Abstract
Cellular signaling activities must be tightly regulated for proper cell fate control and tissue morphogenesis. Here we report that the Drosophila leucine-rich repeat transmembrane glycoprotein Gp150 is required for viability, fertility and development of the eye, wing and sensory organs. In the eye, Gp150 plays a critical role in regulating early ommatidial formation. Gp150 is highly expressed in cells of the morphogenetic furrow (MF) region, where it accumulates exclusively in intracellular vesicles in an endocytosis-independent manner. Loss of gp150 function causes defects in the refinement of photoreceptor R8 cells and recruitment of other cells, which leads to the formation of aberrant ommatidia. Genetic analyses suggest that Gp150 functions to modulate Notch signaling. Consistent with this notion, Gp150 is co-localized with Delta in intracellular vesicles in cells within the MF region and loss of gp150 function causes accumulation of intracellular Delta protein. Therefore, Gp150 might function in intracellular vesicles to modulate Delta–Notch signaling for cell fate control and tissue morphogenesis.
Keywords: Delta/Drosophila/eye/leucine-rich repeat/Notch
Introduction
Development of multicellular organisms requires not only precise cell fate specification, but also proper patterning of differentiating cells. The Drosophila compound eye provides a model system to address how cells are specified and organized. During the third instar larval stage, a dorso-ventral indentation called the morphogenetic furrow (MF) moves from the posterior of the eye tissue to the anterior, and formation of ommatidial clusters is initiated in areas immediately posterior to the MF (for review see Wolff and Ready, 1993). Expression of the proneural basic helix– loop–helix (bHLH) atonal (ato) gene is first induced by Hedgehog signaling in a stripe of cells just anterior to the MF (for review see Heberlein and Moses, 1995; Treisman and Heberlein, 1998). Within the MF, ato expression is restricted to regularly spaced proneural clusters of cells. Only one cell from each proneural cluster continues to express ato and becomes the R8 photoreceptor (R), which then initiates further recruitment of other R cells for ommatidial construction (Jarman et al., 1994, 1995; Baker et al., 1996; Dokucu et al., 1996; Baonza et al., 2001). Notch (N) signaling initially plays a positive role in establishing high levels of ato in cells anterior to the MF (Baker and Yu, 1997; Li and Baker, 2001) by down-regulating Hairy (H) and Extramacrochaetae (Emc), two repressors of ato (Baonza and Freeman, 2001). Subsequently, N-mediated lateral inhibition is required for the refinement of ato expression to regularly spaced individual R8 precursor cells (Parks et al., 1995; Baker et al., 1996; Dokucu et al., 1996). The Drosophila homolog of the vertebrate epidermal growth factor receptor (DER) functions to regulate R8 spacing as well. DER signaling acts non-autonomously to inhibit ato expression in cells anterior and lateral to the proneural clusters to generate regularly spaced proneural clusters (Chen and Chien, 1999; Baonza et al., 2001). Also, a glycoprotein Scabrous (Sca) might be responsible for anterior and lateral repression of Ato, as Sca is produced in proneural clusters and can be secreted (Baker and Zitron, 1995; Lee et al., 1996). Sca associates with N in vivo and can stabilize N proteins at the cell surface (Powell et al., 2001).
In precursor cells located more posterior to the MF, N signaling is required for restricting cellular competence to respond to receptor tyrosine kinase (RTK)-mediated inductive signaling for R cell specification. Supporting this idea, several E(spl) proteins are prominently expressed in the basally located nuclei of the precursor cells (Baker et al., 1996). A deletion of a subset or all of the bHLH genes in the E(spl) complex results in a severe neurogenic phenotype in the posterior region of eye discs (Treisman et al., 1997; Ligoxygakis et al., 1998). Thus, coordinated action of inductive RTK signaling (for review see Freeman, 1997) and N-mediated lateral inhibition (for review see Artavanis-Tsakonas et al., 1999; Mumm and Kopan, 2000) is critical for the sequential recruitment of R cells into the developing ommatidia.
Here we report the identification and isolation of loss-of-function mutations in the Drosophila gp150 gene, which encodes a leucine-rich repeat (LRR) protein that is required for viability, fertility and proper development of the eye, wing and sensory organs. In the eye, removal of gp150 function causes defects in the refinement of R8 cells and recruitment of other cells, which leads to the formation of fused ommatidia, as well as ommatidia containing too many or too few R cells. We show that Gp150 is expressed at high levels in the MF region, which is consistent with a role of gp150 in early ommatidial development. Moreover, genetic analyses suggested that Gp150 functions to modulate Delta (Dl)–N signaling, and immunostaining experiments showed that Gp150 is co-localized with Dl in intracellular vesicles in cells within the MF region. Gp150 might be involved in facilitating lysosomal delivery of the Dl protein and/or Dl transport to the plasma membrane, as loss of gp150 function causes accumulation of intracellular Dl protein. Gp150 appears to be a resident protein of intracellular vesicles and its localization is not affected in endocytosis-defective cells. Based on these observations, we propose that Gp150 might function in subcellular vesicles to control appropriate intracellular levels of Dl to modulate N signaling.
Results
Identification and isolation of loss-of-function mutations in the Drosophila gp150 gene, which encodes a LRR transmembrane protein
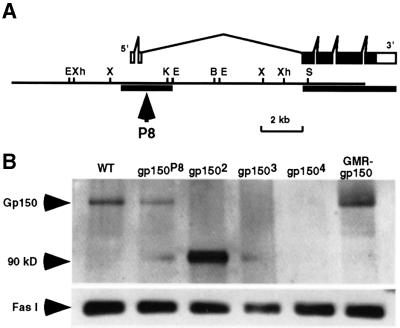
Flies homozygous for two presumably P transposon-induced semi-lethal mutations, l(2)k11107 and l(2)k11120 (Torok et al., 1993), exhibit reduced fertility and abnormal bristle, wing and eye phenotypes. These two allelic recessive mutations were meiotically mapped to the tip of the right arm of the second chromosome. Deficiency mapping further defines the locus of these mutations to the 58D1-5 region. However, no wild-type revertants were recovered by P-element excision (data not shown). Thus, these alleles could have been caused by an imprecise excision of a P-element initially inserted at the locus. To facilitate molecular genetic analysis of this gene, we generated new alleles through ethyl methane sulfonate (EMS) mutagenesis and P-element local hopping (see Materials and methods). Genomic sequences flanking the insertion sites in two of the newly isolated P alleles, P2 and P8, were cloned and used to isolate cDNA clones from a larval eye disc cDNA library. Sequence analysis of full-length cDNAs demonstrated that these cDNA molecules encode a polypeptide identical to the LRR transmembrane glycoprotein Gp150 (Tian and Zinn, 1994).
