Abstract
Using cDNA-based array analysis combined with double-stranded RNA interference (dsRNAi), we have identified yk298h6 as a target gene of Caenorhabditis elegans TGF-β signaling. Worms overexpressing dbl-1, a TGF-β ligand, are 16% longer than wild type. Array analysis shows yk298h6 to be one of several genes suppressed in such worms. Disruption of yk298h6 function by dsRNAi also resulted in long worms, suggesting that it is a negative regulator of body length. yk298h6 was then mapped to, and shown to be identical to, lon-1, a known gene that affects body length. lon-1 encodes a 312 amino acid protein with a motif sequence that is conserved from plants to humans. Expression studies confirm that LON-1 is repressed by DBL-1, suggesting that LON-1 is a novel downstream component of the C.elegans TGF-β growth regulation pathway. Consistent with this, LON-1 is expressed mainly in the larval and adult hypodermis and has dose-dependent effects on body length associated with changes in hypodermal ploidy, but not hypodermal cell proliferation.
Keywords: body length/Caenorhabditis elegans/hypodermis/polyploidization/TGF-β
Introduction
Some of the most fundamental, but least understood, aspects of animal development are the mechanisms by which body size is determined (Conlon and Raff, 1999; Stern and Emlen, 1999; Day and Lawrence, 2000). Body size is also the most obvious way in which animal species differ from each other (Bonner, 1989). This is particularly true for nematodes, which vary in size between 3.0 × 10–1 and 8.0 × 104 mm, but are otherwise quite morphologically uniform (Flemming et al., 2000). Wild-type strains of the nematode Caenorhabditis elegans are 1.2 mm long. Mutations that affect one aspect of body size, length, have long been known in this worm (Brenner, 1974), causing adults to be either long (Lon) or small (Sma). The frequency and viability of these mutants indicate that the worm might be an especially good organism in which to study the molecular regulation of body size.
Recent studies have shown that some C.elegans Sma mutants affect genes that encode components of a transforming growth factor-β (TGF-β) signaling pathway (Patterson and Padgett, 2000). These genes include: sma-6, a homologue of a vertebrate type I Ser/Thr kinase receptor (Krishna et al., 1999), daf-4, a type II Ser/Thr kinase receptor (Estevez et al., 1993) also used in dauer larva formation (Georgi et al., 1990; Ren et al., 1996) and sma-2, sma-3 and sma-4, cytoplasmic SMADs (Massague, 1996; Heldin et al., 1997), that translocate into the nucleus upon signal activation (Savage et al., 1996). Since DAF-4 can function as a type II receptor for BMP signaling in mammalian cells (Estevez et al., 1993) there appears to be substantial functional conservation between nematode and mammalian TGF-β signaling. More recently, DBL-1/CET-1, belonging to the TGF-β superfamily of proteins, has been identified as the ligand that triggers the sma signaling pathway (Morita et al., 1999; Suzuki et al., 1999). Loss-of-function mutations in dbl-1 also have a Sma phenotype, while worms that overexpress it are Lon. DBL-1 is presumed to be a secreted growth factor, but how it actually regulates body length remains largely unknown.
One obvious way in which body length might be controlled by the TGF-β pathway is by regulation of cell proliferation. However, Sma and Lon mutants appear to have wild-type cell numbers (Morita et al., 1999; Suzuki et al., 1999; Flemming et al., 2000). This is not surprising, since ∼50% of the growth of C.elegans occurs during adulthood, when no cell proliferation takes place (Knight et al., 2001). This growth, and even part of larval growth, must then be due to increases in cell size (Flemming et al., 2000). Increases in cell size of at least some tissues such as the hypodermis and intestine are, in C.elegans, associated with increases in somatic ploidy via endoreduplication (Hedgecock and White, 1985). Recently, worms that lack TGF-β activity due to mutations in daf-4, sma-2 and dbl-1 have been shown to have reduced hypodermal ploidy relative to wild type (Flemming et al., 2000; Nyström et al., 2002). This observation also suggests a possible mechanism for body size evolution since the degree of somatic polyploidization appears to be, in part, correlated with evolved differences in body size among nematode species related to C.elegans (Flemming et al., 2000). Identification of further components of this pathway that affect endoreduplication is, therefore, of interest for an understanding not only of the regulation of body size, but also of nematode morphological diversity.
Downstream target genes of dbl-1 signaling might be identified by mutational screens that enhance or suppress the Sma phenotype. However, this kind of genetic screen is often time-consuming and laborious. On the other hand, recent DNA macro/microarray technology (Galitski et al., 1999; Lockhart and Winzeler, 2000; Young, 2000) has permitted the simultaneous screening of thousands of expressed genes. It is even possible, in principle, to examine the expression of all 19 000 C.elegans genes in two different genetic backgrounds or growth conditions (Mochii et al., 1999; Reinke et al., 2000). Another powerful approach recently developed to address gene function is double-stranded RNA interference (dsRNAi), which permits the sequence-specific inactivation of genes (Fire et al., 1998; Bass, 2000; Zamore et al., 2000). dsRNAi has accelerated the high through-put analysis of gene function not only for C.elegans but also for other model organisms (Kennerdell and Carthew, 1998; Ngo et al., 1998; Sanchez Alvarado and Newmark, 1999; Chuang and Meyerowitz, 2000).
Here we report large-scale analyses of gene expression profiles in different genetic backgrounds displaying Sma and Lon phenotypes. We show that one of the genes recovered from this screen has a Lon phenotype when its function is inhibited by dsRNAi. We show that loss-of-function mutations in a known regulator of body length, lon-1, interrupt this gene, and that lon-1 transcription is negatively regulated by dbl-1 signaling. lon-1 encodes an evolutionarily conserved protein that defines the ‘PR-1 related protein superfamily’ from yeast, plant, insect to human, whose biological functions are little known (Szyperski et al., 1998). Moreover, we provide evidence that DBL-1 regulation of hypodermal polyploidization is mediated by LON-1.
Results
Identification of DBL-1-regulated genes by differential hybridization analysis using cDNA-based macroarray
As we reported previously, cDNA array of C.elegans mutants is an efficient way of screening signal-specific target genes (Mochii et al., 1999). To understand the molecular pathway regulating C.elegans body length, we used the same DNA macroarray and screened for genes regulated by dbl-1 signal. In this study, we compared the gene expression profile between dbl-1 null mutants displaying Sma phenotype, dbl-1(nk3) (Morita et al., 1999) and worms overexpressing dbl-1 (which contain a multi-copy of the dbl-1 genomic fragment) with Lon phenotype, ctIs40 (Suzuki et al., 1999). Out of ∼8000 genes arrayed on nylon membranes, 16 genes were found to be up-regulated and seven down-regulated. Gene identification and fold increases of hybridization intensity are summarized in Table I. Altered expression was confirmed for all identified genes by conventional northern blotting. Of the 23 genes identified by cDNA array signals as being regulated by DBL-1, we failed to confirm seven by northern blots.
