Abstract
The soluble Tim9p–Tim10p (Tim, translocase of inner membrane) complex of the mitochondrial intermembrane space mediates the import of the carrier proteins and is a component of the TIM22 import system. The mechanism by which the Tim9p–Tim10p complex assembles and binds the carriers is not well understood, but previous studies have proposed that the conserved cysteine residues in the ‘twin CX3C’ motif coordinate zinc and potentially generate a zinc-finger-like structure that binds to the matrix loops of the carrier proteins. Here we have purified the native and recombinant Tim9p–Tim10p complex, and show that both complexes resemble each other and consist of three Tim9p and three Tim10p. Results from inductively coupled plasma–mass spectrometry studies failed to detect zinc in the Tim9p–Tim10p complex. Instead, the cysteine residues seemingly formed disulfide linkages. The Tim9p–Tim10p complex bound specifically to the transmembrane domains of the ADP/ATP carrier, but had no affinity for Tim23p, an inner membrane protein that is inserted via the TIM22 complex. The chaperone-like Tim9p–Tim10p complex thus may prevent aggregation of the unfolded carrier proteins in the aqueous intermembrane space.
Keywords: mitochondria/mitochondrial protein import/Saccharomyces cerevisiae/TIM complex
Introduction
The mitochondrion has an elaborate set of translocons on the outer and inner membranes to mediate the import of proteins from the cytosol (Ryan and Jensen, 1995; Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner, 1998). Most mitochondrial proteins are synthesized as cytosolic precursors containing a cleavable N-terminal pre-sequence that directs their import into mitochondria via the general protein import pathway. The precursor is escorted through the cytosol by chaperones and then the hetero-oligomeric translocase of the outer membrane (TOM) mediates translocation across the outer membrane. Several TOM components function as receptors, while others form the translocation pore. After passage through the outer membrane, the Tim17p–Tim23p (Tim, translocase of inner membrane) complex of the inner membrane together with the ATP-dependent import motor Hsp70, Tim44p and mGrpE mediates translocation across the inner membrane. Finally, a number of soluble proteins in the matrix involved in the proteolytic maturation and folding of the imported proteins may be required to complete assembly (Ryan and Jensen, 1995; Schatz and Dobberstein, 1996; Neupert, 1997; Pfanner et al., 2001).
The mitochondrion has a separate import pathway for inner membrane proteins, with components residing in the intermembrane space and inner membrane (Koehler et al., 1999b; Bauer et al., 2000; Pfanner et al., 2001). Substrates of this pathway include proteins in the carrier family (Palmieri et al., 1996), such as the ADP/ATP carrier (AAC) and the protein import components Tim22p and Tim23p (Koehler et al., 1998a; Sirrenberg et al., 1998; Leuenberger et al., 1999). Proteins destined for the inner membrane are escorted by cytosolic chaperones and then pass through the TOM complex to the intermembrane space. The intermembrane space complexes, Tim9p– Tim10p and Tim8p–Tim13p, function as putative chaperones to transfer the hydrophobic precursors across the intermembrane space to an inner membrane machinery specialized for the insertion of membrane proteins (Koehler et al., 1998a,b; Sirrenberg et al., 1998; Adam et al., 1999). The inner membrane complex consists of Tim12p, Tim18p, Tim22p, Tim54p and a small fraction of Tim9p and Tim10p, which together form a 300 kDa complex (Kerscher et al., 1997, 2000; Koehler et al., 1998a,b, 2000; Sirrenberg et al., 1998; Adam et al., 1999). Components Tim9p, Tim10p, Tim12p, Tim22p and Tim54p are essential for viability (Kerscher et al., 1997, 2000; Koehler et al., 1998a,b, 2000; Sirrenberg et al., 1998; Adam et al., 1999).
The small Tim proteins (Tim8p, Tim9p, Tim10p, Tim12p and Tim13p) are ∼25% identical and 40–50% similar, yet Tim9p partners exclusively with Tim10p and Tim8p with Tim13p in soluble intermembrane space complexes (Koehler et al., 1998b, 1999a; Adam et al., 1999; Davis et al., 2000). The small Tim family contains the consensus ‘twin CX3C’ motif, in which two cysteines are separated by three amino acids (Koehler et al., 1999b). Neupert and colleagues have shown that this motif might be important for zinc coordination in the monomers (Sirrenberg et al., 1998; Adam et al., 1999; Paschen et al., 2000). The Tim9p–Tim10p complex is 10-fold more abundant than the Tim8p–Tim13p complex (Koehler et al., 1998b, 1999a; Sirrenberg et al., 1998; Adam et al., 1999). Approximately 5% of Tim9p and Tim10p is associated with the 300 kDa Tim18p–Tim22p–Tim54p complex at the inner membrane, whereas 95% is soluble in the 70 kDa intermembrane space complex (Koehler et al., 1998b, 1999a). Tim9p and Tim10p bind to translocation intermediates of the mitochondrial carrier family, Tim17p and Tim22p, whereas Tim8p and Tim13p bind to Tim23p (Endres et al., 1999; Leuenberger et al., 1999; Davis et al., 2000; Paschen et al., 2000), suggesting that the battery of small Tim proteins may have different substrate specificities.
Previous studies have focused on using cross-linking and immunoprecipitation approaches with translocation intermediates to determine the mechanism by which the small Tim proteins mediate protein import. In this study, we have purified the Tim9p–Tim10p complex from the mitochondrial intermembrane space and an Escherichia coli strain expressing both proteins. We show that both native and recombinant complexes have similar properties. The cysteine residues do not coordinate Zn2+ ions, but seemingly are involved in disulfide bonds. Using peptide scans with peptides derived from the AAC sequence, the Tim9p–Tim10p complex binds with highest specificity to the transmembrane domains.
Results
Purification of the native and recombinant Tim9p–Tim10p complex
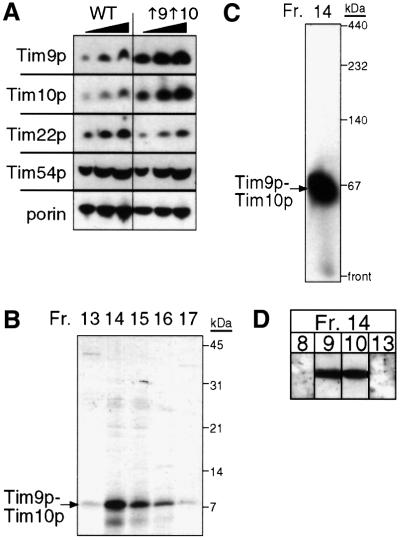
To understand better the mechanism by which the Tim9p–Tim10p complex mediates the import of the carrier proteins, we purified the native complex from yeast for biochemical characterization. A yeast strain, designated ↑9↑10, was constructed in which Tim9p and Tim10p were overexpressed from 2µ plasmids. Strain ↑9↑10 grew at a rate similar to the parental strain. From purified mitochondria, we determined the abundance of components in the TIM22 import pathway by quantitative immunoblot analysis and scanning laser densitometry (Figure 1A). As expected, Tim9p and Tim10p were overexpressed 8-fold in comparison with the parental strain. In contrast, the abundance of Tim8p, Tim13p and membrane components Tim22p and Tim54p was similar between the ↑9↑10 and parental strains (Figure 1A and our unpublished data). Strain ↑9↑10 thus facilitated purification of the Tim9p–Tim10p complex from the mitochondrial intermembrane space.
Fig. 1. Purification of the Tim9p–Tim10p complex from a yeast strain overexpressing Tim9p and Tim10p. (A) Tim9p and Tim10p were overexpressed 6- to 8-fold, but the expression of Tim22p and Tim54p was not changed. Mitochondrial proteins (50, 100 and 150 µg) were separated by SDS–PAGE, followed by immunoblot analysis from the parental strain (WT) and a strain (↑9↑10) co-transformed with two 2µ plasmids expressing TIM9 and TIM10, respectively, and analyzed by immunoblotting with monospecific antisera for Tim9p, Tim10p, Tim22p, Tim54p and porin. Proteins were identified by incubation with [125I]protein A; the amount was quantitated by scanning laser densitometry. (B) The Tim9p–Tim10p complex was purified from the mitochondrial intermembrane space. After successive chromatography steps (see Materials and methods), fractions from the final purification step (gel filtration) were analyzed for Tim9p and Tim10p by SDS–PAGE and Coomassie Blue staining. (C) As in (B), fraction 14 was analyzed on a 6–16% blue native gel followed by immunoblotting with antiserum for Tim10p. (D) As in (B), fraction 14 was separated by SDS–PAGE, followed by immunoblot analysis with monospecific antisera for Tim8p, Tim9p, Tim10p and Tim13p. Proteins were identified by incubation with [125I]protein A.