Our sequence analysis, together with data derived from the Drosophila genome project, indicates that the gp150 gene consists of six exons and five introns (Figure 1A). The gp150 open reading frame (ORF) is restricted to the last four exons, which encode a polypeptide of 1051 amino acids. The following evidence demonstrates that mutant alleles of gp150 have been identified. First, in the P8 allele, a P-element is inserted within the second intron of the gp150 transcription unit, 53 bp downstream of the second exon (Figure 1A). The P8 mutant eye discs exhibited a reduction of gp150 expression, as shown by western blotting (Figure 1B) and immunostaining (data not shown). These experiments revealed that P8 is a partial loss-of-function mutation. Furthermore, mutant phenotypes in four out of six newly isolated P alleles can be frequently reverted to wild type by P-element excision (see Materials and methods), indicating that these mutations were indeed caused by P-element insertions. Secondly, a single base change of G to A was found in codon 94 for tryptophan in an EMS allele (gp1504), which results in a nonsense mutation. Thirdly, western blot and immunostaining analyses revealed that the level of Gp150 is either reduced or eliminated by the mutations (Figures 1B and 5B). Lastly, the mutant eye phenotypes can be effectively rescued by overexpression of a wild-type gp150 gene in the developing eye (Figure 3E and F). These loss-of-function mutations were used in this study to reveal a role of gp150 in eye development.
Fig. 1. Identification of loss-of-function alleles of the gp150 gene. (A) The gp150 gene consists of six exons and five introns with the complete coding sequence restricted within a 4 kb genomic region. The filled boxes represent a complete 3156 bp ORF. Sequences of two genomic regions indicated by the thick horizontal lines were determined to illustrate intron–exon structures of the gp150 transcriptional unit. A PCR method was used to obtain the downstream genomic sequence of gp150 that was not included in the genomic clone. In P8, a P transposon was inserted in the second intron of gp150. Restriction enzymes used for mapping include BamHI (B), EcoRI (E), KpnI (K), SalI (S), XbaI (X) and XhoI (Xh). (B) A western blot was probed with a Gp150 antibody that was made against the N-terminal region (amino acids 5–192) of Gp150. Since no Gp150 protein was detectable in gp1502, gp1503 and gp1504 mutants, these alleles can be considered as molecular nulls. The gp1502 mutation results in the production of a non-functional 90 kDa protein. This is based on observations that gp1502 is recessive and the mutant phenotypes caused by gp1502 are similar to that of gp1503 and gp1504. The 70 kDa Fas I protein served as an internal control and was detected by anti-Fas I (MAb6D8) antibody in all lanes.
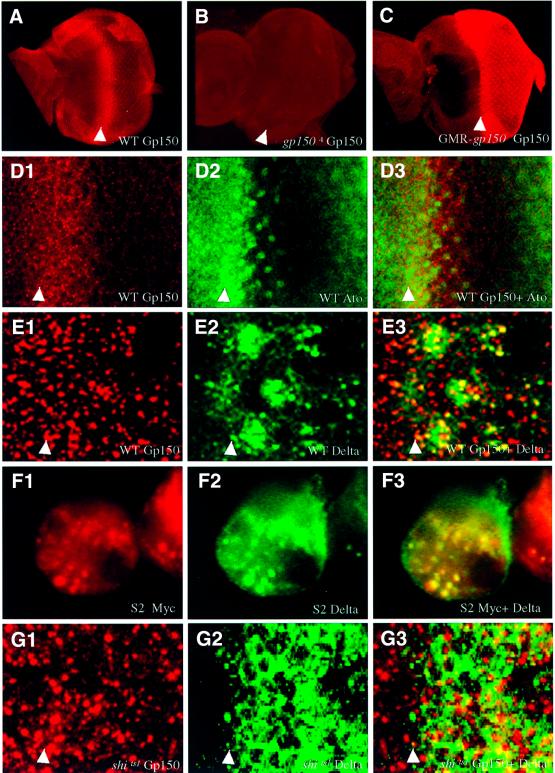
Fig. 5. gp150 is expressed at high levels in the MF region in the developing eye. (A) High levels of Gp150 protein were detected in the MF region of wild-type third instar eye discs. Posterior to the furrow, relatively lower levels of Gp150 can be detected around each individual ommatidium. (B) Gp150 expression was not detectable in gp1504 eye discs. (C) High levels of Gp150 expression were detected in all cells behind the furrow in GMR-gp150 eye discs. (D1–3) Double-staining reveals Gp150 (red, D1) and Ato (green, D2) expression in eye discs. A superimposed image from (D1) and (D2) is shown in (D3). (E1–3) Gp150 and Dl proteins were shown to co-localize in subcellular vesicles in the furrow region. (F1–3) In S2 cells co-transfected with pmt-gp150-myc and pmt-Dl, Gp150 and Dl were also shown to co-localize in subcellular vesicles. (G1–3) Heat-treated shits1 mutant third instar eye discs were double stained with anti-Gp150 (red in G1) and anti-Dl (green in G2) antibodies. A superimposed image is shown in (G3). Arrowheads indicate the location of the MF, and anterior is to the left in all panels.
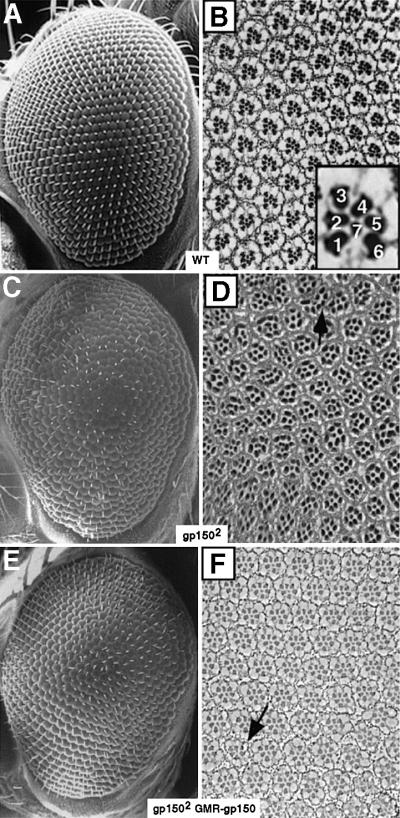
Fig. 3. Loss of gp150 function causes defective eye development. SEM images (A, C and E) and tangential sections (B, D and F) of adult eyes are presented. (A and B) Wild type. The inset highlights seven R cells arranged in a trapezoidal configuration in each ommatidium and the rhabdomere of R8 cannot be seen in this apical section. (C and D) gp1502 mutants. About 35% (n = 522) of the ommatidia contain either too many or too few R cells. The arrow indicates a fused ommatidium. At basal levels, ommatidia with multiple R8-like cells were observed (data not shown). Other gp150 alleles exhibit similar phenotypes. (E and F) In gp1502 GMR-gp150 flies, overexpression of gp150 in the eye effectively rescues the mutant eye phenotype, with >93% (n = 368) of the ommatidia containing a normal complement of R cells. The arrow indicates a mutant ommatidium occasionally seen in this genotype. Anterior is to the left, except (D), in which anterior might be to the top.
gp150 is required for viability, fertility and normal development of the wing and sensory organs
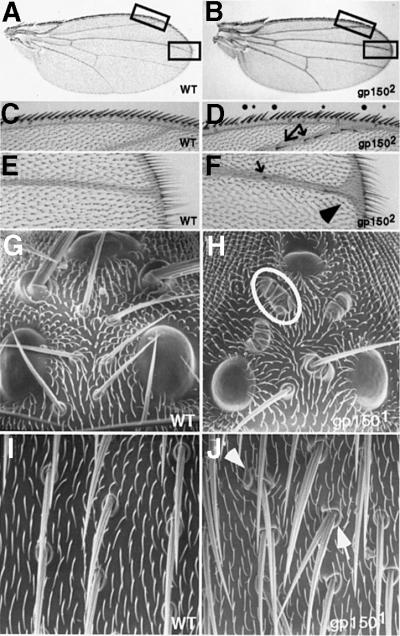
Loss of gp150 function results in a strong reduction in viability. For instance, >60% of gp1502/gp1503 flies fail to develop to the adult stage, with most of them dying during pupal development (Table I). gp150 mutant females are sterile as they fail to lay eggs and the fertility of gp150 mutant males is also strongly reduced (data not shown). In gp150– wings, the size and overall morphology appear normal (Figure 2B). However, closer examination revealed that some dorsal margin bristles are missing or duplicated and ectopic bristles can be found in the distal region of the second vein (Figure 2D). Moreover, the distal end of the third vein is enlarged to form a ‘Delta-like’ structure (Figure 2F).
