Abstract
Insulin-like growth factor II receptor (IGF2R) is a multifunctional cell surface receptor implicated in tumour suppression. Its growth inhibitory activity has been associated with an ability to bind IGF-II. IGF2R contains 15 homologous extracellular domains, with domain 11 primarily responsible for IGF-II binding. We report a 1.4 Å resolution crystal structure of domain 11, solved using the anomalous scattering signal of sulfur. The structure consists of two crossed β-sheets forming a flattened β-barrel. Structural analysis identifies the putative IGF-II binding site at one end of the β-barrel whilst crystal lattice contacts suggest a model for the full-length IGF2R extracellular region. The structure factors and coordinates of IGF2R domain 11 have been deposited in the Protein Data Bank (accession codes 1GP0 and 1GP3).
Keywords: growth factor receptor/IGF-II/protein crystallography/tumour suppression
Introduction
The 300 kDa insulin-like growth factor II receptor (IGF2R, also known as the cation-independent mannose 6-phosphate receptor) is a type I transmembrane glycoprotein expressed ubiquitously in human tissues, with a truncated form of the receptor present in the circulation (Lobel et al., 1988; Oshima et al., 1988). Within IGF2R are distinct binding regions for both IGF-II and phosphomannosyl residues (Oshima et al., 1988; Schmidt et al., 1995), reflecting the multifunctional nature of IGF2R. Major functions include cycling of insulin-like growth factor II (IGF-II), the sorting of newly synthesized lysosomal enzymes and the endocytosis of extracellular lysosomal enzymes (reviewed in Kornfeld, 1992). Additionally, transforming growth factor-β1 (TGF-β1), a growth inhibitor, undergoes activation from its latent precursor via IGF2R binding (Dennis and Rifkin, 1991). IGF2R is also the receptor for the uptake of granzyme B, a serine protease essential for cytotoxic T-cell-induced apoptosis (Motyka et al., 2000).
Loss of IGF2R would be expected to lead to increased levels of circulating IGF-II, reduced levels of TGF-β1 and loss of granzyme B uptake, suggesting that IGF2R may be a growth inhibitor. In the mouse, IGF-II modifies cell survival and proliferation between days 9 and 10 of gestation (Burns and Hassan, 2001), with IGF-II signalling occurring via the IGF-I receptor (IGF1R). Disruption of Igf2r in the mouse results in increased levels of circulating IGF-II, leading to embryonic overgrowth and perinatal lethality. The phenotypic features of overgrowth are similar to defects in human growth, e.g. Beckwith– Weidemann syndrome, and to large offspring syndrome, which occurs following mammalian somatic cell cloning (reviewed in Burns and Hassan, 2001). Such mice are rescued when a second mutation eliminates either IGF1R or IGF-II.
In line with IGF2R functioning as a growth inhibitor, Igf2r may be a tumour suppressor gene, since mutations have been found in several cancers (reviewed in Falls et al., 1999). This hypothesis is supported by evidence of altered ligand binding as a consequence of Igf2r mutations found in hepatocellular and breast cancers (Devi et al., 1999). Whilst IGF-II is a potent regulatory peptide involved in fetal growth, it is only minimally involved in post-natal pathways. However, IGF-II is frequently overexpressed in certain cancers and is implicated in the growth of both human and murine tumours (Toretsky and Helman, 1996; Hassan and Howell, 2000). The Igf-II and Igf2r genes are imprinted, with loss of imprinting being one mechanism by which IGF-II is overexpressed (Falls et al., 1999). Clearly, interruption of IGF-II pathways offers a potential method of tumour control and IGF2R, which appears to be a natural ‘scavenger’ for IGF-II, is an obvious tool for such interruption.
The extracellular region of IGF2R comprises 15 repeating domains that, in addition to being internally homologous, show low sequence similarities (between 14 and 28% identity) to the extracellular domain of cation-dependent mannose 6-phosphate receptor (CDMPR) (Lobel et al., 1988), a nine-stranded β-barrel structure (Roberts et al., 1998). IGF-II binds to domain 11 of IGF2R (Schmidt et al., 1995), with domain 13 proposed to enhance binding by reducing the rate of IGF-II release from the receptor (Devi et al., 1998; Linnell et al., 2001). This enhancement may be associated with a domain 13-specific 43-residue insertion that shares ∼50% sequence identity with type II fibronectin domains (Lobel et al., 1988). Mannosylated proteins are bound by domains 3 and 9 (Dahms et al., 1993). A number of mutations have been characterized, including Ile1572Thr in domain 11 which abolishes IGF-II binding (Garmroudi et al., 1996).
We report here the crystallization of a truncated form of IGF2R comprising domain 11 only (IGF2R-Dom11), and the determination of its three-dimensional structure at a resolution of 1.4 Å based on experimentally derived phases from the anomalous scattering of sulfur. Structure solution based on phases derived from sulfur was originally demonstrated for crambin (Hendrickson and Teeter, 1981), but with modern X-ray sources and detectors it is becoming a convenient phasing method for small proteins.
Results and discussion
The structure of IGF2R-Dom11
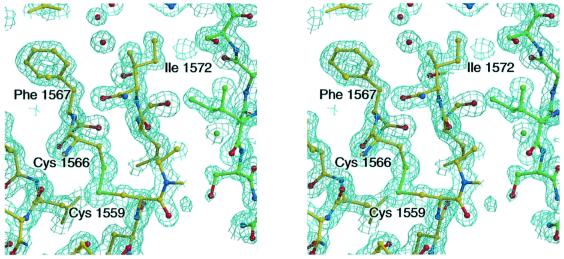
Table I summarizes the data collection and structure refinement statistics for two IGF2R-Dom11 crystal forms (tetragonal and orthorhombic at 1.4 and 1.95 Å resolution, respectively; see Materials and methods). A representative portion of the electron density for the refined 1.4 Å resolution structure is shown in Figure 1.
