Abstract
Caenorhabditis elegans has two heterotrimeric G-protein γ subunits, gpc-1 and gpc-2. Although GPC-1 is specifically expressed in sensory neurons, it is not essential for the detection of odorants or salts. To test whether GPC-1 is involved in sensory plasticity, we developed a water soluble compound adaptation assay. The behaviour of wild-type animals in this assay confirms that prolonged exposure to salts can abolish chemo-attraction to these compounds. This process is time and concentration dependent, partly salt specific and reversible. In contrast, gpc-1 mutant animals show clear deficits in their ability to adapt to NaAc, NaCl and NH4Cl, but normal wild-type adaptation to odorants. Two other loci previously implicated in odorant adaptation, adp-1 and osm-9, are also involved in adaptation to salts. Our finding that G proteins, OSM-9 and ADP-1 are involved in taste adaptation offer the first molecular insight into this process.
Keywords: adaptation/Caenorhabditis elegans/ G proteins/taste
Introduction
Heterotrimeric G proteins form a first intracellular step in many signal transduction cascades (reviewed by Hamm, 1998). Binding of a ligand to a seven-transmembrane receptor results in the activation of the heterotrimeric G protein complex. In the inactive complex, a GDP molecule is bound to the α subunit. Upon activation, GDP is exchanged for GTP, which results in dissociation of the GTP bound α subunit and the βγ dimer. Both entities can activate effector molecules.
Thus far, 20 Gα subunits have been identified in mammals (Hamm, 1998) and in Caenorhabditis elegans (Jansen et al., 1999), yielding a broad repertoire for the transduction of various signals. In mammals, the complexity of this signalling machinery is further increased by six β and 12 γ subunits (Hamm, 1998). These additional levels of complexity are unnecessary in C.elegans, which contains only two β and two γ genes (Jansen et al., 1999).
A first functional characterization of most of the Gα genes in C.elegans has been performed (Lochrie et al., 1991; Mendel et al., 1995; Ségalat et al., 1995; Brundage et al., 1996; Korswagen et al., 1997; Park et al., 1997; Zwaal et al., 1997; Roayaie et al., 1998; Jansen et al., 1999). Based upon alignment of the predicted amino acid sequences, the Gα subunits can be divided into two groups. The first group consists of clear homologues of the four mammalian classes of Gα subunits: Gαo/i (goa-1), Gαs (gsa-1), Gαq (egl-30) and Gα12 (gpa-12). The other 16 Gαs cannot clearly be classified in any of the mammalian classes (gpa-1 to -11, gpa-13 to -16 and odr-3).
Expression patterns and mutant phenotypes also suggest a distinction between the conserved and the new Gαs. The conserved Gα subunits and gpa-7 are involved in regulating muscle and neuron activity (Mendel et al., 1995; Ségalat et al., 1995; Brundage et al., 1996; Korswagen et al., 1997; Berger et al., 1998; Hadju-Cronin et al., 1999; Jansen et al., 1999; Lackner et al., 1999; Miller et al., 1999; Nurrish et al., 1999). These genes are ubiquitously expressed in most neurons and muscle cells and mutations affect locomotion and egg laying. By contrast, 14 of the new Gα genes are likely to function in sensory perception (Zwaal et al., 1997; Roayaie et al., 1998; Jansen et al., 1999). These 14 genes show a restricted expression pattern, almost exclusively in C.elegans sensory neurons in the head (amphid neurons) and the tail (phasmid neurons). Furthermore, mutations in many of these α subunits cause altered behaviour of the animals in various chemotaxis assays (Zwaal et al., 1997; Roayaie et al., 1998; Jansen et al., 1999; G.Jansen and R.H.A.Plasterk, unpublished data).
The two Gβ subunits cannot be classified according to the same functional criteria mentioned above. gpb-1 is ubiquitously expressed and mutations in this gene show that it has an inhibitory function in regulating muscle activity (Zwaal et al., 1996). Maternal gpb-1 expression is essential for the proper orientation of the planes of cell division in early embryogenesis (Zwaal et al., 1996). The second Gβ subunit, gpb-2, shows ubiquitous expression, like gpb-1, and seems to have a stimulatory function in regulating GOA-1 and EGL-30 function at the neuromuscular junction (Chase et al., 2001; Robatzek et al., 2001; Van der Linden et al., 2001). A specific function in sensory perception cannot easily be determined for either of the β subunits because the mutants are either inviable or do not move normally.
The data on the function of the α and β subunits suggest that most of the specificity of G protein mediated signalling in C.elegans is generated via the Gα subunits. In this study we set out to determine the contribution of the Gγ subunits to the potentialities of G protein signalling. We first determined the expression patterns of the two γ subunits. We found one ubiquitously expressed Gγ, gpc-2, and a putative sensory specific γ subunit, gpc-1.
Why would C.elegans need a Gγ subunit specific for sensory perception? gpc-1 could be necessary for the proper migration and development of the sensory neurons, or it could be involved in the detection of several environmental cues. Analysis of loss- and gain-of-function mutants suggests that GPC-1 is not essential for either of these two processes.
Studies in mammals have suggested that Gβγ subunits are involved in receptor adaptation (reviewed by Pitcher et al., 1998). A model has been proposed in which binding of a ligand to a G protein-coupled receptor (GPCR) leads to activation of heterotrimeric G proteins. The free, but membrane-bound Gβγ subunit can now interact with a G protein receptor kinase (GRK), and thereby target the kinase to the GPCR. Subsequent phosphorylation of the GPCR results in the recruitment of arrestin proteins to the receptors, thereby preventing further G protein activation. This mechanism provides a molecular basis for desensitization or adaptation.
There is only one assay to test adaptation of C.elegans to chemosensory cues, an odorant adaptation assay (Colbert and Bargmann, 1995). However, gpc-1 does not seem to be essential for this process, as could be expected from the absence of gpc-1::GFP expression in the olfactory neurons (AWA and AWC). We therefore developed an assay to test the function of GPC-1 in adaptation to water soluble compounds. Our results show that prolonged exposure of wild-type animals to water soluble compounds can abolish chemo-attraction to these same compounds, in a time and concentration dependent, partly salt specific and reversible manner. gpc-1 mutant animals show clear deficits in their ability to adapt to three water-soluble compounds, NaAc, NaCl and NH4Cl. Furthermore, two loci that previously have been implicated in olfactory adaptation, adp-1 and osm-9 (Colbert and Bargmann, 1995; Colbert et al., 1997), are also involved in adaptation to salts.