The Tim9p–Tim10p complex was purified with subsequent chromatographic steps based on a previous purification scheme (Koehler et al., 1998b). We isolated mitochondria from strain ↑9↑10, released the soluble intermembrane space contents by osmotic shock and fractionated the intermembrane space proteins by subsequent chromatography steps: affinity chromatography on a Cibacron blue column, cation-exchange chromatography, anion-exchange chromatography and gel filtration on a Superose 12 column. After each purification step, we monitored Tim9p, Tim10p and the intact 70 kDa complex by SDS–PAGE (Figure 1B) and ‘blue native gel’ electrophoresis followed by immunoblotting. Tim9p and Tim10p co-purified through the entire fractionation procedure as a 70 kDa complex (Figure 1C). Specifically, both flowed directly through the Cibacron blue column, eluted at 60–80 mM NaCl on Mono S and Mono Q columns, and eluted in the 60–70 kDa mass range from the gel filtration column. Because the small Tim proteins may pair differently in the strain overexpressing Tim9p and Tim10p, we confirmed that the 70 kDa complex contained only Tim9p and Tim10p, and not Tim8p and Tim13p by SDS–PAGE followed by immunoblotting with monospecific antisera (Figure 1D).
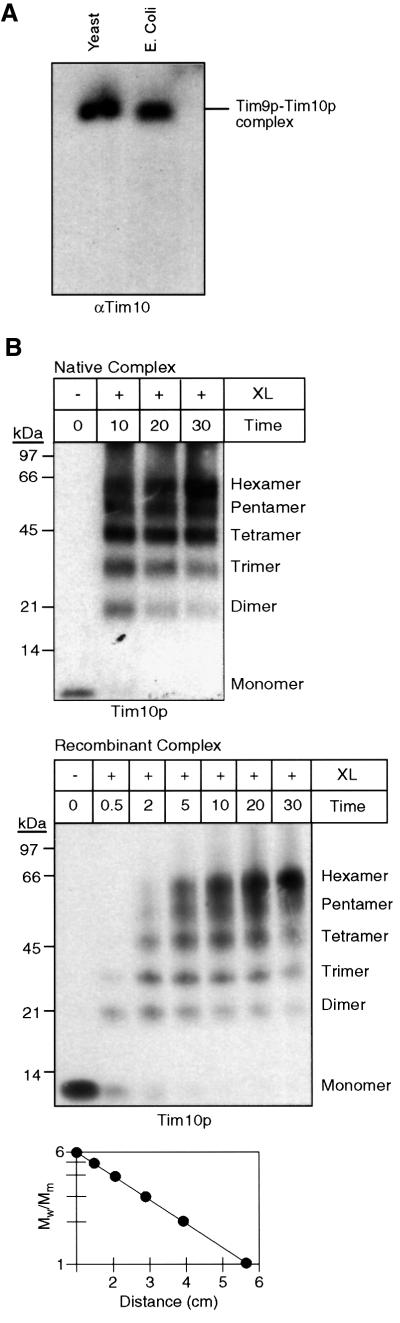
Purification of the native Tim9p–Tim10p complex required large amounts of mitochondria, so we also engineered an E.coli strain to overproduce the Tim9p– Tim10p complex. Because Tim9p and Tim10p have a molecular mass of only 10 kDa and the complex most likely contains three Tim9p and three Tim10p polypeptides, purification tags were not included; the presence of tags could potentially interfere with assembly in E.coli. Our approach was to construct a single plasmid in which both Tim9p and Tim10p were expressed from the same RNA transcript. In brief, TIM9 and TIM10 with their own E.coli ribosomal binding site were cloned in tandem in an expression plasmid and transformed into the E.coli host BL21(DE3). Upon induction, both Tim9p and Tim10p were expressed and assembled efficiently. Analysis of the E.coli lysate on blue native gels showed that Tim9p and Tim10p migrated as a 70 kDa complex; smaller molecular weight complexes were not detected in the dye front (our unpublished data). In comparison with the native Tim9p–Tim10p complex from the mitochondrial intermembrane space, the recombinant complex migrated identically in a native gel, indicating that both complexes are similar with regard to charge and shape (Figure 2A). The purification protocol for recombinant Tim9p–Tim10p complex was similar to that of the native complex. The Tim9p–Tim10p complex was purified to homogeneity from E.coli lysate by subsequent chromatography steps using an anion-exchange column, followed by a cation-exchange column and a gel filtration column. The order of the ion-exchange columns was reversed because contaminating proteins were removed more efficiently. Once again, the Tim9p–Tim10p complex eluted at 60–80 mM NaCl on ion-exchange columns and eluted in the 60–70 kDa mass range from the gel filtration column (our unpublished data). Both recombinant and native Tim9p–Tim10p complex thus displayed similar properties during purification.
Fig. 2. The recombinant and native Tim9p–Tim10p complexes have identical properties. (A) An expression plasmid was constructed in which TIM9 and TIM10 were each placed behind a ribosomal binding site. After transformation into the E.coli host BL21(DE3) and induction, the Tim9p–Tim10p complex (E.coli) was purified (see Materials and methods) and separated on a 6.0% native gel. A mitochondrial intermembrane space fraction (yeast) was also separated. Immunoblot analysis was performed with monospecific antisera against Tim10p (αTim10) and Tim9p (unpublished data). Proteins were identified by incubation with [125I]protein A. (B) For intermolecular cross-linking assays, an intermembrane space fraction (native complex, top blot) and the recombinant Tim9p–Tim10p complex (recombinant complex, bottom blot) were incubated with 0.1% glutaraldehyde (XL) and aliquots were removed at the indicated time points (min). Cross-linked products were separated on 16% tricine gels, followed by immunoblot analysis with monospecific antisera against Tim10p. The number of subunits is indicated on the right. Electrophoretic mobility (graph) of the products from the 10 min time point of cross-linking was plotted against the logarithm of Mw/Mm, where Mw is the relative molecular weight of the cross-linked species and Mm is the molecular mass of the Tim10p or Tim9p monomer (10 kDa).
Recombinant and native Tim9p–Tim10p complex have similar properties
To confirm that the recombinant Tim9p–Tim10p complex assembled identically to the intermembrane space complex and that the number of subunits in the complex was six, we treated the recombinant Tim9p–Tim10p complex and an intermembrane space fraction with 0.1% glutaraldehyde on ice for 30 min (Figure 2B; Azem et al., 1998; van Dijl et al., 1998). The resulting cross-linked intermediates were analyzed by SDS–PAGE and immunoblot analysis with antibodies against Tim10p. We observed six distinct species that we interpreted as monomer to hexamer because their electrophoretic mobility (Mw/Mm) was linearly related to the logarithm of subunit number and the distance each oligomer had migrated from the origin (Figure 2B, graph). As the incubation period approached 30 min, the predominant cross-linked product moved to the hexameric form, but the tetramer was also abundant. The same cross-linking pattern was observed with antibodies against Tim9p (our unpublished data). Results from quantitative amino acid analysis confirmed that the complex was present in equimolar ratios (our unpublished data), as has been shown previously (Koehler et al., 1998b; Adam et al., 1999). We conclude that the complex is a hexamer, most likely comprised of three Tim9p and three Tim10p.
The Tim9p–Tim10p complex does not coordinate zinc
The small Tim proteins contain the conserved ‘twin CX3C’ motif, in which two cysteine residues are separated by three amino acids and the spacing between each CX3C is 11–16 amino acids (Koehler et al., 1998a; Bauer et al., 2000). Previous studies have shown that the individual small Tim proteins fused to the maltose binding protein coordinated zinc in a 1:1 molar ratio, presumably through the cysteine residues (Sirrenberg et al., 1998; Adam et al., 1999; Paschen et al., 2000). We investigated the ‘state’ of the cysteine residues using several techniques, including thiol-trapping assays, reductant and metal ion chelator sensitivity assays, and inductively coupled plasma–mass spectrometry (ICP–MS).