Table I. Data statistics.
| Data set | 2.5 Å (sulfur) | 1.6 Å | 1.4 Å | 1.95 Å |
|---|---|---|---|---|
| Space group | P43212 | P43212 | P43212 | P212121 |
| Wavelength (Å) | 1.771 | 0.934 | 0.933 | 0.934 |
| Resolution range (Å) | 20–2.5 | 20–1.6 | 20–1.4 | 30–1.95 |
| Measurements | 184 517 | 334 004 | 761 896 | 75 027 |
| Unique reflections | 5536 | 20 265 | 29 550 | 9 752 |
| Completeness (%)a | 100 (100) | 99.9 (99.0) | 98.7 (95.6) | 97.2 (92.9) |
| I/σ(I)a | 80.9 (31.9) | 58.2 (14.1) | 35.7 (9.4) | 21.9 (2.4) |
| Rmerge (%)a,b | 4.9 (10.8) | 6.3 (21.4) | 9.7 (45.2) | 9.0 (65.3) |
| Phasing power | 1.79 | |||
| Figure of merit | ||||
| before density modification | 0.36 | |||
| after density modification | 0.92 | |||
| R factor (%)c | 20.9 | 20.7 | ||
| Rfree (%)d | 23.7 | 25.5 | ||
| R.m.s.d. from ideal | ||||
| bonds (Å) | 0.013 | 0.005 | ||
| angles (°) | 1.662 | 1.330 | ||
| Average B factors (Å)2 | 22.49 | 35.31 | ||
| B r.m.s.d. (Å)2 | ||||
| main chain | 2.73 | 1.63 | ||
| side chain | 4.77 | 2.58 | ||
| Total number of non-hydrogen atoms | ||||
| protein | 1006 | 983 | ||
| water | 180 | 79 | ||
| other | 1 SO2–4 | – |
aValues in parentheses correspond to the highest resolution data shell (2.59–2.50 Å for the sulfur data set, 1.66–1.60 Å for the native 1.6 Å data set, 1.45–1.40 Å for the native 1.4 Å data set and 2.02–1.95 Å for the native 1.95 Å data set).
bRmerge = Σ|I – 〈I〉|/Σ〈I〉
cR factor = Σ||Fobs| – |Fcalc||/Σ|Fobs|
dRfree is as for R factor but calculated using a 5% test set of reflections excluded from the refinement.
Fig. 1. Stereodiagram of the final 2Fo–Fc electron density map for the tetragonal crystal form of IGF2R-Dom11, contoured at 1.7σ. Residues Ile1572 and Phe1567 are in the putative IGF-II binding site (hydrophobic patch 1; see text). A crystal contact molecule is shown as green balls-and-sticks demonstrating the packing of hydrophobic patch 1 against hydrophobic patch 2 (around Ile1627).
The model of IGF2R-Dom11 for the tetragonal crystal form comprises residues 1515–1647 (mature IGF2R numbering); seven N-terminal residues of the expressed molecule are disordered as are four C-terminal residues plus the seven residues of the His-tag. The model for the orthorhombic crystal form also comprises residues 1515– 1647, with the exception of loop residues Ala1618– Thr1621 where the electron density is not continuous. The root-mean-squared deviation (r.m.s.d.) between 129 equivalent Cα pairs in the two models is 0.27 Å.
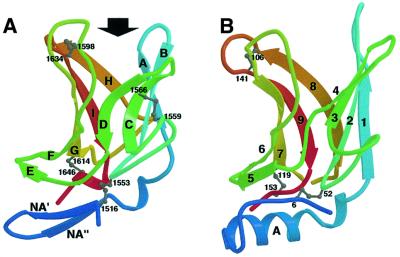
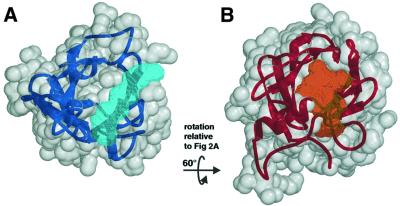
The core structure of IGF2R-Dom11 consists of a flattened β-barrel (Figure 2A), which, as expected from its sequence, is similar to that of CDMPR (Figure 2B). This shared β-barrel topology consists of nine β-strands, here termed βA–βI [equivalent to strands 1–9 in CDMPR as reported by Roberts et al. (1998)], consisting of two crossed β-sheets, the first formed by βA–βD and the second by βE–βI. The most significant structural differences between IGF2R-Dom11 and CDMPR lie in the N-terminal region preceding βA, where IGF2R-Dom11 contains two additional β-strands (βNA′ and βNA′′), which form a β-hairpin capping off the β-barrel. The equivalent region in CDMPR contains an α-helix. A structural superposition between IGF2R-Dom11 and CDMPR (PDB accession code 1C39) yields a r.m.s.d. between 106 equivalenced Cα pairs of 1.61 Å, representing 80 and 70% of the IGF2R-Dom11 and CDMPR structures, respectively. Comparison with the database of known structures using DALI (http://www.ebi.ac.uk/dali/) yields no other significant structural matches, although, as Roberts et al. (1998) note, there is a similarity between the topology of the CDMPR β-barrel and that of the avidin fold.
Fig. 2. Comparison of IGF2R-Dom11 and CDMPR structures. (A) Ribbon diagram of IGF2R-Dom11 showing the flattened β-barrel structure capped off by a β-hairpin. An arrow indicates the putative IGF-II binding region. Ribbons are coloured from blue at the N-terminus to red at the C-terminus and disulfide linkages are represented by grey balls-and-sticks. (B) Ribbon diagram of CDMPR (PDB accession code 1C39) coloured using the same scheme as in (A).
The core of the IGF2R-Dom11 β-barrel is formed primarily by hydrophobic residues (see Figure 3) including Leu1532, Val1548, Met1550, Ile1552, Val1574, Leu1581, Leu1588, Leu1590, Ile1610, Phe1612, Pro1624, Leu1636, Phe1638 and Trp1640, while residues Phe1527 and Leu1529 form the core of the β-hairpin. This hydrophobic nature of core residues is highly conserved in the CDMPR structure as well as in the sequences of the other IGF2R domains.
