Abstract
The DEAD box RNA helicase Dbp5 is essential for nucleocytoplasmic transport of mRNA–protein (mRNP) complexes. Dbp5 is present mainly in the cytoplasm and is enriched at the cytoplasmic side of nuclear pore complexes (NPCs), suggesting that it acts in the late part of mRNP export. Here, we visualize the assembly and transport of a specific mRNP particle, the Balbiani ring mRNP in the dipteran Chironomus tentans, and show that a Dbp5 homologue in C.tentans, Ct-Dbp5, binds to pre-mRNP co-transcriptionally and accompanies the mRNP to and through the nuclear pores and into the cytoplasm. We also demonstrate that Ct-Dbp5 accumulates in the nucleus and partly disappears from the NPC when nuclear export of mRNA is inhibited. The fact that Ct-Dbp5 is present along the exiting mRNP fibril extending from the nuclear pore into the cytoplasm supports the view that Ct-Dbp5 is involved in restructuring the mRNP prior to translation. Finally, the addition of the export factor Dbp5 to the growing transcript highlights the importance of the co-transcriptional loading process in determining the fate of mRNA.
Keywords: Dbp5/pre-mRNP/RNA export/RNA helicase
Introduction
During transport, mRNAs are associated with proteins in ribonucleoprotein (RNP) complexes (Dreyfuss et al., 1993; Daneholt, 1997; Krecic and Swanson, 1999). The proteins, designated hnRNP proteins, are loaded on to pre-mRNA concomitant with transcription. The hnRNP protein population is complex: in mammalian cells, for example, there are ∼20–30 major hnRNP proteins (A–U) and a large number of minor ones. Some of the proteins are confined to the nucleus while others also appear in the cytoplasm (shuttling proteins) (Pinol-Roma and Dreyfuss, 1992). Further analysis of the flow of pre-mRNA/mRNA-associated proteins has shown that some leave the transcript in the nucleoplasm or at the nuclear pore, others are shed subsequent to translocation of the mRNP particle through the nuclear pore, whereas others still accompany the mRNA into polysomes (Visa et al., 1996a; Daneholt, 1997). The pre-mRNP/mRNP complexes change not only in protein composition but also drastically in conformation during the transfer from the genes in the cell nucleus to the polysomes in cytoplasm, perhaps most remarkably during translocation through the nuclear pores (Mehlin et al., 1992). It should be emphasized that the hnRNP proteins not only package the RNA into an RNP particle but also exert a number of specific functions during gene expression; for example, hnRNP proteins can influence splicing of pre-mRNA in the cell nucleus, and translation, stability and localization of mRNA in the cytoplasm (for references, see Krecic and Swanson, 1999).
The transport of mRNA is known to be energy dependent and signal mediated (Nigg, 1997). Several tentative mRNA export factors have been identified (Mattaj and Engelmeier, 1998; Nakielny and Dreyfuss, 1999; Görlich and Kutay, 1999; Cole, 2000; Zenklusen and Stutz, 2001). TAP and Mex67p are particularly intriguing. Mex67p is known to be an essential export factor in yeast (Segref et al., 1997), and TAP, its vertebrate homologue, has also been implicated in mRNA export (Grüter et al., 1998). TAP and Mex67p shuttle between the nucleus and cytoplasm, are associated with poly(A)+ RNA and interact with several FG repeat nucleoporins in the nuclear pore complex (NPC) (Segref et al., 1997; Katahira et al., 1999; Bachi et al., 2000). The binding of TAP and Mex67p to mRNA is likely to be indirect via the Aly/REF proteins and Yra1p, respectively (Strässer and Hurt, 2000; Stutz et al., 2000).
Recently, it has been shown that the mRNP complex is modified concomitant with splicing and that addition of proteins makes the mRNP export competent (Luo and Reed, 1999). A complex of five proteins (SRm160, DEK, RNPS1, Y14 and REF) is formed 20–24 nucleotides upstream of the exon–exon junction (Le Hir et al., 2000). Aly/REF plays a role in mRNP export (Zhou et al., 2000; Rodrigues et al., 2001), presumably by recruiting the export factor TAP to the complex (Le Hir et al., 2001), while Y14 is more likely to function as a marker on exon–exon junctions instrumental in controlling nonsense-mediated decay of mRNA (Kim et al., 2001).
The export of mRNA could be complex with alternative pathways. In fact, recent data suggest that there are multiple TAP pathways in higher eukaryotes (Herold et al., 2000). Furthermore, the shuttling hnRNP proteins A1 and K have nuclear export signals, suggesting a role in mRNA export (Nakielny and Dreyfuss, 1999). Finally, in yeast the export factor Gle2p could participate in the Mex67p pathway, but it is also conceivable that it represents an additional pathway (for references and discussion, see Zenklusen and Stutz, 2001).
Considering the comprehensive and dynamic restructuring of the mRNP complex that takes place during transport, it is interesting that a putative RNA helicase, the DEAD box protein Dbp5, has been shown to be an essential mRNA export factor in both yeast (Snay-Hodge et al., 1998; Tseng et al., 1998) and vertebrates (Schmitt et al., 1999). In general, RNA helicases can affect RNA–RNA as well as RNA–protein interactions in RNP complexes (Luking et al., 1998). The Dbp5 protein is an RNA-dependent ATPase and its activity is essential for its mRNA export function (Schmitt et al., 1999). Dbp5 is located predominantly in the cytoplasm and is associated with the cytoplasmic fibrils of the NPC (Schmitt et al., 1999; Strahm et al., 1999). It is, therefore, believed to function in late steps of mRNA export at the cytoplasmic side of the NPC. Dbp5, however, also enters the nucleus in yeast cells lacking functional CRM1/Xpo1 or Mex67p, indicating that Dbp5 can shuttle in and out of the nucleus (Hodge et al., 1999). These findings could imply that Dbp5 also functions in the nucleus, possibly on the nuclear side of, or within, the NPC (for discussion, see Nakielny and Dreyfuss, 1999).
In the present study we have investigated the appearance and flow of Dbp5 in the larval salivary glands of the dipteran Chironomus tentans. This experimental system allows the visualization of a specific mRNP particle, the Balbiani ring (BR) mRNP, during its assembly on the gene and during transport via nucleoplasm and nuclear pores into the cytoplasm (Daneholt, 1997), and the presence of Dbp5 can be monitored in relation to the drastic conformational changes of the particle taking place during the information transfer. Concomitant with transcription, the growing BR transcript is first packed into an RNP fibril. The fibril forms a spiral structure that is subsequently packed into a short, bent ribbon, which forms the completed product, a well-defined, ring-like structure with a diameter of 50 nm (Skoglund et al., 1983, 1986). At the same time, spliceosomes assemble and disassemble on the growing BR pre-mRNP (Kiseleva et al., 1994), and splicing takes place simultaneously with transcription (Baurén and Wieslander, 1994). The released particle can be readily detected during its transport through the nucleoplasm. Finally, the translocation of the BR particle through the nuclear pore is a multi-step process, including binding of the mRNP particle to the nuclear pore, docking to the entrance of the central channel, unpacking of the particle and passage of the unfolding RNP fibril through the central channel with the 5′ end of the mRNA in the lead. The exiting RNP fibril becomes engaged in polysome formation just outside the nuclear pore (Mehlin et al., 1992; Daneholt, 1997).