Results
Caenorhabditis elegans has two Gγ subunits
The nematode C.elegans has two heterotrimeric G protein γ subunits, gpc-1 and gpc-2 (Jansen et al., 1999). Further screening using BLAST (Altschul et al., 1990) for the presence of Gγ subunits using all 12 mammalian γ subunit sequences (Hamm, 1998) yielded no further γ genes.
The family of Gγ subunits consists of quite divergent members. The percentage identity among the 12 human Gγs ranges from 22 to 79% amino acid identity and the percentage identity between the human and the C.elegans γ subunits ranges from 22 to 37%. Therefore, we cannot exclude the existence of additional, even less conserved, γ subunits in C.elegans. Alignment of the predicted amino acid sequences of the two C.elegans γ subunits with the 12 human Gγ sequences shows that they are not clear homologues of any of the human Gγ subunits (results not shown). GPC-1 shows greatest identity to human Gγ7 and Gγ8olf (35% amino acid identity), GPC-2 shows greatest identity to human Gγ13 (37%).
gpc-1 shows sensory-neuron specific expression
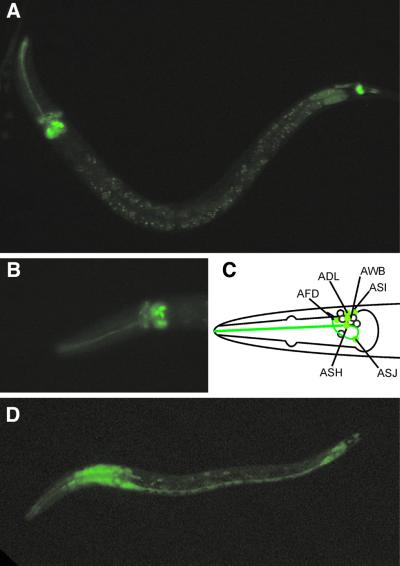
A first indication of the function of the two Gγ subunits was obtained by determining the expression patterns of these genes, using GFP-fusion constructs. gpc-1::GFP expression was found in only 12 cells in the head and two cells in the tail of the animal (Figure 1A). These cells were identified as (putative) chemosensory neurons (White et al., 1986): six pairs of amphid neurons in the head (ADL, ASH, ASJ and faintly in AFD, ASI and AWB; Figure 1B and C) and one pair of phasmid neurons in the tail (PHB). The amphid neurons are involved in sensory perception of the environment of the animal (reviewed in Bargmann and Mori, 1997). ADL and ASH have been implicated in aversion to mechanical and chemical cues. The AWB neurons mediate volatile avoidance. The ASI and ASJ neurons, together with ASG, are involved in the detection of pheromones and food signals that regulate the formation of dauer larvae. Furthermore, the ASI neurons, together with ADF, ASE, ASG and ASK, detect water soluble compounds. Finally, the AFD neurons have a thermosensory function. The function of the phasmid neurons is unclear. The morphology of these cells suggests a sensory function, but so far no experimental evidence exists for such a function.
Fig. 1. gpc-1 shows sensory-neuron specific expression; gpc-2 expression was found in all neurons and muscle cells. Fluorescence micrographs (A and B) and schematic representation of gpc-1::GFP (C) and gpc-2::GFP (D) expressing animals. gpc-1::GFP is expressed in six pairs of amphid neurons in the head: ADL, ASH, ASJ and faintly in AFD, ASI and AWB (A–C) and one pair of phasmid neurons in the tail, PHB (A). gpc-2::GFP expression was found in all neurons and muscle cells (D).
gpc-2::GFP expression was found in all neurons and muscle cells (Figure 1D). This ubiquitous expression suggests that GPC-2 functions in similar processes as the conserved Gαs (Brundage et al., 1995; Mendel et al., 1995; Ségalat et al., 1995; Korswagen et al., 1997; Berger et al., 1998; Hadju-Cronin et al., 1999; Jansen et al., 1999; Lackner et al., 1999; Miller et al., 1999; Nurrish et al., 1999). Analysis of gpc-2 loss-of-function showed that this Gγ subunit is essential for the proper orientation of the planes of cell division during early embryogenesis (Gotta and Ahringer, 2001), as has been found for GPB-1 (Zwaal et al., 1996).
GPC-1 is not essential for olfactory and gustatory perception
Heterotrimeric G proteins function as molecular switches. Consequently, both loss- and gain-of-function mutations of the constituting subunits may affect the signal transduction pathways in which they are involved. In the case of the Gα subunits, gain-of-function alleles were more informative than loss-of-function alleles (Jansen et al., 1999), probably because the effects of loss-of-function mutations were masked by functional redundancy. However, while a phenotype obtained by loss of gene function will always be informative about the function of that gene, one has to be more cautious when interpreting the effects of gain-of-function mutations. We generated a loss-of-function mutant of gpc-1 using Tc1-mediated target-selected gene inactivation (Zwaal et al., 1993). As most of the gene has been deleted, including the translation start, this allele is a molecular null (pk298te). To test the effect of overexpression of gpc-1, we generated transgenic animals that carry additional copies of the wild-type gene, gpc-1XS(pkIs571).
Loss-of-function of gpc-1 has no obvious effects on the nematode. We observed no morphological changes in gpc-1 animals, and they showed wild-type locomotion and egg laying (results not shown). gpc-1 overexpression (gpc-1XS) animals also showed no morphological changes, however, both locomotion and egg laying were slightly reduced. On average, gpc-1XS animals showed 10.8 (± 0.9 SE) body bends/animal/minute, while wild-type animals showed 18.3 (± 0.9) body bends. The uteri of wild-type nematodes contained 14.3 ± 0.6 eggs per animal, while gpc-1XS animal uteri contained 11.2 ± 0.8 eggs. We detected no difference in the developmental stage of newly laid eggs (results not shown).
The specific expression of gpc-1 in the amphid and phasmid neurons suggests a function for this γ subunit in sensory perception, most likely aversion or attraction to water soluble compounds. Alternatively, GPC-1 might be involved in the development of the amphid and phasmid neurons, as has been reported for ODR-3 (Roayaie et al., 1998) and GPA-3 (Zwaal et al., 1997). To determine if the sensory cilia of the amphid and phasmid neurons are in contact with the environment, we exposed the animals to the lipophilic fluorescent dye DiO (Perkins et al., 1986). Neither the loss- nor the gain-of-function mutant of gpc-1 showed reduced dye filling (data not shown).