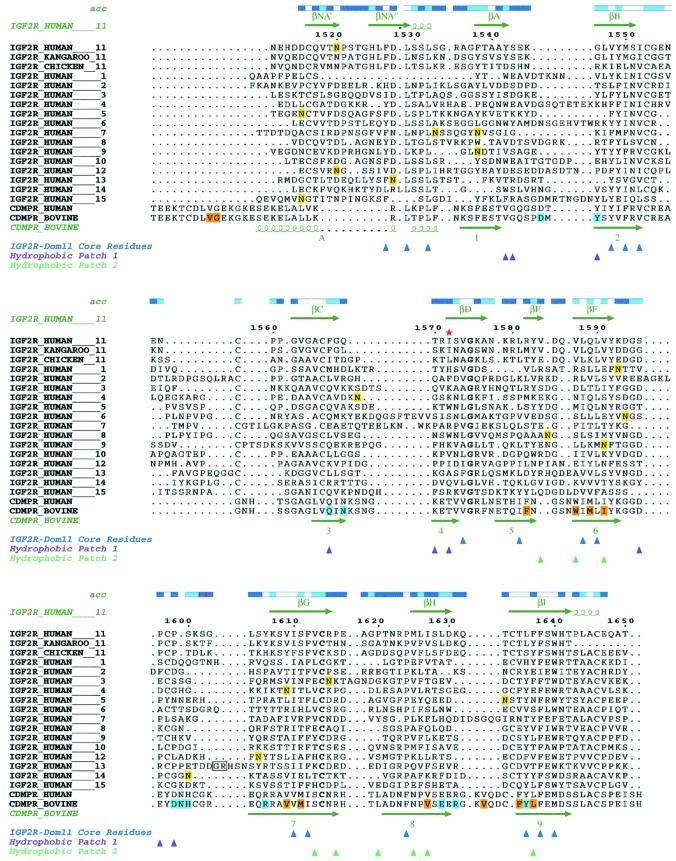
Fig. 3. Alignments of human, kangaroo and chicken IGF2R domain 11 sequences, human IGF2R domains 1–10 and 12–15 and human and bovine CDMPR. The IGF2R sequences were aligned using Clustal_W with manual modifications using Jalview, while the human IGF2R-Dom11 and bovine CDMPR were aligned based on structure. Numbering is that of mature human IGF2R-Dom11. Accessibilities of IGF2R-Dom11 residues are given above the alignments (blue, accessible; cyan, intermediate; white, buried). Secondary structure assignments for IGF2R-Dom11 and bovine CDMPR are displayed above and below the alignments, respectively. Potential glycosylation sites in IGF2R are highlighted in yellow, residues involved in the ligand binding pocket of CDMPR are highlighted in cyan and residues involved in CDMPR dimerization are highlighted in orange. In domain 13, the box indicates the residues between which the fibronection-like sequence is inserted. A red star indicates Ile1572 in the IGF-II binding site. Coloured triangles indicate residues of IGF2R-Dom11 that form the core (blue) or that contribute to the hydrophobic surface patches 1 (purple) and 2 (turquoise).
IGF2R-Dom11 contains four disulfides, three of which (Cys1516–Cys1553, Cys1598–Cys1634 and Cys1614– Cys1646) are in similar, although not identical, positions to the three disulfides in CDMPR (Cys6–Cys52, Cys106–Cys141 and Cys119–Cys153). The fourth disulfide (Cys1559–Cys1566) between the BC loop and βC is unique to the IGF2R-Dom11 structure. From sequence alignment this disulfide is predicted for all the IGF2R domains. The equivalent of Cys1614–Cys1646 is present in all IGF2R domains, while domain 15 lacks the equivalent of Cys1516–Cys1553 and domains 5 and 7 lack the equivalent of Cys1598–Cys1634.
The discontinuous density for the orthorhombic crystal form between Ala1618 and Thr1621 suggests that the GH loop may be flexible, although this region of density is clearly defined in the tetragonal crystal form where it is involved in a crystal contact. In comparison, the most dynamic region of CDMPR, loop A, is disordered in the presence of a small mannose 6-phosphate ligand but becomes more ordered in the presence of pentamannosyl phosphate (Roberts et al., 1998; Olson et al., 1999). The equivalent loop in IGF2R-Dom11 (loop AB) does not demonstrate the same flexibility, being several residues shorter.
The only N-linked glycosylation sequence motif in IGF2R-Dom11 is at the end of βNA′ (Asn1520–Pro1521– Ser1522) but the central proline and the low solvent accessibility of Asn1520 appears to preclude glycosylation.
Interaction surfaces of IGF2R-Dom11 reveal the putative IGF-II binding site
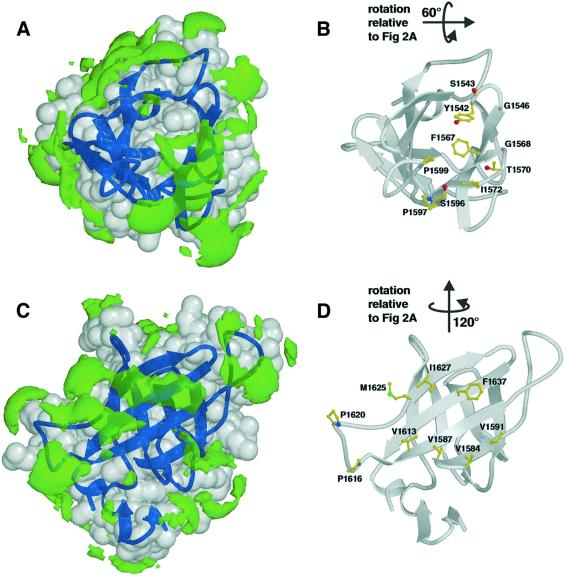
Calculation of an electrostatic potential surface for IGF2R-Dom11 using GRASP (Nicholls et al., 1991) reveals no striking imbalances of charge. In contrast, the use of a hydrophobic probe in GRID (Goodford, 1985) highlights two significant areas of surface hydrophobicity (Figure 4).
Fig. 4. Hydrophobic patches on the surface of IGF2R-Dom11. Views showing (A) hydrophobic patch 1, the putative IGF-II binding region and (B) the residues forming this patch (including Ile1572). Views showing (C) hydrophobic patch 2, an area partially equivalent to the dimerization region of CDMPR, and (D) the residues responsible for this patch. Hydrophobic patches (green) are defined using the program GRID (Goodford, 1985) and are shown here as volumes of pseudo-energies contoured at –2.3 kcal/mol. IGF2R-Dom11 is shown as a grey surface with dark blue secondary structure.