Table I. Gp150 is required for viability.
| 1 | 2 | 3 | 4 | Df(2R)02311 | |
|---|---|---|---|---|---|
| 1 | 61 (n = 633) | 75 (n = 747) | 73 (n = 221) | 61 (n = 334) | |
| 2 | 61 (n = 526) | 43 (n = 319) | 46 (n = 319) | ||
| 3 | 93 (n = 476) | 63 (n = 331) | |||
| 4 | 62 (n = 355) |
1, 2, 3 and 4 correspond to gp1501, gp1502, gp1503 and gp1504, respectively. For example, from a cross between gp1502/CyO and gp1503/CyO, 61% of the predicted gp1502/gp1503 mutants fail to develop to the adult stage. n = total number of adult flies scored.
Fig. 2. Loss of gp150 function results in defective wing and sensory organs. Wild-type (A, C and E) and gp1502 mutant (B, D and F) wings are shown. (C) and (E) are enlarged from boxed areas in (A). (D) and (F) are enlarged from the boxed areas in (B). Some bristles are missing (dots) or duplicated (asterisks) in the wing margin (D). Arrows point to some ectopic bristles in the second and third veins (D and F) and the arrowhead identifies an enlarged ‘Delta-like’ vein structure (F). SEM images of wild-type (G and I) and gp1501 mutant flies (H and J) are shown to illustrate phenotypes in ocellar bristles (G and H) and sensory organs over the notum (I and J). Multiple socket cells are often developed in sensory organs (circled in H and indicated by an arrowhead in J). The external shaft is often missing or duplicated (indicated by an arrow in J). Moreover, there are cases where both socket and shaft cells are absent in some sensory organs (H). All other gp150 alleles exhibit similar phenotypes in the wing and sensory organs.
During normal bristle development, a single sensory organ precursor (SOP) cell divides to produce two daughter cells, IIa and IIb, which divide and generate a trichogen (shaft) and a tormogen (socket), a neuron, a thecogen (sheath) and a glial cell, respectively (for review see Jan and Jan, 1995; Gho et al., 1999). In gp150 mutants, sensory organ development is defective in ocellar, humeral, scutellar and sensory bristles over the notum and dorsal abdomen (Figure 2H and J). In all of the gp1501 mutant flies, one or more ocellars were found to contain multiple socket cells with the external shaft missing (Figure 2H). Such mutant phenotypes implicate cell fate transformation from IIb to IIa and shaft to socket. Interestingly, external cells (socket and shaft) of some sensory organs appeared to be missing (Figure 2H), suggesting a possible cell fate transformation from IIa to IIb. Such mutant phenotypes were observed in other sensory organs as well. For instance, 68% (n = 31) of the anterior scutellars in gp1502/gp1503 flies exhibited no shaft and no socket, 23% with double socket, 3% with one shaft/two sockets and 3% with two shafts/one socket phenotypes. In conclusion, the gp150 function is required for proper development of several distinct tissue types.
Loss of gp150 function results in defective ommatidial development
We have focused on analyzing gp150 function using the eye system. The wild-type compound eye of Drosophila melanogaster represents an organization of 800 precisely arrayed unit eyes called ommatidia (Figure 3A). Each ommatidium contains 12 accessory cells and eight R cells arranged in a trapezoidal configuration (Figure 3B). Compared with the wild type, gp150 mutant eyes are rough. Many ommatidia are irregular in shape and size and contain either too many or too few R cells. Also, fused ommatidia are often visible (Figure 3C and D). These results indicate that gp150 is required for normal ommatidial development. Clonal analysis revealed essentially the same mutant phenotypes as those observed in gp150 homozygous mutants (data not shown).
To reveal the developmental basis of the eye defects, gp150 mutant third instar eye discs were examined. In wild-type eye discs, phalloidin staining outlines apical cell surfaces and highlights the constricted cells within the MF as well as the cells within the evenly spaced developing ommatidial clusters (Figure 4A). In gp150 mutants, ommatidial clusters are often irregularly spaced and the MF appears broader as the cells within the region are no longer constricted in a narrow stripe (Figure 4B). To elucidate early developmental defects that might lead to the aberrant ommatidial organization, ato expression was examined. ato is essential for R8 specification (Jarman et al., 1994) and is initially expressed in cells anterior to the MF. Within the MF, ato expression is first restricted to proneural groups of cells and then to individual R8 precursor cells (Figure 4C; Jarman et al., 1995). In gp150 mutant eye discs, Ato-positive proneural clusters are abnormally patterned and multiple R8 precursor cells are often closely located (Figure 4D). About 10% (n = 72) of the ommatidia contain multiple Ato-positive R8 precursors that become R8 cells, as inferred from their expression of R8-specific Bride-of-sevenless (Boss) protein (Figure 4F; Kramer et al., 1991). To examine how ommatidia assembly can be affected by gp150 mutations, a neural-specific nuclear marker, Elav (Robinow and White, 1991), was used to highlight all R cells. In the wild type, developing ommatidia are evenly patterned with precursor cells sequentially recruited into ommatidia clusters (Figure 4G). In contrast, ommatidia are irregularly organized in gp150 mutant eye discs. Some oversized ommatidia are seen at early stages of ommatidia assembly (Figure 4H). Thus, recruitment of R cells into the clusters appears to occur prematurely in some clusters.
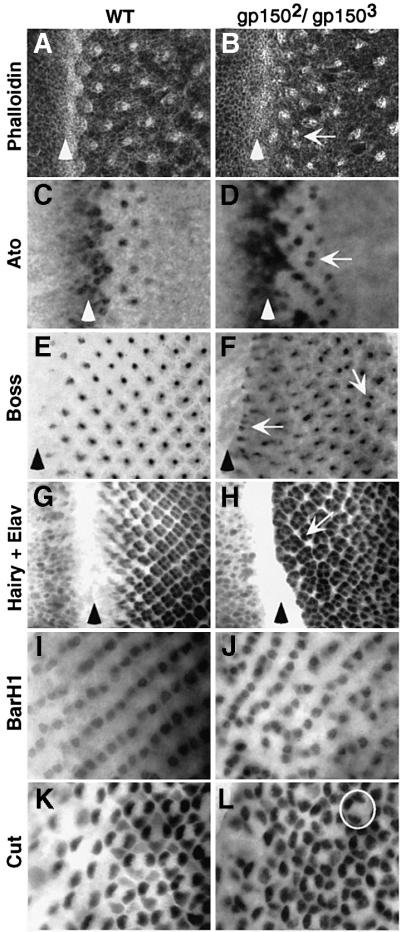
Fig. 4. Loss of gp150 function causes defects in early ommatidial development. Wild-type (A, C, E, G, I and K) and gp1502/gp1503 mutant (B, D, F, H, J and L) third instar eye discs are shown. (A and B) Phalloidin staining. (C and D) Anti-Ato antibody staining. Arrows in (B and D) point to closely located ommatidia. (E and F) Anti-Boss antibody staining. Arrows indicate ommatidia containing more than one Boss-positive R8 cells. (G and H) Double staining for Hairy and Elav. The arrow points to an oversized ommatidium near the furrow. (I and J) Anti-BarH1 antibody staining. (K and L) Anti-Cut antibody staining. The circle indicates an ommatidium missing a cone cell. Arrowheads identify the location of the MF in (A–H). Anterior is to the left in all panels.
