Abstract
The core oscillator generating circadian rhythms in eukaryotes is composed of transcription–translation-based autoregulatory feedback loops in which clock gene products negatively affect their own expression. A key step in this mechanism involves the periodic nuclear accumulation of clock proteins following their mRNA rhythms after ∼6 h delay. Nuclear accumulation of mPER2 is promoted by mCRY proteins. Here, using COS7 cells and mCry1/mCry2 double mutant mouse embryonic fibroblasts transiently expressing GFP-tagged (mutant) mPER2, we show that the protein shuttles between nucleus and cytoplasm using functional nuclear localization and nuclear export sequences. Moreover, we provide evidence that mCRY proteins prevent ubiquitylation of mPER2 and subsequent degradation of the latter protein by the proteasome system. Interestingly, mPER2 in turn prevents ubiquitylation and degradation of mCRY proteins. On the basis of these data we propose a model in which shuttling mPER2 is ubiquitylated and degraded by the proteasome unless it is retained in the nucleus by mCRY proteins.
Keywords: circadian clock/cryptochrome/nuclear–cytoplasmic shuttling/period/ubiquitylation
Introduction
Like most eukaryotes, mammals display circadian rhythms in physiology, metabolism and behavior, which are generated by an intrinsic self-sustaining clockwork with a periodicity of ∼24 h, located in the suprachiasmatic nuclei in the hypothalamus (Klein et al., 1991). The mammalian core oscillator is thought to be composed of interacting transcription–translation-based autoregulatory feedback loops involving a set of clock genes (Dunlap, 1999; Young, 2000; Reppert and Weaver, 2001; Young and Kay, 2001). In mammals, three Period (mPer1, mPer2 and mPer3), two Cryptochrome (mCry1 and mCry2), Clock, Bmal1 and Casein kinase I epsilon (CKIε) genes have been identified (for review see Cermakian and Sassone-Corsi, 2000; Reppert and Weaver, 2000; Young and Kay, 2001). Rhythmic mPer and mCry gene expression is positively driven by heterodimers of the PAS domain-containing CLOCK and BMAL1 transcription factors that act via CACGTG E box enhancer elements (Gekakis et al. 1998) and negatively regulate their own gene products. In vitro and in vivo, mCRY, rather than mPER, proteins are essential to suppress mPer gene expression (Griffin et al., 1999; Kume et al., 1999; Shearman et al., 2000). Bmal1, in contrast to the non-cycling clock gene, is also rhythmically expressed but the phase of oscillations is opposite to that of the mPer genes (Dunlap, 1999). A recent study suggests that the mPER2 protein might be involved in the positive regulation of Bmal1 expression (Shearman et al., 2000). Analysis of mPer mutant mice emphasized the importance of mPER2 in the circadian core oscillator; inactivation of mPer2, but not of mPer1 or mPer3, resulted in blunted clock gene expression (Bae et al., 2001; Zheng et al., 2001).
In addition to cyclic transcription of clock genes, evidence is accumulating that controlled accumulation, localization and degradation of clock proteins constitutes an important feature of circadian clocks in various organisms (Young and Kay, 2001). In view of the controlling function of the mPer2 gene product in the positive and negative loop of the circadian system, the mechanism of nuclear localization of mPER2 as well as its stability are critical features to be elucidated. Transfection studies in COS7 and NIH 3T3 cells have shown that exogenously expressed mPER2 can localize in the nucleus and that co-expression with either mCRY proteins (Kume et al., 1999) or mPER3 (Yagita et al., 2000) can promote its nuclear entry. Despite constitutive high levels of mPer2 mRNA in completely arhythmic mCry1/mCry2-deficient mice (Okamura et al., 1999; van der Horst et al., 1999), mPER2 protein is barely detectable in the cytoplasm and nucleus of suprachiasmatic nucleus (SCN) neurons from these animals (Shearman et al., 2000), which further points to an essential function of mCRY proteins in the nuclear localization of mPER2. In marked contrast, mPER1 protein displays nuclear localization in SCN neurons, hepatocytes and cultured fibroblasts of mCry1/mCry2-deficient mice (Shearman et al., 2000; Yagita et al., 2000). Taken together, these observations suggest the existence of an mCRY-dependent mechanism that specifically controls the subcellular localization and/or stability of mPER2. This prompted us to investigate further the events leading to nuclear accumulation of the mPER2 clock protein.