We have found that the Dbp5 homologue in C.tentans, here called Ct-Dbp5, is located predominantly in the cytoplasm and enriched in the nuclear rim but it is also present within the nucleus. Ct-Dbp5 becomes associated with nascent pre-mRNAs at a large number of active genes, including the BR genes. Immunoelectron microscopy revealed that Ct-Dbp5 is bound to nascent BR pre-mRNP particles and accompanies them through the nucleoplasm and the nuclear pore and into the cytoplasm. Nuclear accumulation of Ct-Dbp5 takes place when synthesis and/or export of mRNA is inhibited. Our results indicate that most or all of the shuttling Ct-Dbp5 exits from the nucleus associated with mRNP. Furthermore, Ct-Dbp5 is present along the mRNP fibril extending into the cytoplasm, supporting the view that Ct-Dbp5 is involved in restructuring the mRNP prior to translation. Finally, the binding of Ct-Dbp5 already to the nascent transcripts suggests that the co-transcriptional loading of pre-mRNA with proteins determines to a large extent both the early and late fate of mRNA.
Results
The C.tentans homologue of Dbp5
Dbp5 homologues exhibit a high degree of similarity in different species (Snay-Hodge et al., 1998; Schmitt et al., 1999). We designed degenerate primers from two conserved regions of Dbp5 by analysing a multiple protein sequence alignment of Dbp5 from six different species. A 400 bp fragment was amplified by RT–PCR, using poly(A)+ RNA from tissue culture cells of C.tentans as a template. The PCR product was significantly related to Dbp5 and was used as a probe to screen a C.tentans λZAP cDNA library. One clone containing a 1858 bp insert was analysed. It contained a 5′ untranslated region of 111 nucleotides, an open reading frame (ORF) of 1419 bp and a 3′ untranslated region of 328 bp.
The open reading frame encodes a 472 amino acid protein with a predicted molecular mass of 52.9 kDa and an isoelectric point of 6.65 (DDBJ/EMBL/GenBank accession No. AJ318937). The protein contains the highly conserved motifs that characterize the well-known members of the DEAD box superfamily (Figure 1). Based on the complete amino acid sequence, it is most closely related to homologues of Dbp5 in different species (Figure 1). The degree of identity to Dbp5 in Drosophila melanogaster is 69%, in human 59%, in mouse 58%, in Xenopus laevis 53%, in Schizosaccharomyces pombe 45% and in Saccharomyces cerevisiae 48%. Moreover, the C.tentans protein also has the conserved six amino acid insertion specific for Dbp5 (Snay-Hodge et al., 1998). Based on the high structural similarity to the Dbp5 group of DEAD box proteins, the C.tentans protein is a member of this subfamily, and we designated the protein as Ct-Dbp5.
Fig. 1. Sequence characteristics of Ct-Dbp5. (A) The amino acid sequence of Ct-Dbp5 compared with Dbp5 in different organisms. Amino acid residues that are identical in at least three species at a given position are shown in white against a black background. Residues that are functionally similar to the predominant one at a position are highlighted against a grey background. The conserved DEAD box helicase motifs are underlined, and the Dbp5-specific insert is boxed. (B) Schematic presentation of Ct-Dbp5. The helicase core domain with its conserved motifs and the Dbp5-specific insert are pointed out. In the insert, the conserved second and fifth positions are marked with a star, in both (A) and (B).
Ct-Dbp5 is abundant in the cytoplasm but also appears in the nucleus
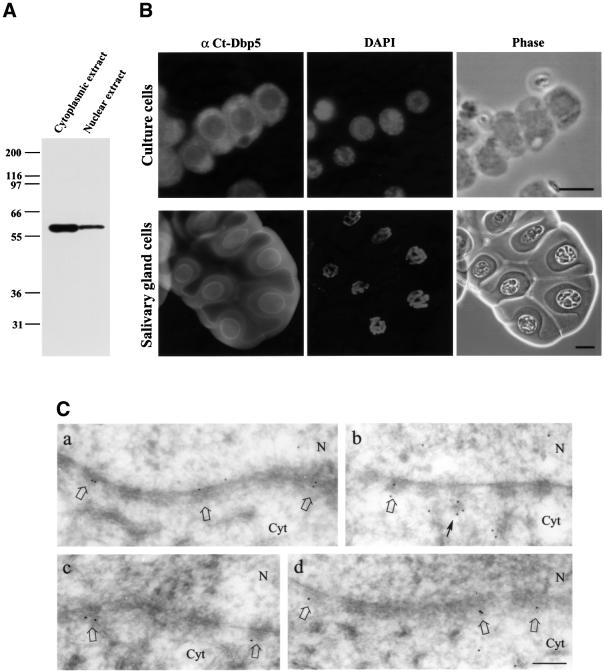
To study the cellular location of Ct-Dbp5, we generated a polyclonal antiserum in rabbits against a His-tagged N-terminal part of Ct-Dbp5. Nuclear and cytoplasmic fractions from tissue culture cells were prepared and analysed by western blot analysis using the affinity-purified polyclonal antibody (Figure 2A). A single polypeptide with a relative mobility of ∼60 kDa was readily found in the cytoplasmic fraction, but it also appeared in the nuclear fraction. The same result was obtained with an affinity-purified polyclonal antibody against the full-length Ct-Dbp5 (data not shown). This is consistent with the location of Ct-Dbp5 homologues in both yeast and mammalian cells, where Dbp5 is present predominantly in the cytoplasm and concentrated at the nuclear envelope (e.g. Snay-Hodge et al., 1998; Tseng et al., 1998; Schmitt et al., 1999).
Fig. 2. Immunological detection of Ct-Dbp5. (A) Western blot analysis of Ct-Dbp5 in cytoplasmic and nuclear extracts from C.tentans tissue culture cells. (B) Localization of Ct-Dbp5 by immunofluorescence microscopy in C.tentans tissue culture cells (top) and salivary gland cells (bottom). The same preparations are also shown stained with DAPI and in phase contrast. (C) Immunoelectron microscopic localization of Ct-Dbp5 in the vicinity of the nuclear envelope. Labelled NPCs are indicated by open arrows. A cytoplasmic cluster of gold particles close to an NPC is indicated by a thin arrow. N, nucleus; Cyt, cytoplasm. Bar equals 10 µm in (B, upper), 20 µm in (B, lower) and 200 nm in (C).