To address whether accumulation of oversized ommatidia close to the MF is due to defects in furrow progression, expression of decapentaplegic (dpp) and a bHLH gene hairy (h) was examined, since both of them are involved in regulating furrow progression (Brown et al., 1995; for review see Treisman and Heberlein, 1998). No discernible effect on either h expression anterior to the furrow or dpp expression within the MF was detected in gp150 mutants (Figure 4G and H and data not shown). Moreover, unlike mutations that disrupt furrow initiation and progression (Heberlein et al., 1993; Ma et al., 1993), gp150 mutations do not cause an apparent size reduction of the eye (Figure 3C). These results suggest that gp150 mutations do not disrupt initiation or progression of the MF.
To find out whether cells recruited at later stages of ommatidial assembly are affected by gp150 mutations, expression of a homeodomain protein BarH1 was examined. In wild-type eye discs, BarH1 is specifically expressed in R1 and R6 cells (Figure 4I; Higashijima et al., 1992). In the mutant, most R1/R6 cells are irregularly positioned (Figure 4J). Their abnormal arrangement further indicates defective ommatidial organization. Moreover, some ommatidia contain additional or fewer R1/R6 cells (Figure 4J) and cone cells (Figure 4L). Thus, phenotypic analysis of gp150 mutant eye discs indicates that gp150 plays a critical role in regulating ommatidial patterning and assembly in the developing eye.
Gp150 is expressed at high levels within cells of the MF region and is localized to intracellular vesicles
To characterize the normal pattern of Gp150 expression in the developing eye, Gp150 antibodies were generated and used for immunostaining experiments. Specificity of the Gp150 antibody was confirmed by the absence of staining in gp1504 eye discs (Figure 5B) and the increased staining in GMR-gp150 eye discs (Figure 5C). In wild-type eye discs, cells immediately anterior to and within the furrow region express high levels of Gp150 protein (Figure 5A and D1). Gp150 proteins accumulate in many small vesicles throughout the apical–basal axis in the furrow region. The area with high levels of Gp150 mostly overlaps with cells expressing the proneural gene ato (Figure 5D1–3). This elevated expression of Gp150 in the furrow region is consistent with the notion that Gp150 is required to regulate early ommatidial development. Posterior to the furrow, lower levels of Gp150 were detected around each individual ommatidium (Figure 5A). The Gp150 protein was not detectable in the nucleus or at the plasma membrane of the eye disc cells.
To examine how Gp150 might distribute in subcellular compartments, we used a Myc-tagged non-functional Fringe protein as a Golgi-specific marker (Munro and Freeman, 2000). Partial overlap between Gp150- and Fringe-positive vesicles was observed in both sca-Gal4/UAS-fringeADD-myc and GMR-Gal4/UAS-fringeADD-myc eye discs (data not shown). Using a green flourescent protein (GFP)-tagged Drosophila Rab7 protein as a marker for late endosomes (Entchev et al., 2000), we also observed that Gp150 can be partially localized to endosomes (data not shown). Thus, these results support the idea that Gp150 is involved in regulating vesicle trafficking.
gp150 genetically interacts with Dl and N
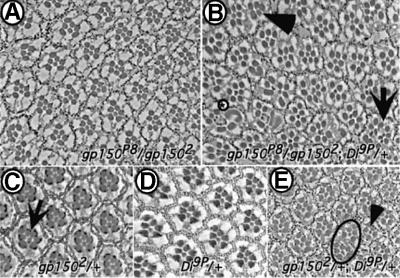
The gp150 mutant eye phenotypes closely mimic those caused by defective Dl–N signaling (Cagan and Ready, 1989; Baker et al., 1990; Parks et al., 1995). Thus, Gp150 might be functionally connected to the N pathway. To investigate this possibility, two assays were devised to test genetic interactions between gp150 and Dl and N. In a genetic sensitization assay, the gp150 function is partially removed in gp150P8/gp1502 flies, which allows us to monitor dominant effects of mutations in other genes on gp150 mutant eye phenotypes. Reduction of Dl function strongly enhanced the gp150 mutant eye phenotypes (Figure 6A and B). There was an ∼2- to 3-fold increase in the number of mutant ommatidia in gp150P8/gp1502; Dl–/+ flies compared with the control (Table II). Although not as effective as Dl, N mutations exhibited a similar effect (Table II). These results suggest that Gp150 functions together with Dl and N to regulate ommatidial development. Using the same assay, mutations such as rlS–135, Ras1e1B and Ras1e2F did not effectively alter the gp150 mutant eye phenotypes (data not shown). Both Ras1 and a MAP kinase, which is encoded by the rolled (rl) gene, are key components of the RTK pathway.
Fig. 6. Genetic interactions between gp150 and Delta. Tangential sections of adult eyes are presented. (A) The gp150P8/gp1502 eye exhibits ∼19% (n = 578) mutant ommatidia. (B) Reduction of Dl function strongly enhances the gp150 mutant phenotypes. Up to 68% (n = 544) abnormal ommatidia, which contain more R cells (arrow), fewer R cells (arrowhead) or apparently fused ommatidial units, were found in gp150P8/gp1502; Dl9P/+ flies. Often rhabdomeres exhibit abnormal morphology (circled asterisk). (C) gp1502/+. The arrow points to an occasionally observed aberrant ommatidium. (D) Dl9P/+ flies, no mutant ommatidia can be found. (E) gp1502/+; Dl9P/+. The circle indicates a fused ommatidium and the arrowhead points to an ommatidium missing an outer R cell.
Table II. gp150 genetically interacts with Notch and Delta.
| Genotype | No. of ommatidia examined | % mutant |
|---|---|---|
| gp150P8/gp1502 | 578 | 19.2 |
| N55e11/+; gp150P8/gp1502 | 629 | 42.3 |
| gp150P8/gp1502; Dl9P/+ | 544 | 67.6 |
| gp150P8/gp1502; Dl6B/+ | 460 | 44.6 |
| Df(2R)02311/+ | 645 | 0.15 |
| gp1502/+ | 1051 | 0.19 |
| N55e11/+ | 599 | 0 |
| N55e11/+; gp1502/+ | 769 | 0.65 |
| Dl9P/+ | 655 | 0 |
| gp1502/+; Dl9P/+ | 852 | 2.70 |
| Dl6B/+ | 534 | 0 |
| gp1502/+; Dl6B/+ | 558 | 0.18 |
| Dl6B/+ (29°C) | 539 | 0.37 |
| gp1502/+; Dl6B/+ (29°C) | 330 | 1.52 |
For each genotype, 5–9 adult eyes were sectioned and scored. Flies were cultured at 25°C, unless indicated otherwise. Ommatidia that were fused or contained too many or too few rhabdomeres were counted as mutant.