Hydrophobic patch 1 (Figure 4A and B) equates spatially to the (rather hydrophilic) mannose 6-phosphate binding site in CDMPR and explains the very different nature of IGF2R-Dom11, which does not bind mannose 6-phosphate. Comparison of these binding site cavities for IGF2R-Dom11 (400 Å3) and CDMPR (640 Å3) reveals a reduction in volume for this cavity in IGF2R-Dom11, which is formed by βA, βB, βC, βD, and loops AB, CD and FG, as a result of reductions in the lengths of both the AB and CD loops and a change in conformation of the FG loop. The cavity in CDMPR partly reflects the shape of its oligosaccharide ligand whereas the IGF2R-Dom11 cavity is elongated (Figure 5). The two cavities overlap but whilst the longer loops of CDMPR constrain its cavity and confer depth, the shorter IGF2R-Dom11 loops result in a shallower cavity.
Fig. 5. Ligand binding cavities for IGF2R-Dom11 and CDMPR. (A) The proposed IGF-II binding cavity (pale blue) of IGF2R-Dom11 (grey surface and dark blue secondary structure) and (B) the mannose 6-phosphate binding cavity (orange) of CDMPR (grey surface and red secondary structure). Cavities were defined by calculating volumes accessible to the centre of a 1.4 Å radius probe but not to a 6 Å probe, selecting the appropriate cavity and expanding these probe centres by spheres of 1.4 Å radius using VOLUMES (R.M.Esnouf, unpublished program).
All residues involved in the hydrogen bonding network of the CDMPR ligand binding site show non-conservative changes (typically from polar to hydrophobic; Figure 3) or are absent in IGF2R-Dom11. Indeed, the residues at the equivalent positions for IGF2R-Dom11 are unique among the domains of human IGF2R whereas, as previously reported, four of the 10 residues postulated to confer sugar binding are conserved for IGF2R domains 3 and 9 (Roberts et al., 1998). Residues Tyr1542, Ser1543, Gly1546, Phe1567, Gly1568, Thr1570, Ile1572, Ser1596, Pro1597 and Pro1599 are significantly exposed to solvent in this region of IGF2R-Dom11 (Figure 3). Of these residues, mutation of Ile1572 to Thr has been shown to abolish IGF-II binding (Garmroudi et al., 1996). Our structure gives no indication that such a mutation would cause major structural perturbation but rather implies that this residue, and hence the associated hydrophobic pocket, is directly involved in IGF-II binding. The ability to bind IGF-II is thought to have evolved in mammalian IGF2R; kangaroo IGF2R binds IGF-II with reduced affinity (Yandell et al., 1999) whilst chicken IGF2R lacks this functionality (Zhou et al., 1995). Sequence alignment between human and kangaroo IGF2R reveals several changes in and around the putative IGF-II binding site, although only three are in the cavity illustrated in Figure 5A (Gln1569Leu, Ser1573Asn and Ser1596Gly). The sequence conservation between human and kangaroo IGF2R domain 11 for residues lining the putative IGF-II binding pocket is high, whilst this is an area of low conservation between human and chicken IGF2R domain 11 and between human IGF2R domain 11 and the other human IGF2R domains. Notably perhaps, loop CD in chicken domain 11 is two residues longer than in human and kangaroo domain 11, a change that potentially deepens the putative binding cavity. However, interspecies differences in IGF-II binding affinities could also be at least partly attributable to differences in domain 13.
Mutational and structural analyses have revealed that the IGF-II residues essential for IGF2R binding (Phe48, Arg49, Ser50, Ala54 and Leu55) form a patch on the surface (Terasawa et al., 1994), suggesting that the IGF2R binding region of IGF-II is also primarily hydrophobic. Structural analyses of other such interactions between cytokine and receptor have shown predominantly hydrophobic interactions (reviewed in Wells and de Vos, 1996). Certainly a similar mechanism of binding between IGF-II and IGF2R would be consistent with the putative ligand binding pocket on IGF2R-Dom11. The interaction sites for IGF2R and the IGF-binding proteins (IGFBPs) on IGF-II are believed to overlap (Terasawa et al., 1994). A structure for the complex of the homologous IGF-I with the N-terminal domain of IGFBP-5 reveals that the principal interactions are between hydrophobic side chains protruding from both IGF-I and the IGFBP-5 fragment (Zeslawski et al., 2001). It is noteworthy that these IGF-I side chains (Glu3, Phe16 and Leu54) are conserved in IGF-II, where they abut the IGF2R binding site.
The second significant area of hydrophobicity on the IGF2R-Dom11 surface covers the upper part of the EFGHI β-sheet (Figure 4C and D). This area partly equates to the hydrophobic dimerization surface in CDMPR, with residues Val1587, Val1591, Ile1627 and Phe1637 (equivalent to Trp91, Ile95, Val131 and Leu144, respectively, in CDMPR) demonstrating some conservation of hydrophobic character. The possible functional significance of this area is uncertain; however, it might be relevant to the higher order structure of the IGF2R extracellular region (see below).
The intrinsic suitability of both hydrophobic patches for protein–protein interactions is underscored by their involvement in crystal packing contacts for both the tetragonal and orthorhombic lattices. For the tetragonal IGF2R-Dom11 model, patch 1 packs against patch 2 burying 699 and 659 Å2 of solvent accessible area, respectively, as calculated by Naccess (Hubbard and Thornton, 1993). The interactions include side chain contacts between Ile1572 and Ile1627 (see Figure 1). In the orthorhombic crystal form, an equivalent packing buries 662 Å2 for patch 1 and 649 Å2 for patch 2.
IGF2R mutations outside the putative IGF-II binding site
In addition to the Ile1572Thr mutation mentioned earlier, several other mutations in IGF2R have been discovered from screening of cancer patients. Of these, three lie in domain 11 (Gly1564Arg, Ala1618Thr and Gly1619Arg; Kong et al., 2000) but their effects on IGF2R function have not yet been assessed. Examining our IGF2R-Dom11 structure, Gly1564 lies on βC and is not solvent accessible, thus the replacement of glycine with a large charged amino acid could significantly disrupt both secondary structure and function. Ala1618 and Gly1619 are in the exposed GH loop, and although these mutations may interfere with the hydrophobicity of patch 2 (and therefore possible higher order structure of IGF2R) their effects might be expected to be small.