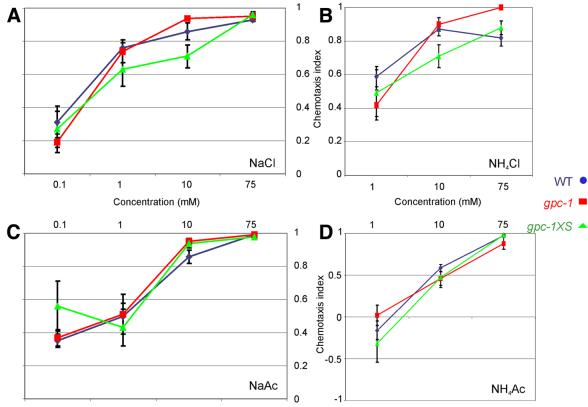
To test if GPC-1 is necessary for the perception of environmental cues, we determined the effect of deletion or overexpression of gpc-1 on the ability to detect aversive compounds (Wicks et al., 2000), water soluble attractants (Wicks et al., 2000) or odorants (Bargmann et al., 1993). We observed no deficits in the detection of these compounds by the gpc-1 mutants (Figure 2 and results not shown). These results indicate that GPC-1 is not essential for the detection of the attractive and repellent compounds tested. Probably GPC-2 or both GPC-1 and -2 together can perform these functions.
Fig. 2. gpc-1 mutant animals show wild-type responses to various salts. Chemotaxis to 0.1, 1, 10 and 75 mM NaCl (A), NH4Cl (B), NaAc (C) and NH4Ac (D) of wild-type (blue circles), gpc-1 (red squares) and gpc-1XS (green triangles) animals.
Water soluble compound adaptation assay
When gpc-1 mutant animals were tested in water soluble compound chemotaxis assays we used 10 min time points as a measure of their ability to detect the compounds tested (Figure 2). At later time points, the chemotaxis index started to decrease. A similar reduction of the animals’ response to a chemical attractant after repeated or continuous exposure to that attractant has previously been observed and been termed desensitization or adaptation (Ward, 1973; Colbert and Bargmann, 1995). This adaptation effect was significantly less prominent after prolonged exposure of gpc-1 mutant animals to certain salts (results not shown). These results suggest a function for gpc-1 in adaptation to water soluble compounds.
To test if GPC-1 is involved in adaptation, we developed a water soluble compound adaptation assay. Detection of tastants by C.elegans is tested after washing well-fed nematodes several times with a low-salt phosphate buffer (Wicks et al., 2000). Animals are then placed on quadrant plates and the distribution of the animals over the quadrants filled with buffered agar, with or without a water soluble compound, is recorded over time. To test whether C.elegans adapts to salts, the animals were exposed to the compound tested during the washing steps.
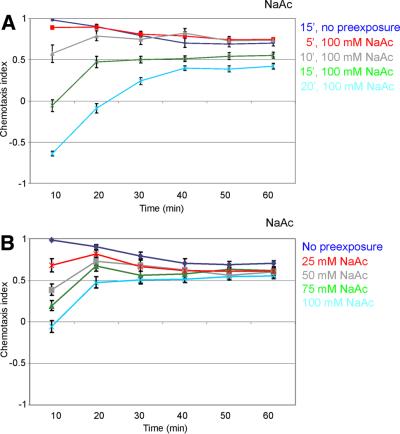
First, we determined the optimal concentration for, and the time course of, adaptation. Wild-type animals were exposed to increasing concentrations of different salts during a 5 to 30 min washing procedure. Subsequently, the animals were tested in the quadrant assay for their attraction to the adapting salt (Figure 3). The optimal pre-exposure time for adaptation to NaAc is 15 min (Figure 3A). Longer exposure increased the effect further, but occasionally seemed toxic. Chemotaxis to 25 mM NaAc was significantly reduced after pre-exposure to 25 mM NaAc (Figure 3B). Increasing salt concentrations up to 100 mM NaAc decreased chemotaxis to NaAc further (Figure 3B). Higher NaAc concentrations did not add to the adaptation (results not shown).
Fig. 3. Pre-exposure to NaAc significantly reduces chemotaxis to that salt, and is time and concentration dependent. (A) Wild-type animals were exposed to 100 mM NaAc for 5–20 min and subsequently tested for chemotaxis to 25 mM NaAc. A 5 min pre-exposure resulted in a significant decrease in chemotaxis to NaAc (p <0.01). Longer pre-exposure resulted in decreasing chemotaxis to NaAc (p <0.001). (B) Wild-type animals were exposed to different concentrations of NaAc (0–100 mM, as indicated) during 15 min, and subsequently tested for chemotaxis to 25 mM NaAc. Pre-exposure to 25 mM NaAc resulted in a significant decrease in chemotaxis (p <0.001). An increase in the salt concentration during the pre-exposure step resulted in a decrease in chemotaxis to NaAc.
Caenorhabditis elegans showed similar responses to several other salts. Adaptation to NaCl and NH4Cl showed similar concentration dependence as adaptation to NaAc (results not shown). In all three cases, optimal adaptation i.e. the strongest effect with the least risk of toxicity, could be obtained after 15 min washing in a 100 mM salt solution. We did not observe significant adaptation to NH4Ac, not even after pre-exposure to 150 mM NH4Ac (results not shown). Thus, C.elegans will adapt to particular chemoattractant salts.
Recovery and specificity of adaptation to salts
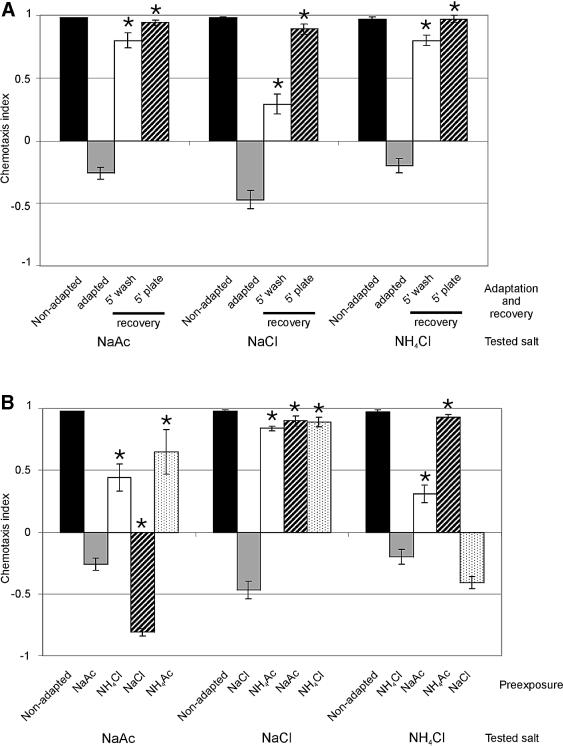
To test for reversibility of adaptation, adapted animals were allowed to recover for 5 min or more on buffered agar plates with no additional salt. Under these conditions, chemotaxis was recovered to non-adapted levels (Figure 4A). When adapted animals were placed on plates with the adapting salt the animals remained fully adapted (results not shown). Chemotaxis could partially, but significantly (p <0.001), be recovered by a 5 min wash step in CTX buffer without salt, immediately after the 15 min pre-exposure (Figure 4A).
