Abstract
The RNA helicase/protease NS3 plays a central role in the RNA replication of hepatitis C virus (HCV), a cytoplasmic RNA virus that represents a major worldwide health problem. NS3 is, therefore, an important drug target in the effort to combat HCV. Most work has focused on the protease, rather than the helicase, activities of the enzyme. In order to further characterize NS3 helicase activity, we evaluated individual stages of duplex unwinding by NS3 alone and in complex with cofactor NS4A. Despite a putative replicative role in RNA unwinding, we found that NS3 alone is a surprisingly poor helicase on RNA, but that RNA activity is promoted by cofactor NS4A. In contrast, NS3 alone is a highly processive helicase on DNA. Phylogenetic analysis suggests that this robust DNA helicase activity is not vestigial and may have specifically evolved in HCV. Given that HCV has no replicative DNA intermediate, these findings suggest that NS3 may have the capacity to affect host DNA.
Keywords: DExH/D/helicase/hepatitis C/NS3/NS4A
Introduction
The RNA virus hepatitis C (HCV) chronically infects an estimated 170 million people worldwide, of whom 10–20% will ultimately develop cirrhosis and 1–5% will develop liver cancer (Di Bisceglie, 1997; Seeff, 1998; Lavanchy, 1999). The HCV genome encodes 10 proteins, one of which is the bifunctional protease/helicase NS3 (Hagedorn and Rice, 2000). The helicase domain of NS3 is essential for viral RNA replication, which is carried out by a complex that includes the NS3 helicase/protease, the NS4A polypeptide and polymerase NS5B (for review see Bartenschlager and Lohmann, 2000).
Despite key roles for NS3 in HCV biology, there are many outstanding questions regarding the helicase activity of this protein: (i) what are the elemental mechanistic features of this RNA helicase (i.e. unwinding rate constants, processivity, etc.), as these parameters are necessary for meaningful structure–function studies and drug screening efforts; (ii) what is the relevance of in vitro studies that have demonstrated NS3 helicase activity on DNA (Tai et al., 1996; Gwack et al., 1997) given that HCV is a cytoplasmically replicating RNA virus (Bartenschlager and Lohmann, 2000; Hagedorn and Rice, 2000); and (iii) what is the mechanistic role of cofactor NS4A, which forms a tight complex with NS3 (Bartenschlager et al., 1995) and anchors it into the endoplasmic reticulum where viral RNA replication is believed to occur (Wölk et al., 2000)?
To answer these questions, we employed in vitro techniques that break down the unwinding mechanism of NS3 into individual, isolated steps. Specifically, the formation of functional complexes (helicase–duplex complexes that are active for unwinding upon addition of ATP) was isolated from the first pass of duplex unwinding, which was in turn dissected from multiple-cycle events in which the helicase can rebind (Figure 1A). Each of these experiments was conducted in a manner that compared cognate RNA and DNA substrates of the same sequence (except for the presence of U versus T) and the relative activities of NS3 and NS3–4A complex. Finally, in parallel with biochemical experiments, we performed a phylogenetic analysis of the helicase family to which NS3 belongs, in order to provide a biological context for our findings.
Fig. 1. Methodological approach. (A) Reaction schematic for dissection of unwinding parameters. Gel shift assay monitoring the single-cycle unwinding of 18 bp RNA (B) and DNA (C) duplex substrates by NS3 (Figure 3A, substrates b and f).
Results and discussion
Functional complex formation
Duplex unwinding is necessarily preceded by the formation of functional helicase–substrate complexes. However, it is problematic to distinguish between non-specifically associated helicase–duplex complexes and those that are properly configured for productive unwinding. This is exemplified by the fact that NS3 readily binds substrates containing either 3′ or 5′ single-strand overhangs (Tai et al., 1996), although it can only unwind substrates containing 3′ overhangs (Gwack et al., 1996). Further more, direct equilibrium binding studies report only minor differences in the affinity of NS3 for single-stranded RNA (ssRNA) (Kd = 11 nM), ssDNA (Kd = 12–36 nM; Preugschat et al., 1996) and double-stranded (dsDNA) substrates with single-stranded overhangs (Kd = 2.2– 6.3 nM; Porter et al., 1998; Tackett et al., 2001).
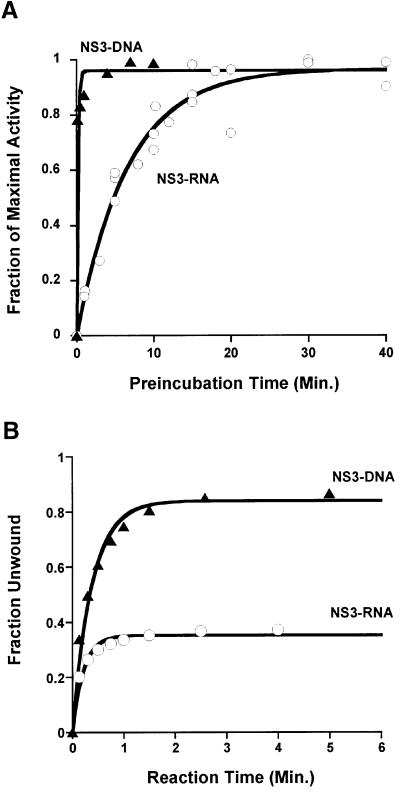
To address this problem, we developed a kinetic approach for monitoring functional complex formation. In this assay, the NS3–substrate incubation time was varied and NS3 was provided in large excess over substrate (ratio of 70:1). Reactions were then initiated by the simultaneous addition of ATP and a trap oligonucleotide (defined as single-cycle conditions, in which dissociated helicase cannot rebind to the substrate; Ali and Lohman, 1997; Jankowsky et al., 2000). Finally, the total amount of unwinding after a constant amount of reaction time (∼5 min) was measured by gel shift (Figure 1). Under these conditions, the resulting extent of unwinding is directly proportional to the fraction of functional NS3–substrate complex. Comparative plots of reaction extent (normalized to the total amount of unwinding at equilibrium) versus pre-incubation time reveal that NS3 forms functional complexes with DNA ∼40 times faster than with RNA (Figure 2A). The relaxation rate constants (kr) are 6 ± 2 min–1 for DNA and 0.15 ± 0.03 min–1 for RNA, which indicate that early stages of reaction initiation are more favorable on DNA and that NS3 has a greater capability for productively binding DNA substrates than RNA substrates of the same sequence. These findings also confirm previous studies showing that measurements of NS3 equilibrium binding affinity and relative extents of ATPase stimulation are not accurate indicators of NS3 helicase function (Tai et al., 1996; Hesson et al., 2000; Paolini et al., 2000).