To investigate the subcellular localization of Ct-Dbp5 further, we stained tissue culture cells and salivary gland cells with the polyclonal antibody against the N-terminal part of Ct-Dbp5. In Figure 2B, it is shown that in both types of cells, Ct-Dbp5 is located mainly in the cytoplasm and concentrated in the nuclear rim. However, a weak but significant signal was also observed in the nucleus. This suggests that the presence of Ct-Dbp5 in the nuclear fraction in the western blot is not due only to association with the NPCs, but that Ct-Dbp5 is also located inside the nucleus.
To study the nuclear rim staining at the ultrastructural level, we carried out immunoelectron microscopy on salivary gland cells. Four examples of segments of the nuclear envelope are displayed in Figure 2C. The gold particles are distributed mainly in the NPCs and predominantly on the cytoplasmic side (labelled NPCs indicated by open arrows; for structural comparison, see e.g. figure 6 in Schmitt et al., 1999). A quantitative analysis showed that 79% of the gold particles were located on the cytoplasmic side (359 gold particles studied; particles within 100 nm from the central plane were recorded). In addition, we noted that there are often clusters of gold particles extending from the nuclear complex into the cytoplasm (Figure 2C, arrow) (see further below). We conclude that Dbp5 is present mainly on the cytoplasmic side of the NPCs, confirming earlier ultrastructural studies of Dbp5 distribution (Schmitt et al., 1999; Strahm et al., 1999). However, there is also some labelling above background on the nuclear side of the NPC.
Ct-Dbp5 is bound to nascent pre-mRNAs in BRs and other chromosomal puffs
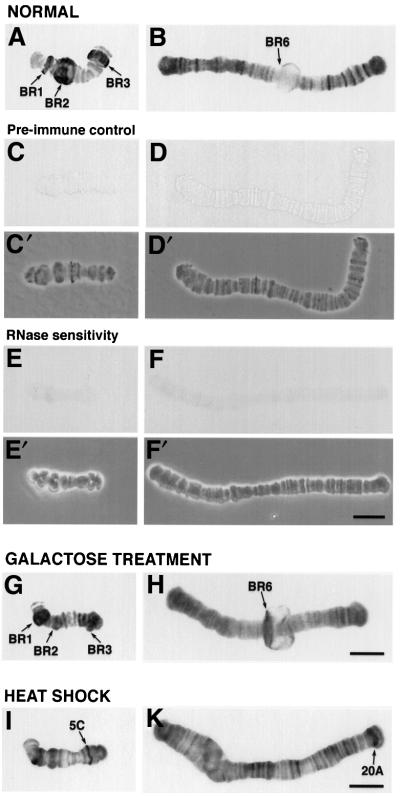
Previously, Dbp5 has been shown to shuttle in and out of the nucleus and to accumulate in the nucleus under certain conditions (Hodge et al., 1999). In Figure 3, we show that Ct-Dbp5 is associated with nascent RNA on polytene chromosomes in salivary gland nuclei. The chromosomes were isolated manually and immunolabelled with polyclonal antibodies specific for Ct-Dbp5. The large BR puffs on chromosome IV were labelled by the anti-Ct-Dbp5 antibodies, and a large number of smaller chromosomal puffs on all chromosomes were also immunostained (Figure 3A and B). No immunolabelling could be found in the control experiment with pre-immune serum (Figure 3C and D). If the isolated chromosomes were digested with RNase A before the immunolabelling, no signal was detected (Figure 3E and F).
Fig. 3. Immunological detection of Ct-Dbp5 in polytene chromosomes. Polytene chromosomes were isolated from untreated (A–F), galactose-treated (G and H) and heat-shocked (I and K) salivary glands and subjected to immunocytology. Some of the untreated chromosomes served as pre-immune controls (C and D), while others were RNase treated (E and F). (C′), (D′), (E′) and (F′), phase contrast images; (A), (C), (E), (G) and (I), chromosome IV; (B) and (H), chromosome III; (D), (F) and (K), chromosome I. After heat shock, the BR genes are switched off and heat shock genes, e.g. in region 5C on chromosome IV (I) and region 20A on chromosome I (K), are turned on. Bar, 20 µm.
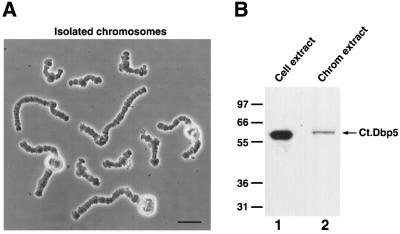
To further confirm that the immunostaining of nascent pre-mRNAs in polytene chromosomal puffs represents Ct-Dbp5, we performed a western blot analysis of the proteins extracted from isolated chromosomes. Approxim ately 1500 polytene chromosomes were isolated from prefixed salivary gland cells (Figure 4A). As expected, Ct-Dbp5 was detected in extracts from the isolated chromosomes (Figure 4B, lane 2).
Fig. 4. The presence of Ct-Dbp5 on isolated polytene chromosomes. (A) Isolated polytene chromosomes from C.tentans salivary gland cells. There are four different chromosomes, two of which carry a nucleolus. Bar, 40 µm. (B) Western blot analysis of extract from the isolated chromosomes. An extract from C.tentans tissue culture cells was used as control.
Taking these results together, we conclude that Ct-Dbp5 is present in the nucleus and that it is associated with nascent transcripts in most transcriptionally active gene loci, including the BR loci.
Ct-Dbp5 is recruited to activated gene loci
To analyse whether the association of Ct-Dbp5 with gene loci is transcription dependent, we specifically induced the transcription of the BR6 gene on chromosome III by galactose treatment as described by Baurén et al. (1996). At the same time, the BR2 genes were turned off at the level of transcription. Before addition of galactose, the active BR2 locus was immunolabelled by the anti-Ct-Dbp5 antibody (Figure 3A), while the silent BR6 locus was unlabelled (Figure 3B). In contrast, after addition of galactose, the immunostaining almost vanished at the BR2 locus (Figure 3G), whereas the active BR6 locus was labelled (Figure 3H).
We also found that Ct-Dbp5 is recruited to major heat shock-inducible puffs (Sass, 1995) when larvae were subjected to a 60 min heat shock at 37°C (Figure 3I and K). The activity of the BR1–3 genes was repressed by heat shock and the immunostaining for Ct-Dbp5 became weak in these gene loci.
