Abstract
Many capsid proteins have peptides that influence their assembly. In hepatitis B virus capsid protein, the peptide STLPETTVV, linking the shell-forming ‘core’ domain and the nucleic acid-binding ‘protamine’ domain, has such a role. We have studied its morphogenic properties by permuting its sequence, substituting it with an extraneous peptide, deleting it to directly fuse the core and protamine domains and assembling core domain dimers with added linker peptides. The peptide was found to be necessary for the assembly of protamine domain-containing capsids, although its size-determining effect tolerates some modifications. Although largely invisible in a capsid crystal structure, we could visualize linker peptides by cryo-EM difference imaging: they emerge on the inner surface and extend from the capsid protein dimer interface towards the adjacent symmetry axis. A closely sequence-similar peptide in cellobiose dehydrogenase, which has an extended conformation, offers a plausible prototype. We propose that linker peptides are attached to the capsid inner surface as hinged struts, forming a mobile array, an arrangement with implications for morphogenesis and the management of encapsidated nucleic acid.
Keywords: core antigen/cryo-electron microscopy/hepatitis B virus/nucleic acid packaging/nucleocapsid morphogenesis
Introduction
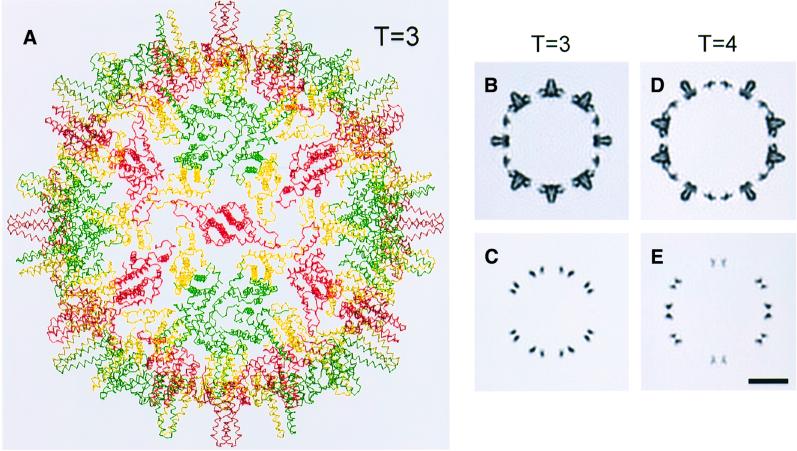
Hepatitis B virus (HBV) is responsible for liver disease on a massive scale (Blumberg, 1997). Its virion consists of a nucleocapsid enclosed within an envelope containing the viral glycoprotein, S-antigen. The capsid constitutes the ‘core’ antigen, and an unassembled variant of the same protein forms another major clinical antigen: e-antigen (Milich et al., 1995; Schodel et al., 1996). Nucleocapsids are fenestrated icosahedral particles of two sizes, with triangulation (T) numbers of 3 (180 subunits) and 4 (240 subunits), respectively (Crowther et al., 1994; Wingfield et al., 1995).
The capsid protein has a ‘core’ domain, and a ‘protamine’ domain (Petit and Pilot, 1985; Nassal, 1992). Polymerized core domains form the capsid shell, while the protamine domain, residues 150–183 and ∼50% arginine, binds nucleic acid, initially the RNA pregenome, and latterly, the DNA genome. Core domain forms dimers (Wingfield et al., 1995) that assemble efficiently into capsids, both in vivo and in vitro (Cohen and Richmond, 1982; Wingfield et al., 1995). The dimer has the overall shape of a capital T whose stem extends outwards as a spike and the tips of whose arms cluster together in five- or six-membered rings around the symmetry axes of the surface lattice (Crowther et al., 1994). The spike is a four-helix bundle of paired helical hairpins (Zlotnick et al., 1996; Conway et al., 1997), and the core domain fold has been determined (Böttcher et al., 1997; Wynne et al., 1999).
Cp140, the shortest core domain shown to form capsids, terminates at residue 140 (Salfeld et al., 1989; Birnbaum and Nassal, 1990), and the first arginine in the protamine domain is at position 150. The intervening nonapeptide is a linker connecting the two domains. The last residue that is sufficiently well ordered to be consistently seen in the crystal structure of the Cp149 capsid (core domain plus linker) is Thr142 (Wynne et al., 1999). However, there are indications that the rest of the linker peptide is not entirely disordered. First, its length and sequence are closely conserved (Bringas, 1997). Secondly, it is a major morphogenic determinant: progressive truncation from the C-terminus reduces the fraction of T = 4 capsids from ∼95% for Cp149 to ∼20% for Cp140 (Zlotnick et al., 1996).
The location of the linker peptide has been controversial. In RNA-free capsids with large moieties appended to the core domain, these moieties are accessible to antibodies (Stahl and Murray, 1989; Yoshikawa et al., 1993), implying that at least some of them are on the outside, and the protamine domain is susceptible to proteolysis by enzymes too large to pass through the holes in the surface lattice (Gallina et al., 1989). On the other hand, encapsidated RNA has been depicted as an inner shell (Crowther et al., 1994; Zlotnick et al., 1997), and it is plausible that protamine domains are bound to it. This consideration places the linker peptides inside the capsid. Moreover, cryo-electron microscopy (cryo-EM) observations of capsids labeled with heavy metal clusters attached to a cysteine appended at position 150 (Zlotnick et al., 1997; Cheng et al., 1999) indicate that this peptide is on the inside.
The goals of the present study were to define the location of the linker peptide and to further explore its properties. To determine the margin of the core domain and conversely, the N-terminal limit of the linker peptide, we examined the assembly properties of Cp138 and Cp139. To investigate the morphogenic effect of the linker, we substituted it with peptides of the same length—both by scrambling the native sequence and by replacing it with an entirely different sequence. In addition, we examined the effect of added linker peptide on in vitro assembly of Cp142, which lacks most of the linker, and the assembly of a linker-deleted construct in which the protamine domain is fused directly to the core domain. To localize the linker in core domain capsids, we used cryo-EM-based difference mapping between Cp149 and Cp140 particles, an approach used previously to visualize a peptide of similar length at the N-terminus of the core domain (Conway et al., 1998b). Finally, we searched the protein database for peptides with similar sequences in proteins whose structures have been solved, and found such a peptide in cellobiose dehydrogenase of Phanerochaete chrysosporium (Hallberg et al., 2000). This peptide, which is surface exposed and has an extended conformation, was docked into our difference densities. These observations are discussed in terms of the multiple functions attributed to the linker peptide: in morphogenesis; in serving as a spacer; and in organizing the interactions between the protein shell and nucleic acid during replication and transcription.