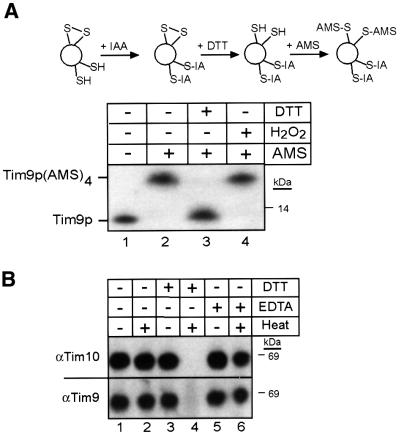
We employed a thiol-trapping method that allowed the separation and visualization of oxidized and reduced species on Tricine gels (Figure 3A, schematic; Jakob et al., 1999). The modification of Tim9p was followed by immunoblot analysis (Figure 3A, blot). First, all accessible thiol groups in the intermembrane space fraction were alkylated (blocked) irreversibly with iodoacetamide (IAA). After removal of excess IAA, disulfide bonds present in the proteins were reduced with dithiothreitol (DTT). In the final step, the thiol groups pre viously involved in disulfide bonds were alkylated with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS). AMS is a thiol-reactive reagent that alkylates cysteine residues, thereby adding 500 Da to the molecular mass of each thiol group. As a control, completely oxidized Tim9p (pre-treated with hydrogen peroxide; Figure 3B, lane 4) showed a slower mobility on tricine gels due to the addition of four AMS molecules to the four cysteine residues in Tim9p; this translated into a 2 kDa change in molecular mass that was easily observed in the 10 kDa protein. In contrast, completely reduced Tim9p (pre-treated with DTT; Figure 3A, lane 3) migrated at the same molecular weight as the untreated complex (Figure 3A, lane 1) because the sulfhydryl residues were blocked with IAA and, therefore, not modified by AMS. When the sample was alkylated with iodoacetamide (avoiding both reduction and oxidation), followed by DTT and AMS treatment (Figure 3A, lane 2), Tim9p migrated at a molecular weight identical to that of the oxidized Tim9p; four AMS molecules covalently bound to Tim9p. This observation thus indicates that the cysteine residues are not reduced and accessible to IAA, but instead are occupied, potentially, in disulfide bonds. Analysis of Tim10p by thiol trapping yielded the same results; the cysteine residues in Tim10p are occupied (our unpublished data). To determine whether disulfide bridges might be intermolecular, the recombinant Tim9p–Tim10p complexes and the intermembrane space fraction were separated on non-reducing denaturing gels. Results from immunoblot analysis showed that both Tim9p and Tim10p migrated as monomers, suggesting that the modification(s) on the cysteine residues were intramolecular (our unpublished results).
Fig. 3. The sulfhydryl groups on the cysteine residues of the ‘twin CX3C’ motif are occupied. (A) To elucidate the state of the cysteine residues in the Tim9p–Tim10p complex, a thiol-trapping method (Jakob et al., 1999) was used (schematic). A mitochondrial intermembrane space fraction (200 µg per lane) was either left untreated (lane 2), reduced with 10 mM DTT (lane 3) or oxidized with 5% H2O2 (lane 4). The samples were then alkylated with IAA to block free sulfhydryl groups, following treatment with DTT to reduce any disulfide bonds. The remaining free sulfhydryl groups were alkylated with AMS, which creates an increase in molecular mass of 0.5 kDa. The samples were separated by SDS–PAGE, followed by immunoblot analysis with antiserum against Tim10p. As a control, an untreated sample is included (lane 1). Tim9p and Tim9p(AMS)4 are denoted. (B) The reductant DTT interferes with refolding of the Tim9p–Tim10p complex. The recombinant Tim9–Tim10p complex was heated to 95°C in the presence of DTT (lane 4) or EDTA (lane 6) and quickly cooled in an ice bath, followed by blue native gel electrophoresis. As a control, samples were untreated (lane 1), heated (lane 2) or treated with DTT (lane 3) or EDTA (lane 5) and incubated on ice. Immunoblot analysis was performed with an antibody against Tim10p and Tim9p.
To distinguish between the possibility that the thiol residues were coordinating a metal versus disulfide bond, the denaturation/renaturation properties of the recombinant Tim9p–Tim10p complex were analyzed in the presence of EDTA and DTT (Figure 3B). The complex was heated to 95°C for 10 min and then immediately placed into an ice bath for another 10 min. Treatment at 95°C resulted in unfolding of the complex, as shown by circular dichroism studies (our unpublished data). Immunoblot analysis on blue native gels showed that Tim9p and Tim10p completely refolded into a 70 kDa complex after this treatment (Figure 3B, lanes 1 and 2), even in the presence of the metal chelator EDTA (Figure 3B, lane 6); recovery was near 100%, as shown by laser scanning densitometry (our unpublished data). However, when DTT was added during the heating process, Tim9p and Tim10p did not refold into a 70 kDa complex (Figure 3B, lane 4). DTT and EDTA treatment did not interfere with the stability of the 70 kDa complex when kept at 4°C (Figure 3B, lanes 3 and 5).
Because the Tim9p–Tim10p complex may bind zinc ions at sites other than the cysteine residues, we used ICP–MS to determine whether the recombinant Tim9p– Tim10p complex coordinated zinc. Three purified samples of the complex from separate bacterial preparations were analyzed. Because the last purification step was gel filtration, the buffer blanks were taken from fractions that were eluted before or after the Tim9p–Tim10p complex. Zinc ions were not detected at levels higher than background in the samples containing the Tim9p– Tim10p complex (Table I). The protein concentration in the Tim9p–Tim10p complex was determined by quantitative amino acid analysis. The amount of zinc ions per Tim9–Tim10p complex was calculated to be 6.8 × 10–3, indicating that zinc was essentially not detectable in the complex. Based on the observation that the Tim9p– Tim10p complex might coordinate one or six zinc ions, we predicted that zinc should be detected at 2.05 and 12.3 µM, respectively, in contrast to the actual measurement of 0.0136 µM (Table I). Furthermore, the presence of other metal ions was not detected at levels higher than that of the buffer blank (our unpublished data). Together, these results suggest that the Tim9p–Tim10p complex does not coordinate Zn2+ ions and the cysteine residues most likely form disulfide bridges.
Table I. Tim9p–Tim10p complex zinc analysis.
| Actual | Predicteda | Predicteda | |
|---|---|---|---|
| (mol zinc:mol complex) |
0:1 |
1:1 |
6:1 |
| Zinc (sample/background, ppb)b | 0.89 (13.5/12.61) | 1.34 × 102 | 8.04 × 102 |
| Zinc (µM)c | 1.36 × 10–2 | 2.05 | 12.3 |
| Protein (mg/ml)d | 0.126 | ||
| Protein (µM)c | 2.05 | ||
| Zinc per complex | 6.82 × 10–3 | 1 | 6 |
aEach ‘twin CX3C’ motif (6) in the Tim9p–Tim10p complex could potentially bind one zinc atom. The predicted levels of zinc are shown (in p.p.b.) for hypothetical Tim9p–Tim10p complexes that bind one zinc per complex or one zinc per monomer (six zinc ions total). Predicted values are based on the same concentration of Tim9p–Tim10p complex used in actual measurements.
bThe average ICP-determined zinc concentration (p.p.b.) is presented along with the average background level of zinc. Background levels of zinc were determined using buffer blanks from the same purification.
cThe micromolar concentrations of both protein and zinc were calculated assuming a molecular weight of 65.38 atomic mass units (a.m.u.) for zinc and 6.15 × 104 a.m.u. for the Tim9p–Tim10p complex.
dThe protein concentration of each sample was determined by quantitative amino acid analysis.