Loss-of-function alleles of gp150 exhibit a very weak dominant eye phenotype as a small number of aberrant ommatidia were found in gp150 heterozygotes (Figure 6C; Table II). This is most likely due to haplo-insufficiency since a deficiency chromosome, in which gp150 is completely deleted, exhibits the same phenotype (Table II). In this second assay, Dl mutations also dominantly enhanced gp150 mutant eye phenotypes, as there was a >10-fold increase in the number of mutant ommatidia in gp1502/+; Dl9P/+ flies (Figure 6C–E; Table II). Compared with Dl, N mutations exhibited a weaker enhancing effect (Table II). Altogether, these genetic data suggest that Gp150 is involved in modulating Dl–N signaling in the eye.
Dl is co-localized with Gp150 in intracellular vesicles within cells of the MF region
To begin to investigate how Gp150 might act to modulate N signaling, we examined whether Dl and Gp150 proteins were co-localized. Cell surface and intracellular expression of Dl within cells of the developing eye has been characterized previously (Parks et al., 1995; Baker and Yu, 1998). Dl protein accumulates to higher levels in endosomal vesicles than on the plasma membrane. Some of these vesicles correspond to multi-vesicular bodies (MVBs) (Parks et al., 1995). Double-labeling experiments showed that Dl was co-localized with Gp150 in intracellular vesicles of cells within the furrow region (Figure 5E1–3). Moreover, Gp150 and Dl proteins were found to be co-localized in 60 out of 66 subcellular vesicles examined in S2 cells that were positive for both Gp150 and Dl expression (Figure 5F1–3). These results suggest that Gp150 may act in late endosomes to modulate the level or activity of Dl protein in the morphogenetic furrow region. Moreover, it is also possible that Gp150 is co-localized with Dl in secretory vesicles or recycling endosomes to facilitate Dl presentation to the plasma membrane.
Gp150 accumulation in intracellular vesicles is independent of Dynamin GTPase-mediated endocytosis
The accumulation of Dl in intracellular vesicles is dependent on Dynamin-mediated endocytosis, which is critical for N signaling (Parks et al., 1995, 2000). The Drosophila Dynamin GTPase is encoded by the shibire (shi) gene, which is required for endocytosis (Chen et al., 1991; Van der Bliek and Meyerowitz, 1991). In shits1 mutants, Dl proteins fail to be endocytosed and, therefore, accumulate at the plasma membrane (compare Figure 5E2 with G2; Parks et al., 1995). To test whether the accumulation of Gp150 in vesicles is also endocytosis dependent, expression of Gp150 was examined in heat-treated shits1 eye discs. Unlike Dl, vesicle localization of Gp150 was not apparently altered when shi function was removed (compare Figure 5E1 with G1). Thus, Gp150 proteins accumulate in intracellular vesicles in an endocytosis-independent manner.
gp150 mutations affect the expression of Dl and N proteins in the MF region
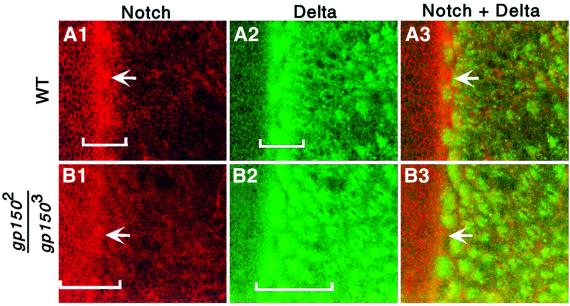
To examine whether loss of gp150 function affects levels and patterns of N and Dl expression in the developing eye, N and Dl antibodies were used for immunostaining experiments. In wild-type eye discs, N is ubiquitously expressed at the cell surface, with some N proteins being detected in subcellular vesicles (Figure 7A1). Cells in the MF accumulate relatively high levels of Notch. Interestingly, cells between Dl-positive clusters in the furrow exhibit even higher levels of N protein (Figure 7A1–3). N expression in the gp150 mutant appeared to be slightly expanded in the MF region and the elevated expression of N in cells between Dl-positive clusters is less apparent (Figure 7B1–3). Posterior to the furrow, high levels of Dl in ommatidial clusters persisted for ∼3–4 rows both at the cell surface and in endosomal vesicles in wild-type eye discs (Figure 7A2 and B3; Parks et al., 1995; Baker and Yu, 1998). However, in gp150 mutants, high levels of Dl were found in clusters distributed over 6–7 rows posterior to the furrow (Figure 7B2 and B3). In addition, regular spacing between Dl-positive clusters was disrupted. Thus, loss of gp150 function alters the expression pattern of Dl and N proteins and, in particular, the level of intracellular Dl appears to be negatively regulated by Gp150 in the furrow region.
Fig. 7. Expression of Delta and Notch in wild-type and gp150 mutant eye discs. Wild-type (A1–3) and gp1502/gp1503 mutant (B1–3) third instar eye discs were stained with anti-N (red in A1 and B1) and anti-Dl (green in A2 and B2) antibodies. Superimposed images are shown in (A3 and B3). Brackets indicate areas that show relatively higher N or Dl levels in the MF region. Arrows point to N-positive cells in between Dl-positive clusters in the MF. Anterior is to the left in all panels.
Discussion
LRR proteins exist in organisms ranging from plants to metazoan animals and constitute an evolutionarily conserved superfamily (for review see Kobe and Deisenhofer, 1994; Kajava, 1998). These proteins are found in different subcellular locations and function in a wide variety of processes such as cell adhesion and signal transduction. Based on our expression analysis using eye discs, Gp150 is exclusively located in intracellular vesicles, some of which are late endosomes and the Golgi apparatus. As Gp150 accumulation in intracellular vesicles does not depend on the Dynamin-mediated endocytic pathway, Gp150 appears to accumulate in the vesicles through an intracellular trafficking route.
A role of Gp150 in modulating Dl–N signaling during early ommatidial development
The development of hundreds of precisely organized ommatidia in the Drosophila compound eye begins with the specification and regular spacing of R8 cells, which become founder cells required for the sequential recruitment of other cell types. Our phenotypic analysis demonstrated that Gp150 plays a critical role during ommatidial development. In the absence of gp150 function, both the selection and patterning of the R8 cells become aberrant and a small number of ectopic R8 cells appear to be specified. The gp150 mutant eye phenotypes mimic what occurs in loss-of-function mutants of Dl and N (Cagan and Ready, 1989; Baker et al., 1990; Parks et al., 1995), suggesting that Gp150 might facilitate Dl–N signaling. Supporting this hypothesis, the reduction of Dl or N function dominantly enhanced the gp150 mutant eye phenotypes.
Clonal analysis was carried out to reveal in which cells the function of Gp150 is required. By examining phenotypically normal but genetically mosaic ommatidia, we found that gp150 could be either wild type or mutant in all R cells (R1–R8; data not shown). Therefore, the function of gp150 does not appear to be required in R cells for normal ommatidial development. Given that R cells are signal-sending cells, which prevent neural differentiation in neighboring precursor cells, this result is consistent with a view that gp150 might be required in the precursor cells, which are presumably Dl–N signal-receiving cells.
