Abstract
We report that in vivo increased acetylation of the repressed Saccharomyces cerevisiae ADH2 promoter chromatin, as obtained by disrupting the genes for the two deacetylases HDA1 and RPD3, destabilizes the structure of the TATA box-containing nucleosome. This acetylation-dependent chromatin remodeling is not sufficient to allow the binding of the TATA box-binding protein, but facilitates the recruitment of the transcriptional activator Adr1 and induces faster kinetics of mRNA accumulation when the cells are shifted to derepressing conditions.
Keywords: ADH2/chromatin remodeling/histone acetylation/Saccharomyces cerevisiae/transcriptional regulation
Introduction
Regulation of gene expression by chromatin structure is well established in eukaryotes. The involvement of ATP-dependent chromatin remodeling complexes and of acetylation/deacetylation in gene activation and/or repression has been clearly shown (Kingston and Narlikar, 1999; Kornberg and Lorch, 1999; Vignali et al., 2000; Wu and Grunstein, 2000). Most of the initial evidence came from genetic studies correlating loss of activation and/or repression with specific mutations in chromatin remodeling complexes and/or acetyltransferases and deacetylases (Hirschhorn et al., 1992; Peterson and Herskowitz, 1992; Braunstein et al., 1993; Rundlett et al., 1996; Kadosh and Struhl, 1998; Kuo et al., 1998; Struhl, 1998). These genetic analyses were followed by biochemical studies showing the in vitro effects of these modifying complexes (Hamiche et al., 1999; Längst et al., 1999; Lorch et al., 1999; Travers, 1999; Whitehouse et al., 1999; Aalfs and Kingston, 2000) on nucleosome structure. Additional studies have added to this matter the analysis of the acetylation levels of specific genomic regions in order to relate the effect of the modification of specific lysine residues to specific patterns of expression. For example, it was shown that transcriptional repression by UME6 involves RPD3-dependent deacetylation of histone H4 (Rundlett et al., 1998; Suka et al., 2001) and that cell cycle-regulated histone acetylation is required for the expression of the yeast HO gene (Krebs et al., 1999), while TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress transcription (Wu et al., 2001). Moreover, it was proposed that targeted histone acetylation by Gcn5 facilitates transcription in a causal fashion (Kuo et al., 2000). In general, promoter histone acetylation is differentially affected by specific activators and repressors (Deckert and Struhl, 2001). A detailed analysis of almost 54 kb of DNA in a search of developmentally regulated patterns of histone acetylation was presented recently (Litt et al., 2001). In addition to targeted histone modification, a background of global acetylation and deacetylation was reported (Kuo et al., 2000; Vogelauer et al., 2000), indicating that the state of acetylation of a genome is in constant flux.
What is still missing in this field is an understanding at the molecular level of the direct effect of in vivo changes in histone acetylation on gene expression. Do changes in histone proteins lead directly to physical changes in nucleosome structure and promoter utilization? An attempt in this direction was recently made in the PHO8 promoter system (Reinke et al., 2001). Interestingly, a transient hyperacetylation peak over the PHO8 promoter, limited to precisely those nucleosomes that are remodeled upon activation, is induced by SAGA; nevertheless, the evidence presented points against a direct effect of acetylation on chromatin accessibility in vivo.
In order to determine whether histone acetylation affects promoter accessibility, we chose as a model system the Saccharomyces cerevisiae ADH2 gene, coding for the enzyme alcohol dehydrogenase II, in its natural chromosomal location. This gene is tightly regulated by glucose and becomes active when the glucose concentration of the medium is lowered or in the presence of non-fermentable carbon sources. An upstream regulatory element (UAS1; Beier and Young, 1982), which binds the transcription factor Adr1 (Denis and Young, 1983), is required for its derepression. We have previously shown that when yeast cells are grown in repressing conditions (3% glucose), two nucleosomes (–1 and +1) occupy the basic promoter elements: the TATA box and the RNA initiation sites (RIS), respectively (Verdone et al., 1996). UAS1 is located in a nucleosome-free region, but one of the two Adr1-binding sites is immediately adjacent to the upstream borders of the TATA box-containing –1 nucleosome. A relevant role for the two nucleosomes in the maintenance of transcriptional repression is shown by the fact that by blocking the production of histone H4 in vivo, and therefore the correct assembly of the nucleosome particles, ADH2 becomes active even in the presence of glucose (Wyrick et al., 1999). When yeast cells are grown in low glucose (0.05%), the two promoter nucleosomes, together with other adjacent particles, undergo ADR1-dependent chromatin remodeling and transcriptional activation (Verdone et al., 1996, 1997; Di Mauro et al., 2000).
In order to understand the role of histone acetylation in ADH2 gene expression, it is necessary to demonstrate that acetylation alters chromatin structure and that the acetylation-dependent nucleosome structural modifications influence the ability of the promoter to be activated.
By genetically altering the steady-state pattern of histone acetylation at the repressed ADH2 promoter, we show that when the histone deacetylases HDA1 and RPD3 are mutated, the structure of the TATA box-containing nucleosome is destabilized, the promoter becomes accessible to Adr1, and, when the cells are shifted to derepressing conditions, the kinetics of mRNA accumulation is faster. We also show that by disrupting the genes for the two acetyltransferases GCN5 and ESA1, the ADH2 promoter structure and function are affected. In particular, in the GCN5 mutant, the chromatin remodeling occurring in derepressing conditions is less pronounced and the kinetics of mRNA accumulation is slower, whereas in the presence of an ESA1 temperature-sensitive mutation, the amount of mRNA is lower even in permissive conditions. Therefore, histone deacetylation/acetylation is directly involved in modulating the accessibility of chromatin at the ADH2 gene.