Fig. 2. NS3 unwinding activity on RNA (open circles) and DNA (filled triangles) substrates. (A) Functional complex formation between NS3 and RNA or DNA substrates (Figure 3A, duplexes b and f, respectively). Unwinding amplitudes were normalized by the total amount of substrate unwound after equilibrium was achieved. (B) Single-cycle unwinding of RNA duplex b (open circles; kunw = 5.5 ± 0.8 min–1) and DNA duplex f (filled triangles; kunw = 2.2 ± 0.7 min–1) by NS3.
Unwinding parameters
A subsequent set of experiments was designed to study the unwinding processes that occur after functional enzyme– substrate complexes are formed. These experiments yielded two particularly informative terms: kunw, the single-cycle unwinding rate constant; and A, the reaction amplitude. Under single-cycle unwinding conditions, kunw excludes kinetic steps involved in helicase association or rebinding and reflects only events involved in unwinding. Values for A are directly proportional to the processivity of the unwinding reaction (as defined previously; Ali and Lohman, 1997; Jankowsky et al., 2000).
These experiments were started by building up a uniform population of functional helicase–substrate complexes. This was accomplished by pre-incubating an excess of NS3 with trace amounts of a duplex substrate such that [So] ∼ [NS3–S]. Single-cycle reactions were then initiated by adding ATP and a trap oligonucleotide. A plot of fraction unwound versus time after initiation fits a single exponential, representing a pseudo-first-order reaction. The single-cycle unwinding rate constants for the DNA and RNA are similar [kunw(DNA) = 2.2 ± 0.7 min–1; kunw(RNA) = 5.5 ± 0.8 min–1; Figures 2B and 3, duplexes b and f] and consistent with single-cycle experiments conducted previously on DNA (Paolini et al., 2000). However, the relative reaction amplitudes for DNA versus RNA are strikingly different. While the DNA substrate was unwound almost to completion, the extent of the reaction on RNA was low (∼38%; Figure 2B). This effect is independent of duplex sequence, as substrates containing a higher A–U content showed the same trend (data not shown). These findings suggest that NS3 is substantially more processive on DNA than RNA.
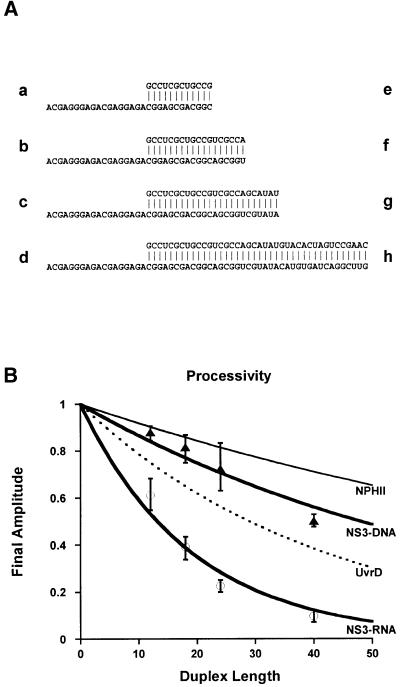
Fig. 3. Processivity on RNA and DNA. (A) The family of comparative helicase substrates. RNA substrates (a–d) contain 12, 18, 24 and 40 bp duplex regions, respectively. DNA substrates (e–h) contain 12, 18, 24 and 40 bp duplex regions, respectively, although the sequences are made of DNA where U has been replaced by T. All substrates contain a 3′ single-stranded overhang region of 18 nucleotides. (B) NS3 processivity measurements (RNA, circles; DNA, triangles). Final unwinding amplitudes were measured under single-cycle conditions for all substrates (A). Error bars denote the variance from a minimum of three different measurements. The processivities of DNA helicase UvrD (Ali and Lohman, 1997; black dotted line) and RNA helicase NPHII (Jankowsky et al., 2000; black thin line) are presented for comparison.
NS3 processivity
To quantitate the relative processivity of NS3 on DNA and RNA (and to correlate it with other helicases), we synthesized a family of eight substrates that contained progressively longer duplex regions (Figure 3A). These substrates control for the effects of overhang length, overhang sequence, duplex sequence and polymer identity (DNA versus RNA). Each of the duplexes was unwound by NS3 under single-cycle conditions and the amplitude for each duplex shown in Figure 3A was quantitated. A plot of final reaction amplitude versus duplex length (Figure 3B) revealed that NS3 is a processive DNA helicase, exceeding even the processivity of DNA helicase UvrD (Ali and Lohman, 1997). However, NS3 is a poorly processive RNA helicase, particularly when compared with the related NPH-II RNA helicase from vaccinia (Jankowsky et al., 2000). Most importantly, the data showed that NS3 was much more processive on DNA than on RNA of the same sequence.