Table I. List of genes identified and dsRNAi phenotype.
| Clone name | Gene | Fold increase (HDF)a | dsRNAi phenotype |
|---|---|---|---|
| yk225d6 | K04E7.1 | 2.3 | Sma |
| yk253f12 | F25H8.5 | 2.2 | no phenotype |
| yk355c4 | C17G1.6 | 3.4 | molting defect |
| yk412e5 | Y38H6C.1 | 1.9 | no phenotype |
| yk608d12 | no gene found | 1.9 | weak Sma |
| yk174b12 | Y57G11C.15 | 2.0 | Let (embryonic lethal) |
| yk479h1 | C07E3.10 | 2.0 | no phenotype |
| yk532c7 | ZK1290.8 | 2.0 | no phenotype |
| yk570a11 | F27E11.3 | 2.5 | no phenotype |
| yk563h1 | C44H4.3 | 1.9 | no phenotype |
| yk604a4 | W04G3.8 | 2.0 | Dpyb |
| yk355e1 | F44E2.4 | 2.8 | no phenotype |
| yk430h5 | F23B2.11 | 3.0 | no phenotype |
| yk479e3 | C44H4.2 | 2.0 | no phenotype |
| yk500g10 | VC5.3 | 6.0 | no phenotype |
| yk44c1 | K10C2.1 | 2.2 | no phenotype |
| yk257f11 | T21C9.5 | 0.3 | Uncc |
| yk361c1 | K10C2.3 | 0.5 | no phenotype |
| yk530e1 | no gene found | 0.4 | no phenotype |
| yk553c5 | no gene found | 0.5 | no phenotype |
| yk71c6 | T21C9.2 | 0.5 | no phenotype |
| yk298h6 | F48E8.1 | 0.5 | Lon |
| yk605h1 | F55A4.2 | 0.5 | no phenotype |
aThe data are the average of four experiments. HDF, high density filter.
bDpy, Dumpy.
cUnc, uncoordinated.
yk298h6 encodes the lon-1 gene
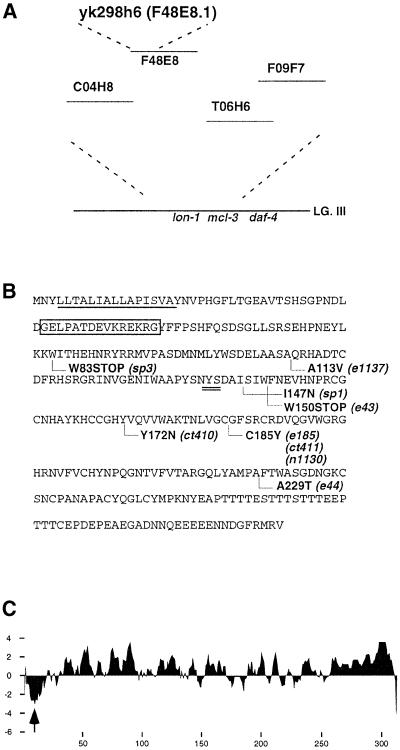
In order to examine the biological relevance of the identified genes to the determination of body length in relation to dbl-1 signaling, we carried out dsRNAi, a method developed recently by which particular genes can be rapidly inactivated (Fire et al., 1998). Among the genes regulated both up and down in our microarray screen, seven showed various dsRNAi phenotypes (Table I), some of which resembled the Sma and Lon phenotypes of known body length mutations. We focused on yk298h6 (F48E8.1), one of the genes suppressed by dbl-1 because its disruption by dsRNAi caused a typical Lon phenotype with a high frequency. Importantly, yk298h6 mapped close to the lon-1 locus (Figure 1A) by database analysis. Expecting that yk298h6 is the gene responsible for lon-1 mutation, we performed rescue experiments using a fragment of cosmid F48E8 harboring the entire yk298h6 sequence. The 8.0 kb genomic fragment was sufficient to rescue the lon-1 body length phenotype, indicating strongly that yk298h6 encodes the lon-1 product (Table II). This was also confirmed by identifying the mutational lesion in nine lon-1 alleles (Figure 1B). Sequence analysis of PCR-amplified lon-1 alleles identified missense and nonsense mutations that cause non-conservative amino acid substitutions in the following alleles: e1137, ct410, e185, ct411, n1130, e44 and sp1 (Figure 1B). Alleles sp3 and e43 each have a stop codon caused by a CG-to-TA transition that converts codon TGG (W83) to the opal terminator TGA and TGG (W150) into the amber terminator TAG, respectively. Based on these findings, we conclude that yk298h6 (F48E8.1 for gene) encodes lon-1.
Fig. 1. yk298h6 encodes the lon-1 gene. (A) The yk298h6 (F48E8.1) gene maps to the lon-1 locus. (B) Primary structure of LON-1 and point mutation site. LON-1 is a 312 amino acid protein. The N-terminal hydrophobic region is underlined. A putative N-glycosylation site is double underlined. The peptide sequence that generates the LON-1 antibody is boxed. Amino acid substitutions in lon-1 alleles are indicated below the sequence. e43 and sp3 each have a nonsense mutation (W150 to stop and W83 to stop, respectively). ct410, ct411, e44, e185, e1137, n1130 and sp1 have missense mutations. (C) The LON-1 Kyte–Doolittle hydrophobicity plot shows a hydrophobic stretch at the N-terminus of the protein (amino acids 4–18, see arrow).
Table II. Effect of lon-1 gene dosage on body length.
| Genotype | Body length (mm)a | N |
|---|---|---|
| N2 | 1.28 ± 0.03 | 85 |
| lon-1(e185) | 1.71 ± 0.01 | 93 |
| lon-1(e43) | 1.43 ± 0.04 | 87 |
| lon-1(ct410) | 1.60 ± 0.01 | 103 |
| lon-1(sp3) | 1.35 ± 0.05 | 19 |
| lon-1(RNAi) | 1.63 ± 0.06 | 35 |
| lon-1(e43)/Dfb | 1.49 ± 0.11 | 8 |
| lon-1(sp3)/Df | 1.29 ± 0.06 | 19 |
| nkIs10c | 1.00 ± 0.03 | 43 |
| lon-1(e185);Exnk51[lon-1(+)] | 1.24 ± 0.11 | 65 |
| lon-1(e185);Exnk52[lon-1(+)] | 1.19 ± 0.09 | 65 |
| lon-1(e185);Exnk53[lon-1(+)] | 0.89 ± 0.10 | 62 |
aMean ± SE. N, measured animals. Body lengths of various mutants were measured using a micrometer at high magnification at 120 h post-hatching.
bDeficiency line.
cLon-1-overexpressed Sma worm.