How might Gp150 modulate Dl–N signaling? So far, Gp150 is the only protein known to co-localize with Dl in intracellular vesicles. In a simple model, Gp150 might facilitate Dl presentation at the cell surface. Gp150 might also promote fusion between Dl-positive MVBs and lysosomes so that the intracellular levels of Dl can be reduced. The latter mechanism might be used to reduce the autonomous inhibitory effect of Dl on N signaling in the signal-receiving cells. In the absence of gp150 function, the levels of intracellular Dl increase, which autonomously blocks the N pathway in N signal-receiving cells and possibly makes these cells more responsive to instructive signals for R cell specification and differentiation, thus resulting in ommatidia with too many R cells. In tissues such as the wing and sensory organs, Dl has been shown to autonomously inhibit N signaling in a signal-receiving cell (Jacobsen et al., 1998; de Celis and Bray, 2000). In some cases, increased intracellular levels of Dl might allow some Dl to be presented at the cell surface to initiate N signaling on adjacent cells. This would increase lateral inhibition and result in the formation of ommatidia with fewer R cells. The fewer R cell phenotype could also be due to an insufficient number of competent retinal precursor cells that are available for the competing ommatidial clusters during R cell recruitment in gp150 mutants. In any event, we do not exclude the possibility that Gp150 might use other mechanisms to modulate Dl–N signaling. For instance, the up-regulation of Dl in gp150 mutant eye discs might be due to impaired N signaling, as N signaling can often cause down-regulation of Dl expression in signal-receiving cells. Further work is needed to distinguish these possibilities.
N signaling is known to be regulated through mechanisms such as site-specific proteolysis and glycosylation (for review see Artavanis-Tsakonas et al., 1999; Mumm and Kopan, 2000). In addition, N is recognized and ubiquitylated by an E3 ubiquitin ligase Itch (Qiu et al., 2000), suggesting that N stability could be regulated through the proteasome-mediated pathway. An Itch-related Drosophila protein, Suppressor of deltex, also negatively regulates N signaling (Cornell et al., 1999). Another negative regulator of N, Numb, has recently been shown to be an endocytic protein (Santolini et al., 2000), suggesting that Numb regulation of N could occur in the endocytic pathway. The regulation of Dl is also complex. For instance, Dl is proteolytically processed so its extracellular domain can be released (Klueg et al., 1998; Qi et al., 1999). From this work, we propose that Gp150 might function in intracellular vesicles, which include endosomes, to adjust appropriate intracellular levels of the Dl protein for modulating N signaling.
As the DER RTK signaling pathway is required for cell specification in the eye, Gp150 might also affect DER-mediated inductive signaling. Our data do not support this model, as mutations in two major components of the DER pathway, Ras1 and MAPK, did not exhibit any apparent dominant effect on the gp150 mutant eye phenotypes. Moreover, reduction of gp150 function did not modify eye phenotypes caused by gain-of-function mutations such as sev-Ras1V12 and sev-raftor (data not shown).
Our functional analysis of Gp150 indicates that both the extracellular and intracellular domains of Gp150 are essential (Dhulkotia et al., 2000 and data not shown). The extracellular domain of Gp150 contains 18 LRR motifs that might provide a scaffold for mediating protein–protein interactions (Kobe and Deisenhofer, 1995). In addition, it has several unique structural features that are conserved during evolution (Dhulkotia et al., 2000). These features include specific potential N-glycosylation sites, cysteine motifs and acidic regions that flank the LRR region. Although the intracellular domain is short, it contains three conserved tyrosine phosphorylation motifs (Dhulkotia et al., 2000) that are potential targets of tyrosine kinases (e.g. Dsrc) and receptor tyrosine phosphatases (e.g. DPTP10D; Fashena and Zinn, 1997). The conserved tyrosine phosphorylation motifs might be used for interaction with SH2 domains of some docking proteins. Further studies are required to reveal the functional significance of these conserved structural motifs, which should help reveal mechanisms of Gp150 action in cell fate control and tissue morphogenesis.
Materials and methods
Genetics and histology
l(2)k11107 and l(2)k11120 (Torok et al., 1993) are referred to as gp1501 and gp1502, respectively. Based on the mutant eye and bristle phenotypes, these mutations were mapped to the 58D position of the second chromosome. All gp150 mutations fail to complement Df(2R)02311 (G.M.Rubin, University of California at Berkeley), a deletion of the 58D1-5 region and several other larger deficiencies (Kerrebrock et al., 1995). On the l(2)k11107 chromosome, there is another recessive rough eye mutation, which is not associated with gp150. We subsequently isolated two EMS alleles, gp1503 and gp1504, by a non-complementation assay from a screen of 6167 F2 lines. Mutant alleles such as gp1502 and gp1503 can be considered as genetic nulls. This is based on the fact that there are ∼35% (n = 522) mutant ommatidia in gp1502/gp1502 flies while the number is 27% (n = 532) in gp1502/Df(2R)02311, and there are ∼45% (n = 484) mutant ommatidia in gp1502/gp1503 flies while the number is 29% (n = 229) in gp1503/Df(2R)02311. In a local P transposon-hopping experiment, we isolated nine gp150 P alleles (P2, P6, P8, P10, P11, P12, P13, P15 and P16) by inducing transposition of P[lacW] inserted at 58E1-2 on the l(2)k08316 chromosome (Torok et al., 1993). The mutant phenotypes can be frequently reverted to wild type by P-element excision in four of the six tested P alleles (P2: 81%, n = 94; P6: 0%, n = 37; P8: 86%, n = 86; P10: 69%, n = 42; P15: 39%, n = 56; P16: 0%, n = 90). P-element transposition was induced by introducing a chromosome carrying a transposase gene (Robertson et al., 1988). FRT42 gp1502 and FRT42 gp1503 chromosomes were generated through meiotic recombination. Mutant clones for gp150– were produced as described (Xu and Rubin, 1993). Other mutations used in this work include N55e11, Dl9P, Dl6B (a temperature-sensitive allele), Ras1e1B, Ras1e2F, rlS-135, shits1, sev-Ras1V12 and sev-raftor. The P[UAS-fngADD-myc] line was a gift from M.Freeman, MRC Laboratory of Molecular Biology, Cambridge, UK and the P[UAS-GFP-DmRab7] line was a gift from M.Gonzalez-Gaitan, Max Planck Institut fur biophysikalische Chemie. Scanning electron microscopy (SEM) and adult eye sections were conducted as described (Lai et al., 1996). Plastic sections were analyzed using a Zeiss Axioplan compound light microscope.
Molecular analysis and germline transformation
Plasmid rescue was performed to isolate the genomic sequence flanking the P insertion site in both P2 and P8 alleles. Three cDNA clones (DDBJ/EMBL/GenBank accession No. AY029705) were isolated after screening 7.2 × 105 recombinants from an eye disc cDNA library (G.M.Rubin). Sequence analyses demonstrated that a deduced full-length protein is identical to Gp150 (Tian and Zinn, 1994). A 16 kb sequence was cloned from a genomic DNA library (K.Moses, Emory University School of Medicine). The gp150-coding region is restricted to a 4 kb sequence containing the third, fourth, fifth and sixth exons. The first (46 bp) and second (72 bp) exons were found 7220 bp upstream of the coding region. To overexpress gp150 in eye discs, a 4.2 kb full-length gp150 cDNA was cloned into pGMR vector (Hay et al., 1994), which drives gene expression in all cells posterior to the MF. Five independent P[GMR-gp150] were produced through germline transformation (Rubin and Spradling, 1982). Standard methods for DNA analysis were used (Sambrook et al., 1989).
