Abstract
The major pathways of mRNA turnover in eukaryotic cells are initiated by shortening of the poly(A) tail. Recent work has identified Ccr4p and Pop2p as components of the major cytoplasmic deadenylase in yeast. We now demonstrate that CCR4 encodes the catalytic subunit of the deadenylase and that Pop2p is dispensable for catalysis. In addition, we demonstrate that at least some of the Ccr4p/Pop2p-associated Not proteins are cytoplasmic, and lesions in some of the NOT genes can lead to defects in mRNA deadenylation rates. The Ccr4p deadenylase is inhibited in vitro by addition of the poly(A) binding protein (Pab1p), suggesting that dissociation of Pab1p from the poly(A) tail may be rate limiting for deadenylation in vivo. In addition, the rapid deadenylation of the COX17 mRNA, which is controlled by a member of the Pumilio family of deadenylation activators Puf3p, requires an active Ccr4p/Pop2p/Not deadenylase. These results define the Ccr4p/Pop2p/Not complex as the cytoplasmic deadenylase in yeast and identify positive and negative regulators of this enzyme complex.
Keywords: deadenylation/decay/mRNA/turnover/yeast
Introduction
The regulation of gene expression is influenced by the control of mRNA turnover. Analysis of mRNA turnover in eukaryotic cells has identified several distinct pathways by which mRNAs are degraded (reviewed in Beelman and Parker, 1995). In most cases, the degradation of the transcript begins with the shortening of the 3′ poly(A) tail (e.g. Shyu et al., 1991; Muhlrad and Parker, 1992). In yeast, shortening of the poly(A) tail primarily leads to removal of the 5′ cap structure (decapping), thereby exposing the transcript to digestion by a 5′ to 3′ exonuclease (Decker and Parker, 1993; Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995). Alternatively, transcripts can also be exonucleolytically degraded in a 3′ to 5′ manner following deadenylation (Muhlrad et al., 1995; Anderson and Parker, 1998).
Several observations identify deadenylation as a key step in mRNA translation and stability. First, many mRNAs are deadenylated prior to decay of the body of the transcript, both in yeast and more complex eukaryotes, indicating the general role of deadenylation in controlling mRNA turnover (Wilson and Treisman, 1988; Shyu et al., 1991; Couttet et al., 1997). Second, deadenylation rates for individual mRNAs are controlled both in cis and in trans by numerous regulatory inputs (e.g. Wilson and Treisman, 1988; Shyu et al., 1991; Muhlrad and Parker, 1992; Caponigro and Parker, 1996). Third, computational modeling indicates that changes in deadenylation rates cause the largest alteration in overall mRNA stability (Cao and Parker, 2001). Finally, because the poly(A) tail can influence translation initiation, control of deadenylation rate can directly influence the translational status of the transcript (for a review see Sachs et al., 1997). Given the pivotal role that deadenylation plays in mRNA function, understanding the mechanism by which mRNA deadenylation occurs and its regulation will be central for our understanding of mRNA decay.
A key step in understanding the control of deadenylation is the identification and characterization of the nuclease(s) that removes the poly(A) tail. Towards this goal, we recently demonstrated that Ccr4p and Pop2p/Caf1p are required for mRNA deadenylation in vivo and co-purify with a deadenylase activity in vitro (Tucker et al., 2001). Although we have previously referred to the POP2 gene as CAF1, we will refer to this gene as POP2, since this is its standard name in the yeast database. This work defined these proteins as components of the major cytoplasmic mRNA deadenylase in yeast.
An important issue is the composition of the Ccr4p/Pop2p deadenylase, in terms of both the catalytic subunit(s) and other factors involved in deadenylation. Both Ccr4p and Pop2p have significant homology to known 3′ to 5′ exonucleases (Moser et al., 1997; Dlakic, 2000). This suggests that either CCR4 and/or POP2 may encode the actual catalytic subunit of the mRNA deadenylase. In addition, Ccr4p and Pop2p have been shown to exist in two large complexes of ∼0.9 and ∼1.9 MDa (Liu et al., 1998). The larger complex includes five Not proteins: Not1p (standard name CDC39), Not2p (standard name CDC36), Not3p, Not4p (standard name SIG1), Not5p and several other proteins (Liu et al., 1998; Bai et al., 1999). As we are discussing their role in a single complex, we will refer to these proteins as Not proteins in this manuscript. The smaller complex contains only Ccr4p, Pop2p, the five Not proteins and two newly identified proteins, Caf40p and Caf130p (C.L.Denis, unpublished observations). Interestingly, the Not proteins have been proposed to be involved in transcription based on genetic and physical interaction with transcription factors (Collart and Struhl, 1993, 1994; Benson et al., 1998; Badarinarayana et al., 2000; Lemaire and Collart, 2000; Denis et al., 2001; Liu et al., 2001). Thus, an unresolved issue is the role of the Ccr4p/Pop2p/Not complex in cytoplasmic deadenylation.
In this work we provide evidence that CCR4 encodes the catalytic subunit of the yeast mRNA deadenylase. This conclusion is based on three observations. First, over-expression of Ccr4p suppresses the deadenylation defects of a pop2Δ mutation and accelerates deadenylation rates in a wild-type strain. Second, isolation of Ccr4p co-purifies with deadenylase activity, and this activity is not dependent on the presence of Pop2p. Third, point mutations in predicted catalytic residues of the exonuclease domain of Ccr4p inhibit mRNA deadenylation in vivo and deadenylase activity in vitro. In addition, we examined the role of the Ccr4p associated NOT proteins in mRNA turnover. We determined that at least some of the NOT1 and NOT2 gene products are localized to the cytoplasm and that lesions in at least some of the Not proteins affect the deadenylation rate of the MFA2 transcript in vivo. These observations suggest that a Ccr4p/Pop2p/Not complex functions together to perform cytoplasmic deadenylation in yeast. These data also imply that the Ccr4p/Pop2p/Not complexes that exist in other eukaryotes will serve similar roles as cytoplasmic deadenylases.
Results
Over-expression of Ccr4p suppresses pop2Δ deadenylation endpoint defects
To examine the relative roles of Ccr4p and Pop2p in mRNA deadenylation, we first determined the effect of over-expression of Ccr4p in the absence of Pop2p. Over-expression of Ccr4p is known to suppress the growth defects associated with pop2Δ strains (Hata et al., 1998). In contrast, over-expression of Pop2p does not suppress the growth defect of a ccr4Δ (Hata et al., 1998). This leads to the simple prediction that Ccr4p is the catalytic subunit of the deadenylase and that over-expression of Ccr4p compensates for the loss of Pop2p in deadenylation. To analyze the effects of Ccr4p over-expression on mRNA deadenylation we examined mRNA poly(A) tail distributions of the PGK1pG mRNA in wild-type, ccr4Δ and pop2Δ strains with or without a Ccr4p over-expression plasmid. The PGK1pG reporter mRNA contains a poly(G) tract in the 3′ untranslated region (UTR) that inhibits 5′ to 3′ exonuclease digestion following decapping. This allows the detection of a decay intermediate that extends from the 5′ side of the poly(G) tract to the 3′ end of the mRNA (Decker and Parker, 1993).