Results
Expression of Cp138 and Cp139
When expressed in Escherichia coli, Cp138 and Cp139 produced insoluble aggregates, unlike Cp140–Cp149, which form capsids and/or dimers. These aggregates resisted extraction with strong chaotropes such as 8 M guanidine hydrochloride but were readily solubilized by the anionic detergent SDS or sarkosyl (N-laurylsarcosine). Cp138 was purified by gel filtration in the presence of anionic detergent followed by reversed phase chromatography. The purified protein was dialyzed against 50 mM Tris–HCl pH 7.5, with 250 mM sodium chloride, giving an optically clear solution. The far-UV CD spectrum indicated ∼40% α-helix, not much less than the 55–60% of Cp144 capsids and dimers (Wingfield et al., 1995). Sedimentation velocity analysis indicated some size heterogeneity, with 80% of the material having a median S value of 6.8, corresponding to a mass of ∼100 kDa, and 10–15% sedimenting at 11.2S. In comparison, Cp149 dimers sediment at 2.5S. These results suggest that Cp138 is capable of folding into a conformation resembling that of native core domain and of oligomerizing, but not of ordered polymerization into capsids.
Assembly and visualization of Cp140 capsids
Expression of Cp140 in E.coli followed the usual pattern, i.e. most of the protein assembled into particles that elute separately from the bulk of soluble bacterial protein during gel filtration (Zlotnick et al., 1996). On shortening the linker, we have observed a progressive increase in the amount of unassembled capsid protein from essentially zero for Cp149 to 10–15% for Cp140. It was our experience that Cp140 is more liable to aggregation than other truncated constructs, e.g. Cp142–Cp149; nevertheless, by not over-concentrating and appropriately adjusting the buffer conditions, especially controlling the pH and including 50 mM Mg2+, we were able to assemble and purify capsids.
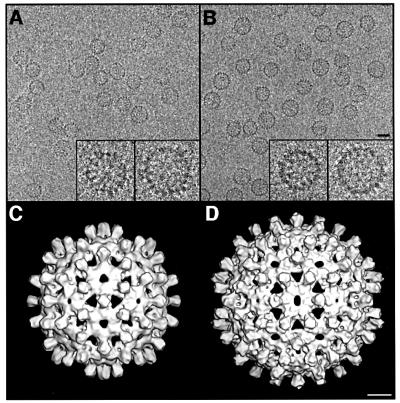
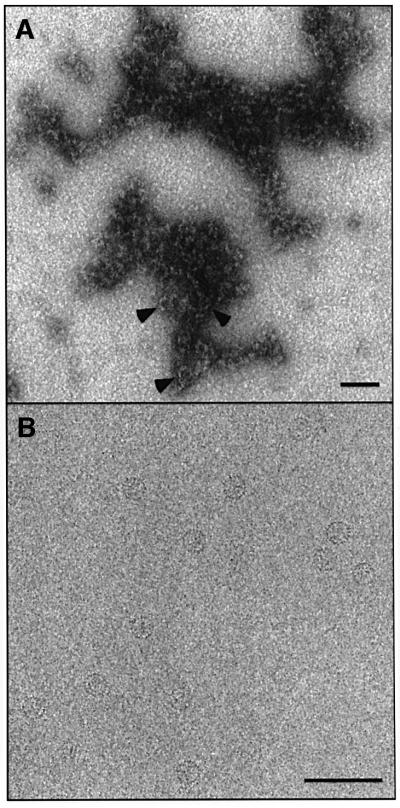
A cryo-micrograph of Cp140 capsids is shown in Figure 1A, and compared with a field of Cp149 capsids (Figure 1B): as expected for Cp140, the smaller T = 3 capsids predominate. Cp140 produces a higher incidence of malformed or incomplete capsids than do other Cp constructs; nevertheless, enough intact capsids of both sizes were imaged to provide adequate data sets for reconstruction. The final density map of the T = 3 capsid (Figure 1C) combined 914 particles. In parallel, we reconstructed the T = 4 capsid (Figure 1D), although fewer particles were available (292 were eventually combined). These density maps were calculated to 10 Å resolution and in both, the four α-helices per spike (center-to-center spacing, ∼10 Å) were resolved in transverse sections (data not shown).
Fig. 1. (A and B) Cryo-electron micrographs of Cp140 and Cp149 capsids. Bar = 250 Å. In each case, examples of a T = 3 and a T = 4 capsid are shown at higher magnification (×3) in the inserts. (C and D) Surface renderings of the outside of Cp140 capsids: (C) T = 3; (D) T = 4. Bar = 50 Å.
Visualization of linker peptides by difference imaging
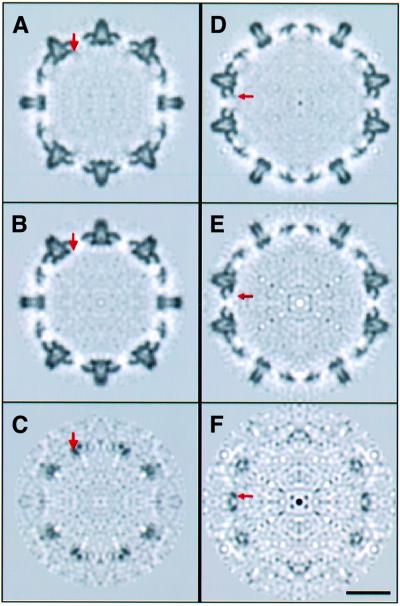
To identify density specifically associated with the linker peptide, we performed difference imaging between Cp149 and Cp140 capsids. Central sections through the two T = 3 maps are compared in Figure 2A and B, and for the T = 4 maps in Figure 2D and E. Because most Cp149 capsids are T = 4, in this case it was the T = 4 density map that had less noise. Each pair of sections matches up well, and corresponding sections through both difference maps consistently show positive density at specific sites on the inner surface (arrows in Figure 2C and F). These additional densities are not so conspicuous in the Cp149 maps because they lie in the interference fringe of ‘negative density’ that persists because of limitations in our procedure for correcting phase contrast effects and the limited resolution (i.e. Fourier truncation effect). In the difference maps, these fringes largely cancel out, making extra densities present on Cp149 more evident; the densities of interest are stronger than the residual noise peaks, and they are consistently observed at the same sites on independently calculated subunits.