LON-1 belongs to the PR-protein superfamily conserved among yeast, plant, insect and human, and may be a type II transmembrane protein
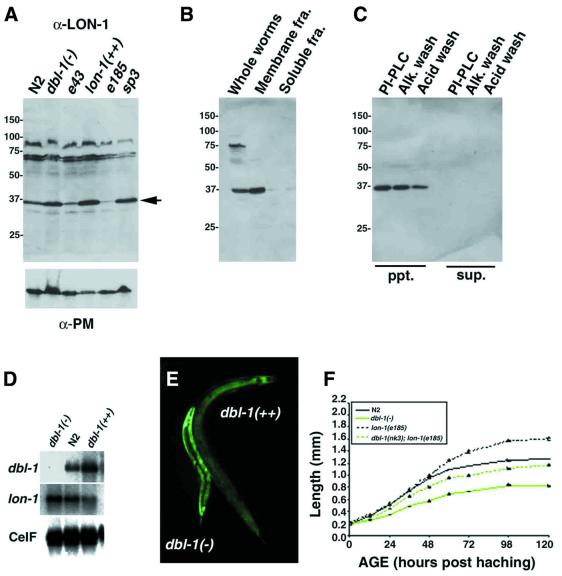
The lon-1 gene encodes a 312 amino acid protein (Figure 1B). By a database search, we have found that genes encoding proteins homologous to lon-1 belong to the PR-protein superfamily and are present in other organisms including yeast, plant, insect and human (Szyperski et al., 1998) (see also Discussion). Although overall sequence similarity between them is not outstanding (>30%), 18 amino acid residues in the stretch of ∼100 amino acids compared are well aligned (Figure 2). Interestingly, almost all the amino acids identified as mutation sites in lon-1 alleles include those conserved between species, indicating that these amino acids are essential for LON-1 function (Figure 2, indicated by asterisk). There are no defined functional motifs within this family, but members of the family are predicted to be secreted or glycosylphosphatidylinositol (GPI)-anchored proteins. Two of the members of this family, pathogenesis related protein-1 (PR-1) in plant and glioma-pathogenesis related protein-1 (GliPR-1) in human, are secreted from cells (Szyperski et al., 1998). Yeast PR-1 like protein-3 (YPR-3) in yeast and cysteine-rich protease inhibitor are predicted to be GPI-anchored proteins (Hamada et al., 1998). However, the analysis of the LON-1 sequence shows that there is a hydrophobic stretch at the N-terminus of the LON-1 protein, indicating that LON-1 might be a transmembrane protein (Figure 1C). To characterize the LON-1 protein, we generated antibodies against a LON-1 peptide corresponding to 42–56 amino acid residues. This antibody (α-LON-1) detected ∼35 kDa of protein prepared from whole worm extract by immunoblotting (Figure 3A). The molecular mass of this protein is the same as that of the predicted LON-1 protein.
Fig. 2. Alignment of LON-1 protein with nine related proteins. The sequences are LON-1 (C.elegans), CG8483 (Drosophila), GliPR-1 (human) (Murphy et al., 1995), PR-1 (Triticum aestivum), SGP28 (human) (Kjeldsen et al., 1996), TPX-1 (testis-specific protein) (pig) (Kasahara et al., 1989), venom allergen 5 (Vespa mandarinia) (Lu et al., 1993), cystein-rich secretory protein (horse), cystein-rich protease inhibitor (mouse), YJH8 (yeast). Asterisks indicate the mutation sites identified in lon-1 alleles. Conserved amino acid residues (at least seven residues) are by bold type.
Fig. 3. LON-1 expression is negatively regulated by dbl-1 signaling. (A–C) Western blot analysis using α-LON-1 antibody. (A) Whole worm extracts of dbl-1(–), N2 wild-type, lon-1(++) and lon-1(–) alleles sp3, e185 and e43. The arrowhead indicates the 35 kDa LON-1 protein. Lower panel is the loading control staining with α-paramyosin antibody. (B) Cryostat sections of N2 worms were homogenized and extracted by Triton X-100 and fractionated by centrifugation (see also Materials and methods). LON-1 protein is detected in the membrane fraction but not in the soluble fraction. (C) The membrane fractions were treated with PI-PLC or alkaline wash (pH 11.5), or acid wash (pH 2.0). LON-1 protein remains associated with the membrane fraction. (D) Northern blot analysis. Poly(A)+ RNA (1.0 µg) prepared from mixed stages of dbl-1(–);dbl-1(nk3), N2, dbl-1(++);ctIs40 worms was blotted and hybridized with dbl-1, lon-1, CeIF (C.elegans initiation factor; for loading control) cDNA. The dbl-1 signal in dbl-1-overexpressing animals increased 3-fold compared with N2, and lon-1 in dbl-1-overexpressing animals decreased 5-fold compared with dbl-1 null animals. (E) lon-1::gfp expression is up- or down-regulated by dbl-1 dosage. The fluorescence indicating lon-1::gfp expression was increased with a dbl-1(–);dbl-1(nk3) background and decreased with a dbl-1(++);ctIs40 background. (F) Growth curve of N2 wild type, dbl-1(nk3), dbl-1(nk3);lon-1(e185) and lon-1(e185). The body length of mutants was measured at various times after hatching.
To address the subcellular localization of LON-1 protein, we fractionated the extract of N2 worms. We detected the LON-1 protein in the membrane fraction, but not the soluble fraction, by western blot analysis (Figure 3B). These results indicate that LON-1 may be a membrane-integrated or -associated protein. Furthermore, the LON-1 protein remains associated with the membrane fraction after treatment with phosphatidylinositol-specific phospholipase C (PI-PLC) or an alkaline wash (Figure 3C). As the sequence following the signal sequence does not closely match with consensus cleavage sequence, we propose that the signal sequence is not cleaved and instead acts as an anchor region. We conclude that LON-1 is likely to be a type II integral membrane protein.
Characterization of lon-1 alleles
To analyze the relationship between the mutation site of lon-1 alleles and their phenotype, we compared the expression level of LON-1 protein and body length in mutants. lon-1(e185) is thought to be the strongest allele because they are the longest worms (Table II). By western blot analysis, LON-1 protein was found to be almost completely undetectable as a result of the point mutation (Figure 3A). Cysteine residues, including C185, are conserved in the PR-protein family and might form disulfide bonds, essential for the structural integrity of the protein. The missense mutation might cause misfolding of LON-1 by creating unpaired cysteine residues and may even prevent LON-1 from being exported from the endoplasmic reticulum, thereby resulting in a severe loss of LON-1 function. Compared with lon-1(e185), nonsense mutation alleles e43 and sp3 display mild phenotypes in terms of body length. LON-1 protein in e43 was weakly detected by western blot analysis (Figure 3A). The weak Lon phenotype of e43 and sp3 may be due to low levels of LON-1 protein produced by translational readthrough. Like other animals, C.elegans has tRNA[Ser]Sec that can insert selenocysteine at UGA codons (Lee et al., 1990). In prokaryotes, substitution of tryptophan at opal stops occurs due to third-position wobble in codon–anticodon recognition. A similar effect in daf-1 alleles is reported by Gunther et al. (2000). Next, we made e43/Df and sp3/Df animals. These animals have a longer phenotype than e43 and sp3 homozygotes. lon-1(e43) and some lon-1 alleles often produce stunted worms and show incomplete penetrance of the Lon phenotype. We cannot rule out that this is due to the secondary induced constriction behind the head that leads to auto-decapitation; nevertheless, dsRNAi of lon-1 seems to cause only the Lon phenotype (data not shown). We speculate that the lon-1 allele with the null or most severe phenotype is lon-1(e185).
lon-1 expression is negatively regulated by dbl-1 signaling
lon-1 was initially identified as being down-regulated in worms overexpressing dbl-1 in our cDNA macroarray (Table I). Northern blot analysis confirmed this, showing that lon-1 transcript levels seem to be regulated in a stepwise manner depending on the gene dosage of dbl-1 (Figure 3D). The change in expression level of lon-1 transcript between ctIs40;dbl-1-overexpressing worms and the dbl-1(–) mutation was estimated as ∼5-fold.