The present study provides evidence that mPER2 can enter the nucleus in a mCRY-independent manner and that the protein shuttles between nucleus and cytoplasm using functional nuclear localization (NLS) and nuclear export (NES) sequences present in the protein. Furthermore, we show that mCRY1 can bind to the C-terminal region of mPER2 (thereby likely preventing the latter protein from leaving the nucleus) and in the presence of mCRY proteins, ubiquitylation and presumably subsequent degradation of mPER2 by the proteasome machinery is inhibited. These findings led us to propose a new working model for the nuclear accumulation and stability of the mPER2 protein that adds another level at which nuclear accumulation of this clock protein can be regulated and may potentially extend to other clock proteins.
Results
mPER2 contains functional nuclear import and export signals
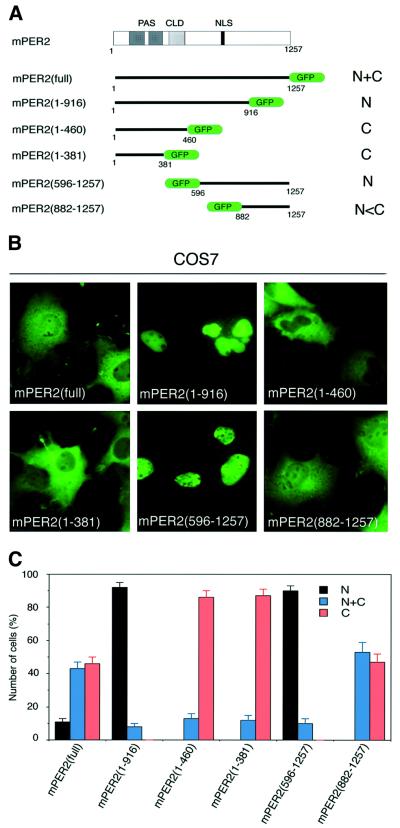
Subcellular localization domains in the mPER2 protein include a sequence reminiscent to a bipartite basic NLS (Albrecht et al., 1997; Shearman et al., 1997). In addition, mPER2 contains a domain (residues 390–450) with 43% sequence identity to the cytoplasmic localization domain (CLD) of the Drosophila PER protein (Saez and Young, 1996; Takumi et al., 1998) but it is not known whether this sequence is functional. To investigate whether mPER2 contains other subcellular localization signals, we have generated a panel of green fluorescent protein (GFP)-tagged full-length and N- or C-terminally truncated mPer2 expression constructs (Figure 1A). The subcellular distribution patterns of these GFP-tagged mPER2 proteins were analyzed in COS7 cells. Figure 1B show representative examples of GFP fluorescence in transiently transfected COS7 cells as well as the ratio between cells with nuclear, nuclear–cytoplasmic and cytoplasmic staining. Whereas full-length mPER2–GFP is observed in both cytoplasm and nucleus, mPER2(1–916)–GFP and mPER2(596– 1257)–GFP localized predominantly in the nucleus (Figure 1B). Apparently, the N- as well as C-terminal regions of mPER2 contain domains that facilitate cytoplasmic localization of the protein. In contrast, mPER2(1– 460)–GFP, mPER2(1–381)–GFP and mPER2(882–1257)– GFP, lacking the putative NLS, tended to be cytoplasmic dominant, which suggests that the putative NLS is functional. On the other hand, regulation of cytoplasmic localization seems to be more complex. The absence of mPER2(596–1257)–GFP in the cytoplasm at first glance might be attributed to the loss of the putative CLD. However, deletion of the CLD from mPER2(1–460)–GFP, as in mPER2(1–381)–GFP, does not instigate an increase in cytoplasmic localization of the protein. Moreover, the prominent nuclear localization of mPER2(1–916)–GFP argues against a potential role for the CLD and can only be explained by assuming that the C-terminal region (residues 916–1257) contains a yet unidentified signal for cytoplasmic localization.
Fig. 1. Different domains of mPER2 influence its subcellular localization. Full-length and truncated GFP-tagged mPER2 proteins were transiently expressed in COS7 cells and analyzed for the subcellular distribution pattern of mPER2 proteins. (A) Schematic diagram of the mPER2 protein, including the position of PAS, CLD and NLS sequences and the six constructs. (B) Representative examples of the subcellular distribution patterns of the various mPER2 proteins, as detected by GFP fluorescence. (C) Percentage of cells showing nuclear (N, black bars), nuclear–cytoplasmic (N+C, blue bars) and cytoplasmic (C, red bars) staining. Three independent experiments were performed in which 100–200 mPER2–GFP-expressing cells were counted. Error bars indicate the SEM.