Ct-Dbp5 is associated with nascent BR pre-mRNP and remains associated with the BR mRNP to and through the nuclear pores
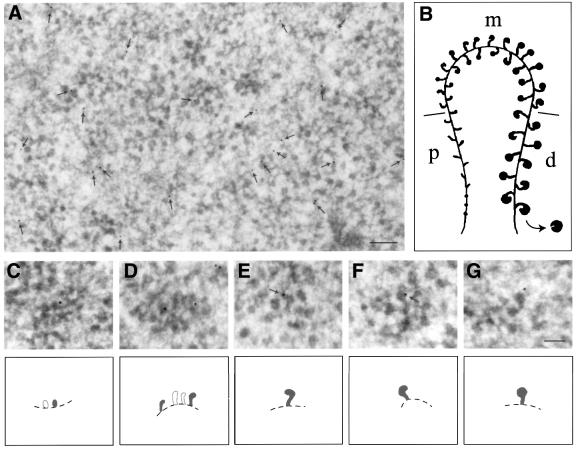
Immunoelectron microscopy of ultrathin cryosections was used to characterize further the behaviour of Ct-Dbp5 in relation to the assembly and transport of gene-specific pre-mRNPs. In a cryosection through a Balbiani ring, a large number of BR gene segments can be seen (Figure 5A), representing proximal, middle and distal portions of the active genes (schematic drawing of a whole BR gene in Figure 5B). The immunosignal can already be seen at low magnification (arrows in Figure 5A); the control specimens are essentially devoid of gold particles (data not shown). Furthermore, as shown at higher magnification, the gold particles are located on both proximal (Figure 5C), middle (Figure 5D) and distal (Figure 5E–G) portions of the BR gene. We conclude that Ct-Dbp5 is associated with the growing pre-mRNP particle, and that it is added to the transcript very early during transcription, maybe even at the start of transcription.
Fig. 5. Immunoelectron microscopic localization of Ct-Dbp5 along transcriptionally active BR genes. (A) Section through a Balbiani ring showing a large number of segments of active genes. Arrows indicate the position of gold particles. (B) Schematic drawing of an active BR gene; growing RNP particles in the promoter-proximal (p), middle (m) and promoter-distal (d) portions of the gene are shown as well as the chromatin axis. (C–G) Immunolabelled gene segments at higher magnification; (C) proximal portion; (D) middle portion; (E and F) distal portions. Schematic drawings are shown below the electron micrographs. Labelled, growing RNP particles are depicted in black and the putative chromatin axis by a broken line. Gold particles at the very tip of growing RNP particles are indicated by arrows in (E) and (F). Bar equals 200 nm in (A) and 100 nm in (G).
The immunosignal is already present in the proximal portion of the active gene but remarkably, it does not get stronger along the active gene. Furthermore, it was noted that the gold particles are often located at the tip of the bent ribbon in the globular portion of the particle (Figure 5E and F, arrows, cf. B), where the 5′ end of the transcript is known to be located (Skoglund et al., 1986). Essentially the same result was obtained in an earlier study of the cap binding protein 20 (CBP20) (cf. figure 5 in Visa et al., 1996b). We conclude that Ct-Dbp 5 is likely to be selectively bound to the 5′ end region of the transcript (see Discussion). It is difficult to rigidly exclude the possibility that Dbp5 is also being added downstream of the 5′ end region but is then not accessible to antibodies. However, as polyclonal antibodies were used and other antibodies can recognize the packaged RNP fibre (cf. e.g. Alzhanova-Ericsson et al., 1996), we regard this latter alternative as less likely.
The BR mRNP particles are released from the gene as globular structures (Figure 5B) and can be readily visualized in the electron microscope as dense RNP granules when in transit through the nucleoplasm to the NPCs. Although the immunosignal is low, BR mRNP particles in the nucleoplasm can be seen labelled by gold markers in our immunoelectron microscopy sections (Figure 6A and B). Thus, we conclude that Ct-Dbp5 still remains bound to BR mRNP particles during intranuclear transport.
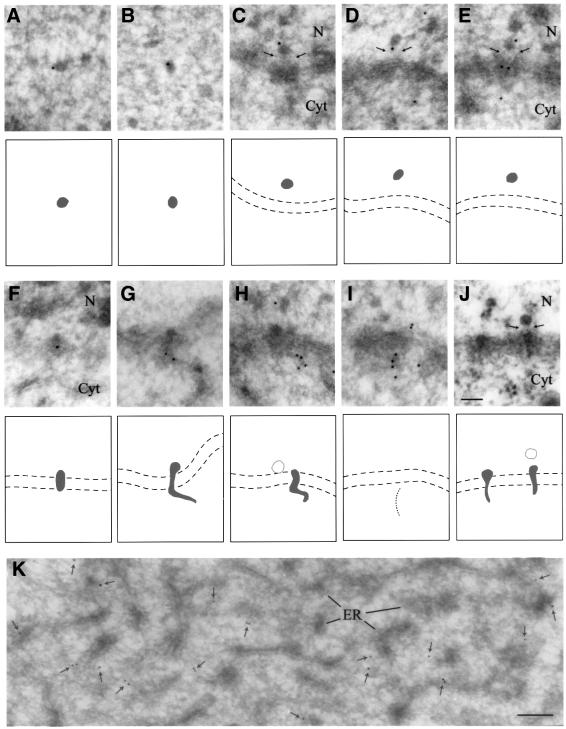
Fig. 6. Immunoelectron microscopic localization of Ct-Dbp5 in nucleoplasmic and translocating BR mRNP particles and in cytoplasm. (A and B) Nucleoplasmic BR RNP particles. (C–E) BR particles bound to the top of the nuclear basket; the basket is indicated by thin arrows. (F–H) BR particles in various stages of translocation through the central channel of the NPC. In (G) and (H) the exiting RNP fibril is extending into the cytoplasm. (I) A line of gold particles extending from an NPC, presumably indicating the presence of an exiting mRNP fibril. (J) Plastic-embedded, translocating BR particles. The arrows indicate the position of the basket. The two membranes of the nuclear envelope are indicated by broken lines in (C–J). (K) Cytoplasm. The gold particles are indicated by small arrows. N, nucleus; Cyt, cytoplasm; ER, endoplasmic reticulum. Bar equals 100 nm in (J) and 200 nm in (K).
When the BR mRNP particle reaches the NPC, it will bind to the nuclear fibres of the NPC, unfold and pass through the channel, and finally exit into the cytoplasm as an extended RNP fibril (Daneholt, 1997). Two examples of translocating, unfolding BR RNP particles can be seen in Figure 6J. Our immunolabelling data show that Ct-Dbp5 is associated with the BR pre-mRNP as it binds to the NPC (Figure 6C–E), as the particle is translocating through the pore (Figure 6F) and, finally, as it appears as an extended RNP fibril on the cytoplasmic side (Figure 6G and H). Furthermore, we recorded in a zone close to the nuclear envelope several examples of linear arrays of gold particles extending into the cytoplasm from the NPC, presumably also representing exiting RNP fibrils, although a distinct RNP particle could not be seen in the NPC (Figure 6I). The association of gold markers with BR mRNP particles at the nuclear side of NPC, during the passage through the NPC and at the cytoplasmic side of the NPC indicates that Ct-Dbp5 is exported to the cytoplasm still associated with the BR mRNP.