Results
The histone acetylation level of the ADH2 promoter changes when the deacetylases HDA1 and RPD3 and the acetyltransferases ESA1 and GCN5 are not functional
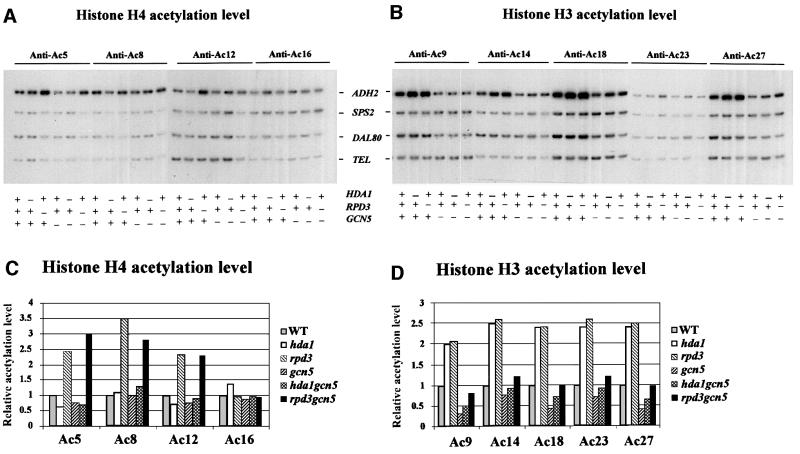
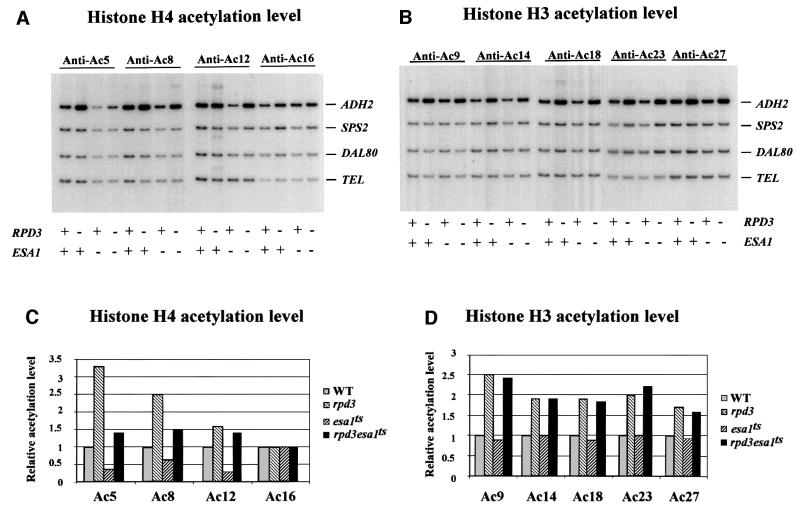
We started our analysis by searching for a direct effect on the ADH2 histone acetylation level of disruptions in the genes coding for the two major S.cerevisiae histone acetyltransferases, GCN5 (a component of both the SAGA and the ADA complexes; Grant et al., 1997) and ESA1 (a member of the NuA4 complex; Smith et al., 1998; Allard et al., 1999; Clarke et al., 1999), and in the genes for the two deacetylases HDA1 and RPD3. The study was performed by chromatin immunoprecipitation (ChIP) analysis of a region encompassing the basic transcription elements, the TATA box and the RNA initiation sites, occupied by nucleosomes –1 and +1, respectively (see the diagram in Figure 3 and Verdone et al., 1997). Figures 1 and 2A and B show the results obtained by PCR amplification of a 337 bp ADH2 fragment relative to a 138 bp fragment, 0.5 kb from the telomere (Tel) of chromosome VI-R, used as an internal control for the quantity of DNA. Nine different antibodies were used (five specific for the histone H3 acetylated lysines AcK9, AcK14, AcK18, AcK23 and AcK27, and four specific for the histone H4 acetylated lysines AcK5, AcK8, AcK12 and AcK16) to immunoprecipitate formaldehyde-crosslinked chromatin of different cell extracts prepared from cultures grown in 3% glucose, i.e. in repressing conditions for ADH2 expression. The quantitative evaluation of these data is reported in Figures 1 and 2C and D; the histone acetylation level for each mutant strain is shown relative to the wild-type level, equivalent to 1. In the case of histone H4, it is clear that RPD3 deacetylates three out of the four lysines (K5, K8 and K12; Figure 1C) and that the same residues are acetylated by ESA1 (Figure 2C). In fact, the hyperacetylation observed in the rpd3 mutant is abolished in the double mutant rpd3/esa1ts (Figure 2C), but not in the double mutant rpd3/gcn5 (Figure 1C). In the case of histone H3, both RPD3 and HDA1 deacetylate the five lysines (K9, K14, K18, K23 and K27; Figure 1D) that are acetylated by GCN5 (Figure 1D). In fact, the hyperacetylation observed in the hda1 and rpd3 single mutants is abolished in the double mutants hda1/gcn5 and rpd3/gcn5 (Figure 1D). Moreover, GCN5 does not acetylate the histone H4 lysines because the hyperacetylation shown in the rpd3 mutant is not abolished in the double mutant rpd3/gcn5 (Figure 1C), and ESA1 does not acetylate the histone H3 lysines because the hyperacetylation shown in a second rpd3 mutant is not abolished in the double mutant rpd3/esa1ts (Figure 2D).
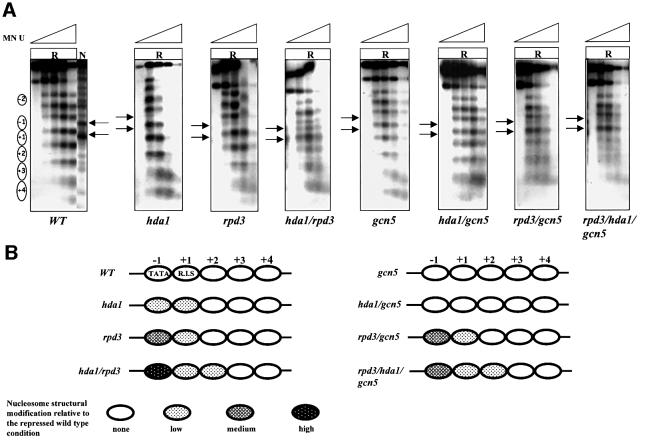
Fig. 3. Effects of histone acetylation on ADH2 chromatin structure. (A) MN analysis of the ADH2 promoter region in repressing conditions (R, 3% glucose). Nystatin-permeabilized spheroplasts were reacted with increasing amounts of MN (U = units/0.25 ml: 0, 0.57, 1.7, 5 and 15), deproteinized, digested with BamHI and HindIII (map positions are –1202 and +760, respectively), and analyzed by indirect end-labeling. Nucleosomes (i.e. protected areas) are represented as ovals. Lane N (naked control) contains deproteinized DNA from plasmid pFA treated in vitro with MN and the same restriction enzymes as the in vivo samples. (B) Summary of nucleosome structural modifications. The chromatin changes, relative to the wild-type YDS2 strain, are summarized; the different shadings indicate the intensity of the changes (none, low, medium and high).
Fig. 1. Effects of histone acetyltransferase GCN5 and histone deacetylases HDA1 and RPD3 on acetylation of histones at the ADH2 promoter. ChIP demonstrating the effects of histone acetyltransferase and histone deacetylase disruptions on the acetylation of (A) histone H4 sites K5, K8, K12 and K16, and (B) histone H3 sites K9, K14, K18, K23 and K27. The ADH2 fragment spans the region from –223 to +114, relative to the ATG. Amplification of a 138 bp fragment 0.5 kb from the telomere (Tel) of chromosome VI-R was used as a reference to ensure equal loading of samples. Yeast strains used for ChIP were wild type (WT) (YDS2), hda1 (WJY111), rpd3 (WJY140), gcn5 (WJY139), hda1/gcn5 (WJY142) and rpd3/gcn5 (WJY143) (Vogelauer et al., 2000). SPS2 and DAL80 were found to be relatively unaffected by these mutations and were used as negative controls. (C and D) Quantification of the increase in H4, H3 acetylation in mutant cells relative to wild-type cells. [α-32P]dATP was added to the PCR mixture, and the PhosphorImager was used to quantitate the intensity of ADH2 PCR bands in these mutants relative to WT after normalizing to the TEL bands.