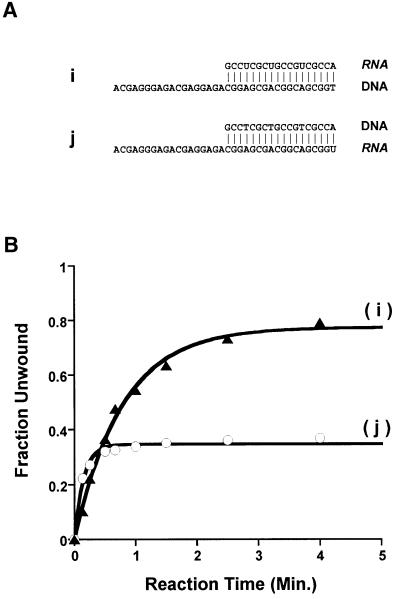
Given these observations, it was of interest to study which of the physicochemical differences between DNA and RNA was responsible for this difference in processivity. DNA lacks 2′-OH groups, has a different physical structure (B- versus A-form, with C2′-endo sugar puckering), has different electrostatic properties and is less thermodynamically stable than RNA of the same sequence (Wang et al., 1982; Sugimoto et al., 1995; Chin et al., 1999; Kankia and Marky, 1999). Because NS3 is essentially a motor, it was of particular interest to determine whether facile DNA unwinding could be traced to a lower overall thermodynamic stability for DNA duplexes. To this end, we created two 18 bp DNA–RNA hybrid duplexes (Figure 4A, duplexes i and j) that are identical in sequence to substrates b and f and identical to each other. The only substantive difference between these substrates is the chemical identity of the two strands, as i and j are both A-form duplexes (C3′-endo sugar pucker) with similar thermodynamic stability (Wang et al., 1982; Sugimoto et al., 1995).
Fig. 4. Unwinding of chimeric duplexes. (A) Hybrid duplexes i and j are analogous in sequence to duplexes b and f in Figure 3A. (B) Single-cycle unwinding of hybrid duplexes i (closed triangles; kunw = 1.3 ± 0.2 min–1) and j (open circles; kunw = 6.7 ± 0.7 min–1) by NS3. In addition to the obvious differences in amplitude (see text), the differences in kunw for i and j are notable. Like the unwinding of RNA duplexes, unwinding of j is somewhat faster than unwinding of DNA and of duplex i. In general, kunw for RNA is 2- to 3-fold faster than for DNA duplexes of the same sequence (see Figures 2B and 5B). While this difference is smaller than unusually considered significant for kinetic studies, it is highly reproducible and may represent an important aspect of RNA unwinding by NS3 that could be explored with techniques for measuring rapid kinetics (such as quench-flow).
A plot of fraction unwound versus time after initiation reveals that the total extent of unwinding (proportional to processivity) was determined entirely by the identity of the bottom strand (Figure 4B). When the bottom strand was composed of DNA (duplex i), the duplex was unwound to the same extent (80%, compare with Figure 2B) as when the entire duplex was composed of DNA, though at a somewhat slower rate (kunw = 1.3 ± 0.2 min–1). When the bottom strand was RNA (duplex j), this substrate was unwound to the same extent (38%, compare with Figure 2B) as if the entire duplex were composed of RNA, with a nearly identical rate constant (kunw = 6.7 ± 0.7 min–1). Other data support the observation that a substantial link does not exist between duplex stability and relative processivity on DNA versus RNA. For example, the 40 bp DNA duplex h was unwound to the same extent as the 12 bp RNA duplex a, despite the fact that duplex h is twice as thermodynamically stable (ΔG = –59.3 versus –27.9 kcal/mol, respectively). Furthermore, RNA duplex b was unwound to the same extent as hybrid duplex j, despite large differences in stability (ΔG = –43.7 versus –32.3 kcal/mol, respectively). By comparing reaction amplitudes for different duplexes while carefully controlling for chemical and conformational identity, these experiments demonstrate that the unusual processivity of NS3 on DNA is not attributable to thermodynamic differences between RNA and DNA duplexes. Furthermore, the experiments using chimeric substrates indicate that NS3 specifically recognizes functionalities on the bottom strand of the substrate (the strand containing the 3′ overhang) and preferentially recognizes the features of DNA.
It is interesting to speculate on the possible mode of discrimination between RNA and DNA strands. Having ruled out the effects of thermostability, helical conformation and electrostatic character (electrostatics is similar for A-form DNA, RNA and chimeras; Chin et al., 1999), it is possible that an important role is played by the identity of the 2′ sugar substituent on the bottom strand. Alternatively, the DNA sugar conformation (predominantly C2′-endo for single-stranded DNA) on the bottom strand may play a role once the strands begin to part. Another possible mechanism for discrimination may be the greater steric hindrance of RNA substituents. This is supported by the fact that RNA duplexes containing bulky 2′-O-methyl substitutions cannot be unwound by NS3 (Hesson et al., 2000). Other types of structural perturbations may also play a role in discrimination by the NS3 helicase (Tackett et al., 2001).
Role of cofactor NS4A
Previous studies have already demonstrated the capacity of NS3 to unwind both RNA and DNA (for review see Hagedorn and Rice, 2000). However, these earlier comparative studies were performed in a manner that allows helicase rebinding, thereby masking amplitude effects (multiple-cycle conditions; Gwack et al., 1997; Levin and Patel, 1999; Tackett et al., 2001). The present study is the first to demonstrate that the in vitro helicase activity of NS3 on RNA is relatively poor even under optimal conditions, primarily due to deficiencies in the processivity of unwinding and the productive binding of RNA duplex substrates. By contrast, we show that the DNA helicase activity of NS3 is superior to RNA helicase activity under reaction conditions optimized for RNA unwinding. These data may explain why previous studies have focused on monitoring the unwinding of DNA, rather than RNA. More importantly, given that viral RNA replication is the putative biological role of NS3 (Bartenschlager and Lohmann, 2000; Hagedorn and Rice, 2000), the results strongly suggest that NS3 requires modulation in order to direct its activity toward RNA substrates. If this is indeed the case, then current drug screening protocols based solely on DNA helicase activity may be misleading, as the ‘true’ RNA helicase may consist of a larger complex with differing characteristics.