Fig. 4. Adaptation to salts is fully reversible and partly salt specific. (A) When adapted animals were allowed to recover for 5 min on low-salt agar plates (5 min plate recovery) chemotaxis to the adapting salt was fully reversed. Recovery in low-salt buffer (5 min wash) had only a partial, but significant effect (p <0.001). Asterisks indicate a statistically significant difference between adapted and recovered animals (p <0.001). (B) Pre-exposure to a salt different from the salt used in the subsequent chemotaxis assay showed that the adaptation effect is partly salt specific. The salts used during pre-exposure and the salts tested for, have been listed below the assays. Asterisks indicate a statistically significant difference between adapted and cross-adapted animals (p <0.001). In all cases, wild-type animals were exposed to 100 mM salt for 15 min and subsequently tested for chemotaxis to 25 mM salt. The chemotaxis index was determined 10 min after the animals were placed on the chemotaxis plates.
To determine the specificity of adaptation, the response of wild-type animals to NaAc was also tested after pre-exposure to NH4Cl (cross-adaptation). Furthermore, since anions and cations are tasted separately (Pierce-Shimomura et al., 2001), we also tested the effects of exposure to NaCl (adapted for Na+) or NH4Ac (adapted for Ac–). Pre-exposure to NH4Cl significantly reduced chemotaxis to NaAc, but clearly less than pre-exposure to NaAc (p <0.001; Figure 4B), indicating the existence of both a salt-specific and an aspecific response. Pre-exposure to NaCl had a very strong effect on chemotaxis to NaAc, suggesting that pre-exposure to NaCl induces a very strong avoidance response, stronger than the attractive signal provided by Ac–. Pre-exposure to NH4Ac had a weak effect on the response to NaAc, although it should be noted that NH4Ac also had a minor effect on chemotaxis to NH4Ac (results not shown). Similar effects were observed on chemotaxis to NH4Cl (Figure 4B). Chemotaxis to NaCl could only be affected after pre-exposure to NaCl, but not the other salts (Figure 4B). Taken together, our results indicate the existence of both a salt-specific and an aspecific adaptation process. Exposure to NaCl induces the avoidance of NaCl, NaAc and NH4Cl. Exposure to NaAc, NH4Cl or NH4Ac had variable effects, probably as a result of a balance between the remaining attractive and repulsive signals.
Our results suggest that adaptation is a specific response rather than the result of toxicity. Although we cannot formally rule out this latter possibility, several observations suggest that this is not the case. First, the adapted animals show no signs of toxic effects of the pre-exposure; they move and disperse normally over the assay plates. Secondly, adaptation is fully reversible. Thirdly, the adaptation effects are at least partially salt specific, and fourthly, salt adapted animals show normal chemotaxis to several different odorants (results not shown). Finally, the identification of mutants with severely hampered adaptation, but no morphological or other behavioural defects (see below) suggests that toxicity of the adapting compound does not play an important role.
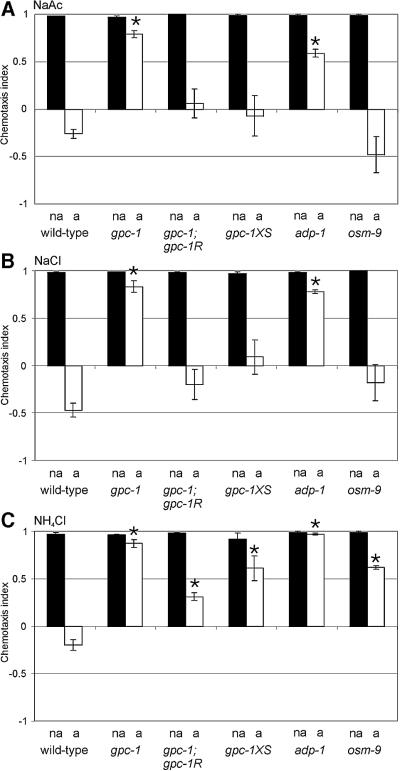
gpc-1 mutants are defective in adaptation to NaAc, NaCl and NH4Cl but not to odorants
gpc-1 mutant animals were tested for adaptation to NaAc, NaCl and NH4Cl. Pre-exposure of gpc-1 animals to 100 mM NaAc for 15 min, had only a minor effect on chemotaxis to NaAc (Figure 5A). Similarly, pre-exposure to NH4Cl or NaCl had almost no effect on chemotaxis to these salts (Figure 5B and C). gpc-1XS animals showed low levels of adaptation to NH4Cl, but almost wild-type levels of adaptation to NaAc and NaCl (Figure 5).
Fig. 5. gpc-1, adp-1 and osm-9 mutant animals are defective in adaptation to salts. Wild-type, gpc-1, gpc-1; gpc-1R, gpc-1XS, adp-1 and osm-9(n1603) animals were tested for chemotaxis to 25 mM NaAc (A), NaCl (B) and NH4Cl (C), after pre-exposure to a low-salt buffer (non-adapted; black bars, na), 100 mM NaAc (A), NaCl (B), or NH4Cl (C) (adapted; open bars, a). The genotypes are indicated below the assays. The chemotaxis index was determined 10 min after the animals were placed on the chemotaxis plates. Asterisks indicate a statistically significant difference between wild-type and mutant adapted animals (p <0.001).
The defect in adaptation to NaCl, NaAc and NH4Cl in gpc-1 animals could be rescued by the introduction of low levels of the wild-type gpc-1 gene in gpc-1 mutant animals (Figure 5). We conclude that although GPC-1 is not essential for the detection of NaCl, NaAc and NH4Cl, it is essential for the subsequent desensitization of the sensory signalling pathway.