Fig. 2. Central sections through T = 3 capsids viewed along a 2-fold axis. The red arrows mark one of the locations at which extra density is present in the Cp149 capsid (A) but not the Cp140 capsid (B). This density is also present at other quasi-equivalent, independently calculated, sites on the icosahedral surface lattice. These densities are more conspicuous in the corresponding section through the difference map (C). Bar = 100 Å. (D–F) show the corresponding sections for T = 4 capsids.
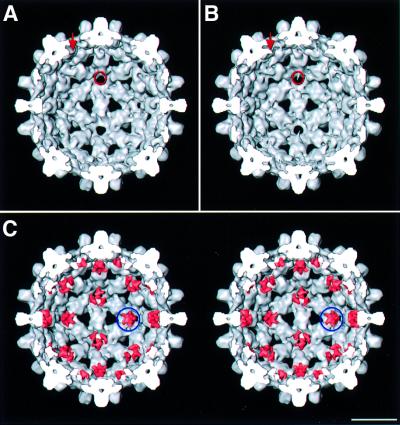
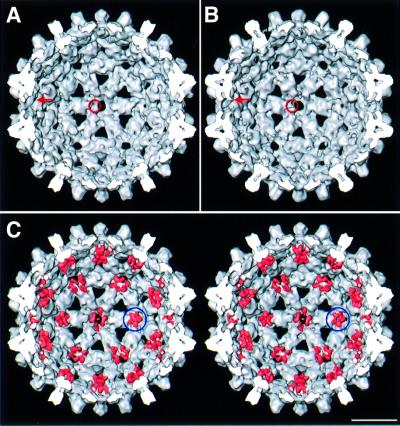
The extra densities on the inner surface of Cp149 capsids are discernible (cf. Figures 3A and B for T = 3 and 4A and B for T = 4), but are seen more readily when the difference densities are juxtaposed on the Cp140 capsids and rendered in a different color (pink). The 5-fold clusters at the vertices of the two capsids are virtually identical in shape and orientation (cf. ringed densities in Figures 3C and 4C), affording further evidence of their authenticity. Similar features are also present around the local 6-fold axes. Here, there is some suggestion of pair-wise interactions between neighboring linker peptides, but this aspect is near the noise level.
Fig. 3. (A and B) Interior surfaces of the T = 3 capsids of Cp140 and Cp149. Additional densities present on Cp149 are marked with a red arrow and ring. (C) Stereo rendering of the Cp140 capsid with the difference densities juxtaposed and contoured at the same level (pink). Difference density at the 5-fold is marked with a blue ring. Bar = 100 Å.
Fig. 4. (A and B) Interior surfaces of the T = 4 capsids of Cp140 and Cp149. Additional densities present on Cp149 are marked with a red arrow and a ring. (C) Stereo rendering of the Cp140 capsid with the difference densities juxtaposed and contoured at the same level (pink). Difference density at the 5-fold is marked with a blue ring. Bar = 100 Å.
Effect on assembly of permuting the linker peptide
One idea for how the linker exerts its morphogenic influence on the assembly of core domain capsids has been molecular crowding of these peptides as dimers congregate around symmetry axes of nascent capsids (Zlotnick et al., 1996): the larger this peptide, the higher the preponderance of 6-fold rings over 5-fold rings and consequently, the higher the proportion of T = 4 capsids. To test this hypothesis, we permuted the last seven amino acids of the linker to ETVPVLT, anticipating that a peptide of the same size and amino acid composition might behave in much the same way, according to this ‘space-filling’ hypothesis. The first two linker residues, having a defined conformation in the crystal structure, were left unchanged. A negatively stained electron micrograph of this protein, Cp142ETVPVLT, as extracted from E.coli, is shown in Figure 5A. There are very few capsids: the protein appears to be aggregating laterally, but not in a manner orderly enough to produce closed shells.
Fig. 5. Effects on assembly of substituting the last seven residues of linker peptide. (A) Cp142ETVPVLT, permuting the native sequence, substitutes a heptapeptide of the same size, net charge and polarity. Under conditions conducive to capsid assembly, most of the material forms aggregates, shown here by negative staining. In places, its appearance suggests a condensate of partially formed capsids. Very few complete capsids are present (arrowheads). (B) Substitution with the N-terminal heptapeptide. This construct, Cp142MDIDPYK, assembled normally, producing almost exclusively T = 4 particles as illustrated here by cryo-EM. Bar = 1000 Å.
Effect on assembly of deleting the linker peptide
Cp140 core domains lack the linker but are assembly competent (see above). To test whether this property is retained in the presence of the protamine domain, we created a construct, Cp183Δ143–149, in which the Cp140 core domain is fused directly to the protamine domain. If this protein were to assemble into capsids when expressed in E.coli, we would expect it to encapsidate bacterial RNA, and if the morphogenic proclivity of Cp140 is retained, these nucleocapsids should be mainly T = 3. However, Cp183Δ143–149 did not produce capsids. It may be that without the linker to serve as a spacer, the protamine domains, possibly with bound RNA, intrude into the inter-dimer interaction surfaces, subverting assembly.