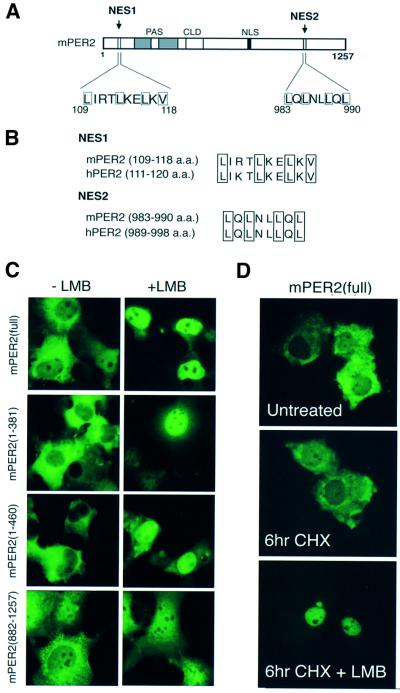
Accumulation of nuclear proteins in the cytoplasm can also be achieved via the CRM1/Exportin1 nuclear export system, acting through NES, composed of the leucine-rich consensus sequence LX(1∼3)LX(2∼4)LXL(V/I/M) (in which X indicates any amino acid; Nigg, 1997; Mattaj and Engelmeier, 1998). Previously, we have reported the presence of a putative NES domain in the N-terminal region of mPER2 (residues 109–118; Takumi et al., 1998), which fits reasonably to the NES consensus sequence and is well conserved between mouse and human PER2 (Figure 2A and B). Closer examination of the C-terminal sequence of mPER2 revealed a second conserved leucine-rich region between residues 983–990 (Figure 2A and B). For simplicity, we refer to NES sequences in the N- and C-terminal region as NES1 and NES2, respectively.
Fig. 2. Pre-existing mPER2 can leave the nucleus via the CRM1/Exportin1 nuclear export machinery. Subcellular localization of transiently expressed full-length and truncated mPER2–GFP proteins in COS7 cells in the absence or presence of an active CRM1/Exportin1 nuclear export system. (A) Schematic representation of the mPER2 protein, indicating putative leucine-rich NES domains in the N-terminal (NES1) and C-terminal (NES2) region of the protein, respectively. (B) Amino acid sequence comparison between mouse and human mPER2 NES domains. (C) Representative examples of the subcellular distribution pattern of mPER2(full)–GFP, mPER2(1–460)–GFP, mPER2(1–381)–GFP and mPER2(882–1257)–GFP in cells cultured in the absence (left) or presence (right) of the nuclear export inhibitor LMB. (D) Subcellular distribution pattern of mPER2(full)–GFP in the absence or presence of LMB under conditions where de novo protein synthesis is blocked by the translation inhibitor CHX.
To provide evidence for the functionality of the NES domains in mPER2, we transfected COS7 cells with full-length or truncated mPER2–GFP expression constructs and studied the effect of leptomycin B (LMB), a potent specific inhibitor of CRM1/Exportin1-mediated nuclear export (Fornerod et al., 1997; Fukuda et al., 1997). After treatment with LMB, mPER2(full)–GFP almost exclusively accumulated in the nucleus, suggesting that after entry, mPER2 can leave the nucleus again via the nuclear export machinery (Figure 2C, top). Inhibition of de novo protein synthesis by cycloheximide (CHX) prior to LMB treatment did not change the subcellular localization of mPER2(full)–GFP observed in the absence or presence of LMB (Figure 2D). This shows that LMB-mediated accumulation of mPER2 in the nucleus is not coupled to de novo protein synthesis, but instead involves already existing mPER2. Somewhat surprisingly, mPER2(1– 460)–GFP, mPER2(1–381)–GFP and mPER2(882–1257)– GFP (all lacking the NLS) are also prominently present in the nucleus when cells are cultured in the presence of LMB, although in comparison with mPER2(full)–GFP considerable amounts of protein are still observed in the cytoplasm (Figure 2C). This indicates that even in the absence of the NLS, mPER2 can still to some extent translocate to the nucleus. More importantly, however, this observation demonstrates that the NES signals can be separately used by the nuclear export machinery. In addition, the distribution patterns of mPER2(1–460)–GFP and mPER2(1–381)–GFP are highly similar, indicating that the CLD does not significantly contribute to cytoplasmic retention of mPER2 and that cytoplasmic localization of mPER2–GFP proteins is regulated by NES sequences instead.