Previously, Colbert and Bargmann (1995) described odorant-specific adaptation in C.elegans. To determine if GPC-1 is also involved in adaptation to volatile compounds, gpc-1 animals were exposed to four different odorants and subsequently tested for chemotaxis to the adapting odorant. The mutant animals showed similar responses to these odorants as the wild-type control animals (results not shown). This result is in agreement with the finding that gpc-1 is not expressed in the AWA and AWC neurons, the cells involved in odorant detection. An alternative explanation would be that gpc-1 is functionally redundant to GPC-2 with regard to odorant adaptation.
The adp-1 and osm-9 mutations affect water-soluble adaptation
Thus far, mutations in three genes have been described that affect chemosensory adaptation: a dominant mutation in adp-1(ky20), several recessive mutations in osm-9 and overexpression of odr-1 (Colbert and Bargmann, 1995; Colbert et al., 1997; L’Etoile and Bargmann, 2000). The osm-9 gene encodes a putative channel protein with limited similarity to the Drosophila TRP phototransduction channel, and is expressed in a subset of sensory neurons (Colbert et al., 1997). Mutations in osm-9 affect chemosensory responses to a subset of odorants mediated by the AWA sensory neurons, and to osmotic and mechanosensory stimuli mediated by the ASH neurons, but does not affect chemotaxis to salts (Colbert et al., 1997). Furthermore, OSM-9 is involved in adaptation to a subset of odorants sensed by AWC. The molecular nature of the adp-1 gene and its mutation are unknown. A mutant in this gene was isolated in a screen for animals defective in adaptation to benzaldehyde (Colbert and Bargmann, 1995). The mutant animals fail to adapt to a subset of AWC-sensed odorants, but show normal chemotaxis to all odorants when not adapted.
To determine if adp-1 and osm-9 are also involved in adaptation to water soluble compounds, we tested these animals in our assay. Before we tested adaptation, we first confirmed that the adp-1(ky20) and osm-9(ky10) animals showed no deficits in chemotaxis to NaAc, NaCl, NH4Ac or NH4Cl (Figure 5; results not shown). Next, we exposed the adp-1(ky20) and osm-9(ky10, n1603 and n2743) animals to 100 mM NaAc, NaCl or NH4Cl for 15 min, and subsequently tested them for their ability to detect these salts. The adp-1 mutants failed to adapt to the salts tested (Figure 5); we observed only very low levels of adaptation. osm-9 animals showed no defects in adaptation to NaAc or NaCl, but low levels of adaptation to NH4Cl (Figure 5). The same behavior was observed for three osm-9 alleles tested (results not shown).
Our results indicate a function for OSM-9 and ADP-1 in adaptation to salts, but not in salt detection. Furthermore, we find a distinction between adaptation to NH4Cl and NaCl or NaAc. The mechanism underlying this difference is unknown. Finally, our results suggest that adaptation to odorants and salts use at least partially overlapping mechanisms.
Discussion
Previous studies indicated that continuous exposure of C.elegans to a water soluble compound results in a decrease in the chemotaxis response to that stimulus (desensitization or adaptation; Ward, 1973; Dusenbery, 1980). When using the recently developed water soluble compound chemotaxis assay (Wicks et al., 2000), we indeed see that animals lose their interest in salts after prolonged exposure. We modified this latter chemotaxis assay so that it can now be used to examine adaptation to water soluble compounds. Using this adaptation assay, we found that chemoattraction to a particular water soluble compound can be abolished by exposing wild-type animals to the water soluble compound before analysis. This adaptation effect is time and concentration dependent, reversible and partly salt specific.
Adaptation mechanisms
Not much is known about the molecular mechanisms used to detect salts or adapt to continuous exposure to salts. The only components that have been identified unequivocally as contributing to sodium salt perception by mammalian cells, are the family of epithelial-type sodium channels (reviewed in Gilbertson et al., 2000). The downstream signalling molecules and the different modes of modulation are largely unknown. Our finding that G proteins, OSM-9 and ADP-1 are involved in taste adaptation offer the first molecular insight into this process.
Adaptation or desensitization in olfaction has been much better characterized. Evidence exists for two mechanisms: a specific (homologous) or a general (heterologous) mechanism (reviewed in Pitcher et al., 1998; Morris and Malbon, 1999). Homologous desensitization is the process in which phosphorylation by a G protein receptor kinase (GRK, targeted by the G protein βγ subunit) and the subsequent binding of arrestins, uncouples the activated receptors from downstream G proteins. This uncoupling process allows cells to specifically decrease their response to continuous stimuli. Heterologous desensitization is less specific, because it occurs by regulation of downstream signalling molecules, e.g. by quenching Gα activity or ion channel regulation or by phosphorylation of signalling molecules by second messenger-dependent kinases and protein kinase C.
The fact that adp-1 and osm-9 function both in odorant and taste adaptation suggests at least a partial overlap between the two mechanisms. Furthermore, we can divide taste adaptation into a specific and an aspecific process (compare homologous and heterologous desensitization). Finally, we found that a G protein γ subunit is involved in taste adaptation. Taken together, these results make it very tempting to speculate that taste adaptation uses a mechanism similar to homologous adaptation. This would imply the existence of G protein-coupled salt sensors, which would function in parallel to, or downstream of, channels. Alternatively, G proteins could be involved in more indirect mechanisms.
What is the function of OSM-9 in adaptation? The OSM-9 protein has been identified as a putative channel protein with limited similarity to the Drosophila TRP phototransduction channel (Colbert et al., 1997). The TRP channel contributes to light adaptation by mediating calcium influx into photoreceptor cells (Peretz et al., 1994; Hardie, 1996). It is unclear how this would fit into a specific salt adaptation mechanism. Finally, what is the molecular nature of the adp-1 gene?
Cellular location of taste adaptation
Based on laser ablation experiments, the ASE cells are considered to be the most important chemosensory cells for chemotaxis to water soluble attractants (Bargmann and Horvitz, 1991). The ADF, ASG and ASI neurons also play a role in this process, possibly depending on salt concentration or how steep the salt gradient is. A recent study by Pierce-Shimomura et al. (2001) revealed that the cellular basis for taste in C.elegans is even more complex. They show that ASEL is primarily sensitive to sodium, whereas ASER is primarily sensitive to chloride and potassium. Caenorhabditis elegans probably uses this functional asymmetry to increase its possibilities to discriminate between different stimuli (Pierce-Shimomura et al., 2001).