Effect on assembly of substituting the linker peptide with the N-terminal peptide
We also performed an assembly experiment in which the linker was substituted by an entirely different peptide of the same size. Confronted with 207 options for residues 143–149, we chose to use the N-terminus of the same protein—MDIDPYK—on the grounds that this peptide does not sabotage assembly when situated at the other end of the polypeptide chain. The resulting construct, Cp142MDIDPYK, assembled adequately, as confirmed by negative staining (not shown). The T = 3:T = 4 ratio of these particles was determined by cryo-EM (e.g. Figure 5B). Essentially all intact capsids (i.e. >99%) were T = 4. When this material was dissociated to dimers with 2 M urea and reassembled in vitro, there were ∼90% T = 4 capsids and ∼10% T = 3 capsids, like Cp149. The latter observation also indicates that, as noted earlier (Zlotnick et al., 1996), in vitro assembly of capsid protein constructs can be affected by their previous history.
Effect on Cp140 assembly of added linker peptide
In principle, the morphogenic effect of linker peptide may involve both linker–linker interactions and interactions between linker and core domain. To test for the latter, we assembled purified Cp140 dimers (which normally produce ∼20% T = 4 capsids) in vitro in the presence of an excess of linker peptide. If linker–core interactions predominate, the peptide would be expected to increase the proportion of T = 4 peptides (>90% are produced by Cp149). However, no such effect was observed. The fraction of T = 4 capsids obtained with this experiment was unchanged according to both negative staining (21%; N = 1375) and cryo-EM (19%; N = 329). This experiment does not rule out linker–core domain interactions, but it implies that the linker has to be appropriately tethered to the core domain for such effects to be expressed.
A similar peptide in the crystal structure of cellobiose dehydrogenase
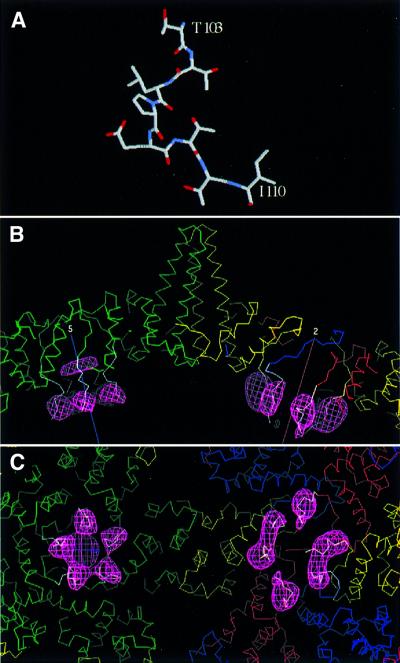
To ascertain whether any similar peptides are present in known structures, we scanned the SwissProt database with FASTA (Pearson and Lipman, 1988) and found such a peptide in residues 103–110 of cellobiose dehydrogenase (Hallberg et al., 2000). When its sequence, TTLPETTI, is compared with the first eight residues of the HBV linker, the central six are identical and the two flanking amino acids are chemically similar, i.e. T for S and I for V. In cellobiose dehydrogenase, this peptide is surface-exposed and has an extended conformation with no regular (i.e. α- or β-) secondary structure (Figure 6A).
Fig. 6. Modeling a peptide structure into linker-associated densities visualized by cryo-EM difference imaging. Residues 103–110 of cellobiose dehydrogenase (Hallberg et al., 2000) are almost completely identical to the linker sequences (A). In (B), this peptide is shown (white α-carbon trace) docked into the difference density (pink mesh) from the T = 4 capsids. Quasi-equivalent core domain subunits are shown in green, yellow, red and blue. Five-fold (at left) and 2-fold (at right) symmetry axes are marked. In (C), the view is from the inside of the capsid. We propose that a semi-flexible hinge allows the linker to pivot, so that it serves as a spacer that prevents the protamine domains from intruding too closely into inter-core domain contacts, and also confers mobility on the protamine domains’ interactions with nucleic acid.
Conformational analysis of linker peptides
We measured the CD spectra of two synthetic peptides corresponding to residues 139–149 and 141–149, respectively. Both spectra were indicative of random coil without detectable content of regular secondary structure (data not shown), emulating the cellobiose dehydrogenase peptide in this respect. We also tried to assess linker conformation in capsids by differential CD analysis between Cp149 and Cp142, but the results were inconclusive because they differ so little in size (5%).
Molecular modeling of difference densities
This peptide was grafted on to quasi-atomic capsid models obtained by fitting the atomic models of the dimers (Wynne et al., 1999) into our cryo-EM density maps of the T = 4 and T = 3 capsids (Figures 6B and C and 7A). The first three residues of the cellobiose dehydrogenase peptide were overlaid on the last three residues seen in the capsid protein (residues 141–143) and fitted well, confirming that they have similar conformations. This operation fixed the position of the rest of the peptide, which matched the observed difference densities quite well at each of the seven sites, i.e. it was at least partially inside or grazing them. The docking was then refined to position the peptides more centrally in the difference densities. Given their limited resolution, there was some ambiguity about how this should be done. The fits shown in Figure 6B and C were obtained by pivoting about residue 143, the putative hinge site (see Discussion), by angles that varied from ∼20° to ∼50°, depending on the quasi-equivalent site in question.
Fig. 7. (A) Quasi-atomic model of the T = 3 HBV Cp142 capsid. The model was created by docking dimers from the crystal structure of the T = 4 capsid into a cryo-EM map of the T = 3 capsid. (B and D) Central sections through quasi-atomic models of the T = 3 (B) and T = 4 (D) capsids. Cp142 capsids, restricted to 10 Å resolution, closely resemble the corresponding sections through the cryo-EM density maps of Cp149 and Cp140 (cf. Figures 2A, B, D and E). Corresponding sections through quasi-atomic models of the icosahedrally coordinated linker peptide (represented by the cellobiose dehydrogenase peptide, appropriately docked) are shown in (C) and (E) (cf. Figure 2C and F). Bar = 100 Å.
Discussion
Some capsid proteins have, in addition to a basic building block or core domain, peptides that regulate assembly and can affect the size of capsid produced. The N-terminus of papillomavirus L1 is one example (Chen et al., 2000). This mechanism is not exclusive to virus capsids and is also operative, for instance, in the assembly of bacterial flagellae (Namba, 2001). Other capsid proteins have positively charged extensions that bind nucleic acid (e.g. Vriend et al., 1986). HBV, with its C-terminal protamine domain connected by the linker peptide to the core domain, has both features. In this study, we have examined structural, conformational and morphogenic properties of its linker peptide.