The Tim9p–Tim10p complex binds to the transmembrane domains of AAC but not Tim23p
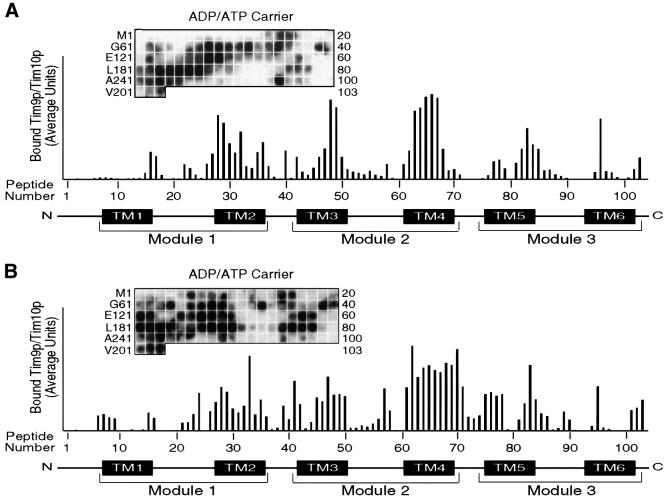
Pfanner and colleagues have used peptide scans to identify sequences in the phosphate carrier and the presequence of CoxIV that mitochondrial import receptors Tom20p, Tom22p and Tom70p bind to (Brix et al., 1999, 2000). Because of the hydrophobic nature of the AAC, we used the same approach to identify sequences that the native and recombinant Tim9p–Tim10p complex might bind. In brief, peptide scans consisted of 13mer peptides overlapping by 10 residues, which were attached covalently to a cellulose membrane. Thus, 103 peptides were synthesized for the 318 amino acids of AAC. The native or recombinant Tim9p–Tim10p complex was incubated to equilibrium with the cellulose-bound peptides, followed by electrotransfer and immunodetection with antibodies against Tim10p and [125I]protein A. Quantitative analysis was performed using laser scanning densitometry (Personal Densitometer SI; Molecular Dynamics) and the ImageQuaNT program, which corrects for the local background around each individual spot. After analysis, the native Tim9p–Tim10p complex bound preferentially to the membrane-spanning domains. Binding was strongest at transmembrane domains 3 and 4, followed by transmembrane domains 2 and 5. The binding affinity was lowest at transmembrane domains 1 and 6, and in the loop regions. The recombinant complex showed a similar binding pattern (Figure 4B), confirming that the properties of the recombinant complex are similar to those of the native complex. Investigation of the amino acid sequences to which the Tim9p–Tim10p complex bound failed to yield a consensus sequence; however, as expected, these sequences were particularly rich in hydrophobic residues. When antibodies against Tim9p were used, a similar pattern of binding was detected, confirming that the intact complex binds to the AAC peptides (our unpublished data).
Fig. 4. Native and recombinant Tim9p–Tim10p complex show a similar binding pattern to a peptide scan derived from the sequence of AAC. (A) The native Tim9p–Tim10p complex (200 nM) purified from the mitochondrial intermembrane space was incubated with a peptide scan consisting of 13mers derived from AAC. The first peptide comprises amino acids 1–13 of the AAC, the second peptide residues 4–16 and the third peptide residues 7–19, etc. The labeling on the left indicates the first amino acid of the left-most peptide of each row. The labeling on the right side indicates the number of the right-most peptide of each row. After washing the membrane, the bound Tim9p–Tim10p complex was transferred to a PVDF membrane, followed by immunoblot analysis with antiserum specific for Tim10p and [125I]protein A. Binding was quantified by scanning laser densitometry from at least three independent experiments and plotted for each peptide. Transmembrane domains and repeat modules of AAC are plotted corresponding to the respective peptides. (B) As in (A), except that 200 nM recombinant Tim9p–Tim10p complex was incubated with the peptide scan.
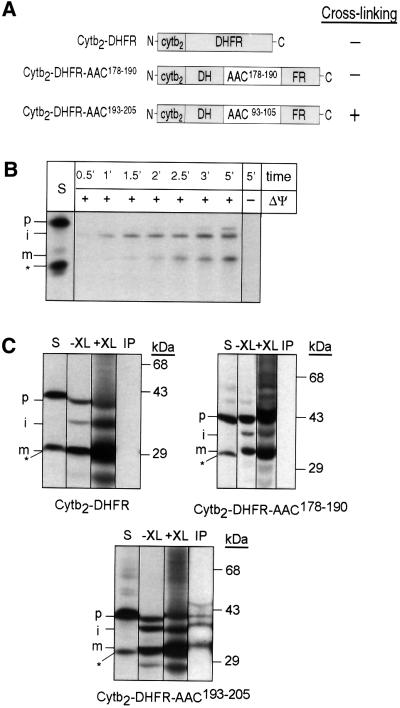
Previous studies have shown that truncations or alterations to the carrier protein sequence can lead to aberrations in the import pathway (Schleiff and McBride, 2000). To confirm that binding on the peptide scans reflects in organello binding of the Tim9p–Tim10p complex, we circumvented this problem by determining whether the Tim9p–Tim10p complex could bind to the peptides when they were placed in the middle of a ‘foreign’ protein and arrested in the intermembrane space (Figure 5A). Specifically, the synthetic precursor that contained the first 167 amino acids of cytochrome b2 was fused to mouse dihydrofolate reductase (DHFR) and synthesized in a reticulocyte lysate system (Figure 5B). Upon import, Cytb2–DHFR is processed by the matrix and intermembrane space proteases, and localizes to the intermembrane space because of the ‘stop-transfer’ sequence (Glick et al., 1992). As shown in Figure 5B, the appearance of the processed intermediate form and mature forms can be followed in an import assay. Note that a non-imported species is synthesized during in vitro transcription/translation; it is most likely that translation is initiated at an internal methionine. The following peptides were placed in the middle of DHFR by inserting annealed oligonucleotides that coded for the following sequences: KKKTLKSDGVAGLYC in construct Cytb2–DFHR– AAC178–190 (no binding, matrix loop between transmembrane domain 3 and 4) and KFLPSVVGIVVYRGC in construct Cytb2–DHFR–AAC193–205 (strong binding, transmembrane domain 4). Lysine and cysteine residues flanked the peptides to provide sites for chemical cross-linking. Because DHFR folds tightly and is protease resistant, we incubated the radiolabeled synthetic precursors with protease and, as expected, found that they were extremely protease sensitive (our unpublished data). The precursor thus should remain unfolded in the intermembrane space after import. We imported the radiolabeled synthetic precursors into energized mitochondria for 20 min at 25°C, reacted the mitochondria with the cross-linker di-succinimidyl suberate (DSS), subjected the solubilized mitochondria to immunoprecipitation with antibodies against Tim9p and Tim10p, and analyzed the immunoprecipitates by SDS–PAGE and fluorography. As expected, cross-links were not observed between Cytb2– DHFR and the Tim9p–Tim10p complex. However, the Tim9p–Tim10p complex was cross-linked to Cytb2– DHFR when it contained peptide AAC193–205 (transmembrane domain 4) but not peptide AAC178–190 (matrix loop between transmembrane domain 3 and 4; Figure 5C). The asterisk marks the non-imported species that is present in the translation reaction. The in organello binding results thus reflect the in vitro results with the peptide scan.
Fig. 5. Tim9p and Tim10p are cross-linked to an AAC peptide placed within a synthetic precursor in the mitochondrial intermembrane space. (A) Diagram of Cytb2–DHFR constructs in which peptides derived from the AAC sequence have been inserted into DHFR. Cytb2–DHFR is a fusion between residues 1–167 of Cytb2 and DHFR. Cytb2–DFHR–AAC178–190 contains AAC residues 178–190 (peptide 60 from Figure 4, which showed no binding). Cytb2–DHFR–AAC193–205 contains AAC residues 193–205 (peptide 65 from Figure 4, which showed binding). (B) The radiolabeled Cytb2–DHFR precursor was synthesized in vitro and incubated for the indicated times at 25°C in the presence and absence of a membrane potential (ΔΨ) with wild-type mitochondria. Samples were treated with protease to remove non-imported precursor and analyzed by SDS–PAGE and fluorography. STD, 25% of the radioactive precursor added to the assay. p, precursor; i, intermediate; m, mature. The asterisk denotes a translation product created by translation initiation from an internal methionine; this product is not imported. (C) The radiolabeled precursor was imported into wild-type mitochondria for 10 min at 25°C (–XL). The chemical cross-linker DSS (+XL) was added at 1 mM. After quenching, the sample was denatured and immunoprecipitated with antibodies against Tim9p and Tim10p (IP). Bound proteins were eluted with SDS-containing sample buffer and analyzed by SDS–PAGE and fluorography. S, 10% of the amount of precursor that was added to the non-cross-linked import assay. p, precursor; i, intermediate; m, mature. The asterisk denotes a translation product created by translation initiation from an internal methionine; this product is not imported.
