0 |
| Rmerge (%) | 5.6 | 7.2 | 6.5 |
| Phasing power acentric (centric) | 1.45 (1.21) | 1.36 (1.06) | 0.90 (0.56–5 Å) |
| Rcullis acentric (centric, anomalous) | 0.73 (0.71, 0.94) | 0.77 (0.81, 0.97) | 0.85 (0.87, 0.95) |
| Overall FOM | 0.46 | ||
H5 crystallized in space group R32 (a = 131.6 Å, c = 254.1 Å) and H9 in space group P63 (a = 108.5 Å, c = 149.3 Å). The statistics for the highest resolution shell of the native data set are in parentheses. Although the H5 crystals diffracted to high resolution, they have high mosaicity (>0.8).
Influenza A virus HA sequence analysis
HA sequences were obtained from the influenza A virus sequence database at Los Alamos National Laboratory. Initial sequence alignment was calculated by PILEUP in GCG (Womble, 2000). Refined sequence alignment among serotypes was based on superposition of H3, H5 and H9 HA crystal structures. Alignment between H3 and influenza C virus HEF sequences was as before (Rosenthal et al., 1998).
Acknowledgments
Acknowledgements
We thank members of our laboratories for helpful discussions, the beamline staff at 14BMC at APS and X25 at BNL-NSLS for assistance in data collection, and Dr Chi-Huey Wong and colleagues for a helpful synthesis. This research is supported by NIH Grant AI-13654, and by a supplement to this grant for Expanded International Research on Emerging and Re-Emerging Diseases, by an International Partnership Research Award in Veterinary Epidemiology of the Wellcome Trust, by the Howard Hughes Medical Institute and by the MRC (U.K.). Y.H. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. D.C.W. is an investigator of the Howard Hughes Medical Institute. Coordinates have been deposited in the Protein Database (1JSM, 1JSD).
References
- Bean W.J., Schell,M., Katz,J., Kawaoka,Y., Naeve,C., Gorman,O. and Webster,R.G. (1992) Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J. Virol., 66, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. et al. (1999) Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997–1998. Virology, 254, 115–123. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 901–921. [DOI] [PubMed] [Google Scholar]
- Bullough P.A., Hughson,F.M., Skehel,J.J. and Wiley,D.C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature, 371, 37–43. [DOI] [PubMed] [Google Scholar]
- Chen J., Lee,K.-H., Steinhauer,D., Stevens,D., Skehel,J.J. and Wiley,D.C. (1998) Structure of the hemagglutinin precursor cleavage site, a major determinant of influenza pathogenicity and the origin of the labile conformation of HA. Cell, 95, 409–417. [DOI] [PubMed] [Google Scholar]
- Claas E.C., Osterhaus,A.D., van Beek,R., De Jong,J.C., Rimmelzwaan,G.F., Seene,D.A., Krauss,S., Shortridge,K.F. and Webster,R.G. (1998) Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet, 351, 472–477. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Connor R.J., Kawaoka,Y., Webster,R.G. and Paulson,J.C. (1994) Receptor specificity in human, avian and equine H2 and H3 influenza virus isolates. Virology, 205, 17–23. [DOI] [PubMed] [Google Scholar]
- Daniels R.S., Downie,J.C., Hay,A.J., Knossow,M., Skehel,J.J., Wang,M.L. and Wiley,D.C. (1985) Fusion mutants of the influenza virus haemagglutinin glycoprotein. Cell, 40, 431–439. [DOI] [PubMed] [Google Scholar]
- Daniels R.S. et al. (1987) The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J., 6, 1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R.W., Gething,M.J., Henneberry,J., White,J. and Helenius,A. (1986) Variant influenza virus hemagglutinin that induces fusion at elevated pH. J. Virol., 57, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–4, 112–13. [DOI] [PubMed] [Google Scholar]
- Francis T. (1940) A new type of virus from epidemic influenza. Science, 91, 405–408. [DOI] [PubMed] [Google Scholar]
- Gambaryan A.S., Matrosovich,M.N., Bender,C.A. and Kilbourne,E.D. (1998) Differences in the biological phenotype of low-yielding (L) and high-yielding (H) variants of swine influenza virus A/NJ/11/76 are associated with their different receptor-binding activity. Virology, 247, 223–231. [DOI] [PubMed] [Google Scholar]
- Gorman O.T., Bean,W.J. and Webster,R.G. (1992) Evolutionary processes in influenza viruses: divergence, rapid evolution and stasis. Curr. Top. Microbiol. Immunol., 176, 75–97. [DOI] [PubMed] [Google Scholar]
- Ha Y., Stevens,D.J., Skehel,J.J. and Wiley,D.C. (2001) X-ray structures of H-5 avian and H9 swine influenza virus haemagglutinins bound to avian and human receptor analogs. Proc. Natl Acad. Sci. USA, 98, 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G., Durkop,I., Becht,H. and Klenk,H.D. (1988) The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol., 69, 839–846. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss,S. and Webster,R.G. (1989) Avian to human transmission of the PB1 gene of influenza A virus in the 1957 and 1968 pandemics. J. Virol., 63, 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk H.D. and Garten,W. (1994) Host cell proteases controlling virus pathogenicity. Trends Microbiol., 2, 39–43. [DOI] [PubMed] [Google Scholar]
- Kraulis J.P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lesk A.M. and Chothia,C. (1980) How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol., 136, 225–270. [DOI] [PubMed] [Google Scholar]
- Lesk A.M. and Chothia,C. (1982) Evolution of proteins formed by β-sheets. II. The core of the immunoglobulin domains. J. Mol. Biol., 160, 325–342. [DOI] [PubMed] [Google Scholar]
- Lin Y.P. et al. (2000) Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl Acad. Sci. USA, 97, 9654–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Tumpey,T.M., Morken,T., Zaki,S.R., Cox,N.J. and Katz,J.M. (1999) A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol., 73, 5903–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macken C., Lu,H., Goodman,J. and Boykin,L. (2001) The value of a database in surveillance and vaccine selection. In Osterhaus,A., Cox,N. and Hampson,A.W. (eds), Options for the Control of Influenza IV. Elsevier, Amsterdam, The Netherlands, pp. 103–106.