Two observations from steady-state mRNA populations in these strains suggested that over-expression of Ccr4p rescues the deadenylation defect in a pop2Δ strain. First, over-expression of Ccr4p corrects the deadenylation endpoint defect seen in pop2Δ strains (Tucker et al., 2001). In wild-type cells the PGK1pG mRNAs have a steady-state poly(A) distribution from 75 to ∼5–10 adenosine residues (Figure 1, lane 2) (Decker and Parker, 1993). In contrast, in ccr4Δ and pop2Δ strains the shortest poly(A) tails seen on the full-length mRNA are 20–26 (Figure 1, lane 4) and 14–20, respectively (lane 6). The key observation was that over-expression of Ccr4p was capable of suppressing the deadenylation endpoint defect seen in a pop2Δ strain with the deadenylation endpoints returning to ∼5–10 adenosine residues (Figure 1, lane 7). Similar results were seen with the MFA2pG mRNA, indicating that this effect was not specific to the PGK1pG mRNA (data not shown). As expected, over-expression of Ccr4p also showed significant complementation of the deadenylation endpoint defect of a ccr4Δ strain, with the shortest poly(A) tails returning to ∼5–12 adenosine residues (Figure 1, lane 5). It should be noted that the ccr4Δ strain complemented by Flag-Ccr4p still showed a small defect in deadenylation, with the distribution of poly(A) tails being shifted to slightly longer lengths. This observation suggests that Flag-Ccr4p has a subtle defect in function, presumably due to the N-terminal Flag epitope. Over-expression of Flag-Ccr4p had little or no effect on the deadenylation endpoint in wild-type cells (Figure 1, lane 3). In contrast, over-expression of Pop2p in a ccr4Δ mutant or wild-type strain had no effect on the deadenylation seen in these strains (data not shown).
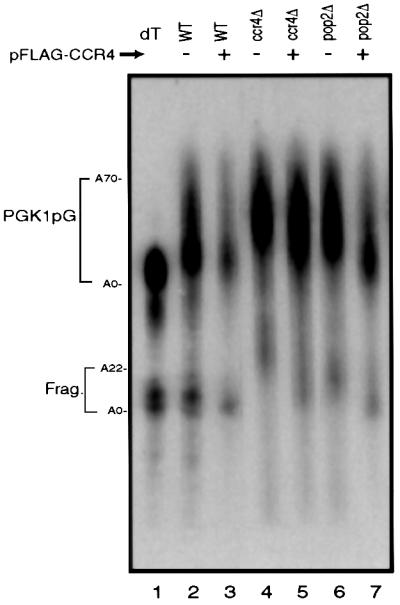
Fig. 1. Comparison of deadenylation end-points. Shown are poly acrylamide northern gels examining the deadenylation end-points for the PGK1pG transcript in wild-type, ccr4Δ and pop2Δ strains containing either a Flag-Ccr4p over-expression plasmid (pRP1045) or vector control as indicated. Here, and in other figures, the full-length (FL) mRNA and decay fragment (Frag.) are indicated on the left. Steady-state mRNA samples were resolved on 6% polyacrylamide/8 M urea northern gels, with or without removal of poly(A) tails with RNase H and oligo(dT) (lane 1). Here, and in other figures, to allow for size resolution of the poly(A) tail the 3′ 319 nucleotides of the 1.4 kb PGK1pG mRNA were cleaved by hybridizing to oRP70 followed by cleavage with RNase H prior to loading on the gel.
Additional evidence that Ccr4p over-expression suppressed the deadenylation endpoint defect came from examining the PGK1pG mRNA fragment produced by decapping and 5′ to 3′ exonucleolytic degradation to the poly(G) tract. In wild-type cells, this decay fragment has a short poly(A) tail of 0–12 residues. This is because decapping and subsequent degradation of the body of the transcript occurs only after significant deadenylation (Figure 1, lanes 1–3) (Decker and Parker, 1993). In contrast, in ccr4Δ and pop2Δ strains these mRNA fragments have poly(A) tails of 20–26 and 14–20 residues, respectively (Figure 1, lanes 4 and 6). This is presumably due to a slow decapping of the partially adenylated full-length mRNAs that accumulate in these strains (Tucker et al., 2001). Over-expression of Ccr4p in pop2Δ strains restores the poly(A) tail on the mRNA fragment to almost the same length as in wild-type cells (Figure 1, lane 7). These two observations indicate that over-expression of Ccr4p corrects the deadenylation endpoint defect seen in the pop2Δ strains, and suggest that over-expression of Ccr4p is capable of bypassing the need for Pop2p in the process of mRNA deadenylation.
Over-expression of Ccr4p accelerates deadenylation rates
To determine whether Ccr4p over-expression suppressed the deadenylation rate defect of a pop2Δ strain, we analyzed the decay of the MFA2pG and PGK1pG reporter mRNAs in wild-type and pop2Δ strains harboring either a vector control or the Ccr4p over-expression plasmid by a transcriptional pulse–chase experiment (Decker and Parker, 1993). In this experiment we utilized the carbon source regulation of the GAL1 upstream activating sequence (GAL UAS) to rapidly induce and then repress transcription of these reporter mRNAs. This produces a pool of newly transcribed mRNAs, the metabolism of which can be followed over time to observe deadenylation and subsequent decay of the mRNA.
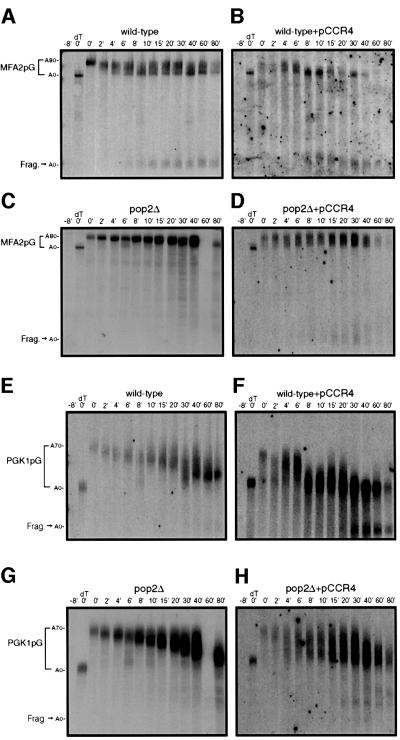
Comparison of transcriptional pulse–chases for the MFA2pG mRNA in wild-type and pop2Δ strains over-expressing Ccr4p with the same strains with a vector control identified a number of significant observations. First, in the wild-type strain over-expressing Ccr4p the MFA2pG mRNA was deadenylated at a rate similar to that of the wild-type control strain, with the appearance of a fully deadenylated mRNA appearing between 2 and 4 min (Figure 2A and B). In this strain background the MFA2pG reporter is not fully repressed by glucose, and for this reason there is some residual transcription. This is the reason for the persistence of a distributed population of adenylated MFA2pG transcripts in wild-type cells. Second, in a pop2Δ strain MFA2pG mRNA was deadenylated slowly over a period >40 min (Figure 2C). Importantly, over-expression of Ccr4p in the pop2Δ strain restored the rapid deadenylation rate of MFA2pG mRNA to that resembling wild type, with some of the transcripts being fully deadenylated by 4 min (Figure 2D, lane 4). This is consistent with the interpretation that over-expression of Ccr4p bypasses the need for Pop2p in the process of mRNA deadenylation.