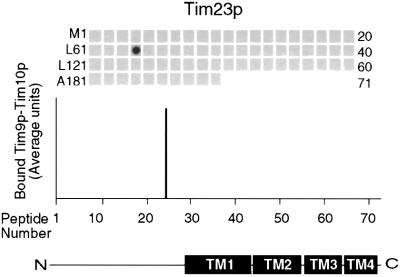
The Tim9p–Tim10p complex interacts specifically with the carrier family, while the Tim8p–Tim13p complex mediates the import of Tim23p (Leuenberger et al., 1999; Davis et al., 2000; Paschen et al., 2000). To confirm the specificity of the AAC-binding interactions, a peptide scan for Tim23p was synthesized and incubated with the native Tim9p–Tim10p complex. The Tim23p peptide scan contained 71 peptides, again overlapping by 10 residues. As expected, after incubation with the Tim9p–Tim10p complex, significant binding to the Tim23p peptides was not detectable (Figure 6). While the Tim9p–Tim10p complex did bind to one peptide, the complex did not bind to the neighboring peptides with 10 residue overlaps, indicating that binding was most likely to be non-specific. This approach provides the first identification of potential motifs that the Tim9p–Tim10p complex may bind. Taken together, the Tim9p–Tim10p complex shows binding specificity for the membrane-spanning domains of AAC and this interaction does not depend on the presence of zinc.
Fig. 6. Native Tim9p–Tim10p complex does not bind to Tim23p. The native Tim9p–Tim10p complex (200 nM) purified from the mitochondrial intermembrane space was incubated with a peptide scan consisting of 13mers derived from Tim23p, as in Figure 4. Binding was quantified by scanning laser densitometry from at least three independent experiments and plotted for each peptide. Transmembrane domains of Tim23p are plotted corresponding to the respective peptides.
Discussion
To understand better the mechanism by which the Tim9p–Tim10p complex mediates the import of inner membrane proteins, we obtained large amounts of the Tim9p–Tim10p complex by purification from a yeast strain overexpressing both proteins and engineering an E.coli strain to overproduce the proteins from one RNA transcript. The recombinant Tim9p–Tim10p complex assembled efficiently in the E.coli cytoplasm because when the E.coli lysate was separated in blue native gels, Tim9p and Tim10p were detected only in a 70 kDa complex. It is possible that unassembled monomers were degraded, but assembly was seemingly efficient. In contrast, Tokatlidis and colleagues used a similar approach but utilized a tagged version of Tim10p (Luciano et al., 2001). In their expression system, the ratio of Tim10p to Tim9p in the E.coli cytosol was 50:1, resulting in the presence of unassembled monomers during purification.
The native and recombinant complexes have similar properties. On native gels that separate by shape and charge, the complexes migrated into the gel identically. Glutaraldehyde cross-linking studies showed that both complexes consisted of hexamers. From previous studies, it was shown that the Tim9p–Tim10p complex contained equimolar ratios of Tim9p and Tim10p (Koehler et al., 1998b; Adam et al., 1999; Luciano et al., 2001). Indeed, results from quantitative amino acid analysis of the recombinant complex revealed that Tim9p and Tim10p were present in equimolar ratios (unpublished data). The complex thus consists of three Tim9p and three Tim10p molecules. The similarity of the native and recombinant Tim9p–Tim10p complex is further confirmed from the thiol-trapping experiments and denaturation experiments in the presence of EDTA and DTT. Finally, both native and recombinant complexes share a similar binding specificity for peptides on the AAC peptide scans. Taken together, these results show that the recombinant and native Tim9p–Tim10p complex are identical.
The Tim9p–Tim10p complex binds to the transmembrane domains of the carrier protein AAC
Cross-linking studies in isolated mitochondria have shown that Tim9p and Tim10p bind directly to translocation intermediates of the carrier proteins (Koehler et al., 1998a,b; Sirrenberg et al., 1998; Adam et al., 1999; Endres et al., 1999). It has been proposed that the ‘twin CX3C’ motif might form a structure similar to a zinc finger and bind to the charged matrix-sided loops of the carrier proteins (Sirrenberg et al., 1998; Endres et al., 1999). However, given the diversity of inner membrane substrates, including Tim17p, Tim22p and the carrier family that can be cross-linked to Tim9p and Tim10p (Leuenberger et al., 1999), the binding sequences might be less specific, much like the diversity of substrates that Tom70p binds (Hines et al., 1990; Schlossmann et al., 1994; Brix et al., 1999). Results from the peptide scan approach indicated that Tim9p–Tim10p bound preferentially to the transmembrane domains, particularly transmembrane domains 2, 3, 4 and 5. In fact, the binding at the regions between the transmembrane domains was the weakest. These results suggest that the Tim9p–Tim10p complex binds preferentially to hydrophobic sequences. Similar experiments were performed with the outer membrane receptors and a peptide scan of the phosphate carrier (Brix et al., 1999, 2000). Pfanner and colleagues reported that Tom70p and Tom20p bind preferentially to multiple segments of the phosphate carrier (Brix et al., 1999, 2000). Tom70p binds to both membrane-spanning domains and charged loops between the membrane-spanning domains, while Tom20p binds to the charged regions between transmembrane domains. Comparison of the binding specificity of the outer membrane receptors and the Tim9p–Tim10p complex thus indicates that the import components recognize different sequences in the carrier proteins.
Pfanner and colleagues have demonstrated that AAC, flanked by DHFR, can cross the TOM complex as a loop and then be cross-linked to Tim9p and Tim10p (Ryan et al., 1999). Similarly, the central matrix loop of the carrier uncoupling protein-1 drives import into mitochondria (Schleiff and McBride, 2000). The results from our peptide scan corroborate these findings. The strongest binding of the Tim9p–Tim10p complex was in the center of AAC at transmembrane domains 3 and 4, while binding was weakest at the N- and C-termini. Although we can not determine the number of Tim9p–Tim10p complexes that bind to one precursor, we propose that ∼4 distinct binding sites are present in the membrane-spanning regions (Figure 4).
Despite the inability to identify a consensus sequence in the AAC polypeptide, the Tim9p–Tim10p complex has specificity for the carrier family. This observation was supported by the in orgenllo cross-linking studies in which peptides were inserted in the middle of DHFR; the Tim9p–Tim10p complex was cross-linked to the peptide derived from transmembrane domain 4. In addition, incubation of the Tim9p–Tim10p complex with the Tim23p peptide scan revealed no specific binding. Previous studies have shown that Tim9p and Tim10p were cross-linked to a Tim23p translocation intermediate (Davis et al., 2000). However, because Tim9p and Tim10p are both in the soluble 70 kDa complex and at the 300 kDa TIM22 translocon, and the TIM22 translocon mediates Tim23p insertion into the membrane, it is feasible that the cross-links were observed with the population of Tim9p and Tim10p in the TIM22 translocon.
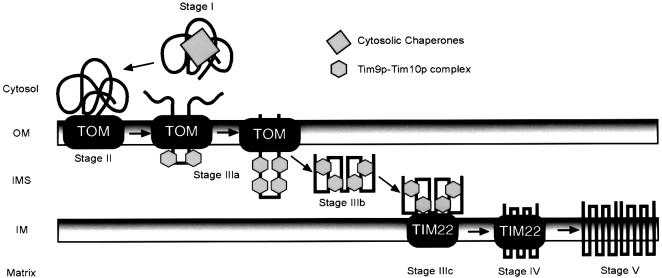
We therefore refine the proposed model by which the Tim9p–Tim10p complex might serve as a putative chaperone to guide the carriers across the intermembrane space (Figure 7; Koehler et al., 1998b; Endres et al., 1999; Pfanner et al., 2001). The import pathway of the carrier family can be dissected into five stages of translocation. At stage I, newly synthesized precursors are escorted through the cytosol by chaperones. The chaperones target the precursor to the TOM complex, where they are handed to a battery of receptors (stage II). In stage IIIa, as the precursor loops through the TOM complex, the Tim9p–Tim10p complex binds to keep the precursor, thereby preventing aggregation in the aqueous intermembrane space. The Tim9p–Tim10p complex then escorts the carrier across the intermembrane space (stage IIIb) to the TIM22 translocon at the inner membrane (stage IIIc). The stage IIIb intermediate is supported by the following observations. First, conditional mutations in tim10 result in an import defect in which the carrier does not cross the TOM complex to reach the TIM22 translocon (Koehler et al., 1998a). Also, cross-linking studies that show a direct interaction between Tim9p and Tim10p and AAC failed to show simultaneous interactions with the TOM translocon, unless AAC was arrested at the TOM complex with DHFR (Ryan et al., 1999; Murphy et al., 2001). Furthermore, mitochondria void of the soluble Tim9p–Tim10p and Tim8p–Tim13p complexes show a decrease in the rate of import for inner membrane substrates by ∼70% (Murphy et al., 2001). This decrease presumably results from the aggregation of these substrates in the intermembrane space and subsequent degradation by proteases such as Yme1p. At stage IV, the precursor is inserted into the inner membrane by the TIM22 translocon and at stage V is assembled into a functional dimer.