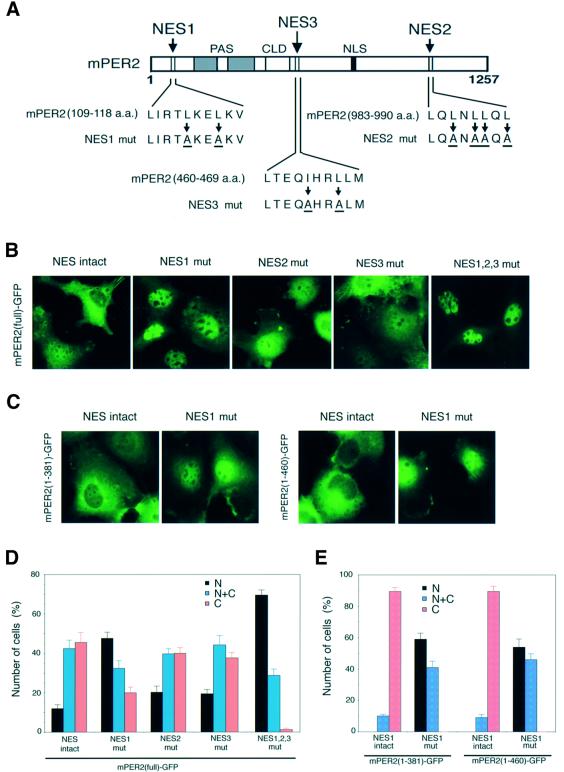
A survey of the complete mPER2 sequence pointed out one additional candidate NES at residues 460–469, referred as NES3 (Figure 3A). To investigate whether NES1, NES2 and NES3 serve as true NES signals and thus contribute to the cytoplasmic localization of mPER2, we made full-length mPER2–GFP expression constructs in which hydrophobic amino acids predicted to be critical in NES functioning had been substituted (Figure 3A). Figure 3 shows representative examples of GFP fluorescence in COS7 cells transfected with mPER2(full)–GFP carrying wild-type or mutant NES sequences (Figure 3B), as well as the ratio between cells with nuclear, nuclear– cytoplasmic and cytoplasmic staining (Figure 3D). In comparison with wild-type mPER2(full)–GFP, mutations in NES1 enhanced the number of cells showing nuclear localization of the protein (p <0.0001). Mutations in NES2 or NES3 caused a relatively small but still significant change in the subcellular localization patterns of mPER2(full)–GFP (p <0.05). These results strongly suggest that all three NES sequences are functional, although NES1 might be more efficiently used by the nuclear export machinery than NES2 and NES3. Importantly, mutations in two out of three NES sequences could not completely abolish nuclear export (data not shown), whereas mutagenesis of all three NES regions in mPER2(full)–GFP induced an almost complete shift from cytoplasmic and nuclear–cytoplasmic to nuclear and nuclear–cytoplasmic localization, which indicates that NES1, NES2 and NES3 additively act in nuclear export of the mPER2 protein. Similarly, mutagenesis of NES1 in mPER2(1–381)–GFP and mPER2(1–460)–GFP caused the subcellular localization patterns of both truncated proteins to shift from almost exclusively cytoplasmic to predominantly nuclear (Figure 3C and E; p <0.0001), indicating once more that cytoplasmic localization of mPER2(1–460)–GFP is regulated by NES rather than CLD sequences.
Fig. 3. NES domains control cytoplasmic distribution of mPER2 in cultured mammalian cells. Subcellular localization of full-length and truncated mPER2–GFP with wild-type and mutant NES domains in transiently transfected COS7 cells. (A) Schematic representation of the three NES domains in mPER2 and the substituted critical hydrophobic amino acids in the mutant NES domains (referred to as NES1 mut, NES2 mut and NES3 mut). (B) Representative examples of the subcellular distribution patterns of mPER2(full)–GFP with intact or mutant NES domains. (C) As (B), except that mPER2(1–381)–GFP and mPER2(1–460)–GFP with an intact or mutant NES1 were used. (D and E) Quantitative analysis of the percentage of cells expressing nuclear (N, black bars), nuclear–cytoplasmic (N+C, blue bars) and cytoplasmic (C, red bars) mPER2–GFP for the experiment shown in (B) and (C), respectively. Data were obtained from three independent experiments and in each experiment 100–200-expressing cells were evaluated. Error bars indicate the SEM.