Further mutations lying outside domain 11 are reported to alter IGF-II binding (Byrd et al., 1999; Devi et al., 1999). One such mutation, Cys1262Ser, lies in domain 9 and is equivalent to Cys1553 in domain 11. Assuming a similar structure to domain 11, the loss of this disulfide bridge would allow the β-hairpin to reposition relative to the β-barrel and potentially disrupt the higher order structure of IGF2R. Another domain 9 mutation, Gly1296Arg (equivalent to Gly1575 in domain 11; Kong et al., 2000) lies buried on βD and is notably the only completely conserved non-cysteine residue in our alignment (Figure 3). The biological effect of this mutation is not yet defined, although for such a non-conservative change it might reasonably be expected to be significant. In domain 10, the Gln1445His and Gly1449Val mutations (equivalent to Arg1582 and Asn1585 in domain 11, respectively; Byrd et al., 1999; Devi et al., 1999) are expected to lie, solvent exposed, around βE. Their effects may arise from distortions of the higher order structure of IGF2R or more directly from proximity to the IGF-II binding site on domain 11.
Comparison of Igf2r from American and Japanese populations has recently revealed nine SNPs which may be useful in characterization of mutations and imprint status in human patients (Killian et al., 2001). Whether these polymorphisms are indicative of population differences in IGF2R function, or are underlying factors relating to physical differences in these populations remains to be seen. Of these mutations, three would give rise to amino acid variations. Leu252Val (equivalent to Arg1580 in domain 11) occurs in domain 2 and is predicted to lie, solvent exposed, on loop DE where a conservative substitution may not have any significant effect. The second amino acid change, Gly1619Arg, has been discussed above. Finally, Asn2020Ser lies in domain 14 according to our alignments (see Figure 3), and not domain 13 as reported (Killian et al., 2001). This mutation would be predicted to lie on the AB loop in a highly variable region where, given the lack of function so far attributable to domain 14, its effects cannot be predicted.
Implications for imprinting and the evolution of IGF-II binding
Recent evidence suggests that the high affinity interaction of IGF-II and IGF2R co-evolved with the reciprocal imprinting of these genes. Imprinting of IGF-II and IGF2R probably occurred during evolution from egg laying to placental embryonic nourishment (John and Surani, 2000), i.e. the protheria (platypus) to metatheria (marsupial) transition 130 million years ago. The latter continue to imprint IGF2R, with silencing of the paternally derived allele but, as stated earlier, do not retain the high affinity binding to IGF-II found in eutherian mammals (Killian et al., 2000). Based on sequence comparison of IGF2R from various species, Killian et al. (2000) have proposed a number of residues to be responsible for low and high affinity interactions with IGF-II. However, no substantive evidence for the functional importance of these residues can be discerned from the IGF2R-Dom11 structure.
Implications for the overall structure and function of IGF2R
Alignments between the 15 domains of IGF2R yield pairwise identities of 10.2–26.6% compared with 10.4– 19.6% between IGF2R domains and the slightly more distantly related CDMPR. The increased identity is limited mainly to the cysteines and to the core β-barrel, with considerable variation in the length and nature of the loop regions (Figure 3). Neither is there any clear bias towards conservation of the dimerization surface seen in CDMPR. However, the dimerization surface does partly overlap with the domain 11 hydrophobic patch 2. Evidence for the dimerization of IGF2R is inconclusive (York et al., 1999; Byrd et al., 2000) and the current structural data contribute little to the argument. Investigation of the positioning of all possible N-linked glycosylation sites on the 15 domains (Figure 3) reveals no favoured regions, although several of the potential sites on other domains map to hydrophobic patch 2 of domain 11, arguing against this feature being a general dimerization face in all domains.
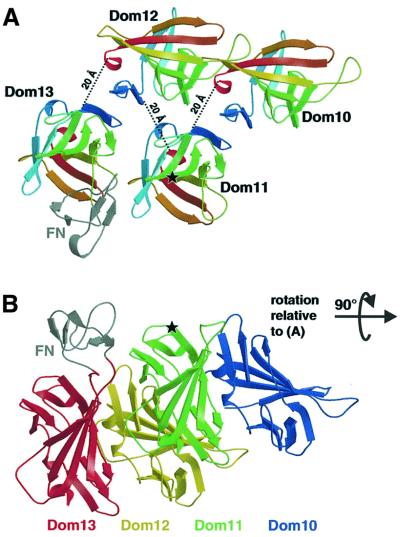
The domain structure may be defined as spanning from the first cysteine to the last (e.g. Cys1516–Cys1646 in IGF2R-Dom11). For domain 11 this implies linkers of nine and five residues to domains 10 and 12, respectively. Between all pairs of IGF2R domains the linker length lies in the range 5–12 residues (typically eight residues), placing considerable constraints on possible arrangements of the domains within the intact IGF2R. These constraints coupled with steric arguments allow us to propose a simple model for the extracellular structure. The packing along a 2-fold screw axis in the orthorhombic crystal form positions N- and C-terminal cysteines from neighbouring IGF2R-Dom11 molecules ∼20 Å apart, a reasonable distance for an eight-residue linker to span. This suggests a model for the full-length IGF2R in which alternate domains have similar orientations such that even- numbered domains all face one direction, whilst odd-numbered domains face the opposite direction (Figure 6). One attractive feature of this model is that the putative IGF-II binding face of domain 11 is adjacent to the region of domain 13 containing the fibronectin type II-like insert (Lobel et al., 1988). It seems plausible that this unique insert contributes to the reported enhancement of IGF-II binding by domain 13 (Devi et al., 1998; Linnell et al., 2001). The model differentiates between even- and odd-numbered domains and it is noteworthy that all the known functional domains of IGF2R have odd numbers (domains 3 and 9 bind mannose 6-phosphate whilst domains 11 and 13 are involved in IGF-II binding). Thus, particular functional importance is attached to a single continuous surface spanning one side of the intact molecule. Structural and functional characterization of multidomain fragments will allow validation and development of this model for the higher order structure.