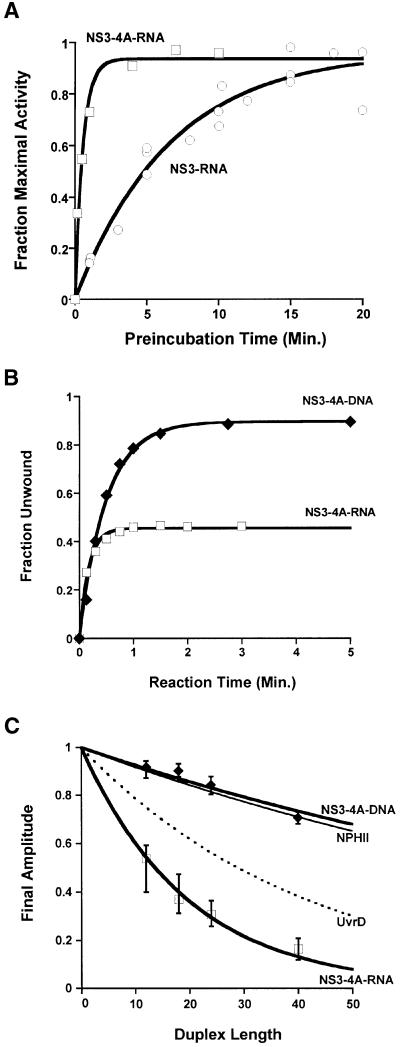
In order to test this ‘modulator’ hypothesis, we examined the role of cofactor NS4A, which is a protein that binds tightly to the protease domain of NS3 (Bartenschlager et al., 1995; Ishido et al., 1998), resulting in the structural reorganization of NS3 (Kim et al., 1996; Love et al., 1996; Yao et al., 1999). The NS3–4A complex was studied with the same kinetic strategies that were applied to the NS3 helicase in isolation (Figures 1, 2 and 3). First, the ability of NS3–4A to form functional complexes was evaluated. The relaxation rate constant for NS3–4A–DNA complex formation was 6 ± 2 min–1 and was nearly identical to that of NS3–DNA complex formation. In contrast, the relaxation rate constant for NS3–4A–RNA complex formation was 1.8 ± 0.2 min–1, which was ∼10 times faster than the binding of NS3 to RNA in the absence of NS4A (Figure 5A). In order to assess whether this rate enhancement was due to structure in the single-stranded overhang region of the RNA substrates, we analyzed the unwinding kinetics of an alternative RNA substrate that contains the same duplex region as b, but which has a 3′ overhang of poly(U). We determined that altering the single-strand overhang sequence does not change the enhanced rate at which NS3–4A binds to RNA (kr = 2.2 ± 0.9 min–1) or the original rate at which NS3 binds to RNA (kr = 0.12 ± 0.08 min–1). This observation supports the conclusion that cofactor NS4A enhances functional NS3–RNA complex formation. Thus, in addition to the known role of NS4A in protease activity (Failla et al., 1994) and membrane anchoring of NS3 (Tanji et al., 1995; Wölk et al., 2000), cofactor NS4A acts as an RNA-specific loading factor, enabling NS3 to form functional protein–RNA complexes.
Fig. 5. NS3–4A activity. (A) Functional complex formation between NS3–4A and RNA (open squares); for comparison, complex formation between NS3 and RNA has been replotted here (open circles). The substrate in both cases was duplex b (see Figure 3A). (B) Single-cycle unwinding of RNA duplex b (open squares; kunw = 5.9 ± 0.5 min–1) and DNA duplex f (closed diamonds; kunw = 2.5 ± 0.9 min–1) by the NS3–4A complex. (C) NS3–4A processivity. Final unwinding amplitudes for the family of duplex substrates (Figure 3A) were measured under single-cycle conditions, as in Figure 3B. Error bars were calculated from the variance in at least three different measurements. The processivities of DNA helicase UvrD (Ali and Lohman, 1997; black dotted line) and RNA helicase NPH-II (Jankowsky et al., 2000; thin black line) are presented for comparison.
Having established a role for NS4A in productive binding to RNA, we then asked whether NS4A might influence other aspects of NS3 unwinding. To address these issues, the unwinding rate constants and processivity for the NS3–4A complex were evaluated under single-cycle conditions. We found that both the single-cycle unwinding rate constant [kunw = 5.9 ± 0.5 min–1 (18 bp duplex b; Figure 5B)] and the processivity (Figure 5C) of NS3 on RNA remained virtually unchanged by the presence of cofactor NS4A (compare with Figure 3B). However, it was possible to detect a slight enhancement in the processivity for DNA unwinding by NS3–4A. Taken together, these observations suggest that NS4A acts prior to unwinding, where it functions to load NS3 onto RNA. In addition, the data indicate that this enhancement of RNA activity is not detrimental to DNA activity, i.e. NS4A binding does not diminish the robust DNA helicase activity of NS3. These experiments assign a mechanistic role to NS4A of enhancing productive binding. They also help resolve previous claims regarding its effect on helicase activity (Morgenstern et al., 1997; Gallinari et al., 1999; Howe et al., 1999). Notably, it has been reported that NS4A enhances ‘unwinding’ under multiple-cycle conditions (Howe et al., 1999). Our results suggest that this effect is due to enhancement of functional complex formation rather than an alteration of the actual unwinding capabilities of NS3.