In conclusion, these data demonstrate that mPER2 can exit the nucleus via the CRM1/Exportin1 nuclear export system, utilizing the NES1, NES2 and NES3 sequences. This finding opens the intriguing possibility that mPER2 proteins shuttle between nucleus and cytoplasm through their NLS and NES sequences.
mPER2 shuttles between the nucleus and the cytoplasm
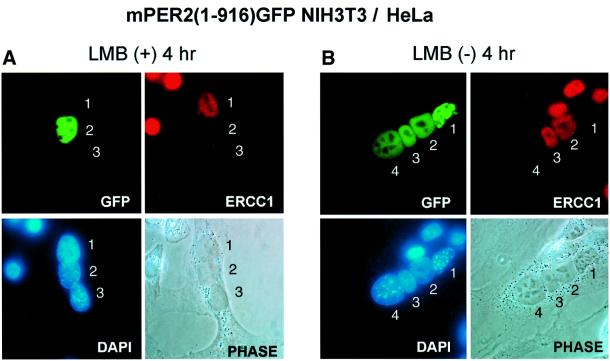
We have shown that under steady-state conditions, mPER2(1–916)–GFP predominantly localizes in the nucleus in spite of the existence of functional NES signals (NES1 and NES3) in this protein. To investigate whether this truncated mPER2 protein can still leave the nucleus through its remaining NES domains and subsequently re-enter the nucleus, we performed a heterokaryon nuclear– cytoplasmic shuttling assay (Borer et al., 1989). The principle of the experiment is that nuclei of cell line X are preloaded with the protein of interest. After fusion with cell line Y, the protein may or may not move to the nucleus originating from the latter. NIH 3T3 cells were transfected with a plasmid encoding mPER2(1–916)–GFP and cultured in the presence of LMB, resulting in nuclear accumulation of truncated mPER2 protein. Twenty-four hours after the transfection, cells were fused with HeLa cells and subcultured in the presence or absence of LMB, while de novo protein synthesis was blocked by CHX. Four hours after cell fusion, the distribution of fluorescent mPER2(1–916)-GFP was analyzed in heterokaryon cells (Figure 4A and B, top left), in which HeLa cell-derived nuclei were identified using antibodies specifically recognizing the human ERCC1 protein (Figure 4A and B, top right). mPER2(1–916)–GFP remained in the NIH 3T3 nuclei when LMB was present in the culture medium (Figure 4A). However, in marked contrast, mPER2(1– 916)–GFP shuttled into HeLa nuclei when the block of nuclear export was released (Figure 4B). These findings indicate that the remaining NES1 and NES3 domains in mPER2(1–916) are functional. Although we anticipated that the heterokaryon shuttling assay might not work for mPER2(full)–GFP because of its prominent nuclear-cytoplasmic localization, we nevertheless could see some relocalization of fluorescence from NIH 3T3 to HeLa nuclei, indicating that intact mPER2 protein is shuttling also. Taken together, these data demonstrate that mPER2 can shuttle between nucleus and cytoplasm.
Fig. 4. Nuclear-cytoplasmic shuttling of truncated mPER2. Heterokaryon shuttling assay using NIH 3T3 cells, transiently expressing nuclear mPER2(1–916)–GFP and (untransfected) HeLa cells. Prior to cell fusion, the nuclear export system and de novo protein synthesis were inhibited by addition of LMB and CHX, respectively (for details, see Materials and methods). After fusion, cell culture was continued either in the presence of LMB (A) or after washing away LMB (B). Under both conditions, translation remained blocked by CHX. Representative examples of the subcellular distribution of the truncated mPER2–GFP proteins are shown (top left), as well as phase contrast photographs of the heterokaryons (bottom right) and their DAPI-stained nuclei (bottom left). HeLa cell nuclei were identified by immunofluorescence using an antibody specifically recognizing human ERCC1 (top right). To facilitate comparison, nuclei in heterokaryon cells are numbered.