Fig. 2. Effects of histone acetyltransferase ESA1 on acetylation of histones at the ADH2 promoter. ChIP demonstrating the effects of histone acetyltranferase esa1ts mutant on the acetylation of (A) histone H4 sites K5, K8, K12 and K16, and (B) histone H3 sites K9, K14, K18, K23 and K27. The ADH2 fragment spans the region from –223 to +114, relative to the ATG. Amplification of a 138 bp fragment 0.5 kb from the telomere (Tel) of chromosome VI-R was used as a reference to ensure equal loading of samples. Yeast strains used for ChIP were WT (LPY3431), rpd3 (NSY164), esa1ts (LPY3430), rpd3/esa1ts (NSY165). SPS2 and DAL80 were found to be relatively unaffected by these mutations and were used as negative controls. (C and D) Quantification of the increase in H4, H3 acetylation in mutant cells relative to wild-type cells. [α-32P]dATP was added to the PCR mixture, and the PhosphorImager was used to quantitate the intensity of ADH2 PCR bands in these mutants relative to WT after normalizing to the TEL bands.
From these data we can conclude that the histone acetylation level of the ADH2 promoter is directly affected by disrupting the two main acetyltransferases GCN5, which is H3 specific, and ESA1, which is H4 specific, and the deacetylases HDA1, which is H3 specific, and RPD3, which is specific for both H3 and H4. How do all these covalent modifications affect the structure of the nucleosomes present at the promoter and therefore the ability of the ADH2 gene to be activated?
Histone deacetylation/acetylation is directly involved in modulating the structure of the TATA box-containing nucleosome
In order to assess the influence of the changes in the acetylation level on the structure of the nucleosomes present in the repressed ADH2 promoter in its natural chromosomal location, we analyzed by indirect end labeling the micrococcal nuclease (MN) digestion pattern in a set of isogenic strains carrying single, double or triple disruptions in the genes coding for the main histone deacetylases and acetyltransferases. The results are shown in Figure 3. Increasing amounts of MN were introduced by nystatin permeabilization of the spheroplasts obtained from cells grown in repressing (R = 3% glucose) conditions. The pattern of in vivo MN cleavage sites is compared with that of an in vitro treated sample (lane N = naked chromosomal DNA) in order to reveal the protection from DNA cleavage due to the presence of nucleosomes. In the case of the wild-type strain, an array of nucleosomes covering the entire area of the promoter and the surrounding regions (with the exception of a nucleosome-free region located between particles –2 and –1; see Verdone et al., 1996) is clearly visible.
When looking at the MN profiles of both the rpd3 and hda1 mutants, we noted a slight modification in the ability of the promoter nucleosomes –1 and +1 to protect the underlying DNA sequence (Figure 3). The loosened structure of these nucleosomes is more clearly seen in the case of the hda1/rpd3 double mutant, suggesting that the hyperacetylation observed on both histones H3 and H4 in the absence of RPD3 and/or HDA1 (caused by the histone acetyltransferase activity of GCN5 and ESA1, respectively; see Figures 1 and 2) is responsible for the partial loss of protection of the ADH2 basic promoter elements.
When the nucleosome structure is analyzed in the gcn5 mutant, the MN profile is very similar to that of the wild type, suggesting that the H3 hypoacetylation characteristic of this mutant (Figure 1C) helps to keep the promoter chromatin in a relatively inaccessible configuration.
When combined with hda1, the gcn5 disruption behaves as a suppressor; in fact, the hda1/gcn5 strain is characterized by a chromatin structure very similar to that of the wild type and of the single gcn5 mutant strain, with an array of tightly associated nucleosomes over the promoter (Figure 3). The implication is that the hypoacetylation of histone H3 in the hda1/gcn5 double mutant is responsible for the suppression of the partial opening of the nucleosome –1 structure, characteristic of the hda1 mutant (Figure 3).
In the double mutant rpd3/gcn5, the –1 nucleosome is characterized by a loosened structure typical of the rpd3 mutant, suggesting that in repressing conditions the gcn5 disruption does not suppress the rpd3 phenotype.
In the case of the triple mutant rpd3/hda1/gcn5, the delicate balance among the various acetyltransferases and deacetylases is such that in repressing conditions the chromatin structure is loosened (Figure 3, last panel); in fact, the gcn5 disruption only partially suppresses the phenotype due to the double hda1/rpd3 disruption.
A diagram showing the results of the chromatin analysis in repressing conditions for the entire set of mutants is presented in Figure 3B.
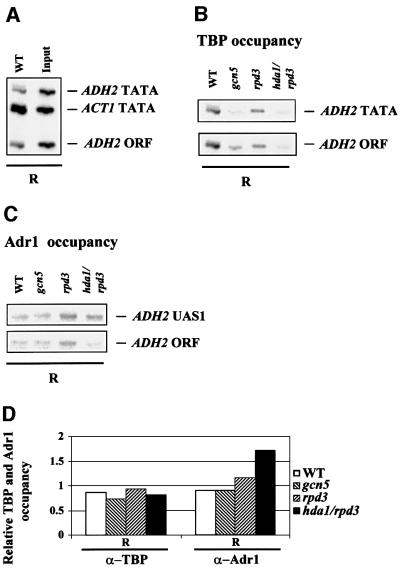
Histone deacetylation/acetylation affects Adr1 but not TATA box-binding protein (TBP) access to the ADH2 promoter in repressing conditions
Adr1 is the major transcription factor binding in the nucleosome-free region upstream of the –1 nucleosome when cells are derepressed. Whether Adr1 is present during repressed growth in a chromatin-bound but transcriptionally inactive form, or is not bound to chromatin, has not been resolved. If the latter were the case, destabilization of nucleosome –1 might allow Adr1 to bind UAS1 even in the presence of glucose, and this could lead to more rapid derepression of ADH2 expression.
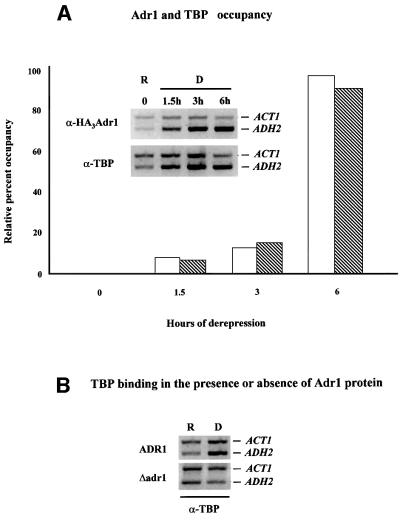
To test this possibility we performed ChIP using a strain containing an HA epitope-tagged version of Adr1 that behaved identically to the wild-type allele. The data shown in Figure 4A indicate that Adr1 is not bound to the ADH2 promoter in glucose-repressed cells containing a full complement of deacetylases. Adr1 can be detected at the ADH2 promoter ∼90 min after the cells are shifted to medium lacking glucose, and the amount of Adr1 at the promoter continues to increase until at least 6 h, as measured by real-time PCR of the immunoprecipitated DNA. When the arrival of TBP at the ADH2 promoter is determined, it appears to coincide with the appearance of Adr1 (Figure 4A), suggesting that its recruitment is ADR1 dependent. Consistent with this interpretation, TBP cannot be detected at the ADH2 promoter in a strain lacking Adr1 (Figure 4B). These results show: (i) that Adr1 is not bound to the ADH2 promoter during glucose repression and recruits TBP upon derepression; and (ii) that Adr1 must be present at the promoter for TBP to bind.