Phylogenetic analysis of NS3
Our in vitro results suggest that a helicase from a cytoplasmically replicating RNA virus prefers DNA substrates. Therefore, it is important to address the biological context of these findings. To do so, we posed two questions: (i) is the ability to unwind both DNA and RNA substrates a general characteristic of DExH/D proteins; and (ii) does the phylogenetic distribution of DExH/D proteins suggest an origin or rationale for the DNA activity of HCV NS3?
Most of the DExH/D proteins characterized to date are implicated in diverse aspects of RNA metabolism (de la Cruz et al., 1999; Jankowsky et al., 2000). However, the DExH/D subgrouping of helicase superfamily 2 (SF2) also contains the well-characterized RecQ group of DNA helicases, which includes the BLM and WRN proteins that are implicated in DNA recombination-associated events (Karow et al., 2000). Of the eight helicases in the DExH/D subfamily for which both RNA and DNA activity has been studied, nearly all show a strong preference for only one type of substrate. For example, the translation initiation factor EIF4a was observed to unwind RNA but not duplex DNA (Rogers et al., 2001). In general, RNA helicases prefer RNA and are either unable to unwind duplex DNA (Lee and Hurwitz, 1992; Warrener and Collett, 1995) or their NTPase activity is not stimulated by DNA (Suzich et al., 1993; Wagner et al., 1998). The RecQ homolog SGS1 was observed to unwind DNA but not duplex RNA (Bennett et al., 1999), which is consistent with the fact that DNA helicases typically prefer DNA and are unable to unwind RNA duplexes or hybrid RNA–DNA duplexes (Matson, 1989). Therefore, it is intriguing that, unlike other helicases studied to date, HCV NS3 possesses a robust in vitro activity (DNA unwinding) that is inconsistent with its putative in vivo function (RNA replication), and it productively binds and unwinds both DNA and RNA substrates.
Phylogenetic analysis was used to investigate the relationship between NS3 and other helicases in the DExH/D family. The first step in this process involved aligning the sequences of known DExH/D proteins. The evolutionary relationships among DExH/D SF2 proteins (of which NS3 is a member) were then determined using a parsimony-based approach that inferred a phylogeny for 154 DExH/D SF2 helicases. The specificity of these helicases for DNA or RNA was then mapped onto the resultant tree (Figure 6).
Fig. 6. Unrooted cladogram of SF2 helicases summarizing the major monophyletic groups as indicated by phylogenetic analysis. All numbers on the branches represent measures of confidence for the node at the end of the branch. Values that are preceded by a ‘d’ are Bremer decay indices. Other numbers (generally to the right of the /) are bootstrap values. Bootstrap values of <50% are not reported. Nucleic acid substrate specificity for each group is indicated.
Analysis of the tree resulted in a number of striking findings. First, HCV NS3 and the DNA helicases typified by RecQ are not close relatives. They are phylogenetically separated by multiple branches that represent groups of proteins demonstrated to have exclusive RNA helicase activity and/or to be involved in well-characterized RNA processes such as splicing (Figure 6). This separation suggests that the DNA helicase activity of NS3 evolved independently from the RecQ family. This conclusion also strongly suggests that DNA unwinding by NS3 is not a vestigial activity that derives from a DNA-specific ancestor.
A second notable feature of the tree is the close evolutionary relationship between NS3 and the p80 protein from bovine viral diarrhea virus (BVDV), a related member of the Flaviviridae family. The protease/helicase p80 is the functional NS3 analog in the BVDV replication complex (Fields et al., 1996). The two proteins have a high sequence similarity (45% similarity, 32% identical). However, unlike HCV NS3, p80 appears unable to unwind dsDNA (Warrener and Collett, 1995) and its NTPase activity is not stimulated by DNA (Suzich et al., 1993). These marked functional differences between HCV NS3 and BVDV p80, particularly given their close sequence similarities, suggest that a fundamental alteration of biopolymer specificity (from RNA to DNA or vice versa) may require only minimal changes in protein sequence or structure. Mutational and biophysical studies are now in progress to identify which residues confer specificity for DNA versus RNA.
Based on these findings, we favor an evolutionary scenario in which NS3 DNA helicase activity evolved recently from a common ancestor of the p80 and HCV NS3 proteins that was an RNA-specific helicase. The robust DNA helicase activity in HCV suggests possible selective pressure for DNA unwinding activity. It is interesting to speculate why DNA activity might be advantageous for HCV. It was recently determined that HCV RNA replication is favored in rapidly dividing cells (Pietschmann et al., 2001) and that NS3 has the capacity to localize in the nuclei of cells transformed with NS3 expression plasmids (Muramatsu et al., 1997; Wölk et al., 2000). While nuclear localization of NS3 is strongly inhibited by complex formation with NS4A (Muramatsu et al., 1997; Wölk et al., 2000), it is difficult to rule out the persistence of small nuclear NS3 populations, even when co-expressed with NS4A (Muramatsu et al., 1997). Unregulated DNA helicase activity is well established to play a role in cellular transformation; for example, overexpression of the SF2 DNA helicase Rad12+ results in the loss of checkpoint control and an increased sensitivity to mutagenic agents (Davey et al., 1998).
As a consequence, DNA helicase activity of HCV NS3 may have direct implications for the pathology of chronically infected patients. HCV has the unusual distinction of being the only known non-retroviral RNA virus associated with the development of cancer in humans (Flint et al., 2000). While this may result from the immune-mediated complications of chronic infection and the deregulation of normal cellular machinery (Sakamuro et al., 1995; Muramatsu et al., 1997), it remains possible that NS3 may act directly on host DNA.