As expected from the dbl-1-dependent repression profiled by cDNA array analysis and northern blotting, green fluorescent protein (GFP) fluorescence confirmed that the lon-1 promoter is negatively regulated by the dbl-1 signal. The fluorescence of lon-1::gfp appeared to be significantly down-regulated in worms overexpressing dbl-1;dbl-1 (++);ctIs40, compared with dbl-1(–) (Figure 3E). These results indicate that lon-1 expression is negatively regulated by dbl-1 signaling at the transcriptional level.
To analyze the genetic relationship of dbl-1 and lon-1, we made double mutants of lon-1 alleles with dbl-1(–) and measured body length (Table IV). The lon-1(ct410);dbl-1 (nk3) double mutant was almost the same length as the lon-1(ct410) single mutant. This result indicates that the lon-1(ct410) mutation is completely epistatic to dbl-1. However, the double mutants of lon-1(e185) and lon-1 (e43) with dbl-1(nk3) had intermediate body lengths. The cause of this incomplete epistacy is unclear.
Table IV. Hypodermal ploidy of mutants and double mutants.
| Mutant | Hypodermal ploidya | Body length (mm) |
|---|---|---|
| N2 | 12.47 ± 0.22 | 1.50 ± 0.02 |
| dbl-1(nk3) | 7.81 ± 0.25 (p < 0.0001) | 1.01 ± 0.02 (p < 0.0001) |
| nkIs10 | 10.73 ± 0.33 (p < 0.003) | 1.00 ± 0.03 (p < 0.0001) |
| lon-1(e185) | 14.36 ± 0.40 (p < 0.0001) | 1.86 ± 0.03 (p < 0.0001) |
| lon-1(e43) | 11.78 ± 0.43 NSb | 1.36 ± 0.06 (p < 0.01) |
| lon-1(ct410) | 10.47 ± 0.40 NS | 1.60 ± 0.02 (p < 0.01) |
| lon-1(sp3) | 11.98 ± 0.78 | 1.35 ± 0.05 |
| lon-1(RNAi) | 13.70 ± 1.34 | 1.63 ± 0.06 |
| lon-1(e43)/Df | 11.63 ± 1.18 | 1.49 ± 0.11 |
| lon-1(sp3)/Df | 13.86 ± 0.72 | 1.29 ± 0.06 |
| dbl-1(nk3);lon-1(e185) | 8.81 ± 0.29 | 1.34 ± 0.02 |
| dbl-1(nk3);lon-1(e43) | 9.64 ± 0.37 | 1.21 ± 0.03 |
| dbl-1(nk3);lon-1(ct410) | 11.48 ± 0.27 | 1.60 ± 0.02 |
aMean ± SE. p values represent the probability of rejecting the null hypothesis of no difference between a given genotype and wild type, as estimated from an F distribution. Preliminary experiments indicate that the Lon phenotype was variably penetrant in some genotypes. In order to circumvent this, we obtained an unbiased estimate of the 80th percentile for length for each genotype. We raised 50 worms (five per plate, 20–30 replicate plates), measured all of them at 120 h of age and then compared only the longest worm of each plate, thus ensuring independence of replicates.
bNot significant.
Worms overexpressing LON-1 have a Sma phenotype
Loss-of-function mutations in dbl-1 cause a Sma phenotype, while worms that overexpress dbl-1 are Lon (Morita et al., 1999; Suzuki et al., 1999). DBL-1 negatively regulates LON-1, and loss-of-function mutations in lon-1 give a Lon worm. If LON-1 is also a dose-dependent regulator of body length, overexpressing lon-1 should give a Sma worm. To address this possibility, we injected high concentrations of lon-1 DNA into lon-1 mutant or N2 worms (see Materials and methods). As predicted, higher dosages of lon-1 caused the Sma phenotype against both lon-1 and N2 backgrounds (Table II; data not shown). The body length of worms overexpressing lon-1;nkIs10 was close to sma mutants such as dbl-1, sma-2, sma-3 and sma-4 but, as discussed below, do not show the male tail phenotype (data not shown).
To determine when mutants become longer than wild type, we measured the length of N2 wild-type, lon-1 (e185), dbl-1(nk3) and dbl-1(nk3);lon-1(e185) hermaphrodites at different times after hatching (Figure 3F). All mutants hatch as L1 larvae indistinguishable in size from wild-type larvae, but the Lon and Sma phenotypes are seen at late L1 stage, and these phenotypes are sustained through to adult stages. These results demonstrate that dbl-1 signaling is effective from all larval stages to adult.
LON-1 is expressed in hypodermal and intestinal cells
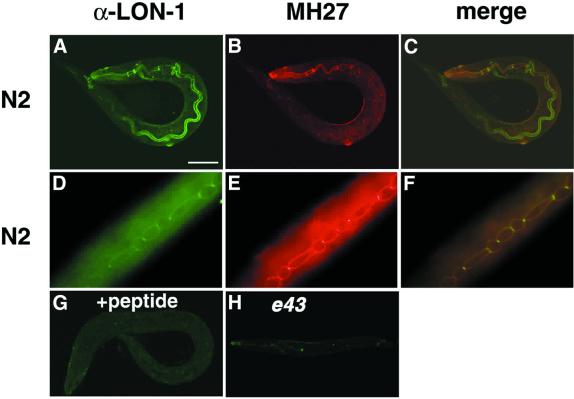
To examine the expression pattern of lon-1 in vivo at the cellular level, we stained the worms with LON-1 antibody (α-LON-1). Fluorescence was detected in almost all hypodermal and intestinal cells (Figure 4A). The signal of hypodermal cells is colocalized with the staining of MH27, which stains the adherens junction of the hypodermal cells (Figure 4A–C). Colocalization of LON-1 and MH27 antigen are also observed around hypodermal seam cells (Figure 4D–F). These expression patterns persisted through the subsequent larval stages to the adult stage. All signals were observed at the surface of the cell membrane. These observations are consistent with the result that LON-1 is a type II transmembrane protein. The signal disappeared completely in the presence of the antigen peptide (Figure 4G). In the lon-1(e43) nonsense mutation allele, LON-1 staining was substantially less (Figure 4F).
Fig. 4. LON-1 proteins are expressed in hypodermal and intestinal cells. All images are stained with purified anti-rabbit LON-1 peptide antibody (1:300 dilution) (green) and MH27 monoclonal antibody (1:100 dilution) (red). (A–C) N2 wild-type L1 stage animal. (A) Staining image of α-LON-1. The fluorescence is observed around the hypodermal cells and in the inner membrane of intestinal cells. (B) MH27. Signal is seen in the hypodermal adherens junction. (C) Merged image of (A) and (B). (D–F) Hypodermal region of N2 wild-type L3 stage animal. (D) α-LON-1, (E) MH27, (F) merged image of (D) and (E), the signals are observed around the seam cells. (G) Staining image of α-LON-1 in the presence of antigen peptide. Staining of α-LON-1 has disappeared completely. N2 wild-type L1 stage animal. (H) Staining image of α-LON-1 to lon-1(e43) mutant L1 stage animal. Weak expression is detected.