mCRY1 interacts with the C-terminus of mPER2 but is not required for nuclear import of mPER2
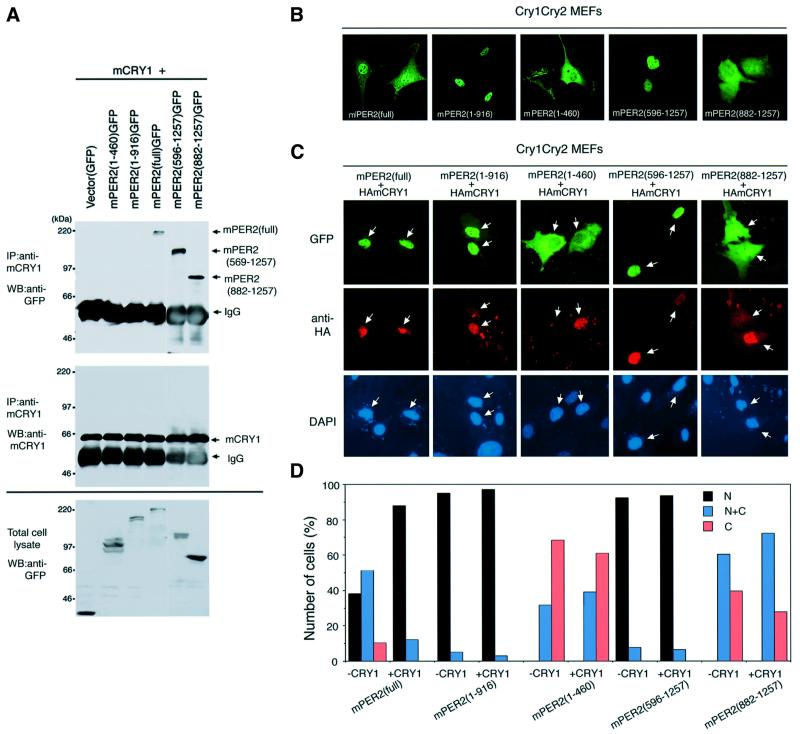
The mPER2 protein is barely detectable in the nucleus of SCN cells from mCry1/mCry2-deficient mice (Shearman et al., 2000) and COS7 cell transfection experiments have shown that co-expression with mCRY proteins resulted in significant accumulation of mPER2 in the nucleus (Kume et al., 1999). These findings suggest that the mCRY proteins are required for nuclear accumulation of mPER2. Yet, cells in the piriform cortex of the brain of mCry-deficient mice constitutively express mPER2 in the nucleus (Shearman et al., 2000), suggesting that mCRY proteins are not absolutely obligatory for nuclear translocation of mPER2. Recently, mPER2 has been shown to physically interact with mCRY1 and mCRY2 (Griffin et al., 1999; Kume et al., 1999). Prior to investigating how mCRY proteins promote nuclear localization of mPER2, we determined the mCRY binding region in mPER2. Figure 5A shows a co-immunoprecipitation experiment using the panel of GFP-tagged mPER2 constructs and mCRY1, co-expressed in COS7 cells. Although all five versions of mPER2–GFP were clearly detectable in the COS7 cell lysates (Figure 5A, bottom left), immunoprecipitation with antibodies against mCRY1 and western blot analysis of the immunoprecipitates with anti-GFP antibodies only revealed mPER2(full)–GFP, mPER2(596– 1257)–GFP and mPER2(882–1257)–GFP. In contrast, mPER2(1–460)–GFP and mPER2(1–916)–GFP did not co-precipitate with mCRY1. A similar result was obtained in the reverse experiment (data not shown), in which anti-GFP and anti-mCRY1 antibodies were used for precipitation of mPER2 proteins and detection of mCRY1 in the precipitate; mCRY1 only co-precipitated with mPER2 (full)–GFP, mPER2(596–1257)–GFP and mPER2(882– 1257)–GFP. These results indicate that the previously reported physical interaction between mPER2 and mCRY1 (and presumably mCRY2; Griffin et al., 1999; Kume et al., 1999) occurs via the C-terminal region of mPER2 (residues 917–1257).