Ultimately, it will be important to test the potential impact of HCV NS3 on the DNA of living cells and phylogenetic analyses will be very helpful in the design of NS3 mutants to facilitate these studies. However, it is important to underscore the serious limitations that currently confront in vivo studies of HCV. The only model systems currently available for studying HCV are the chimpanzee (Hagedorn and Rice, 2000) and immunosuppressed mice transplanted with human liver cells (Mercer et al., 2001). Remarkably, a cell culture system still does not exist for studying viral infection, assembly and impact on cellular metabolism, although specific methods for examining RNA replication in culture appear promising (i.e. the replicon systems; Lohmann et al., 1999; Blight et al., 2000). Consequently, biochemical studies play a vital role in helping to explain the HCV biology that is observed, to define relevant pathways that might be susceptible to inhibitors and to suggest directions for further investigation with the limited in vivo methodologies that are available.
The DNA helicase activity of NS3 is an example of the remarkable functional diversity that can be achieved by the small, streamlined genome of a virus. Our data suggest that HCV has acquired the ability to unwind multiple types of substrates and it employs NS4A to promote the RNA helicase activity necessary for RNA replication. These adaptations suggest new avenues for treatment, as disruption of the NS3–4A interface or of DNA helicase activity could potentially have potent effects on HCV management.
Materials and methods
Protein expression and purification
The proteins NS3 and NS3–4A are both from HCV genotype 1a and were a gift from Dr Hung V.Le (Schering-Plough Research Institute, Kenilworth, NJ). Both constructs contain the ‘full-length’ NS3 protein, which includes both the protease and helicase domains. Briefly, the NS3(1a) protein was expressed in M15 Escherichia coli on a Qiagen pQE40 vector and includes an N-terminal His tag. Cells were grown in Luria–Bertani medium and induced with 1 mM isopropyl-β-d- thiogalactopyranoside. After cell lysis, the supernatant was mixed with Ni-NTA–agarose, from which the protein was subsequently eluted with a solution containing 0.3 M NaCl. NS3–4A(1a) was expressed in insect cells as the NS3–4A polyprotein, which is autocatalytically cleaved between NS3 and 4A by the NS3 protease domain to generate the heterodimeric NS3–4A complex (Sali et al., 1998). When studied at the same concentration under identical reaction conditions (see below), both NS3 and NS3–4A showed almost identical unwinding kinetics and reaction amplitudes (see text), thereby confirming that the protein preparations are of comparable activity. Importantly, the behavior of a full-length NS3 construct that lacks a His tag and derives from genotype 1b was also analyzed (data not shown). This full-length NS3(1b) construct was expressed in a similar manner and its His tag was removed by thrombin cleavage. It showed no kinetic or processivity differences from those reported in the text for the NS3(1a) construct, indicating that the reported results are not affected by the presence of a tag or the specific genotype of the sample.
Nucleic acid substrates
RNA and DNA oligos were chemically synthesized (Wincott et al., 1995) or purchased commercially from Dharmacon, Inc. (Lafayette, CO) and purified by denaturing PAGE. Duplexes were formed and purified as described previously (Jankowsky et al., 2000). Sequences were designed to eliminate intramolecular secondary structural elements (Mathews et al., 1999). To control for sequence effects on kinetic parameters, substrates a–l contain the same sequence at the beginning of the duplex region. A high GC content was necessary to stabilize the 12 bp duplexes that would otherwise dissociate under the conditions used for studying reaction processivity (37°C, 1 nM substrate). This low substrate concentration prevents re-annealing of unwound substrates on the time scale of the reaction. The thermodynamic stabilities of RNA–RNA and DNA–DNA duplexes were calculated using the program RNA structure (Mathews et al., 1999), while the stabilities of RNA–DNA hybrid duplexes were calculated manually using published parameters (Sugimoto et al., 1995).
Single-cycle unwinding and active complex formation
Active complex formation (ES) was measured by incubating NS3 (70 nM) or NS3–4A (70 nM) at 37°C for various times with radiolabeled duplex substrate (1 nM either RNA or DNA duplex, γ-32P-labeled top strand; Figure 3A), in the absence of ATP in a buffer that contained 25 mM MOPS–NH4+ pH 6.5, 3 mM MgCl2, 2 mM DTT, 30 mM NaCl, 1% glycerol, 0.1 mM β-octyl-glucoside and 0.1 mM laurylmaltoside. The buffer conditions were initially based on previously published studies (Hagedorn and Rice, 2000) and then optimized further for maximal helicase activity on RNA. Following substrate–helicase incubation, unwinding reactions were initiated by simultaneous addition of ATP (4 mM) and ‘trap-DNA’ (750 nM single-stranded DNA 60-mer, 5′-CATATGAGTCGTATCGTGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′), thus achieving single-cycle unwinding conditions. Importantly, the presence of the trap-DNA prevents further helicase–substrate association and also prevents reassociation with partially unwound duplexes. Because [E] >> [S], the reaction is effectively first order and the rate constant is equal to the reciprocal relaxation time (1/τ), as described previously (Fersht, 1999). Unwinding amplitudes at varying incubation times were measured by gel shift as described previously (Jankowsky et al., 2000).
Analysis of processivity
Using the same conditions described above, the maximum amount of unwinding for each duplex substrate (Figure 3A) was measured by pre-incubating the helicase with the substrate for ∼25 min before initiating the reaction by the simultaneous addition of 4 mM ATP and 750 nM trap-DNA (Figure 1B and C). The maximum amplitude achieved for each duplex was then plotted against the length of the duplex and this data fit using the equation Afinal = xPN, where x is the initial fraction of functional complex, P is processivity and N is the number of steps made on the duplex, as described previously (Ali and Lohman, 1997; Jankowsky et al., 2000).