Immunocytochemistry and western blot analysis
Mouse polyclonal anti-Gp150 serum was made against the N-terminal region (amino acids 5–192) of Gp150. The serum was diluted 400-fold in a 0.1 M phosphate buffer pH 7.2 with 0.1% Triton X-100 for immunostaining. Third instar eye discs were dissected and fixed in a paraformaldehyde–lysine–phosphate buffer (Tomlinson and Ready, 1987) for 45 min on ice, which was followed by incubation with the anti-Gp150 antibody overnight at 4°C. Texas Red-conjugated anti-mouse IgG was the secondary antibody (Vector Laboratories). For double-labeling experiments, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG and FITC-conjugated anti-guinea pig IgG secondary antibodies were used (Vector Laboratories). Images were collected on a Bio-Rad MRC-1024 confocal laser scanning system. Other primary antibodies include mouse anti-Boss (used in 1:2000 dilution; S.L.Zipursky, University of California at Los Angeles), mouse anti-Cut (1:10) and mouse anti-Elav (1:400; G.M.Rubin), mouse anti-Hairy (1:1; S.Carroll, University of Wisconsin), rabbit anti-Myc (1:500; Santa Cruz Biotechnology, Inc.), mouse anti-Notch (1:1000; C458.2H, the Developmental Studies Hybridoma Bank at the University of Iowa), guinea pig anti-Delta (1:3000; GP581, M.A.T.Muskavitch, Boston College, MA), rabbit anti-Ato (1:10 000; Y.N.Jan, University of California at San Francisco) and rabbit anti-BarH1 (1:100; K.Saigo, University of Tokyo). Biotinylated secondary antibodies for mouse and rabbit IgG and the Vectastain ABC kit (Vector Laboratories) were used for signal detection. FITC-conjugated phalloidin (1.3 µM; Sigma) was used for detecting actin filaments that outline apical cell surface. The shits1 mutants were heat treated at 29°C for 7 h before their third instar eye discs were dissected for antibody staining.
For western analysis, the Gp150 antibody (1:1000) was used to probe a blot containing protein extracts of eight pairs of third instar larval eye discs/brain tissues for each lane. To determine whether equal amounts of proteins were loaded in each lane, the same blot was stripped and reprobed for the 70 kDa Fascilin I (Fas I) protein by anti-Fas I antibody (1:3000; mAb6D8; C.Goodman, University of California at Berkeley). A peroxidase-labeled anti-mouse IgG was the secondary antibody (Boehringer Mannhein) and the BM chemiluminescence blotting kit (Boehringer Mannhein) was used for signal detection.
Cell culture and DNA transfection
Drosophila Schneider S2 cells were grown in Schneider’s medium supplemented with 10% fetal bovine serum (Gibco-BRL). According to the instructions of the Tfx™-20 reagent (Promega), 106 cells were transfected with 2 µg of each expression construct. Both pmt-gp150myc (Fashena and Zinn, 1997) and pmt-Delta (Fehon et al., 1990) were induced via addition of 0.7 mM CuSO4 for 24 h. Cells were fixed and labeled as in Fehon et al. (1990). Primary antibodies include anti-Delta antibody (Gp581; 1:3000) and anti-Myc ascites (1:1000; G.M.Rubin).
Acknowledgments
Acknowledgements
We thank S.Artavanis-Tsakonas, N.Baker, S.Carroll, M.Freeman, M.Gonzalez-Gaitan, C.Goodman, Y.N.Jan, I.Kiss, T.Laverty, K.Moses, R.Ordway, T.L.Orr-Weaver, G.M.Rubin, K.Saigo, J.Treisman, K.Zinn, S.L.Zipursky, the Bloomington Drosophila Stock Center, the Developmental Studies Hybridoma Bank at the University of Iowa and especially M.A.T.Muskavitch for fly stocks and reagents. We also thank A.Unni, D.Hurtado and H.-B.Tang for technical assistance, C.Keller and J.Stains for help with the confocal images, C.Gay for providing the confocal image analysis system, G.Thomas for the fluorescence microscope and much thanks to D.Nguyen for comments on the manuscript and for technical and intellectual discussion. This work was supported by a March of Dimes Basil O’Connor Starter Scholar Research Award and a grant from the National Institute of Health (to Z.-C.L.)
References
- Artavanis-Tsakonas S., Rand,M.D. and Lake,R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- Baker N.E. and Zitron,A.E. (1995) Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenic furrow by scabrous. Mech. Dev., 49, 173–189. [DOI] [PubMed] [Google Scholar]
- Baker N.E. and Yu,S.-Y. (1997) Proneural function of neurogenic genes in the developing Drosophila eye. Curr. Biol., 7, 122–132. [DOI] [PubMed] [Google Scholar]
- Baker N.E. and Yu,S.-Y. (1998) The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech. Dev., 74, 3–14. [DOI] [PubMed] [Google Scholar]
- Baker N.E., Mlodzik,M. and Rubin,G.M. (1990) Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encodes by scabrous. Science, 250, 1370–1377. [DOI] [PubMed] [Google Scholar]
- Baker N.E., Yu,S. and Han,D. (1996) Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol., 6, 1290–1301. [DOI] [PubMed] [Google Scholar]
- Baonza A. and Freeman,M. (2001) Notch signaling and the initiation of neural development in the Drosophila eye. Development, 128, 3889–3898. [DOI] [PubMed] [Google Scholar]
- Baonza A. Casci,T. and Freeman,M. (2001) A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol., 11, 396–404. [DOI] [PubMed] [Google Scholar]
- Brown N.L., Sattler,C.A., Paddock,S.W. and Carroll,S.B. (1995) Hairy and EMC negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell, 80, 879–887. [DOI] [PubMed] [Google Scholar]
- Cagan R.L. and Ready,D.F. (1989) Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev., 3, 1099–1112. [DOI] [PubMed] [Google Scholar]
- Chen C.-K. and Chien,C.-T. (1999) Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc. Natl Acad. Sci. USA, 96, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.S., Obar,R.A., Schroeder,C.C., Austin,T.W., Poodry,C.A., Wadsworth,S.C. and Vallee,R.B. (1991) Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature, 351, 583–586. [DOI] [PubMed] [Google Scholar]
- Cornell M., Evans,D.A.P., Mann,R., Fostier,M., Flasza,M., Monthatong,M., Artavanis-Tsakonas,S. and Baron,M. (1999) The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is a E3 class ubiquitin ligase. Genetics, 152, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J.F. and Bray,S.J. (2000) The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development, 127, 1291–1302. [DOI] [PubMed] [Google Scholar]
- Dhulkotia D., Nguyen,D. and Lai,Z.-C. (2000) Evolutionary conservation of the leucine-rich repeat transmembrane protein Gp150 in Drosophila and Bombyx. Dev. Genes Evol., 210, 145–150. [DOI] [PubMed] [Google Scholar]
- Dokucu M.E., Zipursky,S.L. and Cagan,R.L. (1996) Atonal, Rough and the resolution of proneual clusters in the developing Drosophila retina. Development, 122, 4139–4147. [DOI] [PubMed] [Google Scholar]
- Entchev E.V., Schwabedissen,A. and Gonzalez-Gaitan,M. (2000) Gradient formation of the TGF-β homolog Dpp. Cell, 103, 981–991. [DOI] [PubMed] [Google Scholar]