It should be emphasized that the immunolabelling of the exiting cytoplasmic RNP fibril is stronger than that of the BR particle in the nucleus (Figure 6G–I compared with Figures 5 and 6A and B). Furthermore, the labelling is present along a considerable portion of the mRNA, suggesting a broader distribution along the RNA than just at the very 5′ end. One possible interpretation of the data is that additional Ct-Dbp5 molecules are being added to the BR transcript when it leaves the NPC (see Discussion).
Further out in the cytoplasm, gold particles are associated with the rough endoplasmic reticulum (ER) although they also seem to appear in between the tubuli of the reticulum (Figure 6K). As BR mRNA is known to bind to the ER, it is possible that Ct-Dbp5 remains associated with the BR mRNA also during protein synthesis.
Taking all immunolabeling data together, we con clude that Ct-Dbp5 is added to the 5′ end region of the BR transcript early during transcription, and remains associated with the mRNP during transport through the nucleoplasm, to and through the nuclear pores, and into the cytoplasm. It is also suggested that additional Ct-Dbp5 is added to and along the transcript when the unfolding RNP fibril leaves the NPC and enters the cytoplasm.
Ct-Dbp5 accumulates in the nucleus when synthesis and/or export of mRNA is inhibited
If most or all Ct-Dbp5 leaves the nucleus associated with mRNA, the subcellular distribution of Ct-Dbp5 should be changed if the synthesis and/or export of mRNA is blocked. This was tested in two sets of experiments.
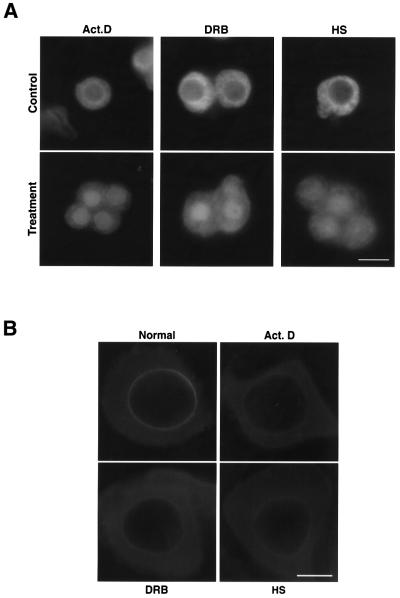
In the first set of experiments, mRNA synthesis was abolished by the RNA polymerase II inhibitors actinomycin D and 5,6-dichloro-1-β-ribofuranosylbenzimidazole (DRB). When C.tentans tissue culture cells were exposed to actinomycin D for 3 h, Ct-Dbp5 accumulated in the nucleus but could still be seen in the cytoplasm (Figure 7A). Ct-Dbp5 also accumulated in the nucleus after a 3 h treatment with DRB (Figure 7A).
Fig. 7. Redistribution of Ct-Dbp5 after treatment with actinomycin D, DRB or heat shock. (A) C.tentans tissue culture cells were exposed to actinomycin D, DRB or heat shock and stained with the anti-Ct-Dbp5 antibodies before and after the treatments. Bar equals 10 µm. (B) C.tentans salivary gland cells were exposed to actinomycin D, DRB or heat shock, treated briefly with detergents to allow antibodies to enter the cytoplasm, but not the nucleus, and stained with the anti-Ct-Dbp5 antibodies. Bar, 20 µm.
In the second set of experiments, we used heat shock treatment to considerably reduce mRNA export (see e.g. Liu et al., 1996). Chironomus tentans tissue culture cells were shifted from 24 to 37°C and kept at the higher temperature for 60 min. Again, Ct-Dbp5 accumulated in the nucleus (Figure 7A).
We conclude that Ct-Dbp5 accumulates in the cell nucleus when the synthesis and/or export of mRNA is inhibited, suggesting that the export of most or all Ct-Dbp5 is closely coupled to the export of mRNA.
The enrichment of Ct-Dbp5 at the NPC is dependent on RNA export
To determine whether the association of Ct-Dbp5 with the NPC is mRNA dependent, we examined whether the protein at the NPC relocated after incubation with the transcriptional inhibitor actinomycin D or DRB and after heat shock treatment. To visualize the nuclear rim-associated fraction of Ct-Dbp5 efficiently, we treated salivary gland cells with Triton X-100 prior to fixation according to Bachi et al. (2000). As shown in Figure 7B, the level of Ct-Dbp5 associated with the nuclear envelope decreased considerably after treatment with actinomycin D or DRB, but fluorescence still remained detectable at the nuclear rim. A similar result was also obtained when C.tentans larvae were subjected to a 60 min heat shock at 37°C (Figure 7B). Thus, the association of Ct-Dbp5 with the NPC is partially sensitive to transcriptional inhibitors and heat shock, and the presence of Ct-Dbp5 at the NPC is at least partly dependent on ongoing mRNA export through the pore.
Discussion
Ct-Dbp5 is likely to be a general mRNA export factor
Ct-Dbp5 is highly similar to its Dbp5 homologues in many species, from yeast to human. It shows the typical conserved domains of the DEAD box superfamily of proteins. In addition, it has a six amino acid insert that is characteristic for the Dbp5 subfamily. It should also be noted that the cellular distribution of Dbp5 in C.tentans cells is similar to that observed in other eukaryotic cells (e.g. Snay-Hodge et al., 1998; Tseng et al., 1998; Schmitt et al., 1999), i.e. by far most of the Dbp5 is in the cytoplasm with a high concentration at the cytoplasmic face of the nuclear envelope. We conclude that Ct-Dbp5 is a typical member of the Dbp5 subfamily of DEAD box proteins.
Dbp5 functions as an mRNA export factor in yeast (Snay-Hodge et al., 1998; Tseng et al., 1998) and in mammals (Schmitt et al., 1999). In the present study, we could establish that Ct-Dbp5 appears associated with RNA at essentially all major transcription sites on the giant chromosomes, implying that Ct-Dbp5 is connected to most or all mRNA transcripts. Furthermore, when specific genes are activated or repressed by galactose treatment or heat shock, the amount of Ct-Dbp5 at the corresponding chromosomal loci follows closely the changes in transcription. Thus, Ct-Dbp5 is likely to be a general mRNA export factor.