Phylogenetic analysis
SF2 DExH/D helicases were identified using the BLAST program (Altschul et al., 1997). The genomes of Caenorhabditis elegans, human, mouse, Arabidopsis and E.coli were searched for proteins similar to NS3 and known yeast DExH/D helicases (de la Cruz et al., 1999). The resulting sequences were inspected for the DExH/D signature and all other SF2 helicase motifs (Jankowsky et al., 2000). Viral helicases of known functional specificity matching the above criteria were added. Sequences that precluded alignment were discarded (although other DNA SF2 helicases are known to exist, none met the above selection criteria; Gorbalenya and Koonin, 1993). Clustal X 1.63 (Thompson et al., 1997) was used to align the sequence region encompassed by helicase motifs I–VI of the 154 helicases. Sequences were aligned in two steps; a first pass alignment forced the anchoring of the six major helicase motifs and emphasized absolute sequence conservation (weight = identity matrix, Gap/Extension = 10/0.1), followed by a second pass that utilized specific alignment parameters (weight = gonnet 250 matrix, Gap/Extension = 10/0.1). The matrix produced was then analyzed using the parsimony algorithms of PAUP*4.0β (Swofford, 1998). Owing to the large size of the data set, a maximum likelihood approach was prohibitive. We performed an aggressive heuristic search with 5000 replicates of random addition of taxa followed by the subtree pruning and regrafting technique of PAUP, saving one tree at each replicate. All characters and state transformations were given equal weight and columns with gaps were retained as phylogenetically informative characters (Phillips et al., 2000). The resulting 4998 trees (ranging in length from 15 391 to 15 471 steps) were then tested with the more rigorous tree–branch reconnection (TBR) function of PAUP. This yielded three parsimonious trees of 15 391 steps each. We calculated the strict consensus of these three trees. To test the support for the consensus topology, we performed 100 bootstrap replicates, each composed of 10 heuristic searches with 100 random addition steps followed by TBR. We also used the program Autodecay in conjunction with PAUP to calculate Bremer supports of confidence for nodes in the tree. Bremer support indices show how many additional evolutionary steps are required to lose the groupings represented by the phylogeny (Bremer, 1995). Ten TBR replicates were performed at each node in the phylogeny to obtain the Bremer index.
Acknowledgments
Acknowledgements
We thank members of the Schering Plough Research Institute for providing NS3 and NS3–4A proteins. We are particularly grateful to Drs Hung Le and Patricia Weber of Schering Plough Research Institute for stimulating our investigations of NS3, for ongoing valuable discussions and for critical review of the manuscript. We are also grateful to Robert Krug, Vincent Racaniello and John Taylor for critical review of the manuscript. Finally, we would like to thank Robert DeSalle and David H.Figurski. This work was supported by grants from the National Institutes of Health (RO1 GM60620 to A.M.P. and 5 T32 GM07367 to P.S.P.) and by the Howard Hughes Medical Institute (A.M.P. is an Assistant Investigator of the HHMI).
References
- Ali J.A. and Lohman,T.M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science, 275, 377–380. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R. and Lohmann,V. (2000) Replication of hepatitis C virus. J. Gen. Virol., 81, 1631–1648. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Lohmann,V., Wilkinson,T. and Koch,J.O. (1995) Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol., 69, 7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- Blight K.J., Kolykhalov,A.A. and Rice,C.M. (2000) Efficient initiation of HCV RNA replication in cell culture. Science, 290, 1972–1974. [DOI] [PubMed] [Google Scholar]
- Bremer K. (1995) Branch support and tree stability. Cladistics, 10, 295–304. [Google Scholar]
- Chin K., Sharp,K.A., Honig,B. and Pyle,A.M. (1999) Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nature Struct. Biol., 6, 1055–1061. [DOI] [PubMed] [Google Scholar]
- Davey S., Han,C.S., Ramer,S.A., Klassen,J.C., Jacobson,A., Eisenberger,A., Hopkins,K.M., Lieberman,H.B. and Freyer,G.A. (1998) Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol. Cell. Biol., 18, 2721–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- DiBisceglie A.M. (1997) Hepatitis C and hepatocellular carcinoma. Hepatology, 26, 34S–38S. [DOI] [PubMed] [Google Scholar]
- Failla C., Tomei,L. and De Francesco,R. (1994) Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol., 68, 3753–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W.H.Freeman, New York, NY.
- Fields B.N., Knipe,D.M. and Howley,P.M. (1996) Fields Virology. Lippincott-Raven, Philadelphia, PA.
- Flint S.J., Enquist,L.W., Krug,R.M., Racaniello,V.R. and Skalka,A.M. (2000) Principles of Virology. ASM Press, Washington, DC.
- Gallinari P., Paolini,C., Brennan,D., Nardi,C., Steinkuhler,C. and De Francesco,R. (1999) Modulation of hepatitis C virus NS3 protease and helicase activities through the interaction with NS4A. Biochemistry, 38, 5620–5632. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- Gwack Y., Kim,D.W., Han,J.H. and Choe,J. (1996) Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun., 225, 654–659. [DOI] [PubMed] [Google Scholar]
- Gwack Y., Kim,D.W., Han,J.H. and Choe,J. (1997) DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur. J. Biochem., 250, 47–54. [DOI] [PubMed] [Google Scholar]
- Hagedorn C.H. and Rice,C.M. (2000) The Hepatitis C Viruses. Springer-Verlag, Berlin, Germany.