Fig. 4. Adr1 and TBP occupancy of the ADH2 promoter. (A) ChIP assays for Adr1-HA and TBP were performed with anti-HA and anti-yTBP, respectively, as described in Materials and methods. The primer pairs for the PCR analysis, followed by gel electrophoresis (inset), are located at –432 and –139 from the ADH2 ATG. The primer pairs for the ACT1 control were located at +418 and +797 in the ACT1 ORF. Real-time PCR analysis was carried out with an Applied Biosystems Model 700 and software as described by the manufacturer. The primer pairs for the real-time PCR analysis are located at –283 and –187 from the ADH2 ATG. The primer pairs for the ACT1 control were located at +448 and +531 in the ACT1 ORF. The real-time PCR data are shown in the graph and are plotted as a percent of the ADH2 product generated using ChIP DNA isolated from repressed cells, after correcting for the amount of ACT1 DNA in each sample (white and hatched bars refer to Adr1 and TBP occupancy, respectively). R, repressed conditions (3% glucose); D, derepressed conditions (0.05% glucose). (B) ChIP assays for TBP occupancy in wild-type and adr1 strains. PCR and electrophoretic analysis were carried out as described in Materials and methods and in (A).
We reasoned that the destabilization of the TATA box-containing nucleosome –1, observed in the deacetylase mutants (Figure 3), could influence the ability of members of the transcription machinery or of the activator itself to bind in vivo their target sites in repressed growth conditions, when neither Adr1 nor TBP was normally bound to the ADH2 promoter. We therefore analyzed by ChIP with anti-Adr1 or anti-yTBP antibodies the occupancy of the ADH2 UAS1 and TATA box region by Adr1 and TBP, respectively, in four different strains: wild type, rpd3, gcn5 and rpd3/hda1. The results are shown in Figure 5. All the extracts were prepared from cells grown in 3% glucose (repressing conditions) and the immunoprecipitated material was amplified by PCR with different pairs of oligonucleotides. As a negative control for unspecifically immunoprecipitated material, we included a fragment (ADH2 ORF) containing a small portion of the ADH2 coding region, located between 800 and 900 bp from the TATA box. The positive control for TBP binding is a fragment (ACT1 TATA) containing the TATA box of the ACT1 gene, which is constitutively expressed in high-glucose conditions (Figure 5A). The results show that there is no change in TBP binding efficiency at the ADH2 promoter when comparing the various mutants with the wild type (Figure 5B). In particular, there is no increase in TBP occupancy for the ADH2 TATA box fragment relative to the ADH2 ORF fragment when comparing the deacetylase mutants with the wild-type strain, suggesting that the hyperacetylation-dependent increased accessibility to MN of the TATA box-containing nucleosome is not affecting the accessibility of the promoter to the transcription machinery. On the other hand, when testing the anti-Adr1 antibody with the same extracts, we observed an increase in the occupancy of the ADH2 promoter by Adr1 in the hda1/rpd3 double mutant (Figure 5C). The quantitative evaluation of these data is presented in Figure 5D.
Fig. 5. Acetylation-dependent Adr1 binding of the repressed ADH2 promoter. (A) Positive control for TBP binding. DNA immuno precipitated with anti-yTBP (α-TBP) from wild-type extract under repressing conditions (R) and input DNA derived from the same extract were amplified with three different pairs of oligonucleotides at the same time: ADH2 TATA fragment (234 bp) spans the region from –223 to +11; ACT1 TATA fragment (182 bp) contains the ACT1 TATA box; ADH2 ORF fragment (108 bp) contains a small portion of the ADH2 coding region from +658 to +778 (negative control). Amplifications were performed using the following amounts of DNA: WT = 1/50; Input = 1/2500. (B) ChIP with anti-yTBP antibody illustrating the occupancy of the ADH2 TATA box region by TBP in the WT, gcn5, rpd3 and hda1/rpd3 strains, in repressing conditions (R). The TBP-immunoprecipitated material was amplified by PCR with two different pairs of oligonucleotides at the same time. (C) ChIP with anti-Adr1 antibody (α-Adr1) demonstrating the occupancy of the ADH2 promoter in the WT, gcn5, rpd3 and hda1/rpd3 strains in repressing conditions (R). The Adr1-immunoprecipitated material was amplified by PCR with two different pairs of oligonucleotides at the same time. The ADH2 UAS1 fragment (217 bp) spans the region from –379 to –162. (D) Densitometric evaluation of the results shown in (B) and (C). The histograms were obtained by dividing the values of the ADH2 TATA and of the ADH2 UAS1 fragments by the value of the ADH2 ORF fragment, used as a negative control.
We conclude that the destabilization of the TATA box-containing –1 nucleosome even in high-glucose conditions (i.e. in the absence of transcription) induces the binding of Adr1 to the promoter, but not the recruitment of TBP, suggesting that the access of the transcriptional activator to its target sites represents an essential but not sufficient regulatory step for ADH2 derepression, since in repressed conditions TBP is still not recruited despite the presence of Adr1.
These data show that the exact pattern of covalent modifications of the promoter nucleosome, and therefore its precise structural organization, has an important role in the control of Adr1 activator chromosomal localization.