Ct-Dbp5 is associated with mRNA from gene to cytoplasm
The most striking result of the present study is the observation that Ct-Dbp5 becomes associated with mRNA as early as the transcription process and accompanies the mRNA to and through the nuclear pores and into the cytoplasm, presumably also into polysomes. Biochemical experiments showing that Ct-Dbp5 co-immunoprecipitates with pre-mRNP particles further support the conclusion that Dbp5 accompanies pre-mRNA/mRNA (our unpublished data). Our results are in agreement with the finding in yeast that Dbp5 can shuttle between the nucleus and cytoplasm (Hodge et al., 1999) and demonstrates that Dbp5 can be recorded also in the nucleus under normal conditions. Furthermore, in previous studies of the export factor Dbp5, it has been emphasized that Dbp5 is present at the cytoplasmic fibrils of the NPC (Schmitt et al., 1999; Strahm et al., 1999), probably associated with CAN/Nup159p (Hodge et al., 1999; Schmitt et al., 1999) and Gle1p (Hodge et al., 1999; Strahm et al., 1999), and it has been assumed that Dbp5 functions in mRNP translocation through the NPC and/or during release into the cytoplasm. Our study implies that the export factor Dbp5 could exert its function anywhere between the gene and the polysome.
The Ct-Dbp5 distribution along the active gene gives information on the location of Dbp5 on the transcript. It was noted that Ct-Dbp5 appears in the most promoter-proximal portion of the gene, that the immunosignal remains approximately the same along the entire gene and that the gold particles seem to bind to the 5′ end region of the growing RNP particles (best seen on the almost finished RNPs). Such a distribution on the active BR gene was established earlier for CBP20 (Visa et al., 1996b), and it was concluded that Ct-Dbp5 is also likely to bind selectively to the BR transcript at or close to its 5′ end. Such a conclusion is strengthened by the observation that during the translocation of BR RNP through the nuclear pores, Ct-Dbp5 is associated with the leading end of the BR transcript, known to be the 5′ end (Mehlin et al., 1992). As proposed below, it is possible that additional Ct-Dbp5 molecules are being added along the transcript when the message enters the cytoplasm.
If a major part of the nuclear Ct-Dbp5 is associated with mRNA and accompanies it during nucleocytoplasmic transport, the distribution of Ct-Dbp5 in the nucleus should be affected if the synthesis and/or export of mRNA is inhibited. This proved also to be the case when mRNA synthesis was blocked by DRB or actinomycin D, and when mRNA export was drastically reduced by heat shock. In all three cases, Ct-Dbp5 accumulates in the nucleus, and the nuclear rim staining is diminished. Thus, the translocation of Ct-Dbp5 from the nucleus to the cytoplasm seems to be closely coupled to ongoing mRNA export. As only a minor part of the cellular Ct-Dbp5 is present inside the nucleus, it seems likely that immediately upon entry into the nucleus, Ct-Dbp5 binds to pre-mRNP and accompanies mRNP to the cytoplasm.
Potential roles of Ct-Dbp5 in assembly and export of mRNP particles
The fact that the RNA helicase Ct-Dbp5 is added to pre-mRNP co-transcriptionally and remains with the mRNA during export into the cytoplasm opens up the possibility that Dbp5 participates in the drastic conformational changes of the BR particle occurring during the information transfer, in particular the stepwise packing of the RNP fibril into the compact ring-like structure concomitant with transcription and the subsequent unpacking of the particle upon passage through the nuclear pores. In addition, some of the proteins are shed from the BR RNP complex just before or during the translocation (Alzhanova-Ericsson et al., 1996; Visa et al., 1996a; Sun et al., 1998; for review, see Daneholt, 1997). It is conceivable that modulation of the structure of the RNA molecule and/or rearrangement of RNA–protein interactions is required to pack and unpack the pre-mRNP complexes properly. It would not be unexpected if an RNA helicase like Dbp5 were involved in one or more of these dynamic processes.
We confirm that Dbp5 is enriched on the cytoplasmic side of the NPC, suggesting that this region is likely to be the main site of action for Dbp5. When the BR mRNA extends into the cytoplasm, proteins are still attached to the RNA (Mehlin et al., 1992; Visa et al., 1996a,b; Daneholt 1997). Some of them are displaced just upon appearance in the cytoplasm like CBP20 (Visa et al., 1996b), while others accompany mRNA into polysomes like the A1-like hnRNP protein hrp36 (Visa et al., 1996a). It has been proposed that Dbp5 could be involved in the dramatic structural changes, including removal and addition of proteins, that are likely to occur when the exiting mRNA is prepared for participation in translation (Snay-Hodge et al., 1998; Tseng et al., 1998; Schmitt et al., 1999). Our demonstration that Dbp5 is associated directly with the exiting mRNA strongly supports this model; an alternative model stating that NPC-anchored Dbp5 is pulling the mRNA through the nuclear pore seems less likely from our data (for discussion, see e.g. Cole, 2000). Furthermore, the fact that Dbp5 is present on at least a substantial portion of the mRNA entering the cytoplasm could imply that Dbp5 operates along the mRNA. It is then of interest that it has recently been shown that a closely related RNA helicase, NPH-II, unwinds RNA substrates in a processive and directional fashion (Jankowsky et al., 2000). Finally, it should be noted that if Dbp5 acts by unloading (e.g. export factors) or remodelling mRNP entering the cytoplasm, Dbp5 could confer directionality to the mRNP export process in the absence of the RanGTPase system (for review, see Görlich and Kutay, 1999).
It is striking that the immunosignal is particularly strong at the more or less extended, exiting mRNP fibril, and that it is distributed along at least a considerable part of the transcript (often a line of several gold particles). This is clearly different from the restricted 5′ end distribution on the nascent transcripts: the growing, still unpacked RNP fibril on the proximal part of the gene does not show the same high and linear labelling as the unfolding RNP fibril in the cytoplasm. One possible interpretation of the data would be that the transcript passes through the central channel with Ct-Dbp5 just bound at the 5′ end region of the transcript, while on the cytoplasmic side of the NPC more Ct-Dbp5 is being added on to the emerging transcript. Since Dbp5 is likely to interact with Can/Nup159p (Hodge et al., 1999; Schmitt et al., 1999) and Gle1p (Hodge et al., 1999; Strahm et al., 1999) present on the cytoplasmic fibrils of the NPC, it has been proposed that Dbp5 gets bound to the cytoplasmic fibrils and is then further loaded on to the exiting mRNA. Such a model would also be in agreement with yeast genetic data showing that the binding of Dbp5 to CAN/Nup159p contributes to efficient mRNA export but is not essential; overexpression of Dbp5 can overcome the defect (Hodge et al., 1999). If some Dbp5 is already present on the mRNA entering the cytoplasm and more can be recruited from a large cytoplasmic pool, it can be readily explained why a loading of Dbp5 on to exiting mRNA via the cytoplasmic fibrils contributes to the export while not being necessary. In conclusion, our data support the hypothesis that the cytoplasmic fibrils of the NPC assist (via CAN/Nup159p and Gle1p) in loading Dbp5 on to the exiting mRNA.