- Fashena S.J. and Zinn,K. (1997) Transmembrane glycoprotein gp150 is a substrate for receptor tyrosine phosphatase DPTP10D in Drosophila cells. Mol. Cell. Biol., 17, 6859–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R.G., Kooh,P.J., Rebay,I., Regan,C.L., Xu,T., Muskavitch,M.A.T. and Artavanis-Tsakonas,S. (1990) Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF homologous genes in Drosophila. Cell, 61, 523–534. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1997) Cell determination strategies in the Drosophila eye. Development, 124, 261–270. [DOI] [PubMed] [Google Scholar]
- Gho M., Bellaiche,Y. and Schweisguth,F. (1999) Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development, 126, 3573–3584. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolf,T. and Rubin,G.M. (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development, 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- Heberlein U. and Moses,K. (1995) Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell, 81, 987–990. [DOI] [PubMed] [Google Scholar]
- Heberlein U., Wolff,T. and Rubin,G.M. (1993) The TGFβ homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell, 75, 913–926. [DOI] [PubMed] [Google Scholar]
- Higashijima S. et al. (1992) Dual Bar homeobox genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev., 6, 50–60. [DOI] [PubMed] [Google Scholar]
- Jacobsen T.L., Brennan,K., Martinez Arias,A. and Muskavitch,M.A.T. (1998) Cis-interactions between Delta and Notch modulate neurogenic signaling in Drosophila. Development, 125, 4531–4540. [DOI] [PubMed] [Google Scholar]
- Jan Y.N. and Jan,L.Y. (1995) Maggot’s hair and bug’s eye: role of cell interactions and intrinsic factors in cell fate specification. Neuron, 14, 1–5. [DOI] [PubMed] [Google Scholar]
- Jarman A.P., Grell,E.H., Ackerman,L., Jan,L.Y. and Jan,Y.N. (1994) atonal is the proneural gene for Drosophila photoreceptors. Nature, 369, 398–400. [DOI] [PubMed] [Google Scholar]
- Jarman A.P., Jan,L.Y. and Jan,Y.N. (1995) Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development, 121, 2019–2030. [DOI] [PubMed] [Google Scholar]
- Kajava A.V. (1998) Structural diversity of leucine-rich repeat proteins. J. Mol. Biol., 277, 519–527. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W., Moore,D.P., Wu,J.S. and Orr-Weaver,T.L. (1995) Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell, 83, 247–256. [DOI] [PubMed] [Google Scholar]
- Klueg K.M., Parody,T.R. and Muskavitch,M.A.T. (1998) Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol. Biol. Cell, 9, 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci., 19, 415–421. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature, 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Kramer H., Cagan,R.L. and Zipursky,S.L. (1991) Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature, 352, 207–212. [DOI] [PubMed] [Google Scholar]
- Lai Z.-C., Harrison,S., Karim,F., Li,Y. and Rubin,G.M. (1996) Loss of tramtrack gene activity results in ectopic R7 cell formation, even in a sina mutant background. Proc. Natl Acad. Sci. USA, 93, 5025–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-C., Hu,X. and Baker,N.E. (1996) The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol. Cell. Biol., 16, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. and Baker,N.E. (2001) Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr. Biol., 11, 330–338. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P., Yu,S.-Y., Delidakis,C. and Baker,N.E. (1998) A subset of Notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development, 125, 2893–2900. [DOI] [PubMed] [Google Scholar]
- Ma C., Zhou,Y., Beachy,P.A. and Moses,K. (1993) The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell, 75, 927–938. [DOI] [PubMed] [Google Scholar]
- Mumm J.S. and Kopan,R. (2000) Notch signaling: from the outside in. Dev. Biol., 228, 151–165. [DOI] [PubMed] [Google Scholar]
- Munro S. and Freeman,M. (2000) The Notch signaling regulator Fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DxD. Curr. Biol., 10, 813–820. [DOI] [PubMed] [Google Scholar]
- Parks A.L., Turner,F.R. and Muskavitch,M.A.T. (1995) Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech. Dev., 50, 201–216. [DOI] [PubMed] [Google Scholar]
- Parks A.L., Klueg,K.M., Stout,J.R. and Muskavitch,M.A.T. (2000) Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development, 127, 1373–1385. [DOI] [PubMed] [Google Scholar]
- Powell P.A., Wesley,C., Spencer,S. and Cagan,R.L. (2001) Scabrous complexes with Notch to mediate boundary formation. Nature, 409, 626–630. [DOI] [PubMed] [Google Scholar]
- Qi H.L., Rand,M.D., Wu,X.H., Sestan,N., Wang,W.Y., Rakic,P., Xu,T. and Artavanis-Tsakonas,S. (1999) Processing of the Notch ligand Delta by the metalloprotease Kuzbanian. Science, 283, 91–94. [DOI] [PubMed] [Google Scholar]
- Qiu L., Joazeiro,C., Fang,N., Wang,H.-Y., Elly,C., Altman,Y., Fang,D., Hunter,T. and Liu,Y.-C. (2000) Recognition and ubiquitination of Notch by Itch, a Hect-type E3 ubiquitin ligase. J. Biol. Chem., 275, 35734–35737. [DOI] [PubMed] [Google Scholar]
- Robinow S. and White,K. (1991) Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol., 22, 443–461. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Preston,C.R., Phillis,R.W., Johnson-Schlitz,D.M., Benz,W.K. and Engels,W.R. (1988) A stable genomic source of P element transposase in Drosophila melanogaster. Genetics, 118, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Santolini E., Puri,C., Salcini,A.E., Gagliani,M.C., Pelicci,P.G., Tacchetti,C. and Di Fiore,P.P. (2000) Numb is an endocytic protein. J. Cell Biol., 151, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S.-S. and Zinn,K. (1994) An adhesion molecule-like protein that interacts with and is a substrate for a Drosophila receptor-linked protein tyrosine phosphatase. J. Biol. Chem., 269, 28478–28486. [PubMed] [Google Scholar]
- Tomlinson A. and Ready,D.F. (1987) Neuronal differentiation in the Drosophila ommatidium. Dev. Biol., 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Torok T., Tick,G., Alvarado,M. and Kiss,I. (1993) P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics, 135, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J.E. and Heberlein,U. (1998) Eye development in Drosophila: formation of the eye field and control of differentiation. Curr. Top. Dev. Biol., 39, 119–159. [DOI] [PubMed] [Google Scholar]
- Treisman J.E., Luk,A., Rubin,G.M. and Heberlein,U. (1997) eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev., 11, 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Bliek A. and Meyerowitz,E.M. (1991) Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature, 351, 411–414. [DOI] [PubMed] [Google Scholar]
- Wolff T. and Ready,D.F. (1993) Pattern formation in the Drosophila retina. In Bate,M. and Martinez-Arias,A. (eds), The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]