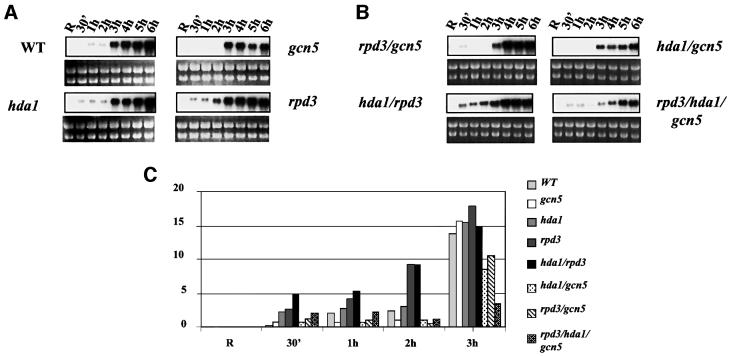
Increased histone acetylation determines faster kinetics of ADH2 promoter transcriptional activation, while decreased histone acetylation causes slower kinetics of derepression
In order to assess the influence of changes in the acetylation level on the expression of our model gene, we analyzed by northern blotting the kinetics of ADH2 mRNA accumulation in the same set of isogenic mutant strains for which the chromatin analysis was performed. Figure 6 shows the results of this analysis; for each mutant, the ADH2 mRNA was tested in the initial repressed conditions (R = 3% glucose) and at different times (from 30 min up to 6 h) after decreasing the amount of glucose in the medium to 0.05%. When comparing the kinetics of both the hda1 and rpd3 mutants with those of the wild type, a faster derepression is observed: the ADH2 mRNA is already present at 30 min after the shift to derepressing conditions, suggesting a role in the transcriptional activation process for the increased histone acetylation caused by the absence of HDA1 or RPD3. The effect of the increased acetylation caused by the absence of the two deacetylases is additive; in fact, when comparing the kinetics of the double mutant hda1/rpd3 with that of each single mutant, the ADH2 mRNA signal at 30 min after the shift is stronger and equivalent to the sum of the signals at 30 min of each single mutant (see also the densitometric evaluation in Figure 6C). This additive effect, due to the hyperacetylation of both histones H3 and H4, correlates with the stronger chromatin modification seen in the case of the hda1/rpd3 double mutant (see Figure 3). We have also analyzed the mRNA accumulation at a shorter time (10 min) after the shift to derepressing conditions: the ADH2 signal is already visible in the rpd3 and hda1 strains, but not in the wild type, and it is even stronger in the hda1/rpd3 strain (data not shown).
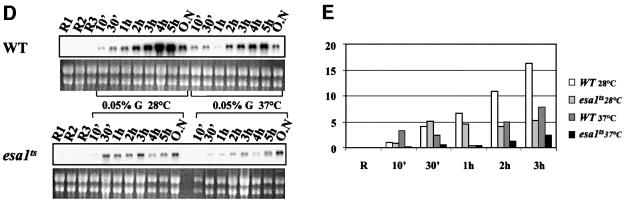
Fig. 6. Effects of histone acetylation on the kinetics of ADH2 mRNA accumulation. (A and B) Northern analysis for the wild-type YDS2 strain and the mutants indicated. Cells were grown overnight in repressing conditions (3% glucose, lanes R), washed, resuspended in fresh medium containing 0.05% glucose and analyzed at the times indicated. (C) Histogram representing the densitometric evaluation of the results shown in (A) and (B), from R (repressing condition) up to 3 h. (D) Northern analysis for the wild-type LPY3430 and its isogenic esa1ts. Cells were grown overnight at 28°C in repressing conditions (R1 lanes). The culture was split into two: one flask was kept at 28°C, the other was shifted to 37°C for 8 h in order to inactivate Esa1 (Clarke et al., 1999). The same amount of cells from each culture was analyzed (R2 = 28°C; R3 = 37°C). After Esa1 inactivation, the cells were washed and resuspended in fresh medium containing 0.05% glucose and analyzed at the times indicated (from 10 min to overnight). (E) Histogram representing the densitometric evaluation of the results shown in (D), from the initial repressing conditions (R = R1, R2, R3) up to 3 h.
On the other hand, when the kinetics of the gcn5 mutant is analyzed, there is no signal of ADH2 mRNA accumulation up to 3 h after the shift, suggesting a role for the GCN5-dependent histone acetyltransferase activity in the initial step of transcriptional activation at this promoter. When combined with the hda1 mutation, the gcn5 disruption behaves as a suppressor; in fact, the kinetics of mRNA accumulation in the hda1/gcn5 strain is very similar to those of the single gcn5 strain, with no signal up to 3 h after the shift. The implication is that the faster kinetics of ADH2 derepression observed in the hda1 mutant is due to GCN5-mediated hyperacetylation. Instead, when combined with the rpd3 mutation, the gcn5 disruption appears to behave as a suppressor only at 1 and 2 h after the shift, whereas at 30 min the faster kinetics due to the absence of RPD3 is the predominant effect. The implication is that the faster kinetics of derepression, observed in the absence of RPD3 only, is due to an increased acetylation level partly mediated by GCN5 and partly by a different histone acetyltransferase, presumably ESA1.
In the triple mutant rpd3/hda1/gcn5, the delicate balance among the various acetyltransferases and deacetylases is such that at 30 min and 1 h after the shift, a low ADH2 mRNA level is visible. However, at 2–3 h after the shift, the mRNA signal is reduced with respect to the double rpd3/gcn5 mutant, and at the same time a reduced extent of chromatin remodeling is observed (data not shown).
We have also studied the effect of an esa1 temperature-sensitive mutant on ADH2 transcription; the results are shown in Figure 6D and E. The effect of the esa1 mutation should be visible only at the restrictive temperature (37°C); nevertheless, it appears that the mutant esa1 protein does not behave normally even at the permissive temperature (28°C). In fact, the ADH2 mRNA level is always lower in the mutant when compared with the wild type both at 28 and 37°C, suggesting a role for the ESA1-mediated promoter acetylation in the transcriptional process.
From this set of data, we conclude that the change in acetylation level, observed at the ADH2 promoter in each mutant relative to the wild type, correlates not only with a defined pattern of nucleosome structural alterations, but also with a change in the kinetics of mRNA accumulation.
The acetyltransferase Gcn5 is required for efficient chromatin remodeling during derepression
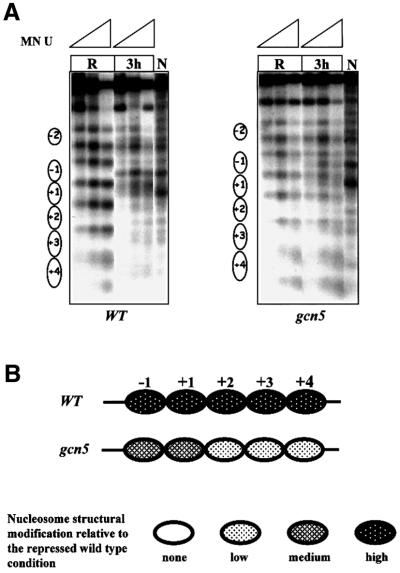
The analysis of the MN profiles of the wild-type and gcn5 mutant strains in repressing conditions did not reveal any difference (Figure 3), even though the histone H3 acetylation level was lower in the mutant when compared with the wild type (Figure 1D). However, the different acetylation pattern is responsible for the slower kinetics of mRNA accumulation observed in the gcn5 strain (Figure 6). We therefore also analyzed the ADH2 promoter chromatin structure in derepressing conditions in the wild-type and gcn5 strains. The results are shown in Figure 7. At 3 h after the shift to derepressing conditions, some differences become evident: (i) the intensity of the MN induced bands appearing inside all particles present on both the promoter and the coding region, very high in the case of the wild type, is reduced in the gcn5 mutant; and (ii) the intensity of the MN induced bands at the borders of all nucleosomes is very similar when going from repressing to derepressing conditions in the gcn5 mutant, whereas it is strongly reduced in the wild type after the metabolic shift.