Fig. 2. Transcriptional pulse–chase analysis of the MFA2pG and PGK1pG transcripts. Shown are polyacrylamide northern gels examining the decay of MFA2pG (A–D) and PGK1pG (E–H) in wild-type, ccr4Δ and pop2Δ strains containing either a Flag-Ccr4p over-expression plasmid (pRP1045) or vector control as indicated. Numbers above the lanes are minutes after transcriptional repression by the addition of glucose following an 8 min induction of transcription (Decker and Parker, 1993). The 0 min time point was treated with RNase H and oligo(dT) to indicate the position of the deadenylated mRNA. Here, and in all subsequent figures, poly(A) tail lengths were determined by comparison of bands to size standards and the poly(A)– mRNA species generated by cleavage of RNase H and oligo(dT) (data not shown).
We also observed that over-expression of Ccr4p accelerated the deadenylation rate of the normally stable PGK1pG mRNA. In wild-type strains, the PGK1pG mRNA was deadenylated at a rate of ∼2 or 3 nucleotides/min (Figure 2E). In wild-type and pop2Δ strains, over-expression of Ccr4p accelerated the deadenylation rate of the PGK1pG mRNA to ∼10–12 nucleotides/min (Figure 2F and H). Interestingly, the pop2Δ strain over-expressing Ccr4p apparently had two populations of PGK1pG mRNA (Figure 2H). One of these populations underwent rapid deadenylation similar to the wild-type strain over-expressing Ccr4p. The other population appeared to be deadenylated at a lower rate, typical of a pop2Δ strain. We do not yet understand the difference between these two populations. One possibility is that these two populations represent pools of cells that have either retained or lost the over-expression plasmid during the course of the experiment. Alternatively, the two pools may reflect a rate-limiting step for interaction of Ccr4p with the mRNA. In either case, the ability of Ccr4p over-expression to at least partly suppress the pop2Δ defects in deadenylation as well as accelerate the rate of deadenylation in an otherwise wild-type strain implies that Ccr4p plays a pivotal role in mRNA deadenylation.
Pop2p is not required for deadenylase activity in vitro
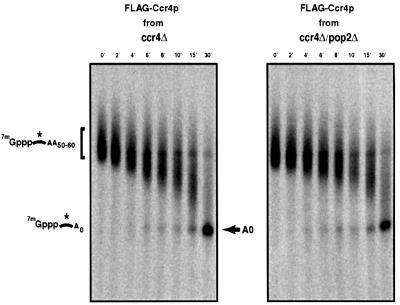
Our analysis indicated that over-expression of Ccr4p was capable of suppressing the deadenylation defects of a pop2Δ strain. In addition, we previously reported that Pop2p co-purifies with deadenylase activity and that this activity is dependent on the presence of Ccr4p (Tucker et al., 2001). Together these observations suggested that the Ccr4p might be sufficient for deadenylase activity independent of Pop2p. Given this, we purified a functional Flag-Ccr4p fusion protein from either a ccr4Δ or a ccr4Δ/pop2Δ strain under conditions that would co-purify endogenous Pop2p.
Flag-Ccr4p fusion protein isolated from ccr4Δ and ccr4Δ/pop2Δ strains was then assayed for its ability to deadenylate a mRNA substrate in vitro. A body labeled, capped mRNA, synthesized in vitro with a 50 nucleotide body and a poly(A) tail length of ∼50–60 adenosines was incubated with purified Flag-Ccr4p extracts and analyzed at various times on 6% polyacrylamide gels. We observed, using Flag-purified preparations with equal amounts of the Ccr4p present from either strain background, that a shorter species of the input RNA appeared over time and a new species accumulated of the correct length to represent a fully deadenylated substrate (Figure 3). In repeated experiments, it was apparent that the relative rates of deadenylation were not significantly different between the two extracts. We interpret these observations to indicate that Pop2p is not required for deadenylation activity. Furthermore, because Pop2p is required for association of Ccr4p with the Not proteins (Bai et al., 1999), this suggests that the Not proteins are also not required for catalytic activity of the deadenylase.
Fig. 3. Flag-Ccr4p co-purifies with deadenylase activity. Analysis of deadenylation activity in Flag-Ccr4p purified fractions from ccr4Δ and ccr4Δ/pop2Δ strains on a capped, polyadenylated substrate. Numbers above the lanes indicate time points taken after addition of substrate to the reaction. The arrow represents the fully deadenylated form of the substrate based on migration of substrate after cleavage with RNase H and oligo(dT) (data not show). The asterisk indicates the position of the radiolabeled phosphates.
The exonuclease domain of Ccr4p is critical for deadenylase activity
The simplest interpretation of the above observations is that Ccr4p is the catalytic subunit of the deadenylase complex. This is also supported by the fact that Ccr4p has a likely exonuclease motif (Dlakic, 2000). Alternatively, it is formally possible that an unknown protein with nuclease activity co-purifies with Flag-Ccr4p. If Ccr4p is the catalytic subunit of the deadenylase, then point mutations in the exonuclease domain of Ccr4p would be predicted to block Ccr4p’s function in deadenylation both in vivo and in vitro. To test this, we constructed four point mutants in amino acids that are absolutely conserved in this family of nucleases and predicted from structural studies to have catalytic roles (Dlakic, 2000). Specifically, in allele ccr4-713 and ccr4-715, Asp713 and Asn715, respectively, were changed to alanine, each predicted to be a structural residue of the catalytic pocket. In allele ccr4-817, Asp817, predicted to be involved in magnesium binding, was changed to alanine. Finally, the allele ccr4-818 changes His818 to alanine; this amino acid is likely to be a key catalytic residue. Each of these point mutants was expressed as Flag-epitope fusions from the GPD1 promoter.