Fig. 7. Proposed model for the role of the Tim9p–Tim10p complex in the import of carrier proteins. Carrier proteins are escorted by cytosolic chaperones (stage I) to receptors of the TOM complex (stage II). The precursor passes through the TOM channel to the intermembrane space and the Tim9p–Tim10p complex binds to hydrophobic residues in the transmembrane domains of the carriers (stage IIIa). In the aqueous intermembrane space, the Tim9p–Tim10p complex maintains the carrier in an import-competent state (stage IIIb) and delivers it to the TIM22 translocon (stage IIIc). The TIM22 translocon mediates insertion into the inner membrane (stage IV) and then the carrier assembles into a functional dimer (stage V).
The ‘twin CX3C’ motif does not coordinate Zn2+
The ‘twin CX3C’ motif is present in all of the small Tim proteins identified so far. The homolog of Tim8p in humans is DDP1/TIMM8a (deafness dystonia protein) and mutations in the DDP1 locus cause Mohr–Tranebjaerg syndrome (MTS; Tranebjaerg et al., 1995; Jin et al., 1996). Mutations reported previously in the DDP1/TIMM8a locus have been frameshift/nonsense mutations or deletions resulting in a truncated or absent protein (Jin et al., 1996). However, the first de novo mutation in DDP1/TIMM8a, C66W, was identified in which a missense mutation changed Cys66 to Trp66 (Tranebjaerg et al., 2000). This mutation occurs in the fourth cysteine of the ‘twin CX3C’ motif, indicating the importance of the cysteine residues. We made a similar mutation in Tim8pC68W, in which Cys68 was changed to Trp68 (K.Roesch and C.Koehler, in preparation). Tim8pC68W was imported into mitochondria, but failed to assemble in a stable 70 kDa complex. This indicates that the cysteine residues might be important structurally for assembly of the 70 kDa complex.
Because the Tim9p and Tim10p monomers have been shown to coordinate Zn2+ ions in an ∼1:1 molar ratio (Sirrenberg et al., 1998; Adam et al., 1999), we investigated whether the Tim9p–Tim10p complex contained Zn2+. We expected that one complex might coordinate up to six Zn2+ ions, but results from ICP–MS studies failed to detect Zn2+ or any other metal ion. While it is plausible that the Tim9p–Tim10p complex could assemble without incorporating Zn2+, a study by Harrison and colleagues suggests that hetero-oligomeric complexes co-expressed in E.coli can coordinate Zn2+ ions if it is requisite for their interaction (Huse et al., 1998). Specifically, Lck (lymphoid-specific, src family protein-tyrosine kinase) and T-cell co-receptor CD4 were co-expressed in E.coli and their interaction was dependent on the presence of a thiol-mediated co-coordination of zinc; purification and analysis of the complex showed that zinc was co-coordinated in a 1:1 molar ratio with the complex. Furthermore, the presence of low concentrations of EDTA at room temperature dissociated the Lck–CD4 complex, whereas high concentrations of DTT and β-mercaptoethanol had no effect. In contrast, our results with the Tim9p–Tim10p complex show that, upon heat treatment to dissociate the complex, the reducing agent DTT, not EDTA, dissociated the complex.
Alternatively, the thiol-trapping experiments with AMS showed that the cysteine residues are not present as free thiols but are probably engaged in disulfide bonds. While it is tempting to model the ‘twin CX3C’ motif as a zinc finger, it differs from the canonical zinc finger in which the two cysteines are separated by two amino acids (Mackay and Crossley, 1998). However, the cysteine residues might be involved in disulfide bonds, based on a similar motif in fractalkine, a novel CX3C chemokine (Mizoue et al., 1999, 2001). The CX3C motif involves Cys8 and Cys12, and is present in the extracellular matrix. Cys8 forms a disulfide bridge with Cys34, while Cys12 forms a disulfide bridge with Cys50, as shown by NMR studies. While the small Tim proteins are located in a different compartment, the CX3C motif of fractalkine indicates that these cysteine residues are capable of forming disulfide bridges.
The thiol residues in the Tim9p–Tim10p complex, therefore, may play a structural role in assembly of the complex or alternatively mediate interactions with the substrate proteins. The importance of thiol–disulfide exchange reactions playing a mechanistic role is already evident in an increasing number of cytoplasmic proteins in bacteria and eukaryotes (Lee et al., 1998; Jakob et al., 1999; Wu et al., 2000; Pomposiello et al., 2001). Examples include the transcription factors OxyR and Flp, anti-σ factor RsrA, the redox-regulated Hsp33, p53 and protein-tyrosine phosphatase 1B. Further studies to determine the structure of the Tim9p–Tim10p complex should provide helpful clues as to the role of the cysteine residues.
Materials and methods
Plasmids and strains
Standard genetic techniques were used for growth, manipulation and transformation of yeast strains (Sikorski and Hieter, 1989; Guthrie and Fink, 1991). The Saccharomyces cerevisiae strain YCK78 used to overexpress Tim9p and Tim10p was designated ↑9↑10 and constructed as follows. Both TIM9 and TIM10 and flanking regions were cloned into pRS426 (2µ, URA3) and pRS425 (2µ, LEU2), respectively. The plasmids were co-transformed into a diploid yeast strain in which one allele of TIM9 and TIM10 were deleted (TIM10/Δtim10::HIS3 TIM9/Δtim9::TRP1 ade8 leu2 ura3). Transformants were sporulated and selected for histidine, tryptophan, leucine and uracil prototrophy.
For expression of the recombinant Tim9p–Tim10p complex in E.coli, the TIM9 was cloned into pET28a (Novagen) as an NcoI–SalI fragment and TIM10 was cloned into pET28a as an NcoI–NdeI fragment; only the open reading frames were cloned and no fusions or tags were constructed. TIM10 with the ribosomal binding site was then removed as a XbaI–NheI fragment and cloned into the XbaI site of the pet28a-Tim9 plasmid. As a result, a single transcript was synthesized in which both Tim9p and Tim10p were translated from their own ribosomal binding site. Expression was induced according to the manufacturer’s protocols (Novagen).
Cytb2–DFHR–AAC178–190 and Cytb2–DHFR–AAC193–205 were constructed as follows. Cytb2–DHFR, a fusion between Cytb21–167 and mouse DHFR, was subcloned into pSP65 (Promega). In brief, oligonucleotide pairs were synthesized so that when annealed and inserted into the SacI site of DHFR, they would generate the following sequences: Cytb2–DFHR–AAC178–190, KKKTLKSDGVAGLYC; and Cytb2–DHFR– AAC193–205, KFLPSVVGIVVYRGC. The peptide was flanked with lysine and cysteine residues to generate a chemical cross-linking site. The constructs were linearized with HindIII for in vitro transcription/translation.
Purification of the Tim9p–Tim10p complex from the mitochondrial intermembrane space and E.coli
Mitochondria (40 mg/ml in 0.6 M sorbitol, 20 mM HEPES–KOH pH 7.4) were hypotonically shocked by dilution with 9 vols of buffer A (1 mM DTT, 20 mM HEPES–KOH pH 7.4) and incubated for 1 h at 4°C. After removing mitoplasts by two centrifugation steps (10 min at 10 000 g), the soluble intermembrane space was loaded onto a Cibacron blue column (1 × 7 cm; Amersham Pharmacia Biotech). The Tim9p–Tim10p complex was washed through the column with buffer A, while contaminating proteins remained bound. The flow through was loaded onto a Mono S cation-exchange column (0.5 × 5 cm; Amersham Pharmacia Biotech). The column was washed with buffer A at a flow rate of 0.5 ml/min and bound proteins were eluted with 30 ml of a linear gradient (0–150 mM NaCl) in buffer A. Fractions (1 ml) were collected and analyzed by SDS–PAGE, followed by immunoblotting or silver staining or by blue native gel electrophoresis. The fractions that contained Tim9p and Tim10p were loaded onto a Mono Q anion-exchange column (0.5 × 5 cm; Amersham Pharmacia Biotech). The column was washed with buffer A at a flow rate of 0.5 ml/min and bound proteins were eluted with 30 ml of a linear gradient (0–200 mM NaCl) in buffer A. Fractions (1 ml) were collected and analyzed as described above. Tim9p and Tim10p co-eluted at 60–80 mM NaCl. These fractions were concentrated to 200 µl in buffer A containing 100 mM NaCl and loaded onto a Superose 12 gel filtration column (1.0 × 30 cm; Amersham Pharmacia Biotech). The column was developed with buffer A containing 100 mM NaCl at a flow rate of 0.2 ml/min. Fractions (1 ml) were collected and analyzed as described above. The molecular mass standards for the Superose 12 column were RNase A (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), bovine serum albumin (BSA; 67 kDa) and aldolase (158 kDa).