Fig. 7. Role of the acetyltransferase Gcn5 in ADH2 promoter chromatin remodeling. (A) MN analysis of the ADH2 promoter region in repressing conditions (R, 3% glucose) and at 3 h after the shift to derepressing conditions (3 h, 0.05% glucose). Nystatin-permeabilized spheroplasts, of both wild-type and gcn5 strains, were reacted with increasing amounts of MN (U = units/0.25 ml: 0.57, 1.7 and 5), deproteinized, digested with BamHI and HindIII (map positions are –1202 and +760, respectively), and analyzed by indirect end-labeling. Nucleosomes (i.e. protected areas) are represented as ovals. Lanes N (naked control) contain deproteinized DNA from plasmid pFA treated in vitro with MN and the same restriction enzymes as the in vivo samples. (B) Summary of nucleosome structural modifications. The chromatin changes in the gcn5 strain, relative to the wild-type YDS2 strain, are summarized; the different shadings indicate the intensity of the changes (none, low, medium and high).
The results are consistent with a reduced ability of the ADH2 nucleosomes to undergo chromatin remodeling in the absence of the GCN5-dependent histone acetyltransferase activity.
Discussion
Histone acetylation state regulates ADH2 promoter chromatin structure and transcription
We have analyzed the in vivo chromatin structure and the kinetics of transcriptional activation of the S.cerevisiae ADH2 promoter as a function of genetically modified histone acetylation levels.
In an exponentially growing wild-type strain in the presence of glucose (repressing conditions for the ADH2 gene), an equilibrium exists among the various histone acetyltransferases and deacetylases. When this equilibrium is altered by abolishing the function of either or both RPD3 and HDA1, an imbalance is created, leading to hyperacetylation of specific lysine residues, thus causing specific modifications of the structure of both nucleosomes –1 and +1 (see Figure 3). The partial loss of protection observed does not induce mRNA synthesis in repressing conditions, but is responsible for inducing a defined timing for the synthesis of the ADH2 mRNA when the cells are shifted to derepressing medium (0.05% glucose). In particular, faster kinetics of mRNA accumulation is obtained when either or both RPD3 and HDA1 genes are disrupted (see Figure 6).
On the other hand, when the equilibrium is altered by abolishing the activity of the acetyltransferase Gcn5, the ability of histone H3 to be correctly acetylated is compromised. In repressing conditions, the structures of nucleosomes –1 and +1 appear very similar to the wild-type situation, whereas in derepressing conditions the reduced acetylation causes a decreased extent of chromatin remodeling, leading to slower kinetics of activation.
When combined with hda1, the gcn5 disruption behaves as a suppressor; in fact, the hda1/gcn5 strain is characterized by a chromatin structure very similar to that of the wild type and of the single gcn5 mutant strain, with an array of tightly associated nucleosomes over the promoter (Figure 3). The implication is that the hypoacetylation of the histone H3 in the hda1/gcn5 double mutant is responsible for the suppression of the partial opening of the nucleosome –1 structure, characteristic of the hda1 mutant (Figure 3). This suppressor effect is also reflected at the functional level: the kinetics of mRNA accumulation, faster in the single hda1 mutant, become slower in the hda1/gcn5 strain (see Figure 6).
The effects of a gcn5 disruption combined with an rpd3 disruption are more complex. Since RPD3 deacetylates both histones H3 and H4 (Figure 1), the most likely interpretation of the data regarding the double mutant rpd3/gcn5 is that the dominant rpd3 mutant effect seen in repressing conditions on the –1 nucleosome structure (Figure 3) and on mRNA synthesis very soon (30 min) after the shift to derepressing conditions (see Figure 6) is due to histone H4 hyperacetylation, whereas the dominant gcn5 mutant effect on both transcription (see Figure 6) and nucleosome structure (data not shown) seen a few hours after the shift is due to histone H3 hypoacetylation.
In the case of the triple mutant rpd3/hda1/gcn5, the delicate balance among the various acetyltransferases and deacetylases is such that in repressing conditions the structure of nucleosomes –1, +1 and +2 is loosened (Figure 3), correlating with the low amount of mRNA signal visible at 30 min and 1 h after the shift (see Figure 6).
Mechanisms of nucleosome modifications by changes in the histone acetylation level
How is the increased accessibility to MN of the two promoter nucleosomes achieved? One of the most common hypotheses deals with the decreased affinity for DNA of the nucleosome particle embedding hyperacetylated histones H3 and H4. In fact, hyperacetylated chromatin adopts a more ‘open’ structure (Garcia-Ramirez et al., 1995) and was shown to be generally sensitive to DNase I in vivo (Hebbes et al., 1994). In recent work, it was shown that hyperacetylated nucleosomes, isolated from HeLa cells grown in butyrate to inhibit all cellular deacetylases, affect the equilibrium constants for site exposure to restriction enzyme cleavage at various positions throughout the nucleosome (Anderson et al., 2001). The effect of histone acetylation on nucleosome–nucleosome interactions has not been defined, although internucleosomal contacts involving the N-terminal tails are likely to have a significant impact on higher order chromatin structure (Annunziato and Hansen, 2000).
Alternatively, the acetylation of specific lysine residues could serve as a marker for the binding of proteins required for a subsequent step in the transcription process (Strahl and Allis, 2000; Marmorstein, 2001). By using purified systems, it was shown that histone acetylation facilitates RNA polymerase II transcription in chromatin (Nightingale et al., 1998). In vitro evidence suggests that the H3/H4 tails are the primary arbiters of transcription factor access to intranucleosomal DNA (Vettese-Dadey et al., 1996; Howe et al., 1998; Vitolo et al., 2000). More recently, histone acetylation by either SAGA or NuA4 HAT complexes was shown to stabilize SWI/SNF binding to promoter nucleosomes (Hassan et al., 2001). The specific timing for the targeting of these complexes in vivo was studied in detail in the case of a cell cycle-regulated S.cerevisiae promoter (Cosma et al., 1999; Krebs et al., 1999).
In the case of the ADH2 promoter, we tested whether recruitment of TBP could be favored when the TATA box-containing nucleosome was remodeled in an rpd3- and hda1-dependent manner. However, we did not observe any increase in the occupancy of the promoter by TBP (Figure 5), suggesting that another factor, capable of inducing faster kinetics of activation in the rpd3, hda1 and rpd3/hda1 strains, is being recruited. This factor turned out to be the Adr1 protein itself, whose binding to the ADH2 UAS1 is inhibited by glucose (Figure 4; Sloan et al., 1999), presumably by means of the TATA box-containing nucleosome. Nevertheless, only after the glucose content of the medium is lowered does the activator recruit the transcription machinery and the mRNA synthesis begin. In this way, the facilitated recruitment of Adr1 in repressing conditions determines faster kinetics of activation soon after the metabolic switch.