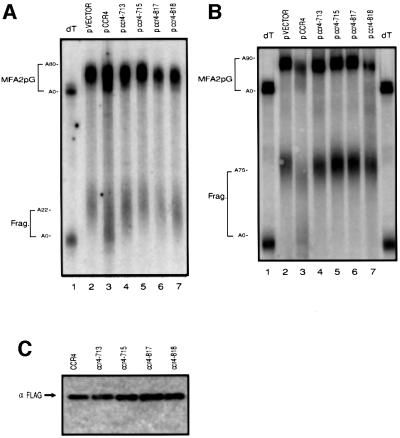
Expression of the wild-type Flag-Ccr4p rescued the slow-growth phenotype (data not shown) and the deadenylation defects of the ccr4Δ mutation for both MFA2pG and PGK1pG mRNAs (Figure 4A; data not shown). In contrast, each of the point mutants failed to complement both the growth (data not shown) and the deadenylation end point defects of the ccr4Δ strain (Figure 4A). In order to confirm the failure of the ccr4 point mutants to complement the deadenylation defect in a ccr4Δ strain, we also examined their phenotype in a ccr4Δ/pan2Δ strain (Figure 4B). This strain, which is also lacking the secondary deadenylase encoded by the PAN2 gene, shows an absolute dependence on Ccr4p for deadenylation. Similar to the results seen in a ccr4Δ strain, we observed that the wild-type Flag-Ccr4p complements the deadenylation defect of a ccr4Δ/pan2Δ strain, although again this strain shows a small defect in deadenylation, consistent with Flag-Ccr4p having a subtle defect in function. In contrast, the point mutants completely failed to complement the deadenylation defect and only accumulated mRNAs with the full poly(A) tail (Figure 4B), which failed to deadenylate appreciably in >1 h (data not shown). In addition, the mRNA decay fragment in the mutants also possess a full-length poly(A) tail, whereas in the complemented strain the fragment is predominantly deadenylated. The presence of a fully adenylated mRNA decay fragment is consistent with decapping occurring without deadenylation in ccr4Δ/pan2Δ strains (Tucker et al., 2001). Consistent with these amino acid residues having a critical role in the function of Ccr4p, each of these mutants was expressed as a stable, full-length protein when assayed by western blot analysis (Figure 4C). These observations all argue that the exonuclease motif of Ccr4p is essential for Ccr4p function in vivo.
Fig. 4. Comparison of deadenylation end-points in ccr4 point mutants. Shown are polyacrylamide northern gels for the MFA2pG transcript in (A) ccr4Δ strains or (B) ccr4Δ/pan2Δ strains, containing either a vector control, Flag-Ccr4p wild-type plasmid (pRP1045) or Flag-Ccr4p plasmids containing specific point mutants (pRP1046–1049) as indicated (see Materials and methods). Steady-state mRNA samples were resolved on 6% polyacrylamide/8 M urea northern gels, with or without removal of poly(A) tails with RNase H and oligo(dT) (lane 1). (C) Western analysis of extracts from ccr4Δ strains harboring both wild-type and mutant forms of the CCR4 plasmid. Probing with anti-Flag antibodies indicates that each mutant produces a full-length Ccr4p.
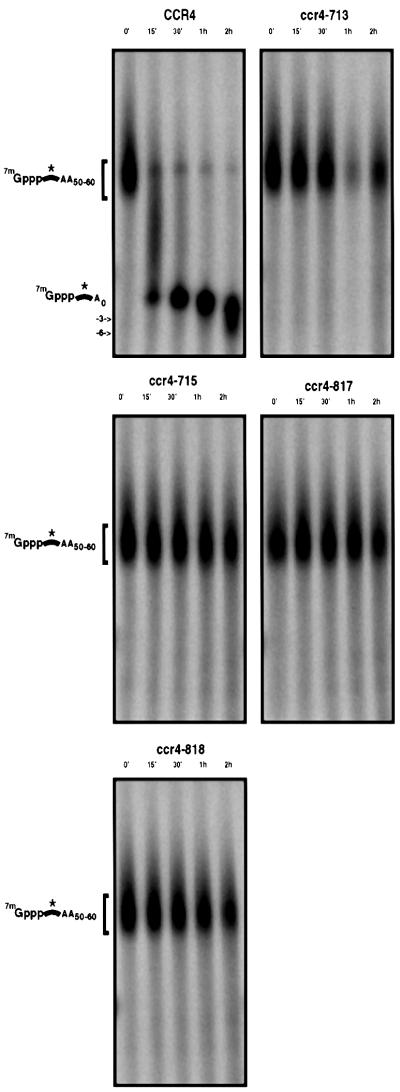
To determine whether the exonuclease domain of the Ccr4p was also required for in vitro deadenylase activity, we purified the Flag-Ccr4p point mutants and assayed their ability to deadenylate an mRNA substrate. In these experiments, we used equal amounts of Ccr4p in the reactions, with concentrations being compared carefully by western blot analysis (data not shown). All four of the mutant CCR4 proteins were essentially completely defective in deadenylation in vitro (Figure 5). Consistent with these results, it has recently been reported that similar mutations in Ccr4p fail to degrade poly(A) RNA in vitro (Chen et al., 2002). These observations indicated that the Ccr4p exonuclease domain is essential for purified Flag-Ccr4p deadenylase activity. Together, the above results indicate that the Ccr4p exonuclease domain is essential for deadenylation, both in vivo and in vitro, and strongly suggests that Ccr4p is the catalytic subunit of the yeast mRNA deadenylase.
Fig. 5. The Ccr4p nuclease motif is required for deadenylase activity. Analysis of deadenylation activity in Flag-Ccr4p purified fractions on a capped, polyadenylated substrate from a ccr4Δ strain containing either a Flag-Ccr4p wild-type plasmid (pRP1045) or Flag-Ccr4p plasmids containing specific point mutants (pRP1046–1049) as indicated (see Materials and methods). Numbers above the lanes indicate time points taken after addition of substrate to the reaction. The asterisk indicates the position of the radiolabeled phosphates.
Pab1p inhibits Ccr4p deadenylation activity in vitro
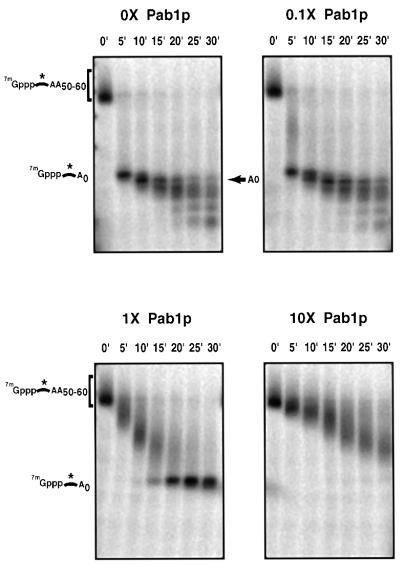
The poly(A) binding protein (Pab1p) has been proposed to both stimulate and inhibit the process of mRNA deadenylation (Sachs and Deardorff, 1992; Ford et al., 1997, 1999). In order to determine how Pab1p interacts with the Ccr4p deadenylase, we examined the effects of recombinant His-tagged Pab1p on Ccr4p deadenylase activity in vitro. The ability of the purified His-Pab1p to bind poly(A) was confirmed by its ability to UV cross-link to a radiolabeled substrate in vitro (data not shown). The purified His-Pab1p was then assayed for its effects on Flag-Ccr4p deadenylation activity. During the course of this investigation, we observed that omission of spermidine from the reaction buffer improved the Flag-Ccr4p deadenylase activity (data not shown). To simplify interpretation of the results, the following reactions were carried out in the absence of spermidine. Addition of increasing amounts of His-Pab1p to the in vitro deadenylation reaction resulted in decreasing rates of deadenylation (Figure 6). At 10-fold excess of His-Pab1p to Ccr4p, deadenylation was substantially slowed. Bovine serum albumin (BSA) was added to all reaction mixtures to maintain a constant protein concentration, but BSA alone had no inhibitory effect on the activity of Ccr4p (data not shown). Addition of His-Pab1p alone had no effect on the integrity of the substrate (data not shown). These data indicate that Pab1p is capable of inhibiting Ccr4p deadenylase activity.