Frozen E.coli cell pellets containing the recombinant Tim9p–Tim10p complex were thawed and lysed by sonication on ice. Cell debris was removed by centrifugation (30 min, 100 000 g, 4°C) and proteins were precipitated by addition of polyethylene glycol (average molecular weight 3350 Da; J.T.Baker, Inc.) to 25% and NaCl to 250 mM. After centrifugation, the pellet was solubilized in buffer A and loaded onto a Q-Sepharose Fast Flow column (5 × 30 cm; Amersham Pharmacia Biotech). The column was washed with buffer A at a flow rate of 5 ml/min and bound proteins were eluted with 500 ml of a linear gradient (0–200 mM NaCl) in buffer A. Fractions (15 ml) were collected and analyzed by SDS–PAGE, followed by immunoblotting, Coomassie Blue staining and blue native gel electrophoresis. After desalting and concentration in buffer A, the fractions containing the Tim9p–Tim10p complex were loaded onto a Resource S column (1.6 × 30 cm; Amersham Pharmacia Biotech). The column was washed with buffer A at a flow rate of 1 ml/min and bound proteins were eluted with a 12 ml gradient (0–100 mM NaCl) in buffer A. Fractions (1 ml) were collected and separated on a Superose 12 column as described for the native Tim9p–Tim10p complex.
Import of radiolabeled proteins into isolated mitochondria and cross-linking studies
Mitochondria were purified from lactate-grown yeast cells (Glick and Pon, 1995) and assayed for in vitro protein import as described (Rospert and Schatz, 1998). Proteins were synthesized in a rabbit reticulocyte lysate in the presence of [35S]methionine after in vitro transcription of the corresponding gene by SP6 polymerase. The reticulocyte lysate containing the radiolabeled precursor was incubated with isolated mitochondria at the indicated temperatures in import buffer (1 mg/ml BSA, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, 20 mM K+–HEPES pH 7.4). Where indicated, the potential across the mitochondrial inner membrane was dissipated with 1 µM valinomycin. Non-imported radiolabeled proteins were removed by treatment with 100 µg/ml trypsin or 50 µg/ml proteinase K for 15–30 min on ice; trypsin was inhibited with 200 µg/ml soybean trypsin inhibitor and proteinase K with 1 mM PMSF, respectively.
The translocation intermediates of Cytb2–DHFR were cross-linked to adjacent proteins with 1 mM DSS. The cross-linking protocol was performed as follows (Koehler et al., 1998b; Leuenberger et al., 1999). After import, mitochondria were washed, suspended at 1 mg/ml in import buffer and incubated with the cross-linker on ice for 30 min followed by a quench with 100 mM Tris–HCl pH 7.5. For immunoprecipitation, solubilized mitochondria were incubated with the corresponding monospecific rabbit IgGs coupled to protein A–Sepharose (Rospert et al., 1994). Glutaraldehyde cross-linking was performed as described previously (van Dijl et al., 1998).
Thiol-trapping studies
Thiol-trapping methods with the thiol-specific reagent AMS (Molecular Probes) were performed according to Jakob et al. (1999). Specifically, mitochondria (10 mg/ml in 0.6 M sorbitol, 20 mM HEPES–KOH pH 7.4) were hypotonically shocked by dilution with 9 vols of 20 mM HEPES–KOH pH 7.4. After removing mitoplasts by centrifugation steps (10 min at 10 000 g), the soluble intermembrane space was divided into four samples. Three of the four samples were controls: one sample was mock treated, the second sample was treated with 10 mM DTT (reduced) and the third sample was treated with 5% H2O2 (oxidized). Half-volume 100 mM IAA (Fluka), 200 mM Tris–HCl pH 8.0 and 10 mM EDTA was added to the samples and the reaction was incubated at 37°C for 5 min to irreversibly block free thiol residues. The samples were acid precipitated and the pellets were resuspended in 50 µl of 10 mM DTT, 100 mM Tris–HCl pH 8.0, 10 mM EDTA and 0.5% SDS to reduce any disulfide linkages, followed by incubation at 44°C for 1 h. After addition of 1/10 vol. 250 mM AMS, the samples were incubated at 25°C for 90 min and stopped with non-reducing Laemmli buffer. Samples were separated on a 16% SDS–tricine gel, followed by immunodetection with antibodies against Tim9p and Tim10p.
ICP–MS analysis
ICP–MS analysis was performed using an ICP–MS Elan 6000 Instrument (Perkin Elmer-Sciex) by the Laboratory for Environmental Analysis, University of Georgia. Seven samples were analyzed for zinc. Three samples contained the Tim9p–Tim10p complex purified from three separate bacterial preparations, along with three blanks taken from the same preparations (buffer that eluted from the column just before or after the complex). The final sample contained the elution buffer that was applied to the sizing column. The concentration of the recombinant Tim9p–Tim10p complex was determined by quantitative amino acid analysis.
Screening of peptide scans with the Tim9p–Tim10p complex
The cellulose-bound peptide scans were prepared by automated spot synthesis (Jerini). Multiple 13mer peptides with a 10 amino acid overlap were synthesized according to the sequence of Tim23p and AAC. The membranes were incubated either with 200 nM native or recombinant Tim9p–Tim10p complex in binding buffer (100 mM KCl, 5% sucrose, 1% BSA, 30 mM Tris–HCl pH 7.4) at 25°C for 2 h as described by Brix et al. (1999). After extensive washing, the bound Tim9p–Tim10p complex was transferred to a polyvinylidene difluoride (PVDF) membrane, followed by detection with antibodies against Tim9p or Tim10p and [125I]protein A. Binding data were acquired by scanning laser densitometry (Personal Densitometer SI; Molecular Dynamics) and quantitated utilizing ImageQuaNT (version 4.2a; Molecular Dynamics). The mean of at least three independent experiments for each peptide spot was used and the local background of each peptide spot was subtracted. Binding data were identical for antibodies against Tim9p and Tim10p.
Blue native gel electrophoresis
Mitochondria (2.5 mg/ml) were solubilized in 20 mM K+–HEPES pH 7.4, 50 mM NaCl, 10% glycerol, 2.5 mM MgCl2, 1 mM EDTA and 0.16% N-dodecylmaltoside (Boehringer Mannheim) for 30 min on ice. Insoluble material was removed by centrifugation at 100 000 g for 10 min and the solubilized proteins were analyzed by blue native gel electrophoresis on a 6–16% linear polyacrylamide gradient (Schägger and von Jagow, 1991; Schägger et al., 1994; Dekker et al., 1996).
Miscellaneous
Mitochondrial proteins were analyzed by SDS–PAGE using a 10 or 16% polyacrylamide gel and a tricine-based running buffer (Schägger and von Jagow, 1987). Proteins were detected by immunoblotting using nitrocellulose or PVDF membranes and visualization of immune complexes with [125I]protein A. Protein concentration was assayed by the bicinchoninic acid method (Pierce) using BSA as the standard.
Acknowledgments
Acknowledgements
We would like to thank David Wong for technical assistance, Dr Hassan and co-workers at the Laboratory for Environmental Analysis, University of Georgia, for ICP–AE analysis, Karl Schmid for purification of the native Tim9p–Tim10p complex, and Drs Einhard Schmidt and Jamil Momand, California State University, Los Angeles, for helpful discussions. C.M.K. is a Damon Runyon–Walter Winchell Scholar. This work was supported by the Damon Runyon–Walter Winchell Cancer Research Foundation (DRS18), the American Heart Association (0030147N), Burroughs Wellcome Fund New Investigator Award in the Toxicological Sciences (1001120), Research Corporation (RI0459) and the National Institutes of Health (1R01GM61721-01). S.P.C. is funded by the USPHS National Research Service Award (GM07185).