Fig. 6. Tentative model for the relative arrangement of IGF2R domains 10–13. (A) View looking down on to the putative IGF-II binding site around residue Ile1572 (indicated by a star), showing each domain coloured from blue at the N-terminus to red at the C-terminus. The fibronectin-like insert in domain 13 (coloured grey and labelled FN) was obtained from the Protein Data Bank (accession code 1J7M), edited and positioned approximately at the insertion site using O (Jones et al., 1991). Dotted lines give the distances between the final cysteine of one domain and the first cysteine of the subsequent domain. (B) View at 90° from that in (A) showing the proximity between our proposed IGF-II binding site (again indicated by a star) and the fibronectin-like insert (coloured grey and labelled FN). Domains are coloured from blue (domain 10) to red (domain 13).
Materials and methods
Cloning
The region of Igf2r encoding domain 11 (nucleotides 4522–4954) was PCR amplified, using specific primers to incorporate restriction sites and a carboxypeptidase A-cleavable C-terminal histidine tag [Lys-(His)6-STOP]. The PCR fragment was ligated using NdeI and BamHI sites into pET-22b (Novagen) under the control of the T7 promotor.
Protein expression, refolding and purification
A truncated version of IGF2R comprising only domain 11 (amino acids 1508–1666; IGF2R-Dom11) was produced in Escherichia coli Origami strain (Novagen) under the control of the T7 promoter. Inclusion bodies were prepared and solubilized in reducing and denaturing buffer [6 M guanidine hydrochloride, 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM EDTA and 10 mM dithiothreitol (DTT)]. Aliquots were frozen at –70°C. Prior to refolding, a 0.8 ml aliquot of ∼20 mg resolubilized protein was diluted up to 1.6 ml using fresh denaturant buffer and incubated for 1 h at 4°C. The fully reduced and denatured IGF2R-Dom11 was then diluted into conditions permitting formation of native protein [200 mM Tris–HCl pH 8, 10 mM EDTA, 1 M l-arginine, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 6.5 mM cysteamine and 3.7 mM cystamine] (Gao et al., 1998). After refolding for 48 h at 4°C, soluble protein was concentrated for analysis and purification by gel filtration. The elution profile of concentrated, soluble IGF2R-Dom11 from a Superdex 75 column (Amersham-Pharmacia Biotech) gave a major peak of ∼16 kDa consistent with a monomeric form of the molecule. A high molecular weight aggregate peak was observed to elute from the column before the 16 kDa peak. Gel analysis of the major peak showed only one band and the expected molecular weight of the peak protein was confirmed by electrospray mass spectrometry. The IGF-II binding ability of purified IGF2R-Dom11 was assessed by surface plasmon resonance on a BIAcore biosensor 2000 at 37°C in HBS buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v surfactant P20). Briefly, biotinylated IGF-II (Gropep) was immobilized on to a streptavidin-coated sensor chip (type SA) and a 15 µM solution of IGF2R-Dom11 was passed over the chip for 1 min at a flow rate of 40 µl/min. Affinities calculated using the BIAevaluation software indicated that immobilized IGF-II bound the purified IGF2R-Dom11 with an apparent Kd of 4.1 × 10–8 M, similar to results obtained with eukaryotically expressed IGF2R-Dom11 (unpublished data). Wild type and Ile1572Thr mutant forms of domains 10–13 were used as positive and negative controls (Linnell et al., 2001).
Crystallization
Purified IGF2R-Dom11 was concentrated to 12 mg/ml in 100 mM Tris–HCl pH 8.0, 100 mM NaCl. Crystallization trials were performed at room temperature by vapour diffusion from sitting drops using microbridges. Drops contained 1 µl of protein solution plus 1 µl of reservoir solution from Crystal Screen kits I and II (Hampton Research, CA). Crystals appeared in several conditions after ∼4 days. Orthorhombic crystals grew from 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.5 and 30% PEG 4000, and these were subsequently shown to diffract to a resolution of 1.95 Å. Tetragonal crystals were grown from 0.2 M lithium sulfate, 0.1 M Tris–HCl pH 8.5 and 30% PEG 4000. This crystal form diffracted to a high resolution limit of 1.4 Å.
Data collection
All X-ray diffraction data were collected using synchrotron radiation at the European Synchrotron Radiation Facility (ESRF; Grenoble, France) and were autoindexed, integrated and scaled with the HKL program package (Otwinowski and Minor, 1997). An initial 1.6 Å resolution data set was collected using X-rays of wavelength 0.934 Å on beamline ID-14.1 using a tetragonal crystal flash cooled to 100 K in liquid nitrogen. The crystals belonged to space group P43212 with unit cell dimensions a = b = 49.5 Å, c = 118.5 Å and contained one molecule per asymmetric unit (estimated solvent content 43%). Under the same experimental conditions, a 1.95 Å resolution data set was collected using an orthorhombic crystal. These crystals belonged to space group P212121 with unit cell dimensions a = 31.4 Å, b = 49.8 Å and c = 83.0 Å again with one molecule per asymmetric unit (estimated solvent content 36%). A third data set, to 2.5 Å resolution was subsequently collected on BM14, using a tetragonal crystal and longer wavelength X-rays (1.77 Å) to enhance the sulfur anomalous scattering signal. A final 1.4 Å resolution data set was also collected at this time using a tetragonal crystal on ID-14.2 using X-rays of wavelength 0.933 Å. Representative statistics for these data sets are shown (Table I).