Expression of lon-1 cDNA in hypodermal but not in intestinal cells rescued Lon phenotype
We show that LON-1 protein is expressed in the intestinal inner membrane and hypodermal adherens junction where MH27 antigen is also expressed (Figure 4). To address the functional sites of LON-1 protein in the regulation of body length, we expressed a lon-1 cDNA under region-specific promoters. We tested four cell type-specific promoters: Pdpy-7, Pyk92e8, Punc-54 and Pdbl-1. Pdpy-7, the promoter for the collagen gene dpy-7, is specific for hypodermal cells. Pyk92e8 is expressed only in intestinal cells (Mochii et al., 1999; Yoshida et al., 2001). Punc-54, the promoter for the myosin heavy chain, is expressed in major body wall muscle. Pdbl-1, the promoter for dbl-1, the ligand of the Sma pathway, is expressed almost exclusively in neuronal cells (Morita et al., 1999; Suzuki et al., 1999). Using these promoters, we expressed the lon-1 cDNA. The cell type specificity of these promoters was confirmed by GFP fluorescence by the injection of a GFP cDNA with each of above tissue-specific promoters (data not shown). Hypodermal cell-specific expression of LON-1 (Pdpy-7 lon-1) rescued the Lon phenotype, but the intestinal cell-specific expression (Pyk92e8 lon-1) did not (Table III). Two other promoters, Punc-54 and Pdbl-1 also could not rescue the Lon phenotype. These results indicate that expression of LON-1 protein in the hypodermal cells is sufficient to regulate body length.
Table III. Expression of lon-1 cDNA in hypodermal, but not intestinal, cells rescued a lon-1 phenotype.
| Genotype | Transgene | Expression sites | Body length (mm)a | N |
|---|---|---|---|---|
| lon-1(e185) | – | – | 1.71 ± 0.01 | 93 |
| lon-1(e185) | Pdpy-7 lon-1 | hypodermal cells | 1.12 ± 0.12 | 108 |
| lon-1(e185) | Pyk92e8 lon-1 | intestinal cells | 1.59 ± 0.01 | 60 |
| lon-1(e185) | Punc-54 lon-1 | body wall muscle | 1.60 ± 0.01 | 47 |
| lon-1(e185) | Pdbl-1 lon-1 | neural cells | 1.57 ± 0.01 | 37 |
| lon-1(e185) | Plon-1 lon-1 | hypodermal and intestinal cells | 1.30 ± 0.02 | 68 |
aMean ± SE.
LON-1 regulates the polyploidization of hypodermal nuclei
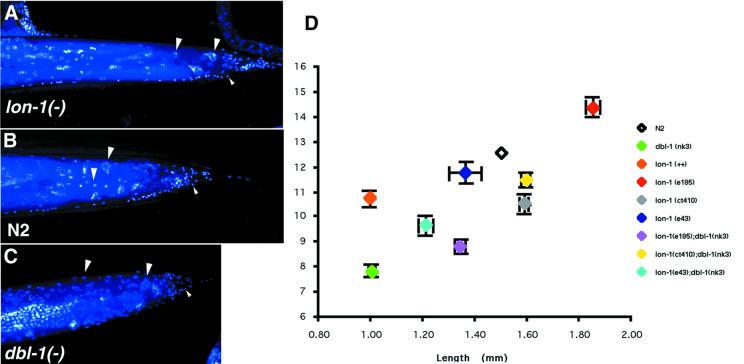
Recently, loss-of-function mutations in daf-4 and sma-2, both of which encode signaling components downstream of dbl-1, have been reported to decrease polyploidy of the hypodermal nuclei (Flemming et al., 2000). The C.elegans TGF-β growth pathway is thus implicated in the regulation of endoreduplication. If lon-1 is a downstream target of TGF-β signaling, it may also be a dose-dependent regulator of hypodermal ploidy. To address this possibility, we estimated the hypodermal ploidy of loss-of-function mutations of lon-1 and worms that overexpress lon-1. We found that endoreduplication of hypodermal nuclei was indeed increased in lon-1(e185) and lon-1 (RNAi) worms by 19 and 13%, respectively, compared with wild type (Figure 5 and Table IV). Importantly, lon-1-overexpressing Sma worms (nkIs10) have reduced hypodermal ploidy, like dbl-1, daf-4 and sma-2 mutants (Table IV; Flemming et al., 2000; Nyström et al., 2002). It is intriguing to note that Nyström et al. (2002) failed to detect increased hypodermal ploidy in worms that overexpress dbl-1, despite the general phenotypic similarity of these worms to lon-1(–). Furthermore, dpy-2(–) and lon-3(–) mutants both have altered body size but wild-type ploidy (Flemming et al., 2000; Nyström et al., 2002). Indeed, among the many mutations that affect body size in C.elegans, so far only the TGF-β signal mutants and lon-1 loss-of-function and overexpression mutants have increased or decreased ploidy in hypodermal cells.
Fig. 5. LON-1 regulates the polyploidization of hypodermal nuclei in a dose-dependent manner. (A–C) DAPI staining of the tail region of an adult hermaphrodite. (A) lon-1(e185), (B) N2 wild type, (C) dbl-1(nk3). Large arrowheads indicate the nuclei of hypodermal cells (polyploid nuclei) and small arrowheads diploid nuclei. Anterior to the left and posterior to the right. (D) The relationship between hypodermal ploidy and body length in lon-1(–);dbl-1(0) double mutant worms.
Discussion
lon-1 regulates body length of C.elegans and is negatively regulated by TGF-β signaling
In this study, we first show that DNA array technology combined with dsRNAi is capable of identifying a biologically important gene. Although not all genes identified displayed a phenotype when inactivated by dsRNAi, one of the genes that did was found to encode lon-1 whose inactivation causes a Lon worm. Our finding that lon-1 expression is negatively regulated by dbl-1 provides further insight into the important pathway regulating nematode body size yet identified. The lon-1 expression profile suggests that it most likely acts as a downstream component of the C.elegans TGF-β growth pathway.
Although lon-1 controls body length acting downstream of dbl-1, it is not likely be involved in all processes of dbl-1 function. Another phenotype commonly found in dbl-1, daf-4, sma-6 and sma-2, sma-3, sma-4 mutants is a change in male tail ray identity (Estevez et al., 1993; Savage et al., 1996; Krishna et al., 1999; Morita et al., 1999; Suzuki et al., 1999). mab-21, a gene that also regulates male tail identity, has been shown to be downstream of dbl-1 signaling (Morita et al., 1999). Since mab-21 is thought not to regulate body length (Chow et al., 1995; Morita et al., 1999), the TGF-β signaling pathway must bifurcate upstream of mab-21, one branch regulating body length and the other regulating tail ray patterning. Consistent with this, we find no evidence of altered ray identities in lon-1(–) worms, although they do have an elongated male tail. Similarly, worms overexpressing LON-1 (nkIs10) have wild-type tails (data not shown). This suggests that lon-1 functions downstream of the bifurcation point and is involved exclusively in body length regulation.