References
- Adam A., Endres,M., Sirrenberg,C., Lottspeich,F., Neupert,W. and Brunner,M. (1999) Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J., 18, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azem A., Weiss,C. and Goloubinoff,P. (1998) Structural analysis of GroE chaperonin complexes using chemical cross-linking. Methods Enzymol., 290, 253–268. [DOI] [PubMed] [Google Scholar]
- Bauer M.F., Hofmann,S., Neupert,W. and Brunner,M. (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol., 10, 25–31. [DOI] [PubMed] [Google Scholar]
- Brix J., Rudiger,S., Bukau,B., Schneider-Mergener,J. and Pfanner,N. (1999) Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22 and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem., 274, 16522–16530. [DOI] [PubMed] [Google Scholar]
- Brix J., Ziegler,G.A., Dietmeier,K., Schneider-Mergener,J., Schulz,G.E. and Pfanner,N. (2000) The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol., 303, 479–488. [DOI] [PubMed] [Google Scholar]
- Davis A.J., Sepuri,N.B., Holder,J., Johnson,A.E. and Jensen,R.E. (2000) Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol., 150, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J., Muller,H., Rassow,J. and Pfanner,N. (1996) Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol. Chem., 377, 535–538. [PubMed] [Google Scholar]
- Endres M., Neupert,W. and Brunner,M. (1999) Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J., 18, 3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S. and Pon,L. (1995) Isolation of highly purified mitochondria from S. cerevisiae. Methods Enzymol., 260, 213–233. [DOI] [PubMed] [Google Scholar]
- Glick B.S., Brandt,A., Cunningham,K., Muller,S., Hallberg,R.L. and Schatz,G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell, 69, 809–822. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA.
- Hines V., Brandt,A., Griffiths,G., Horstmann,H., Brutsch,H. and Schatz,G. (1990) Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J., 9, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M., Eck,M.J., Harrison,S.C. (1998) A Zn2+ ion links the cytoplasmic tail of CD4 and the N-terminal region of Lck. J. Biol. Chem., 273, 18729–18733. [DOI] [PubMed] [Google Scholar]
- Jakob U., Muse,W., Eser,M. and Bardwell,J.C. (1999) Chaperone activity with a redox switch. Cell, 96, 341–352. [DOI] [PubMed] [Google Scholar]
- Jin H. et al. (1996) A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nature Genet., 14, 177–180. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Holder,J., Srinivasan,M., Leung,R.S. and Jensen,R.E. (1997) The Tim54p–Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol., 139, 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Sepuri,N.B. and Jensen,R.E. (2000) Tim18p is a new component of the Tim54p–Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell, 11, 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Jarosch,E., Tokatlidis,K., Schmid,K., Schweyen,R.J. and Schatz,G. (1998a) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science, 279, 369–373. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S., Oppliger,W., Schmid,K., Jarosch,E., Dolfini,L., Junne,T., Schatz,G. and Tokatlidis,K. (1998b) Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J., 17, 6477–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Leuenberger,D., Merchant,S., Renold,A., Junne,T. and Schatz,G. (1999a) Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl Acad. Sci. USA, 96, 2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S. and Schatz,G. (1999b) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci., 24, 428–432. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Murphy,M.P., Bally,N., Leuenberger,D., Oppliger,W., Dolfini,L., Junne,T., Schatz,G. and Or,E. (2000) Tim18p, a novel subunit of the inner membrane complex that mediates protein import into the yeast mitochondrial inner membrane. Mol. Cell. Biol., 20, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.R., Kwon,K.S., Kim,S.R. and Rhee,S.G. (1998) Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem., 273, 15366–15372. [DOI] [PubMed] [Google Scholar]
- Leuenberger D., Bally,N.A., Schatz,G. and Koehler,C.M. (1999) Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J., 17, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P., Vial,S., Vergnolle,M.A., Dyall,S.D., Robinson,D.R. and Tokatlidis,K. (2001) Functional reconstitution of the import of the yeast ADP/ATP carrier mediated by the TIM10 complex. EMBO J., 20, 4099–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- Mizoue L.S., Bazan,J.F., Johnson,E.C. and Handel,T.M. (1999) Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry, 38, 1402–1414. [DOI] [PubMed] [Google Scholar]
- Mizoue L.S., Sullivan,S.K., King,D.S., Kledal,T.N., Schwartz,T.W., Bacon,K.B. and Handel,T.M. (2001) Molecular determinants of receptor binding and signaling by the CX3C chemokine fractalkine. J. Biol. Chem., 276, 33906–33914. [DOI] [PubMed] [Google Scholar]
- Murphy M.P., Leuenberger,D., Curran,S.P., Oppliger,W. and Koehler,C.M. (2001) The essential function of the small Tim proteins in the TIM22 import pathway does not depend on formation of the soluble 70-kilodalton complex. Mol. Cell. Biol., 21, 6132–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Palmieri F., Bisaccia,F., Capobianco,L., Dolce,V., Fiermonte,G., Iacobazzi,V., Indiveri,C. and Palmieri,L. (1996) Mitochondrial metabolite transporters. Biochim. Biophys. Acta, 1275, 127–132. [DOI] [PubMed] [Google Scholar]
- Paschen S.A., Rothbauer,U., Kaldi,K., Bauer,M.F., Neupert,W. and Brunner,M. (2000) The role of the TIM8–13 complex in the import of Tim23 into mitochondria. EMBO J., 19, 6392–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. (1998) Mitochondrial import: crossing the aqueous intermembrane space. Curr. Biol., 8, R262–R265. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Geissler,A., Schleiff,E. and McBride,H. (2001) Versatility of the mitochondrial protein import machinery. Nature Rev. Mol. Cell. Biol., 2, 339–349. [DOI] [PubMed] [Google Scholar]
- Pomposiello P.J., Demple,B., Bacon,K.B. and Handel,T.M. (2001) Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol., 19, 109–114. [DOI] [PubMed] [Google Scholar]
- Rospert S. and Schatz,G. (1998) Protein translocation into mitochondria. In Celis,J.E. (ed.), Cell Biology: A Laboratory Handbook. Academic Press, San Diego, CA, pp. 277–285.
- Rospert S., Muller,S., Schatz,G. and Glick,B.S. (1994) Fusion proteins containing the cytochrome b2 presequence are sorted to the mitochondrial intermembrane space independently of hsp60. J. Biol. Chem., 269, 17279–17288. [PubMed] [Google Scholar]
- Ryan K.R. and Jensen,R.E. (1995) Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell, 83, 517–519. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Muller,H. and Pfanner,N. (1999) Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem., 274, 20619–20627. [DOI] [PubMed] [Google Scholar]
- Schägger H. and von Jagow,G. (1987) Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schägger H. and von Jagow,G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem., 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer,W.A. and von Jagow,G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem., 217, 220–230. [DOI] [PubMed] [Google Scholar]
- Schatz G. and Dobberstein,B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Schleiff E. and McBride,H. (2000) The central matrix loop drives import of uncoupling protein 1 into mitochondria. J. Cell Sci., 113, 2267–2272. [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Dietmeier,K., Pfanner,N. and Neupert,W. (1994) Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J. Biol. Chem., 269, 11893–11901. [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Folsch,H., Stuart,R.A., Neupert,W. and Brunner,M. (1998) Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature, 391, 912–915. [DOI] [PubMed] [Google Scholar]
- Tranebjaerg L. et al. (1995) A new X linked recessive deafness syndrome with blindness, dystonia, fractures and mental deficiency is linked to Xq22. J. Med. Genet., 32, 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L., Hamel,B.C., Gabreels,F.J., Renier,W.O. and Van Ghelue,M. (2000) A de novo missense mutation in a critical domain of the X-linked DDP gene causes the typical deafness–dystonia–optic atrophy syndrome. Eur. J. Hum. Genet., 8, 464–467. [DOI] [PubMed] [Google Scholar]
- van Dijl J.M., Kutejova,E., Suda,K., Perecko,D., Schatz,G. and Suzuki,C.K. (1998) The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc. Natl Acad. Sci. USA, 95, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.H., Thomas,J.A. and Momand,J. (2000) p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem. J., 351, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]