- Hesson T., Mannarino,A. and Cable,M. (2000) Probing the relationship between RNA-stimulated ATPase and helicase activities of HCV NS3 using 2′-O-methyl RNA substrates. Biochemistry, 39, 2619–2625. [DOI] [PubMed] [Google Scholar]
- Howe A.Y., Chase,R., Taremi,S.S., Risano,C., Beyer,B., Malcolm,B. and Lau,J.Y. (1999) A novel recombinant single-chain hepatitis C virus NS3–NS4A protein with improved helicase activity. Protein Sci., 8, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido S., Fujita,T. and Hotta,H. (1998) Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun., 244, 35–40. [DOI] [PubMed] [Google Scholar]
- Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2000) The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature, 403, 447–451. [DOI] [PubMed] [Google Scholar]
- Kankia B.I. and Marky,L.A. (1999) DNA, RNA and DNA/RNA oligomer duplexes: a comparative study of their stability, heat, hydration and Mg2+ binding properties. J. Phys. Chem. B, 103, 8759–8767. [Google Scholar]
- Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- Kim J.L. et al. (1996) Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor. Cell, 87, 343–355. [DOI] [PubMed] [Google Scholar]
- Lavanchy D. (1999) Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat., 6, 35–47. [PubMed] [Google Scholar]
- Lee C.G. and Hurwitz,J. (1992) A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem., 267, 4398–4407. [PubMed] [Google Scholar]
- Levin M.K. and Patel,S.S. (1999) The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem., 274, 31839–31846. [DOI] [PubMed] [Google Scholar]
- Lohmann V., Korner,F., Koch,J., Herian,U., Theilmann,L. and Bartenschlager,R. (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science, 285, 110–113. [DOI] [PubMed] [Google Scholar]
- Love R.A., Parge,H.E., Wickersham,J.A., Hostomsky,Z., Habuka,N., Moomaw,E.W., Adachi,T. and Hostomska,Z. (1996) The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell, 87, 331–342. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- Matson S.W. (1989) Escherichia coli DNA helicase II (uvrD gene product) catalyzes the unwinding of DNA.RNA hybrids in vitro. Proc. Natl Acad. Sci. USA, 86, 4430–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer D.F. et al. (2001) Hepatitis C virus replication in mice with chimeric human livers. Nature Med., 7, 927–933. [DOI] [PubMed] [Google Scholar]
- Morgenstern K.A., Landro,J.A., Hsiao,K., Lin,C., Gu,Y., Su,M.S. and Thomson,J.A. (1997) Polynucleotide modulation of the protease, nucleoside triphosphatase and helicase activities of a hepatitis C virus NS3–NS4A complex isolated from transfected COS cells. J. Virol., 71, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S., Ishido,S., Fujita,T., Itoh,M. and Hotta,H. (1997) Nuclear localization of the NS3 protein of hepatitis C virus and factors affecting the localization. J. Virol., 71, 4954–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini C., De Francesco,R. and Gallinari,P. (2000) Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol., 81, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Phillips A., Janies,D. and Wheeler,W. (2000) Multiple sequence alignment in phylogenetic analysis. Mol. Phylogenet. Evol., 16, 317–330. [DOI] [PubMed] [Google Scholar]
- Pietschmann T., Lohmann,V., Rutter,G., Kurpanek,K. and Bartenschlager,R. (2001) Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol., 75, 1252–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D.J., Short,S.A., Hanlon,M.H., Preugschat,F., Wilson,J.E., Willard,D.H. and Consler,T.G. (1998) Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J. Biol. Chem., 273, 18906–18914. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Averett,D.R., Clarke,B.E. and Porter,D.J. (1996) A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem., 271, 24449–24457. [DOI] [PubMed] [Google Scholar]
- Rogers G.W. Jr, Lima,W.F. and Merrick,W.C. (2001) Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem., 276, 12598–12608. [DOI] [PubMed] [Google Scholar]
- Sakamuro D., Furukawa,T. and Takegami,T. (1995) Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J. Virol., 69, 3893–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali D.L. et al. (1998) Serine protease of hepatitis C virus expressed in insect cells as the NS3/4A complex. Biochemistry, 37, 3392–3401. [DOI] [PubMed] [Google Scholar]
- Seeff L.B. (1998) The natural history of hepatitis C—a quandary. Hepatology, 28, 1710–1712. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA–DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- Suzich J.A., Tamura,J.K., Palmer-Hill,F., Warrener,P., Grakoui,A., Rice,C.M., Feinstone,S.M. and Collett,M.S. (1993) Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol., 67, 6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. (1998) PAUP*: Phylogenetic Analysis using Parsimony (*and Other Methods). Sinauer, Sunderland, MA.
- Tackett A.J., Wei,L., Cameron,C.E. and Raney,K.D. (2001) Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res., 29, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai C.L., Chi,W.K., Chen,D.S. and Hwang,L.H. (1996) The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol., 70, 8477–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y., Hijikata,M., Satoh,S., Kaneko,T. and Shimotohno,K. (1995) Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol., 69, 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1997) CLUSTAL X Multiple Sequence Alignment Program. European Molecular Biology Organization, Hamburg, Germany.
- Wagner J.D., Jankowsky,E., Company,M., Pyle,A.M. and Abelson,J.N. (1998) The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J., 17, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.H., Fujii,S., van Boom,J.H., van der Marel,G.A., van Boeckel,S.A. and Rich,A. (1982) Molecular structure of r(GCG)d(TATACGC): a DNA–RNA hybrid helix joined to double helical DNA. Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- Warrener P. and Collett,M.S. (1995) Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol., 69, 1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wincott F. et al. (1995) Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res., 23, 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölk B., Sansonno,D., Kräusslich,H.-G., Dammacco,F., Rice,C.M., Blum,H.E. and Moradpour,D. (2000) Subcellular localization, stability and trans cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J. Virol., 74, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N., Reichert,P., Taremi,S.S., Prosise,W.W. and Weber,P.C. (1999) Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease– helicase. Structure Fold Des., 7, 1353–1363. [DOI] [PubMed] [Google Scholar]