The BR mRNP fibril that extends into the cytoplasm is rapidly engaged in translation. In fact, occasionally ribosomes can be seen on the exiting BR mRNP fibril while the 3′ end is still on the nuclear side of the NPC (Mehlin et al., 1992). Subsequently, the BR mRNA-containing polysomes anchor to the ER, and secretory proteins are produced for export. As Ct-Dbp5 is abundant in the cytoplasm and appears at least partly at the ER, it is possible that Dbp5 also remains associated with mRNA during protein synthesis.
Co-transcriptional programming of the fate of mRNA
During the assembly of the pre-mRNP particle on the gene, various proteins are being added to the growing transcript. It was early believed that these proteins, in particular the hnRNP proteins, bind to the transcript and package the often very large RNA molecules into a manageable RNP parcel. However, it now seems more attractive to widen the scope and look upon a pre-mRNP complex formed as a substrate for molecular machineries responsible for processing of pre-mRNA, and nucleocytoplasmic transport, translation and turnover of mRNA. Co-transcriptional programming of the pre-mRNP particle as well as subsequent regulatory modifications of the particle become key issues. The recently discovered exon–exon junction complex formed concomitant with splicing and located just upstream of exon–exon junctions, represents a striking example of a post-transcriptional regulatory modification affecting downstream events: nucleocytoplasmic transport and most likely also nonsense-mediated decay of mRNA in the cytoplasm (see Introduction). The main observation in the present study is that the RNA helicase Dbp5 appears to bind to the 5′ end region of the transcript already co-transcriptionally and remains bound to the transcript during nucleocytoplasmic transfer and presumably also into polysomes. It could be that the mRNA export factor Dbp5 exerts multiple roles both in the nucleus and cytoplasm, or alternatively, that it acts selectively at the exit of mRNA into the cytoplasm. In either case, our results on Ct-Dbp5 highlight the importance of the co-transcriptional loading of pre-mRNA with proteins, and support the view that these proteins to a considerable extent decide the fate of the mRNA not only in the nucleus but also in the cytoplasm.
Materials and methods
Animals and tissue culture cells
Chironomus tentans was raised in the laboratory. Salivary glands were isolated from fourth instar larvae. Chironomus tentans tissue culture cells (the Wyss cell line) were cultivated at room temperature.
Treatments with drugs and heat shock
Larvae were treated with galactose according to Baurén et al. (1996). In the DRB and actinomycin D experiments, tissue culture cells or isolated salivary glands were kept in cell cultivation medium containing 90 µM DRB (Sigma) or 5 µg/ml of actinomycin D for 3 h at 18°C. In all cases, control cells were incubated in parallel in medium without drugs. In the heat shock experiments, tissue culture cells were incubated for 60 min at 37°C. Larvae were heat shocked in water at 37°C for 60 min.
Antibodies
Polyclonal antisera were raised in rabbits against the His-tagged N-terminal part (amino acid residues 1–123) of Ct-Dbp5 (Ct-Dbp5-N) and against full-length Ct-Dbp5. The antibodies were affinity-purified by binding to recombinant, glutathione S-transferase-tagged, full-length Ct-Dbp5 immobilized on NHS-activated Sepharose beads (Pharmacia Biotech).
cDNA cloning and sequencing
The full-length Dbp5p protein from S.cerevisiae (accession number P20449), was used for database searches. Putative homologues were detected in various organisms including human (Q9UMR2), mouse (Q61655), fission yeast (Q09747), Xenopus (AAF99574) and Drosophila (O61305). Degenerate PCR primers were designed based on the two highly conserved regions GFNTPSKIQE and DEADVMIATQ and used for RT–PCR. Poly(A)+ RNA isolated from tissue culture cells was used for cDNA synthesis, and touchdown PCR was performed on a Perkin Elmer Thermocycler. The obtained PCR product was subcloned into the pCR 2.1 vector (Invitrogen), and several PCR clones were sequenced and analysed. A cloned fragment with high homology to Dbp5 was used as a probe to screen a λ ZAP cDNA library from the salivary glands of C.tentans. The DNA sequencing kit (Dye Terminator Cycle Sequencing; Applied Biosystems) was used for DNA sequencing and the reactions were analysed on a 373A automated DNA sequencer (Applied Biosystems). The DNA sequences were analysed by the University of Wisconsin Genetics Computer Group (GCG) sequence analysis programs and EGCG extensions to the Wisconsin Package sequence analysis programs.
Gel electrophoresis and western blot analysis
Nuclear and cytoplasmic extracts were prepared from C.tentans tissue culture cells as described by Sun et al. (1998). For analysis of chromosomal proteins, polytene chromosomes were isolated from salivary glands according to Sun et al. (1998). The specimens were boiled in sample buffer and the proteins separated by electrophoresis in 10% polyacrylamide–SDS gels. After electrophoresis, the proteins were blotted on to transfer membranes (Immobilon PVDF; Millipore) using a Trans-Blot semi-dry electrophoretic transfer apparatus (Bio-Rad). Membranes were blocked for 1 h in phosphate-buffered saline (PBS), containing 10% dried milk powder, and incubated for 1 h with affinity-purified polyclonal antibodies against Ct-Dbp5 (1:3000 dilution) in PBS, containing 1% non-fat dried milk and 0.05% Tween 20. The membranes were washed in PBS containing 0.05% Tween 20 and incubated with peroxidase-conjugated secondary antibody (Dako). After washing, the position of the antibodies was detected by chemiluminescence using the ECL detection system (Amersham Pharmacia).
Immunofluorescence microscopy of tissue culture cells and salivary glands
Chironomus tentans culture cells were centrifuged (Cytospin; Shandon Astmoor, Runcorn, UK) on to the slides. The cells were fixed in 4% paraformadehyde in PBS for 15 min at room temperature. After washing in PBS, the cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min and washed again in PBS. The cells were blocked in 2% bovine serum albumin (BSA) in PBS for 1 h and incubated for 2 h at room temperature with 100 µl of affinity-purified anti-Ct-Dbp5-N antibody (1:20 dilution in PBS with 2% BSA) or pre-immune serum (1:600 dilution in TKM with 2% BSA) as a negative control. The specimens were then incubated with FITC-conjugated Fab′ fragment of anti-rabbit immunoglobulins (Dako), washed with PBS, mounted in a medium with 4′-6-diamidine-2-phenylindole (DAPI) (Vector Laboratories) and examined in a Zeiss fluorescence microscope.
Salivary glands were dissected from fourth instar larvae and fixed in 4% paraformadehyde in PBS for 15 min at room temperature. After washing in PBS the cells were permeabilized with 0.5% Triton X-100 in PBS for 60 min and washed again in PBS. The immunolabelling was performed as above for tissue culture cells, the only difference being that the glands were incubated with the anti-Ct-Dbp5-N antibody overnight at 4°C.