Our results also permit insight into an unsolved problem of C.elegans growth regulation, namely, which tissue is most critical in determining the size of a worm. Obvious candidates include the hypodermis which covers the entire worm and synthesizes the cuticle, and the body wall muscle that underlies the hypodermis. The TGF-β ligand DBL-1 is expressed mainly in neuronal cells such as ventral nerve cord and AFD (Morita et al., 1999; Suzuki et al., 1999), but although the ventral cord may the source of important growth factors, it is not likely to be a critical structural component of worm length. Expression of another TGF-β component, daf-4, the type II receptor, has shed little light on this question since it is almost ubiquitously expressed (Gunther et al., 2000). More interestingly, we have found recently that the type I receptor sma-6 is highly expressed in hypodermis (Yoshida et al., 2001) and that expressing sma-6 exclusively in the hypodermis by a hypodermal-specific promoter is sufficient to rescue the sma-6(–) Sma phenotype (Yoshida et al., 2001). We also show that LON-1 expression in hypodermal cells is essential for body length regulation (Table III). Taken together with these results, we suggest that the hypodermis is the target tissue of dbl-1 in the determination of body length.
LON-1 may control polyploidization of hypodermal nuclei
In the development of almost all plants and animals, specialized polyploid and polytene cell types arise through endocycles, cell cycles lacking cell division (Brodsky and Uryvaeva, 1985; Royzman and Orr-Weaver, 1998). Endoreduplication is, therefore, a normal and important component of cell proliferation and growth in many organisms. However, endoreduplication also occurs in pathological conditions such as tumorigenesis (Holland et al., 1998; Lengauer et al., 1998). Nevertheless, the molecular mechanism of the regulation of polyploidy has remained largely unknown (Reed and Orr-Weaver, 1997; Royzman and Orr-Weaver, 1998; Galitski et al., 1999), although Britton and Edgar (1998) have recently shown that the Drosophila larval endoreduplication cycle is dependent on nutrition as well as cyclin E and E2F transcription factor expression. Here, we show that LON-1 regulates the polyploidy of hypodermal nuclei from almost 8C to 16C depending on its gene dosage [dbl-1(0), lon-1-overexpressing worms and loss-of-function mutant lon-1 (e185)] (Figure 5 and Table IV). How LON-1 does this is not known, but it could function as an inhibitor of an as yet unknown factor that promotes endoreduplication. Interestingly, lon-1 is also expressed at all stages in the intestine, another tissue that undergoes endoreduplication in C.elegans (Hedgecock and White, 1985), therefore, we are currently examining the ploidy of intestinal cells of lon-1 mutants.
Regulation of endoreduplication and body length by DBL-1
The observation that polyploid cells (be they due to somatic polyploidization or germ-line polyploidy) are typically larger than diploid cells has often led to the suggestion that genome content directly regulates cell size (Brodsky and Uryvaeva, 1985; Day and Lawrence, 2000). In the absence of other levels of body size regulation, changes in ploidy might, then, have a direct effect upon body size. Flemming et al. (2000) proposed that TGF-β mutants might be Sma because they were endoreduplication deficient. They noted, however, that there was no evidence in C.elegans for a direct causal connection between ploidy and body length. Our findings that lon-1 is simultaneously a dose-dependent regulator of body length and of endoreduplication adds strength to the idea that ploidy regulates body length in worms. However, not all evidence from the TGF-β pathway in C.elegans tends to confirm this idea. While worms that are DBL-1 defective are Sma and have reduced hypodermal ploidy, worms that overexpress DBL-1 have a Lon phenotype but appear to have a wild-type ploidy (Nyström et al., 2002). One possible explanation for this discrepancy may be that DBL-1 does not suppress lon-1 activity to the same extent as the lon-1 null mutant; indeed, our northern blot analysis shows that DBL-1 overexpression only reduces the level of lon-1 transcript by ∼50%. This explanation also suggests that relative to the regulation of ploidy, body length determination is more sensitive to a change in DBL-1 activity, since partial suppression of lon-1 is sufficient to promote the increase in body size seen in lon-1(–) mutants. Alternatively, lon-1 may be able to overcome exogenous DBL-1 activity to block hyperendoreduplication to some extent. It is also noted that hypodermal polyploidy is unchanged in some alleles such as lon-1(e43) and lon-1(ct410) in which some residual LON-1 protein exists. It is possible that an interaction of LON-1 protein with an as yet unknown protein(s) may contribute to the suppression of hypodermal ploidy.
We propose here a model for the regulation of body length and hypodermal ploidy by DBL-1. dbl-1 is expressed in the neuronal cells including those of the AFD amphid neuron and ventral nerve cord (Morita et al., 1999; Suzuki et al., 1999). DBL-1 might be secreted by nervous cells and delivered to hypodermal cells that express the type I receptor sma-6 (Yoshida et al., 2001). At the hypodermal syncytium, dbl-1 signaling is activated and negatively regulates the expression of LON-1 through SMA-2, SMA-3 and SMA-4. LON-1 inhibits the endo cell cycle (Royzman and Orr-Weaver, 1998) and regulates the hypodermal ploidy.
It remains possible that the TGF-β signaling pathway and lon-1 regulate endoreduplication as a result of body size change. LON-1 might, for example, regulate one or more of the many collagens of which the C.elegans cuticle is composed. Loss-of-function mutations in many of these collagens give short worms with a characteristic Dumpy (Dpy) phenotype: short and fat worms as distinct from the short and thin Sma phenotype. Indeed, another Lon mutant, lon-3, has recently been found to encode a collagen and, like lon-1, is a dose-dependent regulator of body length (Nyström et al., 2002). lon-3(–) worms, as well as worms overexpressing LON-3, have, however, a wild-type ploidy and are thought to affect body length via changes in cuticle elasticity (Nyström et al., 2002; Y.Suzuki and W.B.Wood, manuscript in preparation). Therefore, it is unlikely that body size change is the cause for change in hypodermal ploidy. Also, if lon-1 regulates collagens, it probably does not regulate lon-3, since analysis of double mutants suggests that these genes act largely additively (Nyström et al., 2002).
LON-1 belongs to the PR-protein superfamily, which is conserved from yeast, plant, insect to human
Searches of databases revealed that LON-1 has conserved amino acid motifs found in proteins in yeast, plants, insects and humans. One of these proteins, the human GliPR protein, was found to be highly expressed in brain tumors, suggesting that GliPR plays an important role in tumor growth (Murphy et al., 1995). LON-1 also has sequence homology with the plant PR-1 protein, which plays a central role in the defense system of plants, for example, during the manifestation of systematic acquired resistance (SAR) (Uknes et al., 1992, 1993). PR-1-related proteins from tobacco and tomato have in vitro activity against Phytophthora infestans, but the underlying molecular mechanism for the action of these proteins is not known (Szyperski et al., 1998). The characterization of the three-dimensional structure of GliPR (human) and PR-1 (plant) revealed that the putative active site residues of these proteins is strictly conserved. This suggests strongly that human GliPR and plant PR-1 proteins operate according to the same molecular mechanism, which establishes a possible functional link between the human immune system and a plant defense system (Szyperski et al., 1998).
In C.elegans, we show that LON-1 regulates the endoreduplication of hypodermal nuclei by demonstrating that loss of function of lon-1 causes hyperendoreduplication of hypodermal nuclei, and overexpression of lon-1 causes hypoendoreduplication (Figure 5 and Table IV). Members of the PR-protein superfamily are secreted, GPI-anchored and transmembrane proteins, raising the possibility that LON-1 might function as an extracellular ligand. Alternatively, the PR-protein superfamily, including LON-1, might function as enzymes because putative enzyme active sites are located in the largest surface cleft of the three-dimensional structure of GliPR, PR-1 and LON-1 (Szyperski et al., 1998). LON-1 might also function as an inhibitor of enzyme or protease that regulates the endocycle. We are tempted to speculate further that other members of the PR-protein superfamily might also regulate the endo cell cycle under various pathological conditions of plants and animals. Intriguingly, endoreduplication is a common response of plants to the saliva of endoparasitic nematodes (Goverse et al., 2000). Future analysis of the molecular mechanism of LON-1 in regulating the polyploidization of hypodermal nuclei may, then, shed light on a diversity of biological problems as well as the functional sites of the PR-protein superfamily.