Minimum-sized core domain for correct folding
It has previously been reported that Cp138 is not assembly competent (Birnbaum and Nassal, 1990; Beames and Lanford, 1993; Metzger and Bringas, 1998), and we have found that Cp139 shares this property. Moreover, a monoclonal antibody that bound to Cp140 failed to react with Cp138 (Salfeld et al., 1989), suggesting a different conformation. Our experiments were intended to determine whether this breakdown in assembly reflects a failure to produce functional building blocks or a failure of building blocks to form capsids. We found that Cp138 could assemble into soluble multimers, rich in α-helix, but not into capsids. This observation suggests that with Cp138, the dimer interface, the four-helix bundle, may be formed correctly, but incompleteness of its arms, which engage in the inter-dimer contacts, precludes assembly into capsids. Wynne et al. (1999) noted a snug interaction involving Leu140 in their crystal structure of Cp149 T = 4 capsids, and inferred that this was a key interaction to stabilizing the protein fold. Our data with the Cp138 and 139 are consistent with this proposal, which is further supported by the observation that the mutant Cp140.L140A is unstable and also produces insoluble aggregates when expressed in E.coli (P.T.Wingfield and S.J.Stahl, unpublished data).
Expression characteristics and capsid stability
The various HBV capsid protein constructs that we have expressed exhibit three physical states: capsids, dimers and aggregates. Whereas Cp183 (full-length protein) assembles efficiently into capsids, most shorter constructs leave some unassembled dimers, ranging up to 10–15% for Cp140. We use gel filtration to resolve capsids from dimers and soluble bacterial proteins. In general, the amount of unassembled protein correlates with instability of the parent capsid (P.T.Wingfield and N.R.Watts, unpublished results). Thus, Cp140 capsids are somewhat fragile and aggregation prone compared with the very stable Cp149 capsids. Instead of capsids, Cp138, Cp142ETVPVLT and Cp183Δ143–149 produce insoluble aggregates, typical of the inclusion bodies formed when many proteins are overexpressed in E.coli.
Location of the linker peptide
Prior evidence is strong that the linker is on the inside when attached to the protamine domain–RNA complex or metal clusters (see Introduction), but in the absence of such constraints, the data are less conclusive. Uncon strained linkers may, in principle, fluctuate between internal and external positions as in the ‘breathing’ motions attributed to capsid protein peptides of rhinovirus (Smith et al., 1993) and poliovirus (Li et al., 1994). In this context, our difference mapping visualizes extended, linker-associated, densities at specific sites on the inner surface. These linkers are in essentially the same position as when bound to undecagold (Zlotnick et al., 1997) or tetrairidium (Cheng et al., 1999), indicating that their placement reflects properties of the linker per se and not of the ligands. While these data do not rule out the possibility that linkers fluctuate between the inner and outer surfaces, they establish that the linkers spend most of the time in more-or-less defined positions on the inner surface.
Visualizing peptides in cryo-EM density maps at moderate resolution
The resolution of the density maps used in this study is ∼10 Å, and the resulting difference maps disclosed the linker peptide at all seven quasi-equivalent sites: three in the T = 3 capsid and four in the T = 4 capsid. The detail seen is limited, as expected for this resolution: positive connected densities are seen, with the correct attachment points at the C-termini of core domains; they are elongated and the direction of their long axes is apparent (Figures 4 and 6B).
Recalling that an octapeptide appended to the N-terminus of HBV core domain was detected by the same approach (Conway et al., 1998b; Böttcher et al., 1998), we suppose that peptides of similar size may, in general, be detected in cryo-EM difference maps of this resolution and quality (signal-to-noise ratio). This capability may be exploited to localize peptides in large complexes, either by comparing natural variants (e.g. species-specific variants), or by engineering in peptides at selected sites, provided that the recombinant proteins assemble correctly and are otherwise isomorphous. These requirements are experimentally testable, i.e. the difference map should be empty apart from the appended peptide.
Disorder and visibility
At first sight, it is paradoxical that a peptide may be visible, albeit in limited detail, in a cryo-EM density map but invisible in a crystal structure (Stewart et al., 1993). However, in cases of limited disorder, well-defined low resolution data offer an advantage because, if a structural element is sampling multiple conformations that occupy essentially the same space, it should be discernible in a cryo-EM map, but perhaps not in the corresponding crystal structure. The HBV linker peptide represents a case in point: it was detected in cryo-EM experiments, but most of it was not seen in the 3.3 Å crystal structure of the Cp149 capsid (Wynne et al., 1999), although data collection extended to relatively low resolution (39 Å) in that study. This does not necessarily mean that the peptide is disordered. An alternative possibility, discussed further below, is that the peptide has a defined structure and is connected to the core domain by a semi-flexible hinge.
Conformation of the linker peptide
Several observations suggest that the linker peptide has an extended conformation. First, it is nearly sequence-identical to a cellobiose dehydrogenase peptide that lies on the surface of that molecule and has low temperature factors, reflecting a well-defined conformation (Hallberg et al., 2000). The cellobiose dehydrogenase peptide has an extended structure with no α-helix or β-sheet content. Our CD spectra showed that isolated linker peptide also lacks regular secondary structure, with the caveat that a peptide in solution may adopt a different conformation from when it is bound to a folded protein. Secondly, the cellobiose dehydrogenase peptide fits well into our difference density maps calculated between Cp149 and Cp140 capsids. Optimal docking required only that the peptides be shifted slightly from their initial conformation specified by the two or three residues at which the crystal structures of the peptide and the capsid protein overlap.
To explain current data, we propose that there is a semi-flexible hinge in the linker peptide but it has an otherwise defined conformation. The resolving power of our difference maps is insufficient to pinpoint the hinge(s), although a good candidate is position 143 beyond which the linker is invisible in the crystal structure. Trans–cis isomerization of Pro144 is another possibility. The fact that visible difference densities extend as far as the docked peptides imply that the rest of the linker is structured and its mobility about the hinge is limited. The optimized fits also suggest that the linker may have different conformations at distinct quasi-equivalent sites, consistent with the ‘hinge’ hypothesis.