Of the four cells involved in salt detection only the ASI neurons express gpc-1. If GPC-1 would mediate salt adaptation in ASI, this would mean that adaptation is uncoupled from detection (via ASE). It is interesting to note that there is a direct connection from the ASI to the ASE neurons, made via a chemical synapse (White et al., 1986). This connection would enable transmission of the adaptation signal from ASI to ASE, resulting in desensitization of the sensory signal in ASE. Alternatively, the chemosensory signals (detecting and modulating) from all four pairs of chemosensory neurons involved in salt detection could be collected and integrated in interneurons.
GPC-1 is also expressed in the ASH and ADL neurons, which mediate aversion. Occasionally we see that pre-exposure to high salt concentrations results in aversion. This could be an indication that adaptation is mediated by the aversion neurons ASH and ADL, while ADF, ASE, ASG and ASI mediate attraction. These stimulatory and inhibitory signals would generate a balance between attraction and aversion, integrated in interneurons.
Our hypotheses on the cellular location of GPC-1-mediated salt adaptation are based on the gpc-1 expression pattern, determined using a gpc-1::GFP fusion construct, and laser ablation experiments to identify the cells involved in salt perception using another salt chemotaxis assay (Bargmann and Horvitz, 1991). Therefore, definitive conclusions can only be drawn after confirmation of the function of GPC-1 in the different amphid neurons and the identification of the cells involved in salt detection and adaptation in the quadrant assay.
In C.elegans, several non-associative learning processes have been demonstrated, including habituation to mechanosensory stimuli (Rose and Rankin, 2001) and olfactory stimuli (Bernhard and van der Kooy, 2000). Furthermore, the animals show altered responses to specific stimuli such as temperature (Bargmann and Mori, 1997), odorants (Colbert and Bargmann, 1997) and food (Sawin et al., 2000), as a result of changes in their environment or past experience. Recently, associative learning has also been reported (Wen et al., 1997). Despite extensive progress in these behavioural, cellular and genetic studies, little is known at the molecular level of these complex processes, or which neurons are involved. In contrast, the analysis of olfactory adaptation, in combination with the detailed characterization of olfactory signalling, has resulted in the identification of the first molecules involved in olfactory adaptation (Colbert et al., 1997; L’Etoile and Bargmann, 2000). The development of the water soluble compound adaptation assay and the identification of the first molecules involved have given us the tools to dissect this adaptation process.
Materials and methods
Molecular biology methods
PCR was performed as described previously (Zwaal et al., 1993). Sequence comparisons were performed using BLAST (Altschul et al., 1990) and the Wisconsin Sequence Analysis Package (Genetics Computing Group, Version 9.0).
GFP fusion constructs
The promoter fusion for gpc-1 was generated by subcloning a 5.0 kbp XbaI–ScaI fragment from cosmid K02A4 into the expression vector pPD95.77 (a gift from A.Fire, J.Ahn, G.Seydoux and S.Xu). The resulting construct, pRP1434, contains 4.2 kbp of upstream sequences, and most of the gpc-1 coding region, fused in-frame to the GFP gene (Chalfie et al., 1994). The gpc-2::GFP construct, pRP2090, was made by fusing a 2.5 kbp PCR fragment generated with gpc2-1 (5′-TCTGCAGCACGACGATAATC, extended with a SphI site) and gpc2-2 (5′-GTCGATTGGGTTCACAAGTG, extended with a BamHI site) into vector pPD95.77. This construct contains 2.3 kbp of upstream sequence and most of the gpc-2 open reading frame. At least two independent transgenic lines were generated for each construct. Except for variation in expression level, the transgenic lines generated for the Gγ subunits showed the same expression pattern. The transgenic array was integrated for one of the lines of each of the fusion constructs.
Cell identifications were made by comparing the fluorescence image with Nomarski images of the same animal. At least 20 animals were examined for each fusion gene. Particular cells were identified by using a combination of their position and morphology (White et al., 1986).
Strains and genetics
All animals were maintained according to standard methods (Brenner, 1974). Strains used in this study were: Bristol N2; CB1282 dpy-20(e1282) IV; CX10 osm-9(ky10) IV; CX20 adp-1(ky20) II; MT3642 osm-9(n1603) IV; MT6317 osm-9(n2743) IV; NL828 mut-2(r459) I gpc-1(pk243::Tc1) X; NL792 gpc-1(pk298) X; NL1575 dpy-20(e1282) IV pkIs575[gpc-1::GFP dpy-20(+)]; NL1598 dpy-20(e1282) IV pkIs571[gpc-1(+) dpy-20(+)]; NL2336 dpy-20(e1282) IV pkIs1275[gpc-2::GFP dpy-20(+)].
A gpc-1::Tc1 insertion mutant (pk243) was isolated as described (Zwaal et al., 1993) using gpc1-1 (5′-CTGCTGCTTCACCTA in the sequence GATATTTACGTATAAATTATAT. A loss-of-function mutant (pk298) was derived from this strain using primers gpc1-1 and gpc1-2 (AAACTAGTCTACTCTGCCTAGTG), nested with gpc1-3 and gpc1-4 (CCTTTCATAATAACGTCTGATG). The deletion removes the first exon, and runs from 0.6 kbp upstream of the predicted ATG (ATATTTACGT are the first 10 deleted bp), to 36 bp into the first intron (GATTTACATA are the last 10 bp of the deletion). The gpc-1 mutant strain was outcrossed six times before phenotypic analysis.
A gpc-1 overexpression/rescue construct was generated by subcloning a 6.4 kbp XbaI fragment of cosmid K02A4 into the plasmid vector pGEM. This construct contains the complete predicted open reading frame of the gene and 4.2 and 1.4 kbp of upstream and downstream sequence, respectively.
Germline transformation was carried out as described (Mello et al., 1991). Marker dpy-20 DNA (pMH86; Han and Sternberg, 1991) was used at a concentration of 100 µg/ml and test DNA at a concentration of 10 (for rescue of the adaptation phenotype of gpc-1 animals) or 50 µg/ml. Transgenic animals were identified by rescue of the dpy-20(e1282) phenotype. The transgenic arrays were integrated by irradiating transgenic animals with 40 Gy of γ radiation from a 137Cs source. Before phenotypic analysis all transgenic strains were outcrossed at least twice. Only one gpc-1 overexpression strain was generated: gpc-1XS(pkIs571). Rescue of the adaptation phenotype was obtained with four strains, gpc-1R(gjIs4), gjIs5, gjIs6 and gjIs8.