A model for the control of Adr1 chromosomal localization
The position of both UAS1 and UAS2 in the ADH2 promoter is such that apparently there is no need to invoke a reorganization of the chromatin structure in order to allow the access of the activator to the promoter. They both are, in fact, located in a nucleosome-free region (Verdone et al., 1996). However, if one analyzes the exact location of the more upstream borders of the TATA box-containing –1 nucleosome (Verdone et al., 1996), it turns out that the left Adr1 consensus site in the UAS1 palindromic sequence is more accessible than the right one. Considering that the protein is quite large (1323 amino acids), one could expect some kind of steric hindrance exerted by the nucleosome particle on the binding of the second Adr1 molecule at the right consensus site. The present finding that Adr1 is not present on the wild-type promoter in high-glucose medium (Figure 4), but can bind in the same conditions in a strain carrying a disruption in both HDA1 and RPD3 histone deacetylases (Figure 5), points to the possibility that the repressive glucose effect is exerted, at least in part, at the DNA binding domain level by the presence of a defined nucleosome structure. When the TATA box-containing nucleosome is covalently modified by GCN5- and/or ESA1-mediated hyperacetylation, it becomes destabilized (as proposed; see Chiang et al., 1996), already allowing Adr1 to be correctly localized on the chromosome under repressing conditions. Nevertheless, TBP is not recruited in these conditions, suggesting that the access of the transcriptional activator to its target sites represents an essential but not sufficient regulatory step for ADH2 derepression.
Histone deacetylation/acetylation is, therefore, directly involved in altering the chromatin structure at the ADH2 promoter, influencing the binding of the major transcriptional activator with a concomitant effect on the kinetics of mRNA accumulation.
Materials and methods
Yeast strains and media
Saccharomyces cerevisiae strains used in this work are WJY139 (MATa trp1-1 his3-11,15 ade2-1 leu2-1 can1-100 gcn5::URA3), WJY140 (MATa trp1-1 his3-11,15 ade2-1 ura3-52 can1-100 rpd3::LEU2), WJY111 (MATa ura3-52 his3-11,15 ade2-1 leu2-1 can1-100 hda1::TRP1), WJY141 (MATa ura3-52 his3-11,15 ade2-1 can1-100 rpd3::LEU2, hda1::TRP1), WJY142 (MATa his3-11,15 ade2-1 leu2-1 can1-100 hda1::TRP1, gcn5::URA3), WJY143 (MATa trp1-1 his3-11,15 ade2-1 can1-100 rpd3::LEU2, gcn5::URA3) and WJY149 (MATa his3-11,15 ade2-1 can1-100 rpd3::LEU2, hda1::TRP1, gcn5::URA3). All these mutants are isogenic to YDS2 (Laman et al., 1995).
LPY3430 (MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1::HIS3, esa1-L327S::URA3) and LPY3431 (MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52, ESA1) were kindly provided by L.Pillus (Clarke et al., 1999). NSY164 (rpd3) and NSY165 (rpd3/esa1ts) were constructed by deleting RPD3 in LPY3431 and LPY3430, respectively (Suka et al., 2001).
W303-1a (MATa ade2 CAN1-100 his3-11,15 leu 2-13,112 trp1-1 ura3-1), TYY202 (W303-1a adr1::LEU2) and KVRY9 (W303-1b MATα ADR1-HAX3 KANr ade2 CAN1-100 his3-11,15 leu2-13,112 trp1-1 ura3-1) were used to test Adr1 and TBP occupancy of the ADH2 promoter.
Yeast strains were grown in YPD medium (1% yeast extract, 2% bacto peptone, 3% glucose). To obtain ADH2 derepression, the cells were collected by centrifugation, washed once with water, and resuspended in the same volume of fresh YP medium containing 0.05% glucose for the appropriate time.
Chromatin analysis
The analysis of nucleosome position and/or structure was performed using MN digestion of spheroplasts coupled with the indirect end-labeling procedure (Wu, 1980). Cells growing exponentially (A600 = 0.3 OD/ml) in ADH2 repressing (3% glucose) or derepressing (0.05% glucose) conditions were washed once with water and then resuspended in zymolyase buffer (1 M sorbitol, 50 mM Tris–HCl pH 7.5, 10 mM β-mercaptoethanol). Incubation with zymolyase (0.01 mg/OD) was for 20 min at room temperature. The resulting spheroplasts were collected by centrifugation and resuspended in nystatin buffer (1 M sorbitol, 20 mM Tris–HCl pH 8.0, 1.5 mM CaCl2, 50 mM NaCl, 100 µg/ml nystatin) in order to permeabilize cell membranes for the subsequent treatment with MN (Venditti and Camilloni, 1994). Incubation with MN was for 15 min at 37°C and the reaction was stopped with 5 mM EDTA, 1% SDS (final concentrations). The samples were then treated with proteinase K for 2 h at 56°C and purified by phenol–chloroform extraction and ethanol precipitation.
After secondary digestion with the appropriate restriction endonuclease, the samples were run on 1.5% agarose gels in TBE buffer and transferred to nitrocellulose filters. Southern blotting and hybridization were performed by standard procedures.
pFA plasmid DNA (Verdone et al., 1997) was used to prepare the probe for the indirect end-labeling analysis and as deproteinized material for control reactions with MN.
RNA analysis
Aliquots containing the same number of cells were collected by centrifugation, and total RNA was prepared as described previously (Schmitt et al., 1990). After spectrophotometric determination of the amount of RNA present in each aliquot, 10 µg of RNA were loaded onto 1.2% agarose–MOPS gels containing formaldehyde and ethidium bromide.
Northern blot analysis was performed by standard procedures. For hybridization, a 5′-end-labeled oligonucleotide (5′-GTTGGTAGCCTTAACGACTGCGCTAAC-3′), specific for the ADH2 gene (from +710 to +684 of the coding region), was used.
ChIP with anti-acetylated histone antibodies and PCR analyses
Highly specific antibodies raised against individual sites of acetylation were as described previously (Suka et al., 2001). ChIP and PCR reactions were carried out as described previously (Rundlett et al., 1998; Hecht and Grunstein, 1999) with some modifications (Suka et al., 2001). All the primers were designed as 24mers with ∼50% GC content.
[α-32P]dATP was added to the PCR reaction (1 µCi/12.5 µl) for quantification by PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
ChIP with anti-Adr1 and anti-TBP antibodies and PCR analyses
ADR1 was tagged with a triple HA epitope using pUG6 (KAN) in strain W303-1b to create KVRY9 (the anti-HA3Adr1 antibody was used for the experiment shown in Figure 4A). The anti-Adr1 antibody (used for the experiment shown in Figure 5C) is described in Dombek et al. (1993); the antibody against yeast TBP was kindly provided by S.Hahn. Chromatin was prepared as described previously (Kuras and Struhl, 1999; Li et al., 1999). Glycine was added to a final concentration of 330 mM. The concentration of NaCl was adjusted from 150 to 275 mM during incubation with the anti-TBP antibody. Immunoprecipitated DNA was analyzed by PCR using primer pairs for specific regions. Multiple PCR reactions were performed in order to check the linear range of amplification for each primer set and DNA sample. Reactions were carried out in 25 µl and contained 70 pmol of each primer, 200 µM dNTPs and 1 µCi of [α-32P]dATP (sp. act. 3000 Ci/mmol). Cycling was for 5 min at 94°C, followed by 21 cycles with 30 s at 94°C, 30 s at 56°C, 1 min at 72°C, then 4 min at 72°C. PCR products were quantified using PhosphorImager and ImageQuant software (Molecular Dynamics).