Structure determination
It was initially hoped that the CDMPR structure (Roberts et al., 1998) could be used for molecular replacement with the first two data sets but this strategy proved unsuccessful. An alternative strategy was made possible by collecting a data set at 1.77 Å: determination of the sulfur substructure from its anomalous scattering (for sulfur at 1.77 Å, f′ = 0.35 and f′′ = 0.72). The program Shake ‘N’ Bake (Weeks and Miller, 1999) was used to locate up to seven sulfur sites (assumed to be three Met Sδ plus four disulfide bridges; at 2.5 Å resolution it was not expected that individual Cys Sγ atoms would be resolved). Four clear sites were found and their positions and B factors were refined anisotropically using SHARP (de La Fortelle and Bricogne, 1997). Three further sites could then be located and the four original major sites showed strong anisotropy, indicating that they were the locations of the disulfide bridges. The individual Cys Sγ atoms were then positioned at appropriate separations along the long axes of the thermal ellipsoids. These 11 sites were then refined using CNS (Brünger et al., 1998), and after density modification an interpretable electron density map was obtained. The experimental phases from the solvent-flattened map were combined with the 1.6 Å resolution data set and successive rounds of GAP (D.I.Stuart and J.Grimes, unpublished program) were used to simultaneously phase extend and solvent flatten. An initial model (114 residues) was built automatically into the resultant map using ARP/wARP in mode warpNtrace (Perrakis et al., 1999).
Refinement
The initial model was refined against the 1.6 Å data set using energy minimization and individual B-factor refinement in CNS. Rigid-body refinement of the model against the 1.4 Å data then allowed subsequent rounds of energy minimization and individual B-factor refinement to be performed using these data. Water molecules were automatically picked in CNS. Between each refinement round, the model was manually checked and rebuilt using O (Jones et al., 1991), with reference to 2Fo–Fc and Fo–Fc electron density maps generated using phase information calculated from the model. Of the 11 anomalous sites identified in phasing, eight were Cys Sγ involved in disulfide bridges, two were methionine Sδ and one was from a well ordered sulfate ion. The stereochemistry of the final structure was checked using PROCHECK (Laskowski et al., 1993), which showed no residues in disallowed regions of the Ramachandran plot. Final refinement statistics are given in Table I.
Molecular replacement solution of the orthorhombic data set
In order to determine the structure of IGF2R-Dom11 in the P212121 crystal, molecular replacement was carried out using a partially refined model for the P43212 data. Using EPMR (Kissinger et al., 1999), a solution with a correlation coefficient of 77.0% and an R-factor of 39.9% was found. The solution was defined by Eulerian rotation angles of 180.4°, 90.2° and 89.1° and a translation of 18.1, 37.8 and 14.3 Å. Further refinement and picking of water molecules was performed according to standard protocols in CNS. The final structure was checked using PROCHECK (Laskowski et al., 1993), which showed no residues in disallowed regions of the Ramachandran plot. Final refinement statistics are given in Table I.
Structural analyses and visualization
Secondary structure motifs were confirmed using Promotif (Hutchinson and Thornton, 1996) and surface accessibility was evaluated with the program Naccess (Hubbard and Thornton, 1993). Structural superpositions were performed with SHP (Stuart et al., 1979) and sequence alignments used Clustal_W (Thompson et al., 1994) with manual modifications carried out using Jalview (http://circinus.ebi.ac.uk:6543/jalview). Crystal contacts were assessed using the program CONTACTS (R.Esnouf, unpublished program) and cavity volumes and surfaces were calculated using VOLUMES (R.Esnouf, unpublished program). Figure 3 was created using ESPript (Gouet et al., 1999). Other figures were produced using Bobscript (Esnouf, 1999) and rendered with Raster3D (Merritt and Bacon, 1997). All calculations and figures, unless otherwise stated, are based on the more complete and higher resolution tetragonal model.
Coordinates
The coordinates from both the tetragonal and orthorhombic crystal forms have been deposited in the Protein Data Bank, with accession codes 1GP0 and 1GP3, respectively.
Acknowledgments
Acknowledgements
We thank R.Aplin for mass spectrometry, N.Riffell and the staff of ID-14.1, ID-14.2 and BM14 for help during data collection, J.Dong, J.Grimes, S.Ikemizu and N.Zaccai for assistance with computation, and D.Stuart, C.Love and K.Drickamer for helpful discussions. This work was supported by Cancer Research UK.
Note added in proof
An independently determined set of coordinates for IGF2R-Dom11 has been released in the PDB subsequent to the preparation of this manuscript (accession code 1E6F). This unpublished structure shows no significant differences from our structure.
References
- Brünger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burns J.L. and Hassan,A.B. (2001) Cell survival and proliferation are modified by insulin-like growth factor 2 between days 9 and 10 of mouse gestation. Development, 128, 3819–3830. [DOI] [PubMed] [Google Scholar]
- Byrd J.C., Devi,G.R., de Souza,A.T., Jirtle,R.L. and MacDonald,R.G. (1999) Disruption of ligand binding to the insulin-like growth factor II/mannose 6-phosphate receptor by cancer-associated missense mutations. J. Biol. Chem., 274, 24408–24416. [DOI] [PubMed] [Google Scholar]
- Byrd J.C., Park,J.H., Schaffer,B.S., Garmroudi,F. and MacDonald,R.G. (2000) Dimerization of the insulin-like growth factor II/mannose 6-phosphate receptor. J. Biol. Chem., 275, 18647–18656. [DOI] [PubMed] [Google Scholar]
- Dahms N.M., Rose,P.A., Molkentin,J.D., Zhang,Y. and Brzycki,M.A. (1993) The bovine mannose 6-phosphate/insulin-like growth factor II receptor. The role of arginine residues in mannose 6-phosphate binding. J. Biol. Chem., 268, 5457–5463. [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dennis P.A. and Rifkin,D.B. (1991) Cellular activation of latent transforming growth factor β requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc. Natl Acad. Sci. USA, 88, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi G.R., Byrd,J.C., Slentz,D.H. and MacDonald,R.G. (1998) An insulin-like growth factor II (IGF-II) affinity-enhancing domain localized within extracytoplasmic repeat 13 of the IGF-II/mannose 6-phosphate receptor. Mol. Endocrinol., 12, 1661–1672. [DOI] [PubMed] [Google Scholar]
- Devi G.R., De Souza,A.T., Byrd,J.C., Jirtle,R.L. and MacDonald,R.G. (1999) Altered ligand binding by insulin-like growth factor II/mannose 6-phosphate receptors bearing missense mutations in human cancers. Cancer Res., 59, 4314–4319. [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Falls J.G., Pulford,D.J., Wylie,A.A. and Jirtle,R.L. (1999) Genomic imprinting: implications for human disease. Am. J. Pathol., 154, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.F., Gerth,U.C., Wyer,J.R., Willcox,B.E., O’Callaghan,C.A., Zhang,Z., Jones,E.Y., Bell,J.I. and Jakobsen,B.K. (1998) Assembly and crystallization of the complex between the human T cell coreceptor CD8α homodimer and HLA-A2. Protein Sci., 7, 1245–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmroudi F., Devi,G., Slentz,D.H., Schaffer,B.S. and MacDonald,R.G. (1996) Truncated forms of the insulin-like growth factor II (IGF-II)/mannose 6-phosphate receptor encompassing the IGF-II binding site: characterization of a point mutation that abolishes IGF-II binding. Mol. Endocrinol., 10, 642–651. [DOI] [PubMed] [Google Scholar]
- Goodford P.J. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem., 28, 849–857. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Hassan A.B. and Howell,J.A. (2000) Insulin-like growth factor II supply modifies growth of intestinal adenoma in Apc(Min/+) mice. Cancer Res., 60, 1070–1076. [PubMed] [Google Scholar]
- Hendrickson W.A. and Teeter,M.M. (1981) Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulfur. Nature, 290, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.J. and Thornton,J.M. (1993) Naccess Computer Program. University College, London.