Chemotaxis assays
Volatile chemotaxis assays were performed as described (Bargmann et al., 1993). Attractants used were 2,4,5-trimethylthiazole, pyrazine (10 and 100 mg/ml), diacetyl, isoamylalcohol, benzaldehyde and 2,3-pentanedione. Aversive odorants used were 2-nonanone and 1-octanol. All attractants were tested in a 10 or 100× dilution (unless otherwise indicated) in ethanol. The aversive odorants were tested undiluted or 10× diluted in ethanol. Odorants were obtained from Sigma Chemie, Acros Organics, Fluka Chemie, Merck, or Pyrazine Specialties (Atlanta, GA).
Soluble compound avoidance assays were performed as described (Wicks et al., 2000). We used 150 mM and 15 mM CuSO4, 4% and 0.4% SDS and 4 M fructose as aversive compounds.
Chemotaxis to water-soluble compounds was assessed as described (Wicks et al., 2000). Briefly, pairs of opposite quadrants of four-quadrant Petri plates (Falcon X plate, Becton Dickinson Labware) were filled with buffered agar (2% agar, 5 mM K2HPO4/KH2PO4 pH 6.6, 1 mM CaCl2 and 1 mM MgSO4), either containing a dissolved attractant or no attractant. As attractants we used 100 ìM, 1, 10, 25 and 75 mM NaAc, NaCl, NH4Ac and NH4Cl. Adjacent quadrants were connected with a thin layer of molten agar. A population of well fed, young adult nematodes was washed three times with CTX buffer (5 mM KH2PO4/K2HPO4 pH 6.6, 1 mM CaCl2 and 1 mM MgSO4) and 100–200 worms were placed at the intersection of the four quadrants. The distribution of the worms over the four quadrants was determined at 10, 20, 30, 40, 50 and 60 minutes. A chemotaxis index [CI = (A – C)/A + C; where A is the number of worms over quadrants 1 and 3, C is the number of worms over quadrants 2 and 4] was calculated at each time point.
To test whether C.elegans adapts to water soluble compounds, the animals were pre-exposed to the compound tested during the washing steps. We determined the optimal concentration for adaptation and the time course of adaptation by exposing wild-type animals to increasing concentrations of NaAc, NaCl, NH4Ac or NH4Cl during a 5 to 30 min washing procedure. Subsequently the animals were tested in a standard water soluble compound chemotaxis assay for their attraction to the adapting salt (10 or 25 mM). Recovery of adaptation was determined in two ways. Animals pre-exposed to 100 mM adapting salt for 15 min were washed for an additional 5 min with CTX buffer without the adapting salt. This 20 min washing procedure did not seem toxic to wild-type animals, however, longer washing could not be tested due to toxicity. In the alternative recovery procedure the adapted animals were put on an agar plate containing only CTX buffered agar. After 5 min these animals were washed off this plate with CTX buffer, allowed to sediment for 1–2 min and subsequently tested in a standard chemotaxis assay. To determine the specificity of adaptation we exposed wild-type animals to 100 mM NaAc, NaCl, NH4Ac or NH4Cl and tested them for chemotaxis to NH4Cl, NaCl or NaAc.
All chemotaxis assays were performed at least four times.
Other assays
Living animals were stained with DiO (Molecular Probes) as described by Perkins et al. (1986). Locomotion and egg laying were assayed as described by Korswagen et al. (1997).
Statistical analysis
Statistical analysis of behavioural data was done with SPSS 8.0 for Windows. Depending on the assay, a paired t-test, a one way ANOVA, and Dunnett post-hoc, or factorial ANOVA, and Tuckey post-hoc comparisons, were used. An α level of 0.05 was used in all tests. All results are given as mean ± SE.
Acknowledgments
Acknowledgements
We thank Pia Werner, Henri van Luenen, Karin van der Linden, Alberto de la Fuente, Karen Thijssen and Marieke van der Horst for technical assistance, Frank Grosveld and Chris de Zeeuw for comments on the manuscript, the Caenorhabditis elegans Stock Center and Cori Bargmann for strains, the Sanger Center for providing cosmids and Andy Fire for GFP vectors. This work was supported by ALW/NWO grant 805.48.009 to G.J. and a Human Frontier Science Program Long Term Fellowship to D.W.
References
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I. and Horvitz,H.R. (1991) Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron, 7, 729–742. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I. and Mori,I. (1997) Chemotaxis and thermotaxis. In C. elegans II, Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R., (eds.) CSH Press, New York, NY, pp. 717–737. [PubMed]
- Bargmann C.I., Hartwieg,E. and Horvitz,H.R. (1993) Odorants-selective genes and neurons mediate olfaction in C. elegans. Cell, 74, 515–527. [DOI] [PubMed] [Google Scholar]
- Berger A.J., Hart,A.C. and Kaplan,J.M. (1998) Gαs-induced neurodegeneration in Caenorhabditis elegans. J. Neurosci., 18, 2871–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard N. and van der Kooy,D. (2000) A behavioral and genetic dissection of two forms of olfactory plasticity in Caenorhabditis elegans: Adaptation and habituation. Learn. Mem., 7, 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage L., Avery,L., Katz,A., Kim,U.J., Mendel,J.E., Sternberg,P.W. and Simon,M.I. (1996) Mutations in a C. elegans Gqα gene disrupt movement, egg-laying and viability. Neuron, 16, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Tu,Y., Euskirchen,G., Ward,W. and Prasher,D. (1994) Green fluorescent protein as a marker for gene expression. Science, 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Chase D.L., Patikoglou,G.A. and Koelle,M.R. (2001) Two RGS proteins that inhibit Go and Gq signaling in C. elegans neurons require a Gβ5-like subunit for function. Curr. Biol., 11, 222–231. [DOI] [PubMed] [Google Scholar]
- Colbert H.A. and Bargmann,C.I. (1995) Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron, 14, 803–812. [DOI] [PubMed] [Google Scholar]
- Colbert H.A. and Bargmann,C.I. (1997) Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learn. Mem., 4, 179–191. [DOI] [PubMed] [Google Scholar]
- Colbert H.A., Smith,T.L. and Bargmann C.I. (1997) OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabitis elegans. J. Neurosci., 17, 8259–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenbery D.B. (1980) Responses of the nematode Caenorhabditis elegans to controlled chemical stimulation. J. Comp. Physiol., 136, 327–331. [Google Scholar]