Acknowledgments
Acknowledgements
We thank S.Venditti and G.Camilloni for helpful discussions, and C.Presutti for critical reading of the manuscript. This work was supported by grants from CNR TP on Biotechnology, MURST 40% 2001, MURST 5% ‘BSU’, NIH GM 26079 to E.T.Y. and NIH GM 23674 to M.G.
References
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- Allard S., Utley,R.T., Savard,J., Clarke,A., Grant,P., Brandl,C.J., Pillus,L., Workman,J.L. and Côté,J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.D., Lowary,P.T. and Widom,J. (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 307, 977–985. [DOI] [PubMed] [Google Scholar]
- Annunziato A.T. and Hansen,J.C. (2000) Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr., 9, 37–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D.R. and Young,E.T. (1982) Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature, 300, 724–728. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Chiang Y.-C., Komarnitsky,P., Chase,D. and Denis,C.L. (1996) ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J. Biol. Chem., 271, 32359–32365. [DOI] [PubMed] [Google Scholar]
- Clarke A.S., Lowell,J.E., Jacobson,S.J. and Pillus,L. (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol., 19, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Deckert J. and Struhl,K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol., 21, 2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L. and Young,E.T. (1983) Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol. Cell. Biol., 3, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mauro E., Kendrew,S.G. and Caserta,M. (2000) Two distinct nucleosome alterations characterize chromatin remodeling at the Saccharomyces cerevisiae ADH2 promoter. J. Biol. Chem., 275, 7612–7618. [DOI] [PubMed] [Google Scholar]
- Dombek K.M., Camier,S. and Young,E.T. (1993) ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol. Cell. Biol., 13, 4391–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Rocchini,C. and Ausio,J. (1995) Modulation of chromatin folding by histone acetylation. J. Biol. Chem., 270, 17923–17928. [DOI] [PubMed] [Google Scholar]
- Grant P.A. et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Hassan H., Neely,K.E. and Workman,J.L. (2001) Histone acetyl transferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell, 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone acetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J.N., Brown,S.A., Clark,C.D. and Winston,F. (1992) Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev., 6, 2288–2298. [DOI] [PubMed] [Google Scholar]
- Howe L., Ranalli,T.A., Allis,C.D. and Ausio,J. (1998) Transcriptionally active Xenopus laevis somatic 5 S ribosomal RNA genes are packaged with hyperacetylated histone H4, whereas transcriptionally silent oocyte genes are not. J. Biol. Chem., 273, 20693–20696. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998) Histone deacetylase activity of RPD3 is important for transcriptional repression in vivo. Genes Dev., 12, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. and Lorch,Y. (1999) Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev., 9, 148–151. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Kuo,M.-H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M., Zhou,J., Jambeck,P., Churchill,M.E.A. and Allis,C.D. (1998) Histone acetyltransferase activity of Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.-H., vom Baur,E., Struhl,K. and Allis,C.D. (2000) Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell, 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- Laman H., Balderes,D. and Shore,D. (1995) Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 3608–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F.V. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Li X.-Y., Virbasius,A., Zhu,X. and Green,M. (1999) Enhancement of TBP binding by activators and general transcription factors. Nature, 399, 605–609. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Recillas-Targa,F., Prioleau,M.-N. and Felsenfeld,G. (2001) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J., 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., Zhang,M. and Kornberg,R.D. (1999) Histone octamer transfer by a chromatin-remodeling complex. Cell, 96, 389–392. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. (2001) Protein modules that manipulate histone tails for chromatin regulation. Nature Rev. Mol. Cell. Biol., 2, 422–432. [DOI] [PubMed] [Google Scholar]
- Nightingale K.P., Wellinger,R.E., Sogo,J.M. and Becker,P.B. (1998) Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J., 17, 2865–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. and Herskowitz,I. (1992) Characterization of the yeast SWI1, SWI2 and SWI3 genes, which encode a global activator of transcription. Cell, 68, 573–583. [DOI] [PubMed] [Google Scholar]
- Reinke H., Gregory,P.D. and Hörz,W. (2001) A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell, 7, 529–536. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J.S., Dombeck,K.M. and Young,E.T (1999) Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J. Biol. Chem., 274, 37575–37582. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Eisen,A., Gu,W., Sattah,M., Pannuti,A., Zhou,J., Cook,R.G., Lucchesi,J.C. and Allis,C.D. (1998) ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl Acad. Sci. USA, 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka,Y., Carmen,A.A., Wu,J. and Grunstein,M. (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell, 8, 473–479. [DOI] [PubMed] [Google Scholar]
- Travers A.A. (1999) An engine for nucleosome remodeling. Cell, 96, 311–314. [DOI] [PubMed] [Google Scholar]
- Venditti S. and Camilloni,G. (1994) In vivo analysis of chromatin following nystatin-mediated import of active enzymes into Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1219, 677–689. [DOI] [PubMed] [Google Scholar]
- Verdone L., Camilloni,G., Di Mauro,E. and Caserta,M. (1996) Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol., 16, 1978–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L., Cesari,F., Denis,C.L., Di Mauro,E. and Caserta,M. (1997) Factors affecting Saccharomyces cerevisiae ADH2 chromatin remodeling and transcription. J. Biol. Chem., 272, 30828–30834. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M., Grant,P.A., Hebbes,T.R., Craine-Robinson,C., Allis,C.D. and Workman,J.L. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J., 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Hassan,A.H. and Workman,J.L (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo J.M., Thiriet,C. and Hayes,J.J. (2000) The H3–H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5 S DNA within a nucleosome. Mol. Cell. Biol., 20, 2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Flaus,A., Cairns,B.R., White,M.F., Workman,J.L. and Owen-Hughes,T. (1999) Nucleosome mobilization catalyzed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Wu C. (1980) The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature, 286, 854–860. [DOI] [PubMed] [Google Scholar]
- Wu J. and Grunstein,M. (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci., 25, 619–623. [DOI] [PubMed] [Google Scholar]
- Wu J., Suka,N., Carlson,M. and Grunstein,M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell, 7, 117–126. [DOI] [PubMed] [Google Scholar]
- Wyrick J.J., Holstege,F.C., Jennings,E.G., Causton,H.C., Shore,D., Grunstein,M., Lander,E.S. and Young,R.A. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature, 402, 418–421. [DOI] [PubMed] [Google Scholar]