Fig. 6. Pab1p inhibits Ccr4p deadenylase activity in vitro. Analysis of deadenylation activity in Flag-Ccr4p purified fractions with addition of increasing amounts of purified Pab1p. Purified Pab1p was added to each time course in molar amounts relative to Flag-Ccr4p as indicated. Numbers above the lanes indicate time points taken after addition of substrate to the reaction. The asterisk indicates the position of the radiolabeled phosphate.
The Ccr4p/Pop2p/Not complex localizes to the cytoplasm
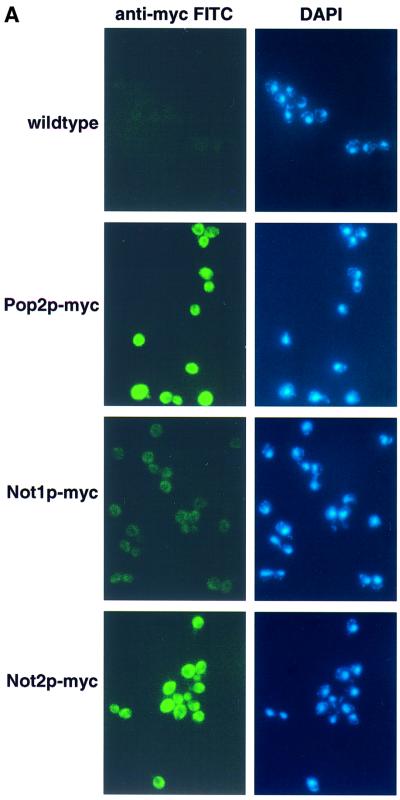
Previous experiments indicated that the Ccr4p and Pop2p were components of a larger complex including the Not proteins (Not1p–5p). This complex of proteins has been implicated to function in transcription (Collart and Struhl, 1993, 1994; Benson et al., 1998; Badarinarayana et al., 2000; Lemaire and Collart, 2000; Liu et al., 2001). However, because the majority of Ccr4p and Pop2p localizes to the cytoplasm (Tucker et al., 2001) and is found in a complex with the Not proteins (Liu et al., 1998), we hypothesized that the Ccr4p/Pop2p/Not complex might represent a cytoplasmic deadenylase complex. A critical prediction of this hypothesis is that the Not proteins should be present in the cytoplasm. In order to determine the subcellular distribution of the Not proteins we tagged the NOT1 and NOT2 genes (as markers for the complex) with multiple myc-epitopes on their C-termini and located these proteins by immunofluorescence (see Materials and methods). These epitope-tagged proteins were functional as assessed by their ability to complement the growth phenotypes associated with deletion of either gene (data not shown). Importantly, detection of the fusion proteins by immunofluorescence indicated that the majority of Not1p-myc and Not2p-myc are present in the cytoplasm similar to Pop2p-myc (Figure 7A). These data are consistent with at least a significant percentage of the Ccr4p/Pop2p/Not complex localizing to the cytoplasm.
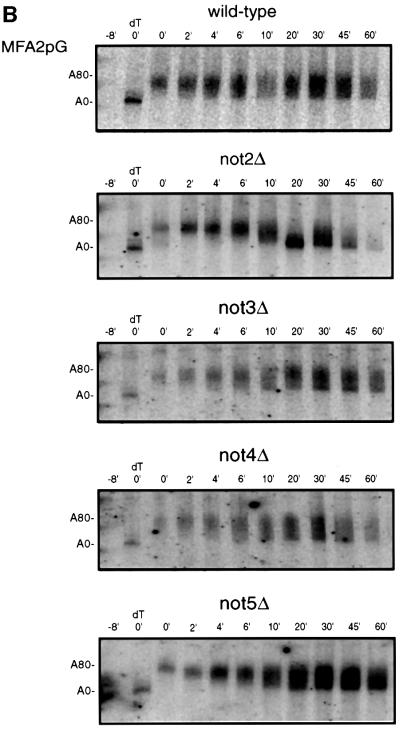
Fig. 7. The Not protein complex functions in deadenylation. (A) Localization of Not1p and Not2p by indirect immunofluorescence. Strains expressing myc-tagged Not1p (yRP1668), myc-tagged Not2p (yRP1669), myc-tagged Pop2p (yRP1622), or wild-type control (yRP841), were stained with anti-myc antibodies and anti-mouse (FITC) secondary antibody and DAPI as indicated. It is critical to note that all photographs were taken under identical conditions. The localization of myc-tagged Not1p was consistently cytoplasmic; however, the signal often appeared weaker. This observation is presumably a result of the Not1p myc-epitope being less accessible to the anti-myc antibodies. (B) Transcriptional pulse–chase analysis of the MFA2pG in not2Δ, not3Δ, not4Δ and not5Δ strains. Shown are polyacrylamide northern gels examining the decay of MFA2pG in the indicated strains. Numbers above the lanes are minutes after transcriptional repression by the addition of glucose following an 8 min induction of transcription (Decker and Parker, 1993). The 0 min time points were treated with RNase H and oligo(dT) to indicate the position of the deadenylated mRNA.
Ccr4p-associated Not proteins affect the MFA2 deadenylation rate
To examine the role the Not proteins in mRNA turnover, we determined whether mRNA turnover was altered in strains lacking Not2p, Not3p, Not4p or Not5p. Not1p is essential and its role in deadenylation is currently being assessed by the generation of conditional alleles. In these experiments we analyzed the decay of the MFA2pG and PGK1pG mRNAs in the not2Δ, not3Δ, not4Δ and not5Δ backgrounds using the transcriptional pulse–chase procedure (Decker and Parker, 1993).