Materials and methods
General method and strains
Caenorhabditis elegans strains were cultured as described by Brenner (1974). Genetic crosses were performed as described by Wood (1988). The following strains were used: wild-type C.elegans variety Bristol strain N2, lon-1(e43)III, lon-1(e44)III, lon-1(e185)III, lon-1(e1137)III, lon-1(ct410)III, lon-1(ct411)III, lon-1(n1130)III, lon-1(sp1)III, lon-1 (sp3)III, dbl-1(–);dbl-1(nk3)V, dbl-1(++);dbl-1(ctIS40), him-5(e1492)V.
cDNA array and differential hybridization
Caenorhabditis elegans cDNA array was prepared as described by Mochii et al. (1999). Total RNA was extracted from synchronously cultured dbl-1(nk3);dbl-1(–) or ctIS40;dbl-1(++) L3 stage with Trizol reagent (Gibco-BRL), and poly(A)+ RNA was purified by using Oligotex-dT 30 (Roche Molecular Biochemicals). For probe labeling, the oligo(dT)-depleted RNA was incubated with random hexamer, Superscript reverse transcriptase (Gibco-BRL) and [33P]dCTP at 37°C for 2 h. Hybridization was performed as described. Hybridized signals were detected using the Fuji BAS system and quantified with HDG Analyzer software (Genomic Solutions). Experiments were carried out four times with independently prepared RNAs. Northern analyses were performed using poly(A)+ RNA and results were quantified using the Fuji BAS system.
dsRNAi analysis
Each template was amplified by PCR from YK clones, using the primers: CMO24: 5′-TTGTAAAACGACGGCCAG-3′ and CMO422: 5′-GCGTAATACGACTCACTATAGGGAACAAAAGCTGGAGCT-3′. dsRNAs were synthesized from these templates using MEGAscript T7 kit (Ambion). dsRNAs were injected into the gonadal syncytia of adult hermaphrodites, and the phenotypes of F1 progeny were analyzed. For RNAi feeding, full-length lon-1 cDNA was inserted in a pPD129.36 vector.
Generation of antibody against LON-1, and western analysis
Antisera LP1-1 was raised against a synthetic peptide with the sequence CGELPATDEVKREKRG (corresponding to amino acids 42–56 in LON-1). The peptide was coupled to keyhole limpet hemocyanin (KLH). Two rabbits were injected with the conjugated peptide. The antiserum was affinity purified on a Sepharose peptide affinity column. Whole worm extracts were prepared by suspending washed worms in 10 vol of SDS sample buffer and immediately boiling for 3 min before loading onto a 10% SDS–polyacrylamide gel. Extracts resolved by SDS–PAGE were transferred to nitrocellulose and the filter was blocked in 5% non-fat dried milk before incubation in a 1:300 dilution of affinity-purified LON-1 peptide antibodies. Anti-paramyosin antibody (5-23) was used in a 1:1000 dilution.
Membrane extracts were prepared as described previously (Epstein et al., 1988). One gram of packed worms mixed with 2.0 ml of OTC compound was frozen in liquid N2. Cryostat sections were dissolved in 10 ml relaxing buffer and washed twice for centrifugation with relaxing buffer. The pellet was resuspended in 1% Triton X-100 in relaxing buffer and homogenized with Teflon homogenizer. Homogenized solutions were centrifuged at 5000 g for 10 min and the resulting supernatant was centrifuged at 15 000 g for 30 min. The resulting pellet was used as membrane fraction and the supernatant as soluble fraction.
Immunofluorescent staining
Mixed stage populations of each strain were fixed and stained according to the protocol of Finney and Ruvkun (1990). Affinity-purified LON-1 anti-rabbit antibody was used at a concentration of 1:200 and MH27 anti-mouse monoclonal antibody at 1:300. The secondary antibody was FITC-conjugated anti-rabbit antibody (Jackson) and Texas-Red- conjugated anti-mouse antibody (Amersham) diluted 1:100.
Plasmids and germ-line transformation
A lon-1 PstI–ScaI genomic fragment (8.2 kb) from cosmid F48E8 was subcloned into the PstI–ScaI site of pBluescript KS– to generate pYS1. In rescue and overexpression experiments, pYS1 was injected at 50 or 200 ng/µl as described (Mello et al., 1991) with myo-3::gfp as the co-injection marker. lon-1::gfp was generated by PCR amplification of 3.0 kb upstream of the lon-1 ORF from cosmid F48E8, and subcloned into pPD95,69 (a gift from A.Fire) in-frame. Extrachromosomal arrays were integrated into chromosomes using UV irradiation, followed by at least two backcrosses with N2 or him-5.
Analysis of polyploidization of hypodermal nuclei
Hypodermal DNA content was determined using videomicrodensitometry as described by Flemming et al. (2000). On average, 20 nuclei were scored per worm at late adulthood (>120 h after hatching).
Acknowledgments
Acknowledgements
We thank J.Hodgkin for providing the lon-1 e43, e44 and e1137 alleles. Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, National Center for Research Resources. This research was supported by a grant from the Biotechnology and Biological Sciences Research Council (UK) to A.M.L., a NERC studentship to A.J.F. and a grant from the ‘Research for the Future’ program of Japan Society for the Promotion of Science to N.U.
References
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Bonner J.T. (1989) The Evolution of Complexity by Means of Natural Selection. Princeton University Press, Princeton, NJ.
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J.S. and Edgar,B.A. (1998) Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development, 125, 2149–2158. [DOI] [PubMed] [Google Scholar]
- Brodsky V.Y. and Uryvaeva,I.V. (1985) Genome Multiplication in Growth and Development. Cambridge University Press, Cambridge, UK.