The nuclear rim-associated fraction of Ct-Dbp5 was observed after drug treatments and heat shock as described by Bachi et al. (2000). Salivary glands were permeabilized with 0.5% Triton X-100 in PBS for 3 min and fixed in 4% paraformadehyde in PBS for 15 min at room temperature, washed again in PBS and immunolabelled as above for salivary glands.
Immunocytology of isolated polytene chromosomes
Polytene chromosomes were isolated as described above. When RNase treated, the isolated chromosomes were incubated with 100 µg/ml RNase A for 60 min at room temperature immediately after isolation. Subsequently, the chromosomes were blocked with 50 µl of 2% BSA in TKM (100 mM KCl, 1 mM MgCl2, 10 mM triethanolamine pH 7.0) for 60 min at room temperature and incubated for 2 h with 80 µl of the affinity-purified anti-Ct-Dbp5-N antibody (1:20 dilution) or pre-immune serum (1:600 dilution in TKM with 2% BSA) as a negative control. The slides were washed in 0.1% Tween 20 in TKM and then incubated for 60 min with 80 µl of the gold-conjugated anti-rabbit antibody (6 nm gold; Jackson Immuno-Research Labs, West Grave, PA) diluted 1:50 in TKM buffer containing 2% BSA. The specimens were washed in TKM and with distilled water. Immunogold silver enhancement solution (IntenSEM; Amersham) was added for 8–15 min at room temperature. The preparations were rinsed in distilled water, mounted in 30% glycerol and photographed in a Zeiss light microscope.
Immunoelectron microscopy of salivary glands
Immunoelectron microscopy staining of ultrathin cryosections was performed according to Visa et al. (1996a). Isolated salivary glands were fixed in 4% formaldehyde and 0.1% glutaraldehyde in 0.1 M cacodylate buffer pH 7.2, for 20–25 min at room temperature. After rinsing, the fixed glands were cryoprotected with 2.3 M sucrose and frozen by immersion in liquid nitrogen. Ultrathin cryosections were prepared and deposited on to nickel grids coated with formvar and carbon. The grids were blocked on drops of PBSG (0.1 M glycine in PBS) containing 10% newborn calf serum for 20 min, and incubated with the anti-Ct-Dbp5-N antibody (diluted 1:10) for 60 min, and then with anti-rabbit immunoglobulin conjugated with 12 nm gold particles (Jackson Immuno-Research Labs) for 60 min at room temperature. In control experiments, a polyclonal, affinity-purified anti-SCP3 antibody (diluted 1:10) replaced the anti-Ct-Dbp5 antibody. After immunolabelling, the sections were stained with 2% aqueous uranyl acetate and embedded in polyvinyl alcohol (9–10 kDa; Aldrich). The specimens were examined in a Philips CM 120 microscope.
Acknowledgments
Acknowledgements
We are grateful to Kerstin Bernholm and Lise-Marie Fjelkestam for technical assistance and to Sergej Masich for computer work. This work was supported by the Swedish Research Council (Natural and Engineering Sciences), Human Frontier Science Program Organisation, Knut and Alice Wallenberg Foundation, and Ingabritt and Arne Lundberg Foundation. J.Z. was a recipient of fellowships from the Wenner-Gren Foundation and the Swedish Cancer Fund.
References
- Alzhanova-Ericsson A.T., Sun X., Visa,N., Kiseleva,E., Wurtz,T. and Daneholt,B. (1996) A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev., 10, 2881–2893. [DOI] [PubMed] [Google Scholar]
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G. and Wieslander,L. (1994) Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell, 76, 183–192. [DOI] [PubMed] [Google Scholar]
- Baurén G., Jiang,W.-Q., Bernholm,K., Gu,F. and Wieslander,L. (1996) Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J. Cell Biol., 133, 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C.N. (2000) mRNA export: the long and winding road. Nature Cell Biol., 2, E55–E58. [DOI] [PubMed] [Google Scholar]
- Daneholt B. (1997) A look at messenger RNP moving through the nuclear pore. Cell, 88, 585–588. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Herold A., Suyama,M., Rodrigues,J.P., Braun,I.C., Kutay,U., Carmo-Fonseca,M., Bork,P. and Izaurralde,E. (2000) TAP/NXF1 belongs to a multigene family of putative RNA export factors with a conserved modular structure. Mol. Cell. Biol., 20, 8996–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.A., Colot ,H.V., Stafford,P. and Cole,C.N. (1999) Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J., 18, 5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2000) The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature, 403, 447–451. [DOI] [PubMed] [Google Scholar]
- Katahira J., Strasser,K., Podtelejnikov,A., Mann,M., Jung,J.U. and Hurt,E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E., Wurtz,T., Visa,N. and Daneholt,B. (1994) Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J., 13, 6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and non-sense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liang,S. and Tartakoff,A.M. (1996) Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J., 15, 6750–6757. [PMC free article] [PubMed] [Google Scholar]
- Luking A., Stahl,U. and Schmidt,U. (1998) The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol., 33, 259–296. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mehlin H., Daneholt,B. and Skoglund,U. (1992) Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscopy tomography. Cell, 69, 605–613. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanics and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B.J., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass H. (1995) Transcription of heat shock gene loci versus non-heat shock loci in Chironomus tentans polytene chromosomes: evidence for heat-induced formation of novel putative ribonucleoprotein particles (hsRNPs) in the major heat shock puffs. Chromosoma, 103, 528–538. [DOI] [PubMed] [Google Scholar]
- Schmitt C. et al. (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J., 18, 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma,K., Doye,V., Hellwig,A., Huber,J., Lührmann,R. and Hurt,E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund U., Andersson,K., Björkroth,B., Lamb,M.M. and Daneholt,B. (1983) Visualization of the formation and transport of a specific hnRNP particle. Cell, 34, 847–855. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Andersson,K., Strandberg,B. and Daneholt,B. (1986) Three-dimensional structure of a specific pre-messenger RNP particle established by electron microscope tomography. Nature, 319, 560–564. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot,H.V., Goldstein,A.L. and Cole,C.N. (1998) Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J., 17, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog,B., Zenklusen,D., Rychner,E., Kantor,J., Rosbach,M. and Stutz,F. (1999) The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J., 18, 5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K. and Hurt,E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Alzhanova-Ericsson,A.T., Visa,N., Aissouni,Y., Zhao,J. and Daneholt,B. (1998) The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J. Cell Biol., 142, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.S.-I., Weaver,P.L., Liu,Y., Hitomi,M., Tartakoff,A.M. and Chang,T.-H. (1998) Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J., 17, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson,A.T., Sun,X., Kiseleva,E., Björkroth,B., Wurtz,T. and Daneholt,B. (1996a) A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell, 84, 253–264. [DOI] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996b) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D. and Stutz,F. (2001) Nuclear export of mRNA. FEBS Lett., 498, 150–156. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]