- Gilbertson T.A., Damak,S. and Margolskee,R.F. (2000) The molecular physiology of taste transduction. Curr. Opin. Neurobiol., 10, 519–527. [DOI] [PubMed] [Google Scholar]
- Gotta M. and Ahringer,J. (2001) Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nature Cell Biol., 3, 297–300. [DOI] [PubMed] [Google Scholar]
- Hadju-Cronin Y., Chen,W., Patikoglou,G., Koelle,M. and Sternberg P.W. (1999) Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev., 14, 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm H.E. (1998) The many faces of G protein signaling. J. Biol. Chem., 273, 669–672. [DOI] [PubMed] [Google Scholar]
- Han M. and Sternberg,P.W. (1991) Analysis of dominant negative mutations of the Caenorhabditis elegans let-60 ras gene. Genes Dev., 5, 2188–2198. [DOI] [PubMed] [Google Scholar]
- Hardie R.C. (1996) INDO-1 measurements of absolute resting and light-induced Ca2+ concentrations in Drosophila photoreceptors. J. Neurosci., 16, 2924–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Thijssen,K.L., Werner,P., van der Horst,M., Hazendonk,E. and Plasterk,R.H.A. (1999) The complete family of genes encoding G proteins of Caenorhabditis elegans. Nature Genet., 21, 414–419. [DOI] [PubMed] [Google Scholar]
- Korswagen H.C., Park, J-H., Ohshima,Y. and Plasterk,R.H.A. (1997) An activating mutation in a Caenorhabditis elegans Gs protein induces neuronal degeneration. Genes Dev., 11, 1493–1503. [DOI] [PubMed] [Google Scholar]
- Lackner M., Nurrish,S. and Kaplan,J. (1999) Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron, 24, 335–346. [DOI] [PubMed] [Google Scholar]
- L’Etoile N.D. and Bargmann,C.I. (2000) Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron, 25, 575–586. [DOI] [PubMed] [Google Scholar]
- Lochrie M.A., Mendel,J.E., Sternberg,P.W. and Simon,M.I. (1991) Homologous and unique G protein α subunits in the nematode Caenorhabditis elegans. Cell Regul., 2, 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel J.E., Korswagen,H.C., Liu,K.S., Hajdu-Cronin,Y.M., Simon,M.I., Plasterk,R.H.A. and Sternberg,P.W. (1995) Participation of the Go protein in multiple aspects of behavior in C. elegans. Science, 267, 1652–1655. [DOI] [PubMed] [Google Scholar]
- Miller K., Emerson,M. and Rand,J. (1999) Goα and diaceylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron, 24, 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.J. and Malbon,C.C. (1999) Physiological regulation of G protein-linked signaling. Physiol. Rev., 79, 1373–1430. [DOI] [PubMed] [Google Scholar]
- Nurrish S., Segalat,L. and Kaplan,J. (1999) Serotonin inhibition of synaptic transmission: Gαo decreases the abundance of UNC-13 at release sites. Neuron, 24, 231–242. [DOI] [PubMed] [Google Scholar]
- Park J-H., Ohshima,S., Tani,T. and Ohshima,Y. (1997) Structure and expression of the gsa-1 gene encoding a G protein α(s) subunit in C. elegans. Gene, 194, 183–190. [DOI] [PubMed] [Google Scholar]
- Peretz A., Sandler,C., Kirschfeld,K., Hardie,R.C. and Minke,B. (1994) Genetic dissection of light-induced Ca2+ influx into Drosophila photoreceptors. J. Gen. Physiol., 104, 1057–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock,E.M., Thomson,J.N. and Culotti,J.G. (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol., 117, 456–487. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J.T., Faumont,S., Gaston,M.R., Pearson,B.J. and Lockery,S.R. (2001) The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature, 410, 694–698. [DOI] [PubMed] [Google Scholar]
- Pitcher J.A., Freedman,N.J. and Lefkowitz,R.J. (1998) G protein-coupled receptor kinases. Annu. Rev. Biochem., 67, 653–692. [DOI] [PubMed] [Google Scholar]
- Roayaie K., Gage Crump,J., Sagasti,A. and Bargmann,C.I. (1998) The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron, 20, 55–67. [DOI] [PubMed] [Google Scholar]
- Robatzek M., Niacaris,T., Steger,K., Avery,L. and Thomas,J.H. (2001) eat-11 encodes GPB-2, a Gβ5 ortholog that interacts with Go and Gq to regulate C. elegans behavior. Curr. Biol., 11, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J.K. and Rankin,C.C. (2001) Analysis of habituation in Caenorhabditis elegans. Learn. Mem., 8, 63–39. [DOI] [PubMed] [Google Scholar]
- Sawin E.R., Ranganathan,R. and Horvitz H.R. (2000) C. elegans locomotory rate is modulated by environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron, 26, 619–631. [DOI] [PubMed] [Google Scholar]
- Ségalat L., Elkes,D.A. and Kaplan,J.M. (1995) Modulation of serotonin controlled behaviors by Go in Caenorhabditis elegans. Science, 267, 1648–1651. [DOI] [PubMed] [Google Scholar]
- Van der Linden A.M., Simmer,F., Cuppen,E. and Plasterk,H.A. (2001) The G protein β subunit GPB-2 in Caenorhabditis elegans regulates the Goα-Gqα signaling network through interactions with the RGS proteins EGL-10 and EAT-16. Genetics, 158, 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. (1973) Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl Acad. Sci. USA, 70, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Kumar,N., Morrison,G.E., Runciman,S., Rambaldini,G., Rousseau,J. and van der Kooy,D. (1997). Mutations that prevent associative learning in C. elegans. Behav. Neurosci., 111, 342–353. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate,E., Thomson,J.N. and Brenner,F.R.S. (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond., 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Wicks S.R., de Vries,C.J., van Luenen,H.G.A.M. and Plasterk,R.H.A. (2000) CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol., 221, 295–307. [DOI] [PubMed] [Google Scholar]
- Zwaal R.R., Broeks,A., van Meurs,J., Groenen,J.T.M. and Plasterk,R.H.A. (1993) Target selected gene inactivation by using a frozen transposon insertion mutant bank. Proc. Natl Acad. Sci. USA, 90, 7431–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal R.R., Ahringer,J., van Luenen,H.G.A.M., Rushforth,A., Anderson,P. and Plasterk,R.H.A. (1996) G proteins are required for the spatial orientation of early cell cleavage in C. elegans embryos. Cell, 86, 619–629. [DOI] [PubMed] [Google Scholar]
- Zwaal R.R., Mendel,J.E., Sternberg,P.W. and Plasterk,R.H.A. (1997) Two neuronal G proteins are involved in the chemosensation of dauer inducing pheromone by C. elegans. Genetics, 145, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]