- Chow K.L., Hall,D.H. and Emmons,S.W. (1995) The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates. Development, 121, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Chuang C.F. and Meyerowitz,E.M. (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon I. and Raff,M. (1999) Size control in animal development. Cell, 96, 235–244. [DOI] [PubMed] [Google Scholar]
- Day S.J. and Lawrence,P.A. (2000) Measuring dimensions: the regulation of size and shape. Development, 127, 2977–2987. [DOI] [PubMed] [Google Scholar]
- Epstein H.F., Berliner,G.C., Casey,D.L. and Ortiz,I. (1988) Purified thick filaments from the nematode Caenorhabditis elegans: evidence for multiple proteins associated with core structures. J. Cell Biol., 106, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M., Attisano,L., Wrana,J.L., Albert,P.S., Massague,J. and Riddle,D.L. (1993) The daf-4 gene encodes a bone morphogenetic protein receptor controlling C.elegans dauer larva development. Nature, 365, 644–649. [DOI] [PubMed] [Google Scholar]
- Finney M. and Ruvkun,G. (1990) The unc-86 gene product couples cell lineage and cell identity in C.elegans. Cell, 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flemming A.J., Shen,Z.Z., Cunha,A., Emmons,S.W. and Leroi,A.M. (2000) Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc. Natl Acad. Sci. USA, 97, 5285–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard,A.L., Fowler,S. and Lazarow,P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol., 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T., Saldanha,A.J., Styles,C.A., Lander,E.S. and Fink,G.R. (1999) Ploidy regulation of gene expression. Science, 285, 251–254. [DOI] [PubMed] [Google Scholar]
- Georgi L.L., Albert,P.S. and Riddle,D.L. (1990) daf-1, a C.elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell, 61, 635–645. [DOI] [PubMed] [Google Scholar]
- Goverse A.J., de Engler,A.J., Verhees,J., van der Krol,S., Helder,H.H. and Gheysen,G. (2000) Cell cycle activation by plant parasitic nematodes. Plant Mol. Biol., 43, 747–761. [DOI] [PubMed] [Google Scholar]
- Gunther C.V., Georgi,L.L. and Riddle,D.L. (2000) A Caenorhabditis elegans type I TGFβ receptor can function in the absence of type II kinase to promote larval development. Development, 127, 3337–3347. [DOI] [PubMed] [Google Scholar]
- Hamada K., Fukuchi,S., Arisawa,M., Baba,M. and Kitada,K. (1998) Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet., 258, 53–59. [DOI] [PubMed] [Google Scholar]
- Hedgecock E.M. and White,J.G. (1985) Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol., 107, 128–133. [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hogan B.L. (1996) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev., 10, 1580–1594. [DOI] [PubMed] [Google Scholar]
- Holland E.C., Hively,W.P., Gallo,V. and Varmus,H.E. (1998) Modeling mutations in the G1 arrest pathway in human gliomas: overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev., 12, 3644–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Gutknecht,J., Brew,K., Spurr,N. and Goodfellow,P.N. (1989) Cloning and mapping of a testis-specific gene with sequence similarity to a sperm-coating glycoprotein gene. Genomics, 5, 527–534. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew,R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Kjeldsen L., Cowland,J.B., Johnsen,A.H. and Borregaard,N. (1996) SGP28, a novel matrix glycoprotein in specific granules of human neutrophils with similarity to a human testis-specific gene product and a rodent sperm-coating glycoprotein. FEBS Lett., 380, 246–250. [DOI] [PubMed] [Google Scholar]
- Knight C., Patel,M.N., Azevedo,R.B.R., Flemming,A. and Leroi,A.M. (2001) A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol. Dev., in press. [DOI] [PubMed]
- Krishna S., Maduzia,L.L. and Padgett,R.W. (1999) Specificity of TGFβ signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development, 126, 251–260. [DOI] [PubMed] [Google Scholar]
- Lee B.J., Rajagopalan,M., Kim,Y.S., You,K.H., Jacobson,K.B. and Hatfield,D. (1990) Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol., 10, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Lockhart D.J. and Winzeler,E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- Lu G., Villalba,M., Coscia,M.R., Hoffman,D.R. and King,T.P. (1993) Sequence analysis and antigenic cross-reactivity of a venom allergen, antigen 5, from hornets, wasps and yellow jackets. J. Immunol., 150, 2823–2830. [PubMed] [Google Scholar]
- Massague J. (1996) TGFβ signaling: receptors, transducers and Mad proteins. Cell, 85, 947–950. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochii M., Yoshida,S., Morita,K., Kohara,Y. and Ueno,N. (1999) Identification of transforming growth factor-β-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl Acad. Sci. USA, 96, 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Chow,K.L. and Ueno,N. (1999) Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-β family. Development, 126, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Murphy E.V., Zhang,Y., Zhu,W. and Biggs,J. (1995) The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene, 159, 131–135. [DOI] [PubMed] [Google Scholar]
- Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström J., Aili,M., Shen,Z.Z., Flemming,A., Leroi,A.M. and Tuck,S. (2002) lon-3 is a dose-dependent regulator of body size in C.elegans. Genetics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.I. and Padgett,R.W. (2000) TGF β-related pathways. Roles in Caenorhabditis elegans development. Trends Genet., 16, 27–33. [DOI] [PubMed] [Google Scholar]
- Patterson G.I., Koweek,A., Wong,A., Liu,Y. and Ruvkun,G. (1997) The DAF-3 smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev., 11, 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed B.H. and Orr-Weaver,T.L. (1997) The Drosophila gene morula inhibits mitotic functions in the endo cell cycle and the mitotic cell cycle. Development, 124, 3543–3553. [DOI] [PubMed] [Google Scholar]
- Reinke V. et al. (2000) A global profile of germline gene expression in C.elegans. Mol. Cell, 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Ren P., Lim,C.S., Johnsen,R., Albert,P.S., Pilgrim,D. and Riddle,D.L. (1996) Control of C.elegans larval development by neuronal expression of a TGF-β homolog. Science, 274, 1389–1391. [DOI] [PubMed] [Google Scholar]
- Royzman I. and Orr-Weaver,T.L. (1998) S phase and differential DNA replication during Drosophila oogenesis. Genes Cells, 3, 767–776. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A. and Newmark,P.A. (1999) Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl Acad. Sci. USA, 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C., Das,P., Finelli,A.L., Townsend,S.R., Sun,C.Y., Baird,S.E. and Padgett,R.W. (1996) Caenorhabditis elegans genes sma-2, sma-3 and sma-4 define a conserved family of transforming growth factor β pathway components. Proc. Natl Acad. Sci. USA, 93, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.L. and Emlen,D.J. (1999) The developmental basis for allometry in insects. Development, 126, 1091–1101. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yandell,M.D., Roy,P.J., Krishna,S., Savage-Dunn,C., Ross,R.M., Padgett,R.W. and Wood,W.B. (1999) A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development, 126, 241–250. [DOI] [PubMed] [Google Scholar]
- Szyperski T., Fernandez,C., Mumenthaler,C. and Wuthrich,K. (1998) Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Natl Acad. Sci. USA, 95, 2262–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S., Mauch-Mani,B., Moyer,M., Potter,S., Williams,S., Dincher,S., Chandler,D., Slusarenko,A., Ward,E. and Ryals,J. (1992) Acquired resistance in Arabidopsis. Plant Cell, 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S., Dincher,S., Friedrich,L., Negrotto,D., Williams,S., Thompson-Taylor,H., Potter,S., Ward,E. and Ryals,J. (1993) Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell, 5, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.B. (1988) The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Yamakawa T., Miyata,S., Ogawa,N., Koshikawa,N., Yasumitsu,H., Kanamori,T. and Miyazaki,K. (1998) cDNA cloning of a novel trypsin inhibitor with similarity to pathogenesis-related proteins and its frequent expression in human brain cancer cells. Biochim. Biophys. Acta, 1395, 202–208. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Morita,K., Mochii,M. and Ueno,N. (2001) Hypodermal expression of a C.elegans TGF-β type I receptor SMA-6 for the growth and maintenance of body length. Dev. Biol., 240, 32–45. [DOI] [PubMed] [Google Scholar]
- Young R.A. (2000) Biomedical discovery with DNA arrays. Cell, 102, 9–15. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]