Capsid modeling shows an apparent enhanced mobility of linker peptide
To test this hypothesis, we performed a modeling experiment aimed to assess whether linker peptide has a higher temperature factor than the core domain and hence, greater mobility. To do so, we first docked the crystal structure of the core domain dimers (Wynne et al., 1999) into our cryo-EM maps (see Materials and methods). For the T = 4 capsid, this required movements of only a few Å from their positions in the capsid crystal structure. The resulting quasi-atomic model of the T = 3 capsid is shown in Figure 7A. We then restricted the resolution to 10 Å according to the procedure of Belnap et al. (1999), applying a temperature factor to optimize the models’ agreement with the cryo-EM density maps (Figure 7B and C, cf. Figure 2). The purpose of this temperature factor was to simulate the cumulative effect of several factors that cause cryo-EM amplitudes to fall off more rapidly than diffraction data towards higher resolution (e.g. Henderson, 1992; Conway and Steven, 1999). The same operation was applied to maps of the cellobiose dehydrogenase peptides in their docked positions. For them, a separate temperature factor was chosen to optimize their match with the cryo-EM difference maps (cf. Figures 7D and E, 2C and F). Best agreement was obtained by assigning a peptide temperature factor that was ∼3-fold higher than for the capsid, suggesting local disorder, consistent with the ‘hinge’ hypothesis.
Summary of morphogenic observations: linker peptide as a hinged strut
The linker peptide sequence tolerates some sequence changes, as attested by assembly of T = 4 capsids with the N-terminal peptide substituted as linker, but is not wholly undiscriminating, to judge by the assembly-negative experiment with the permuted linker. To influence assembly, the linker has to be tethered to the core domain, as the failure of added linker to influence Cp142 assembly implies. This observation also suggests that, apart from covalent tethering, the linker associates only weakly with the core domain, an inference that is consistent with its imputed mobility (see above). Inside nucleocapsids, hinge motions of the linker may facilitate transcription and retrotranscription by affording the protamine domains room for maneuver in organizing the pregenome and genome. With linkers hinging about their pivot points, protamine domains would form a mobile platform on which retrotranscription and transcription take place inside the capsid.
We note that a linker is required for intracellular assembly of core domains with the protamine domain attached, since the linker-deleted mutant, Cp183Δ143–149, failed to assemble. It is plausible that the linker facilitates assembly by ensuring that the protamine domain does not interfere with polymerization of dimers, i.e. it serves as a spacer.
Regulation of assembly
Unlike Cp142, Cp183Δ143–149 did not produce capsids, suggesting that assembly proceeds differently when the protamine domain and RNA are present. In addition to the linker serving as a spacer, it appears likely that the initiation step, i.e. formation of the first stable intermediate, is altered. For core domain assembly, kinetic data suggest that this complex is a trimer of dimers (Zlotnick et al., 1999). For HBV nucleocapsid assembly, on the other hand, a polymerase–RNA complex has been implicated as the initiator (Bartenschlager and Schaller, 1992; Pollack and Ganem, 1993). Such a mechanism has the advantage that it prevents the fruitless (for virus proliferation) expenditure of capsid protein in empty shells by lowering the critical concentration of protein required for initiation. On the other hand, just as most Cp149 capsids are T = 4, so are the majority of HBV nucleocapsids in infected human liver (Kenney et al., 1995), and of those obtained upon expressing the full-length protein in E.coli (Crowther et al., 1994). These observations imply that once assembly is initiated, the core domain and linker determine the size of shell that will be completed.
Materials and methods
Construction of HBV capsid protein genes
Variants of the HBV capsid proteins were produced in E.coli using plasmid pET11a and host bacterial strain BL21 (DE3) as described (Zlotnick et al., 1996). The DNAs encoding Cp138, Cp142 and Cp149 were prepared as NdeI–BamHI fragments using PCR as described by Scharf et al. (1986) and inserted between these same restriction sites into the expression vector pET11a. Cp138 refers to amino acids 1–138, etc. Plasmids directing the expression of Cp142MDIDPYK and Cp142ETVPVLT were constructed by replacing the SalI–BamHI fragment encoding amino acids 143–149 of wild-type Cp149 with synthetic DNA SalI–BamHI fragments encoding the variants above. The deletion (residues 143–149) in the DNA encoding Cp183 was introduced using PCR as described by Vallette et al. (1989). This mutant is referred to as Cp183Δ143–149.
Protein purification and capsid assembly
The various constructs were expressed in E.coli strain BL21 (DE3) using the vector pET11a (Zlotnick et al., 1996). Capsids were recovered from bacterial extracts by gel filtration on Sepharose 4B; these partially purified capsids were dissociated into dimeric protein using 1.5 M urea at pH 9.5 and further purified by a second round of gel filtration using Superdex 200 equilibrated with 100 mM sodium carbonate, 2 mM dithiothreitol pH 9.5 (Zlotnick et al., 1996). Capsid purification was also carried out without the alkaline pH dissociation step: here a second gel filtration step was performed using Sepharose 4B. Dimeric protein (stored in the above pH 9.5 buffer at –80°C) was reassembled into capsids by mixing protein (1 mg/ml) with 0.2 vol of 0.5 M mM HEPES, 2 M sodium chloride, 300 mM magnesium chloride, 10 mM dithiothreitol pH 5.0. Lowering the pH to neutrality and increasing the ionic strength induces assembly, while the presence of magnesium ion helps to alleviate the particle aggregation to which Cp140 is prone. Assembly was allowed to proceed for 1 h at 21°C. Particles were concentrated by ultrafiltration (Ultrafree-15; Millipore, Bedford, MA), then separated from unassembled protein by gel filtration on a 1 × 6 cm column of Sepharose CL-6B (Amersham Pharmacia Biotech, Piscataway, NJ) equilibrated with 20 mM HEPES, 50 mM sodium chloride, 50 mM 50 magnesium chloride pH 8.0. Protein concentration was determined by absorbance at 280 nm (using ε280 = 29 500 M–1 cm–1) corrected for light scattering. Peptides corresponding to HBV Cp residues 139–149 and 141–149 supplied by Multiple Peptide Systems (San Diego, CA) were >96% pure by RP-HPLC. The peptides were dissolved directly at 5 mg/ml in 50 mM sodium phosphate pH 7.2.