Comparison of transcriptional pulse–chases of each of the mutant strains tested indicated that the rates of deadenylation for MFA2pG were slowed in not2Δ and not5Δ strains relative to wild type, and may be marginally slower than normal in not3Δ and not4Δ strains (Figure 7B). The poly(A) tail of the MFA2pG mRNA in wild type shortened at a rate of ∼13 nucleotides/min, with a significant percentage of the population reaching an oligo(A) length of 10–12 nucleotides by 4 min (Figure 7B). In contrast, in both the not2Δ and not5Δ deletion strains, the MFA2pG mRNA was deadenylated at a rate of ∼4 nucleotides/min, with a significant percentage of the population reaching an oligo(A) length of 10–12 nucleotides only after 10–20 min (Figure 7B). Likewise, both the not3Δ and not4Δ deletion strains also showed slight decrease in the MFA2pG mRNA deadenylation rate. These observations, together with the data presented above, suggest that the Ccr4p/Pop2p/Not complex functions in the cytoplasm as the mRNA deadenylase. However, because the affects on deadenylation are relatively minor we cannot rule out the possibility that these are indirect effects of Not protein function in other aspects of mRNA metabolism.
COX17 mRNA is deadenylated via the Ccr4p/Pop2p/Not deadenylase
The identification of the Ccr4p/Pop2p/Not complex as the major yeast deadenylase allows us to address how specific mRNA binding proteins regulate deadenylation. One mRNA-specific activator of deadenylation in yeast is the mRNA-binding protein Puf3p. This protein is a member of the Puf or Pumilio-HD protein family, whose members bind to 3′ UTR sequences and promote deadenylation rates on specific mRNAs (for a review see M.Wickens, D.S.Bernstein, J.Kimble and R.Parker, submitted). Puf3p binds to the 3′ UTR of the COX17 mRNA and enhances deadenylation and subsequent decapping of this mRNA (Olivas and Parker, 2000). To determine whether Puf3p enhances deadenylation by affecting the Ccr4p deadenylase, we analyzed the decay of the COX17 mRNA by transcriptional pulse–chase in ccr4Δ, pop2Δ and ccr4Δ/pan2Δ strains harboring a plasmid expressing the COX17 mRNA under the control of the GAL1 UAS.
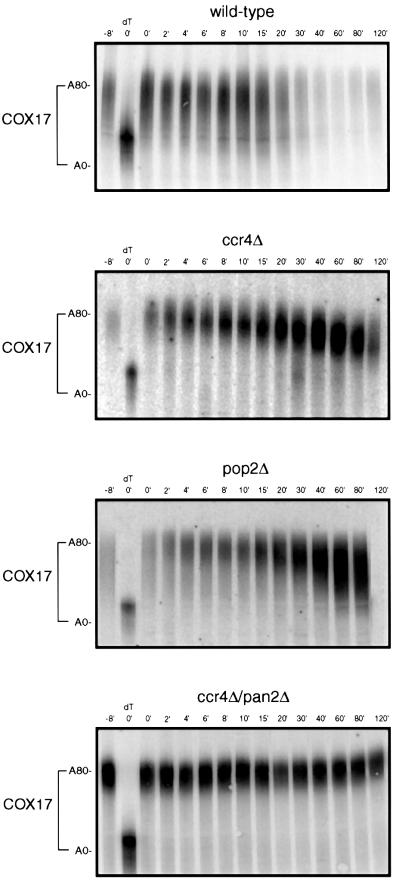
Comparison of transcriptional pulse–chases for the COX17 mRNA in ccr4Δ, pop2Δ and ccr4Δ/pan2Δ strains to wild type indicated that the rates of deadenylation for COX17 were greatly impaired in ccr4Δ, pop2Δ, or the double mutant (Figure 8). In wild-type strains, the COX17 mRNA has a distribution of poly(A) tails skewed to longer tails (Figure 8). This is consistent with the COX17 mRNA having a rate-limiting step in the initiation of deadenylation, which is then followed by very rapid deadenylation and decapping (Olivas and Parker, 2000). In contrast, in both the ccr4Δ and pop2Δ mutant strains, COX17 mRNA was deadenylated at a rate of ∼0.5 nucleotides/min (Figure 8). We also observed that the COX17 mRNA failed to deadenylate over an extended time course in the ccr4Δ/pan2Δ strain. These data indicate that the rapid deadenylation of the COX17 mRNA requires components of the Ccr4p/Pop2p/Not complex.
Fig. 8. Transcriptional pulse–chase analysis of the COX17 transcript. Shown are polyacrylamide northern gels examining the decay of COX17 in wild-type, ccr4Δ and ccr4Δ/pan2Δ strains. Numbers above the lanes are minutes after transcriptional repression by the addition of glucose following an 8 min induction of transcription (Decker and Parker, 1993). The 0 min time point was treated with RNase H and oligo(dT) to indicate the position of the deadenylated mRNA. To allow for size resolution of the poly(A) tail the 3′ 160 nucleotides of the 390 nucleotide COX17 mRNA were cleaved by hybridizing to oRP808 followed by treatment with RNase H prior to loading on the gel.
Discussion
Ccr4p is the catalytic subunit of the cytoplasmic deadenylase
Several observations indicate that Ccr4p is the catalytic subunit of the Ccr4p/Pop2p/Not deadenylase complex. First, we have previously reported that Ccr4p is required in vivo and in vitro for deadenylase activity (Tucker et al., 2001). Second, over-expression of Ccr4p both suppresses the deadenylation defects of a pop2Δ strain and increases the rate of deadenylation in an otherwise wild-type background (Figures 1 and 2). Third, Ccr4p co-purifies with a deadenylase activity that is independent of Pop2p (Figure 3). Furthermore, we have been unable to detect any nuclease activity from full-length Pop2p purified from Escherichia coli, or from yeast in the absence of Ccr4p under the range of conditions we have examined (Tucker et al., 2001; data not shown). Finally, mutations in conserved amino acid residues in the exonuclease motif of Ccr4p inhibit Ccr4p function in vivo and deadenylase activity in vitro (Figures 4 and 5). Together these observations strongly suggest that Ccr4p is the catalytic subunit of the mRNA deadenylase. Similarly, Chen and colleagues (J.Chen, Y.Chiang and C.L.Denis, submitted) also report that the Ccr4p functions as a 3′ to 5′ exonuclease. Given the substantial conservation of Ccr4p in other eukaryotes it is likely that it will also function as a deadenylase in other eukaryotes. Consistent with this idea, it has recently been shown that the human CCR4 protein demonstrates a 3′ to 5′ exonuclease activity in vitro (Chen et al., 2002).