- Hutchinson E.G. and Thornton,J.M. (1996) PROMOTIF—a program to identify and analyze structural motifs in proteins. Protein Sci., 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John R.M. and Surani,M.A. (2000) Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell, 101, 585–588. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Killian J.K., Byrd,J.C., Jirtle,J.V., Munday,B.L., Stoskopf,M.K., MacDonald,R.G. and Jirtle,R.L. (2000) M6P/IGF2R imprinting evolution in mammals. Mol. Cell, 5, 707–716. [DOI] [PubMed] [Google Scholar]
- Killian J.K., Oka,Y., Jang,H.S., Fu,X., Waterland,R.A., Sohda,T., Sakaguchi,S. and Jirtle,R.L. (2001) Mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) variants in American and Japanese populations. Hum. Mutat., 18, 25–31. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R., Gehlhaar,D.K. and Fogel,D.B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D, 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Kong F.M., Anscher,M.S., Washington,M.K., Killian,J.K. and Jirtle,R.L. (2000) M6P/IGF2R is mutated in squamous cell carcinoma of the lung. Oncogene, 19, 1572–1578. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. (1992) Structure and function of the mannose 6-phosphate/insulin-like growth factor II receptors. Annu. Rev. Biochem., 61, 307–330. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Linnell J., Groeger,G. and Hassan,A.B. (2001) Real time kinetics of insulin-like growth factor II (IGF-II) interaction with the IGF-II/mannose 6-phosphate receptor: the effects of domain 13 and pH. J. Biol. Chem., 276, 23986–23991. [DOI] [PubMed] [Google Scholar]
- Lobel P., Dahms,N.M. and Kornfeld,S. (1988) Cloning and sequence analysis of the cation-independent mannose 6-phosphate receptor. J. Biol. Chem., 263, 2563–2570. [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: Photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Motyka B. et al. (2000) Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell, 103, 491–500. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association—insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Olson L.J., Zhang,J., Lee,Y.C., Dahms,N.M. and Kim,J.J. (1999) Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor. J. Biol. Chem., 274, 29889–29896. [DOI] [PubMed] [Google Scholar]
- Oshima A., Nolan,C.M., Kyle,J.W., Grubb,J.H. and Sly,W.S. (1988) The human cation-independent mannose 6-phosphate receptor. Cloning and sequence of the full-length cDNA and expression of functional receptor in COS cells. J. Biol. Chem., 263, 2553–2562. [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Roberts D.L., Weix,D.J., Dahms,N.M. and Kim,J.J. (1998) Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor. Cell, 93, 639–648. [DOI] [PubMed] [Google Scholar]
- Schmidt B., Kiecke-Siemsen,C., Waheed,A., Braulke,T. and von Figura,K. (1995) Localization of the insulin-like growth factor II binding site to amino acids 1508–1566 in repeat 11 of the mannose 6-phosphate/insulin-like growth factor II receptor. J. Biol. Chem., 270, 14975–14982. [DOI] [PubMed] [Google Scholar]
- Stuart D.I., Levine,M., Muirhead,H. and Stammers,D.K. (1979) Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol., 134, 109–142. [DOI] [PubMed] [Google Scholar]
- Terasawa H., Kohda,D., Hatanaka,H., Nagata,K., Higashihashi,N., Fujiwara,H., Sakano,K. and Inagaki,F. (1994) Solution structure of human insulin-like growth factor II: recognition sites for receptors and binding proteins. EMBO J., 13, 5590–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky J.A. and Helman,L.J. (1996) Involvement of IGF-II in human cancer. J. Endocrinol., 149, 367–372. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB version 2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Wells J.A. and de Vos,A.M. (1996) Hematopoietic receptor complexes. Annu. Rev. Biochem., 65, 609–634. [DOI] [PubMed] [Google Scholar]
- Yandell C.A.,Dunbar,A.J., Wheldrake,J.F. and Upton,Z. (1999) The kangaroo cation-independent mannose 6-phosphate receptor binds insulin-like growth factor II with low affinity. J. Biol. Chem., 274, 27076–27082. [DOI] [PubMed] [Google Scholar]
- York S.J., Arneson,L.S., Gregory,W.T., Dahms,N.M. and Kornfeld,S. (1999) The rate of internalization of the mannose 6-phosphate/insulin-like growth factor II receptor is enhanced by multivalent ligand binding. J. Biol. Chem., 274, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Zeslawski W., Beisel,H.G., Kamionka,M., Kalus,W., Engh,R.A., Huber,R., Lang,K. and Holak,T.A. (2001) The interaction of insulin-like growth factor-I with the N-terminal domain of IGFBP-5. EMBO J., 20, 3638–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Ma,Z. and Sly,W.S. (1995) Cloning and expression of the cDNA of chicken cation-independent mannose-6-phosphate receptor. Proc. Natl Acad. Sci. USA, 92, 9762–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]