Circular dichroism
HBV capsid proteins were exchanged into 50 mM sodium phosphate pH 7.2 by gel filtration using Sephadex G 25 M (PD-10 columns, Amersham Pharmacia Biotech). Spectra of capsids (1 mg/ml) and peptides (2.5 mg/ml) were recorded using cells of path length 0.01 cm and a Jasco J-720 spectropolarimeter as described by Wingfield et al. (1995).
Analytical ultracentrifugation
Sedimentation equilibrium and velocity determinations were performed as described previously (Wingfield et al., 1995).
Cryo-EM and image reconstruction
Samples at 0.25 mg/ml were frozen rapidly in thin films of buffer suspended over holey carbon films and imaged with a CM200-FEG electron microscope (FEI, Mahwah, NJ) equipped with a Model 626 cryoholder (Gatan, Pleasanton, CA), as described (Cheng et al., 1999). Focal pairs of micrographs were recorded at 38 000× magnification, using low-dose techniques. Negatives were screened by optical diffraction. Four pairs of well stigmated and drift-free micrographs whose diffractograms of particle fields (not including carbon films) showed visible Thon fringes extending beyond (10 Å)–1 were digitized on a SCAI scanner (Z/I Imaging, Huntsville, AL) on a 7 µm raster (1.8 Å/pixel). These data were processed according to Conway and Steven (1999). Particles were picked with the semi-automatic X3D procedure: 2629 were T = 3 sized and 664 were T = 4 sized. The positions of the first contrast transfer function (CTF) zeros ranged from (16.6 Å)–1 to (18.7 Å)–1 in the first exposures and from (21.0 Å)–1 to (22.9 Å)–1 in the second exposures. CTF effects were deconvolved and focal pairs combined with CTFMIX. Particle orientations and origins were determined by iterative model-based refinement with the PFT algorithm (Baker and Cheng, 1996), using earlier reconstructions (Conway et al., 1997) as starting models. Density maps were calculated by Fourier–Bessel methods (Fuller et al., 1996), including data to 10 Å resolution. The Cp149 reconstructions have been described (Cheng et al., 1999). In calculating difference maps, the radial scaling and normalization of maps under comparison were matched according to Conway et al. (1998a).
Modeling
Crystal structure coordinates were fitted manually into electron density maps using the program O (Jones et al., 1991). First, dimers from the Cp149 crystal structure (1QGT in the Protein Data Bank) were fitted into the Cp147 T = 4 density map of Conway et al. (1997), with small shifts (root mean square deviation = 3.3 Å) to achieve an optimal fit. We also fitted the 1QGT dimers into a T = 3 Cp147 cryo-EM map at 10 Å resolution (Conway et al., 1998b). Next, Cα coordinates of residues 103–104 or 103–105 (with residues up to 110 attached) of the cytochrome domain of cellobiose dehydrogenase (1D7B in the Protein Data Bank) were overlaid onto residues 141–142 and 141–143, respectively, of the capsid protein. Where possible, the 1D7B peptide was moved as a rigid body to achieve maximal overlap with the Cp149–Cp140 difference densities. However, to optimize the fit of the linker peptides into the difference density, some pivoting was necessary. This could be done about any of several sites: in the fits shown in Figure 6, 1D7B residues 105–110 were pivoted about the Cα coordinates of residue 105. Renditions of these models at 10 Å resolution were calculated according to Belnap et al. (1999), using a (pseudo)temperature factor of 375 Å2 for the capsid model (beyond this value, the low-density region at the center of the spike filled in) and 1000 Å2 for the peptides. The latter value was chosen by visual matching of corresponding sections from the models and the difference maps for both T = 3 and T = 4.
Acknowledgments
Acknowledgements
We are grateful for the expert assistance of Ira Palmer and Joshua Kaufmann in the preparation of protein constructs. This work was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
References
- Baker T.S. and Cheng,R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol., 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R. and Schaller,H. (1992) Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J., 11, 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beames B. and Lanford,R.E. (1993) Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology, 194, 597–607. [DOI] [PubMed] [Google Scholar]
- Belnap D.M., Kumar,A., Folk,J.T., Smith,T.J. and Baker,T.S. (1999) Low-resolution density maps from atomic models: how stepping ‘back’ can be a step ‘forward’. J. Struct. Biol., 125, 166–175. [DOI] [PubMed] [Google Scholar]
- Birnbaum F. and Nassal,M. (1990) Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol., 64, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B.S. (1997) Hepatitis B virus, the vaccine and the control of primary cancer of the liver. Proc. Natl Acad. Sci. USA, 94, 7121–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B., Wynne,S.A. and Crowther,R.A. (1997) Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature, 386, 88–91. [DOI] [PubMed] [Google Scholar]
- Böttcher B., Dyson,M.R. and Crowther,R.A. (1998) Finding the small difference: A nine amino acid extension to the hepatitis B core protein. In Calderon Benevides,H. and Yacaman,M.J. (eds), Proceedings of the 14th International Congress on Electron Microscopy, Cancun. Vol. 1. Institute of Physics Publishing, Bristol, UK, pp. 737–738.