The role of Pop2p
The identification of Ccr4p as the catalytic subunit of the Ccr4p/Pop2p/Not deadenylase complex raises the issue as to the role of Pop2p. Pop2p has a significant role in deadenylation, since ccr4Δ and pop2Δ strains possess almost identical defects in deadenylation in vivo (Tucker et al., 2001; Figure 2). However, Pop2p is dispensable for Ccr4p catalytic activity in vitro (Figure 3). Currently, there are two possible functions for Pop2p. In one model, Pop2p might be a second catalytic subunit of the deadenylase. This possibility was originally suggested by the observation that Pop2p has homology to the RNase D family of 3′ to 5′ exonucleases (Moser et al., 1997). In addition, there is a recent report that a GST–Pop2p fusion protein from E.coli co-purifies with nuclease activity (Daugeron et al., 2001). However, several lines of evidence suggest that, although Pop2p may have some nuclease activity under certain conditions, the yeast Pop2p does not primarily function as a nuclease. First, over-expression of Pop2p does not suppress the deadenylation defects of a ccr4Δ strain, nor does it have any effect on mRNA deadenylation in an otherwise wild-type strain (data not shown). Second, sequence analysis indicates that the yeast Pop2p is missing residues thought to be essential for catalysis among this RNase D family of nucleases (Moser et al., 1997). Third, point mutations in the remaining conserved residues predicted to be required for catalysis complement the growth defect of pop2Δ strains (J.Chen and C.L.Denis, unpublished observation). Fourth, purified Pop2p isolated from a ccr4Δ strain fails to show any nuclease activity in vitro (Tucker et al., 2001). However, since the Pop2p homologs in higher eukaryotes contain intact nuclease domains, it is likely that Pop2p will function as a nuclease in other organisms (Moser et al., 1997).
An alternative model for Pop2p function in yeast is that Pop2p acts to increase the activity of Ccr4p, either directly or through additional protein–protein interactions. This possibility is supported by the observation that Pop2p is required for interaction of the Ccr4p with the Not proteins (Liu et al., 1998; Bai et al., 1999; Liu et al., 2001), which may serve as activators of deadenylation (see below). In addition, in pop2Δ strains Ccr4p is unable to remove the last 20–26 adenylate residues; however, when Ccr4p is over-expressed in pop2Δ strains deadenylation is now completed (Figures 1 and 2). This is consistent with the idea that the Ccr4p/Pop2p complex has a higher affinity for the mRNA substrate than Ccr4p alone. A principal goal in future work will be to determine the specific function of Pop2p in deadenylation and investigate the possibility that Pop2p may play a key role in determining mRNA-specific rates of deadenylation by governing Ccr4p–substrate interactions.
The Not proteins are components of the cytoplasmic mRNA deadenylase
Three observations indicate that the Ccr4p/Pop2p/Not complex as a whole functions as the cytoplasmic mRNA deadenylase. First, essentially all Ccr4p and Pop2p, which are known to be required for deadenylation (Tucker et al., 2001), are found in a complex with the Not proteins (Liu et al., 1998). Second, Not1p and Not2p are found localized to the cytoplasm (Figure 7A). This is consistent with the cytoplasmic localization of Ccr4p, Pop2p (Tucker et al., 2001) and the deadenylation process. Moreover, since only a small fraction of the Not proteins are found as free subunits (Collart and Struhl, 1994; Oberholzer and Collart, 1998), this suggests that at least a substantial amount of the Ccr4p/Pop2p/Not protein complex is also cytoplasmic. Finally, deletion analysis of the NOT genes indicate some defects in deadenylation rate for the MFA2pG mRNA (Figure 7B). Together we interpret these observations to indicate that a complex consisting minimally of Ccr4p, Pop2p and the Not proteins is the cytoplasmic deadenylase.
The suggestion that the Not proteins are components of a deadenylase holoenzyme has two implications. First, it suggests that some of the phenotypic changes associated with defects in Not proteins, such as the G1 arrest seen in not1 and not2 mutants (Breter et al., 1983), may be due to changes in the deadenylation and decay rates of specific mRNAs. Second, it strengthens the link between proteins involved in deadenylation and transcription. For example, Ccr4p has been identified in a Paf1p-containing RNA polymerase II transcription complex (Chang et al., 1999). It is also apparent that some of the Not proteins display physical interactions with TFIID and components of SAGA (Benson et al., 1998; Badarinarayana et al., 2000; Lemaire and Collart, 2000) and components of the 1.9 MDa Ccr4–Not complex that interacts with the SRB9-11 proteins of the RNA polymerase II holoenzyme (Liu et al., 2001). The interactions between deadenylation and transcription suggest an important, and yet to be explained, connection between these two fundamental steps in gene expression.
Specific mRNA binding proteins regulate the Ccr4p/Pop2p/Not mRNA deadenylase
Several observations indicate that the Ccr4p/Pop2p/Not complex is the target of mRNA-binding proteins that control mRNA-specific rates of deadenylation. This was first suggested by the observation that the rapid deadenylation of the MFA2 mRNA requires Ccr4p deadenylase (Tucker et al., 2001). Although the mRNA-specific binding proteins responsible for the rapid deadenylation of the MFA2 mRNA are unknown, specific 3′ UTR sequences promote deadenylation of this mRNA (Muhlrad and Parker, 1992). In another example, we have now shown that Ccr4p and Pop2p are required for the rapid deadenylation of the COX17 mRNA (Figure 8). The rapid deadenylation of this mRNA is promoted by the binding of Puf3p to sequence elements in the 3′ UTR (Olivas and Parker, 2000). In principle, Puf3p could enhance deadenylation by the Ccr4p/Pop2p/Not deadenylase, either by recruiting this enzyme complex to the mRNA, or by altering the mRNP structure in a manner that makes the substrate more accessible to the deadenylase. One interesting possibility, suggested by the observation that Pab1p inhibits Ccr4p in vitro (Figure 6), is that mRNA-specific activators of deadenylation, such as Puf3p, may function by promoting the dissociation of Pab1p from the poly(A) tail. This idea is consistent with the observation that yeast strains lacking Pab1p show uniform rates of deadenylation (Caponigro and Parker, 1995). An essential field for future research will be to determine how specific mRNA-binding proteins dictate the deadenylation rates of individual mRNAs via the Ccr4p/Pop2p/Not deadenylase.
Materials and methods
Strains
All strains used in this study are trp1 ura3-52 leu2-3, cup1Δ::LEU2PM. Strains differ as follows: yRP840 (Hatfield et al., 1996) MATa his4-539; yRP841 (Hatfield et al., 1996) MATα lys2-20; yRP1616 (Tucker et al., 2001) MATa his4-539 ccr4Δ::NEO; yRP1617 (Tucker et al., 2001) MATα pop2Δ::URA3; yRP1618 (Tucker et al., 2001) MATa his4-539 ccr4Δ::NEO pop2Δ::URA3; yRP1620 (Tucker et al., 2001) MATa his4-539 ccr4Δ::NEO pan2Δ::URA3; yRP1622 MATα his4-539 POP2-myc::NEO; yRP1668 MATα lys2-20 NOT1-myc::NEO; yRP1669 MATα lys2-20 CDC36-myc::NEO; yRP1670 MATα lys2-20 not2Δ:: NEO; yRP1671 MATα lys2-20 not3Δ::NEO; yRP1672 MATα lys2-20 not4Δ::NEO; and yRP1673 MATα not5Δ::NEO. myc-tagged versions of Not1p and Not2p were generated as described previously (Longtine et al., 1998) and confirmed both by Southern and western blots.