- Matrosovich M., Tuzikov,A., Bovin,N., Gambaryan,A., Kilmov,A., Castrucci,M.R., Donatelli,I. and Kawaoka,Y. (2000) Early alterations of the receptor-binding properties of H1, H2 and H3 avian influenza virus hemagglutinins after their introductions into mammals. J. Virol., 74, 8502–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Zhou,N., Kawaoka,Y. and Webster,R. (1999) The surface glycoproteins of H5 influenza viruses isolated from humans, chickens and wild aquatic birds have distinguishable properties. J. Virol., 73, 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–96. [DOI] [PubMed] [Google Scholar]
- Nobusawa E., Aoyama,T., Kato,H., Suzuki,Y., Tateno,Y. and Nakajima,K. (1991) Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology, 182, 475–485. [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D., Rimmelzwaan,G.F., Martina,B.E., Bestebroer,T.M. and Fouchier,R.A. (2000) Influenza B virus in seals. Science, 288, 1051–1053. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Reid A.H., Fanning,T.G., Hultin,J.V. and Taubenberger,J.K. (1999) Origin and evolution of the 1918 ‘Spanish’ influenza virus hemagglutinin gene. Proc. Natl Acad. Sci. USA, 96, 1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohm C., Zhou,N., Suss,J., Mackenzie,J. and Webster,R. (1996) Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology, 217, 508–516. [DOI] [PubMed] [Google Scholar]
- Rosenthal P.B., Zhang,X., Formanowski,F., Fitz,W., Wong,C.-W., Meier-Ewert,H., Skehel,J.J. and Wiley,D.C. (1998) Three dimensional structure of the haemagglutinin–esterase-fusion glycoprotein of influenza C virus. Nature, 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. and Wiley,D. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem., 69, 531–569. [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A., Martín,J., Lin,Y.-P., Wharton,S.A., Oldstone,M.B.A., Skehel,J.J. and Wiley,D.C. (1996) Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc. Natl Acad. Sci. USA, 93, 12873–12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez D.L., Perdue,M.L., Cox,N., Rowe,T., Bender,C., Huang,J. and Swayne,D.E. (1998) Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J. Virol., 72, 6678–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. et al. (1998) Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science, 279, 393–395. [DOI] [PubMed] [Google Scholar]
- Taylor R.M. (1949) Studies on survival of influenza virus between epidemics and antigenic variants of the virus. Am. J. Publ. Health, 39, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean,W.J., Gorman,O.T., Chambers,T.M. and Kawaoka,Y. (1992) Evolution and ecology of influenza A viruses. Microbiol. Rev., 56, 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C. and Skehel,J.J. (1987) The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem., 56, 365–394. [DOI] [PubMed] [Google Scholar]
- Wiley D.C., Wilson,I.A. and Skehel,J.J. (1981) Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature, 289, 373–378. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Skehel,J.J. and Wiley,D.C. (1981) Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature, 289, 366–373. [DOI] [PubMed] [Google Scholar]
- World Health Organization (1980) A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull. World Health Organ., 58, 585–591. [PMC free article] [PubMed] [Google Scholar]
- Womble D.D. (2000) GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol., 132, 3–22. [DOI] [PubMed] [Google Scholar]
- Yuanji G. and Desselberger,U. (1984) Genome analysis of influenza C viruses isolated in 1981/82 from pigs in China. J. Gen. Virol., 65, 1857–1872. [DOI] [PubMed] [Google Scholar]
- Zhang X., Rosenthal,P.B., Formanowski,F., Fitz,W., Wong,C.H., Meier-Ewert,H., Skehel,J.J. and Wiley,D.C. (1999) X-ray crystallographic determination of the structure of the influenza C virus haemagglutinin–esterase–fusion glycoprotein. Acta Crystallogr. D, 55, 945–961. [DOI] [PMC free article] [PubMed] [Google Scholar]