- Bringas R. (1997) Folding and assembly of hepatitis B virus core protein: A new model proposal. J. Struct. Biol., 118, 189–196. [DOI] [PubMed] [Google Scholar]
- Chen X.S., Garcea,R.L., Goldberg,I., Casini,G. and Harrison,S.C. (2000) Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell, 5, 557–567. [DOI] [PubMed] [Google Scholar]
- Cheng N., Conway,J.F., Watts,N.R., Hainfeld,J.F., Joshi,V., Powell,R.D., Stahl,S.J., Wingfield,P.T. and Steven,A.C. (1999) Tetrairidium, a four-atom cluster, is readily visible as a density label in three-dimensional cryo-EM maps of proteins at 10–25 Å resolution. J. Struct. Biol., 127, 169–176. [DOI] [PubMed] [Google Scholar]
- Cohen B.J. and Richmond,J.E. (1982) Electron microscopy of hepatitis B core antigen synthesized in E.coli. Nature, 296, 677–679. [DOI] [PubMed] [Google Scholar]
- Conway J.F. and Steven,A.C. (1999) Methods for reconstructing density maps of ‘single particles’ from cryoelectron micrographs to sub nanometer resolution. J. Struct. Biol., 128, 106–118. [DOI] [PubMed] [Google Scholar]
- Conway J.F., Cheng,N., Zlotnick,A., Wingfield,P.T., Stahl,S.J. and Steven,A.C. (1997) Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature, 386, 91–94. [DOI] [PubMed] [Google Scholar]
- Conway J.F., Cheng,N., Zlotnick,A., Stahl,S.J., Wingfield,P.T., Belnap,D.M., Kanngiesser,U., Noah,M. and Steven,A.C. (1998a) Hepatitis B virus capsid: localization of the putative immuno dominant loop (residues 78 to 83) on the capsid surface and implications for the distinction between c and e-antigens. J. Mol. Biol., 279, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Conway J.F., Cheng,N., Zlotnick,A., Stahl,S.J., Wingfield,P.T. and Steven,A.C. (1998b) Localization of the N terminus of hepatitis B virus capsid protein by peptide-based difference mapping from cryoelectron microscopy. Proc. Natl Acad. Sci. USA, 95, 14622–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R.A., Kiselev,N.A., Böttcher,B., Berriman,J.A., Borisova,G.P., Ose,V. and Pumpens,P. (1994) Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell, 77, 943–950. [DOI] [PubMed] [Google Scholar]
- Fuller S.D., Butcher,S.J., Cheng,R.H. and Baker,T.S. (1996) Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol., 116, 48–55. [DOI] [PubMed] [Google Scholar]
- Gallina A., Bonelli,F., Zentilin,L., Rindi,G., Muttini,M. and Milanesi,G. (1989) A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol., 63, 4645–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg B.M., Bergfors,T., Backbro,K., Pettersson,G., Henriksson,G. and Divne,C. (2000) A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Struct. Fold. Des., 8, 79–88. [DOI] [PubMed] [Google Scholar]
- Henderson R. (1992) Image contrast in high-resolution electron microscopy of biological macromolecules: TMV in ice. Ultramicroscopy, 46, 1–18. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kenney J.M., von Bonsdorff,C.H., Nassal,M. and Fuller,S.D. (1995) Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure, 3, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Li Q., Yafai,A.G., Lee,Y.M.-H., Hogle,J. and Chow,M. (1994) Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol., 68, 3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger K. and Bringas,R. (1998) Proline-138 is essential for the assembly of hepatitis B virus core protein. J. Gen. Virol., 79, 587–590. [DOI] [PubMed] [Google Scholar]
- Milich D.R., Sallberg,M. and Maruyama,T. (1995) The humoral immune response in acute and chronic hepatitis B virus infection. Springer Semin. Immunopathol., 17, 149–166. [DOI] [PubMed] [Google Scholar]
- Namba K. (2001) Poles of partly unfolded conformations in macromolecular self-assembly. Genes Cells, 6, 1–12. [DOI] [PubMed] [Google Scholar]
- Nassal M. (1992) The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol., 66, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.A. and Pilot,J. (1985) HBc and HBe antigenicity and DNA-binding activity of major core protein P22 in hepatitis B virus core particles isolated from the cytoplasm of human liver cells. J. Virol., 53, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J.R. and Ganem,D. (1993) An RNA stem–loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol., 67, 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salfeld J., Pfaff,E., Noah,M. and Schaller,H. (1989) Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol., 63, 798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S.J., Horn,G.T. and Erlich,H.A. (1986) Direct cloning and sequence analysis of enzymically amplified genomic sequences. Science, 233, 1076–1078. [DOI] [PubMed] [Google Scholar]
- Schodel F., Peterson,D. and Milich,D. (1996) Hepatitis B virus core and e antigen: Immune recognition and use as a vaccine carrier moiety. Intervirology, 39, 104–110. [DOI] [PubMed] [Google Scholar]
- Smith T.J., Olson,N.H., Cheng,R.H., Chase,E.S. and Baker,T.S. (1993) Structure of a human rhinovirus–bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc. Natl Acad. Sci. USA, 90, 7015–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S.J. and Murray,K. (1989) Immunogenicity of peptide fusions to hepatitis B virus core antigen. Proc. Natl Acad. Sci. USA, 86, 6283–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P.L., Fuller,S.D. and Burnett,R.M. (1993) Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J., 12, 2589–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallette F., Mege,E., Reiss,A. and Adesnik,M. (1989) Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res., 17, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G., Verduin,B.J. and Hemminga,M.A. (1986) Role of the N-terminal part of the coat protein in the assembly of cowpea chlorotic mottle virus. A 500 MHz proton nuclear magnetic resonance study and structural calculations. J. Mol. Biol., 191, 453–460. [DOI] [PubMed] [Google Scholar]
- Wingfield P.T., Stahl,S.J., Williams,R.W. and Steven,A.C. (1995) Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis and in vitro capsid assembly. Biochemistry, 34, 4919–4932. [DOI] [PubMed] [Google Scholar]
- Wynne S.A., Crowther,R.A. and Leslie,A.G. (1999) The crystal structure of the human hepatitis B virus capsid. Mol. Cell, 3, 771–780. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A. et al. (1993) Chimeric hepatitis B virus core particles with parts or copies of the hepatitis C virus core protein. J. Virol., 67, 6064–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A., Cheng,N., Conway,J.F., Booy,F.P., Steven,A.C., Stahl,S.J. and Wingfield,P.T. (1996) Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry, 35, 7412–7421. [DOI] [PubMed] [Google Scholar]
- Zlotnick A., Cheng,N., Stahl,S.J., Conway,J.F., Steven,A.C. and Wingfield,P.T. (1997) Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl Acad. Sci. USA, 94, 9556–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A., Johnson,J.M., Wingfield,P.W., Stahl,S.J. and Endres,D. (1999) A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry, 38, 14644–14652. [DOI] [PubMed] [Google Scholar]