Plasmids
The Flag-CCR4 plasmid (pRP1045) was created by PCR amplification of the CCR4 gene from yRP840, placing the Flag epitope 5′ of the CCR4 gene, and ligated into the yeast expression vector pG-1 (Schena et al., 1991) and confirmed by sequencing. Point mutants in Flag-Ccr4 (pRP1046–1049 are ccr4 alleles 713, 715, 817 and 818, respectively) were generated from pRP1045 using QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. The COX17 plasmid under the control of the GAL1 UAS (pG74/ST30) was as described previously (Beers et al., 1997).
RNA isolation and analysis
All procedures with RNA were carried out as previously described: transcriptional shut-off experiments and RNA isolations and normalization (Caponigro et al., 1993), transcriptional pulse–chase experiments (with the exception that all growth media was supplemented with 1% sucrose and 2% raffinose or, in the case of Figure 7B, 2% sucrose only) (Decker and Parker, 1993) and RNase H reactions (Muhlrad and Parker, 1992).
Localization of epitope-tagged Not1p, Not2p and Pop2p
yRP841, yRP1622, yRP1668 and yRP1669 were grown to mid-log, fixed and analyzed by standard methods using anti-myc-FITC 9E10 antibody (CoVance) at a dilution of 1:200 and goat anti-mouse IgG-FITC (Roche) was used at a dilution of 1:300.
In vitro deadenylation assays
Flag-Ccr4 proteins were purified from ccr4Δ and ccr4Δ/pop2Δ strains (yRP1616 and yRP1618) harboring the appropriate plasmid following similar procedures (Tharun and Parker, 1999).
Recombinant Pab1p was isolated from 500 ml of E.coli (BL21) harboring an inducible His-Pab1p plasmid (Amrani et al., 1997). Cells were induced for 1 h in 100 mM IPTG and lysed in 3 ml/g YPER (Pierce). Lysate was bound to 1 ml Talon metal affinity resin (Clontech), washed (0.5 M NaCl, 20 mM Tris pH 7.5, 4 mM MgCl2, 10% glycerol, 0.5 mM β-mercaptoethanol) and eluted (0.5 M NaCl, 20 mM Tris pH 7.5, 4 mM MgCl2, 10% glycerol, 0.5 mM β-me, 100 mM imidazole pH 7.7). Eluted protein was dialysed twice (first: 50 mM KCl, 20 mM HEPES pH 7.6, 5 mM EDTA; second: 50 mM KCl, 20 mM HEPES pH 7.6).
To prepare substrate, capped RNA was transcribed, as previously described (LaGrandeur and Parker, 1998), from plasmid pRP1038 that was digested with HindIII and mung bean nuclease. This produced a body-labeled, 50 nucleotide transcript with a 50 nucleotide adenosine tail that lacked any additional 3′ nucleotides. Capped labeled substrate was prepared cold using the same template and cap labeled as described previously (LaGrandeur and Parker, 1998).
Flag-Ccr4p elution fractions were assayed for deadenylation activity as previously described (Tucker et al., 2001). Pab1p inhibition of Flag-Ccr4p was assayed as above, utilizing 0.98 pmol Flag-Ccr4p, 24 fmol cap-labeled substrate and concentrations of recombinant Pab1p as indicated. Reactions were performed in 20 mM HEPES, pH 7.0, 1 mM MgOAc, 1 mM DTT, 0.2 µg/µl BSA, 0.2% NP-40 and 1 U/µl RNasin.
Acknowledgments
Acknowledgements
We thank the Parker and Denis laboratories for comments on the manuscript. This work was supported by NIH grant GM45443 and funds from the Howard Hughes Medical Institute to R.P. M.A.V-S. recieved additional support from the Fulbright/CONACYT commission.
References
- Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.S.J. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V., Chiang,Y.C. and Denis,C.L. (2000) Functional interaction of CCR4–NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics, 155, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Salvadore,C., Chiang,Y.C., Collart,M.A., Liu,H.Y. and Denis,C.L. (1999) The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separated from NOT2, NOT4 and NOT5. Mol. Cell. Biol., 19, 6642–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- Beers J., Glerum,D.M. and Tzagoloff,A. (1997) Purification, characteriz ation and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem., 272, 33191–33196. [DOI] [PubMed] [Google Scholar]
- Benson J.D., Benson,M., Howley,P.M. and Struhl,K. (1998) Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J., 17, 6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breter H.J., Ferguson,J., Peterson,T.A. and Reed,S.I. (1983) Isolation and transcriptional characterization of three genes which function at start, the controlling event of the Saccharomyces cerevisiae cell division cycle: CDC36, CDC37 and CDC39. Mol. Cell. Biol., 3, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. and Parker,R. (2001) Computational modeling of eukaryotic mRNA turnover. RNA, 7, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1995) Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev., 9, 2421–2432. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) mRNA turnover in yeast promoted by the MATα1 instability element. Nucleic Acids Res., 24, 4304–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad,D. and Parker,R. (1993) A small segment of the MAT α1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol., 13, 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., French-Cornay,D., Fan,H.Y., Klein,H., Denis,C.L. and Jaehning,J.A. (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol., 19, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chiang,Y.-C. and Denis,C.L. (2002) CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J., 21, 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J., 12, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1994) NOT1(CDC39), NOT2(CDC36), NOT3 and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev., 8, 525–537. [DOI] [PubMed] [Google Scholar]
- Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M., Mauzion,F. and Seraphin,B. (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res., 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- Denis C.L., Chiang,Y.C., Cui,Y. and Chen,J. (2001) Genetic evidence supports a role for the yeast CCR4–NOT complex in transcriptional elongation. Genetics, 158, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. (2000) Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci., 25, 272–273. [DOI] [PubMed] [Google Scholar]
- Ford L.P., Bagga,P.S. and Wilusz,J. (1997) The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol., 17, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H., Mitsui,H., Liu,H., Bai,Y., Denis,C.L., Shimizu,Y. and Sakai,A. (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics, 148, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman,C.A., Stevens,A. and Parker,R. (1996) Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribo nuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Grandeur T.E. and Parker,R. (1998) Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J., 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M. and Collart,M.A. (2000) The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4–Not complex and physically interacts with the Not5 subunit. J. Biol. Chem., 275, 26925–26934. [DOI] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana,V., Audino,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.Y. et al. (2001) Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4–NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem., 276, 7541–7548. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1992) Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev., 6, 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U. and Collart,M.A. (1998) Characterization of NOT5 that encodes a new component of the Not protein complex. Gene, 207, 61–69. [DOI] [PubMed] [Google Scholar]
- Olivas W.M. and Parker,R. (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J., 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B. and Deardorff,J.A. (1992) Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell, 70, 961–973. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Schena M., Picard,D. and Yamamoto,K.R. (1991) Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol., 194, 389–398. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Tharun S. and Parker,R. (1999) Analysis of mutations in the yeast mRNA decapping enzyme. Genetics, 151, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Wilson T. and Treisman,R. (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature, 336, 396–399. [DOI] [PubMed] [Google Scholar]