Fig. 5. mCRY1 physically interacts with the C-terminus of mPER2 and promotes nuclear accumulation rather than translocation of full-length mPER2. (A) Identification of the mCRY1 binding region in mPER2. mCRY1 was immunoprecipitated from cell lysates of COS7 cells transiently expressing mCRY1 and either wild-type or truncated GFP-tagged mPER2 and the precipitate was analyzed for the presence of mPER2. Top, immunoblot analysis of co-precipitated mPER2–GFP proteins, detected with anti-GFP antibodies; middle, immunoblot analysis of precipitated mCRY1 protein, detected with anti-mCRY1 (TD1119) antibody and bottom, immunoblot analysis of total cell lysates, confirming the presence of the various mPER2 proteins. (B) Subcellular localization of full-length and truncated mPER2–GFP proteins in transiently transfected mCry1/mCry2 double-mutant MEFs. Representative examples of fluorescent cells are shown. Note that the distribution patterns of full-length and truncated mPER2 proteins in mCRY-deficient cells are not essentially different from that in COS7 cells (see Figure 1). (C) As (B), except that cells were co-transfected with a HA-tagged mCry1 cDNA expression construct. Representative examples of the subcellular distribution patterns of mPER2 proteins, as detected by GFP fluorescence are shown (top). HA-mCRY1 protein is visualized by immunocytochemistry using anti-HA antibodies (middle) and nuclei were stained by DAPI (bottom). Note the difference in the amount of co-expressed mCRY1, as indicated by the white arrows. (D) Quantitative analysis of the subcellular localization of mPER2–GFP fusion proteins in the absence or presence of coexpressed m HA-mCRY1 as shown in (C) and (D). Data represent the mean of three independent experiments. In each experiment, 100–200 mPER2-expressing cells were evaluated for nuclear (N, black bars), nuclear–cytoplasmic (N+C, blue bars) and cytoplasmic (C, red bars) fluorescence.
Next, we investigated how the various subcellular localization signals of mPER2 influence the distribution of the protein over the cytoplasm and nucleus in the absence of mCRY proteins. To this end, we expressed the panel of full-length and truncated mPER2–GFP proteins in mCry1/mCry2 double-deficient mouse embryonic fibroblasts (MEFs; Figure 5B). The majority of cells show nuclear–cytoplasmic or nuclear localization of mPER2(full)–GFP, indicating that mCRY proteins are not essential for nuclear translocation of mPER2. mPER2 (1–460)–GFP and mPER2(882–1257)–GFP fluorescence is predominantly cytoplasmic and mPER2(1–916)–GFP and mPER2(596–1257)–GFP staining appears almost exclusively nuclear (Figure 5B). Thus, the subcellular distribution pattern of exogenously expressed wild-type and truncated mPER2-GFP proteins in mCry-deficient MEFs does not significantly differ from that in COS7 cells (compare Figures 1 and 5B).
We then examined the effect of co-expression of HA-tagged CRY proteins on the subcellular localization of wild-type and mutant PER2-GFP proteins in mCry1/mCry2 MEFs. As shown in Figure 5C, exogenously expressed HA-mCRY1 protein localizes in the nucleus of mCry1/mCry2 mutant MEFs. In line with a previous study in COS7 cells (Shearman et al., 2000), simultaneous expression of HA-mCRY1 with mPER2(full)–GFP significantly increased the percentage of cells with nuclear mPER2(full)–GFP (p <0.0001; Figure 5D). A similar result was obtained when HA-mCRY1 was replaced by untagged mCRY1 or untagged hCRY2 (data not shown). The subcellular localization pattern of nuclear dominant mPER2(1–916)–GFP and cytoplasmic dominant mPER2 (1–460) did not change after co-expression of HA-mCRY1 (p = 0.570 and 0559, respectively; Figure 5D), which would be in agreement with the absence of the mCRY binding region in these C-terminally truncated mPER2 versions. mPER2(596–1257)–GFP, on the other hand, can bind mCRY through its C-terminus but remained exclusively localized in the nucleus after co-expression of HA-mCRY1 (p = 0.815; Figure 5D). Interestingly, co-expression of mCRY1 appeared not to change the subcellular localization of mPER2(882–1257)–GFP (p = 0.231; Figure 5D), despite the ability of mCRY1 to interact with this protein.
Taken together, these results suggest that the nuclear translocation of mPER2 does not depend on mCRY proteins and that the primary factor for determining subcellular localization exists in the mPER2 protein itself (NLS, NES, PAS and binding domains for other proteins that can co-transport mPER2 in the nucleus). Accordingly, the stimulating effect of mCRY1 on the nuclear accumulation of mPER2 must have its origin at another level rather than promoting nuclear import.
mCRYs inhibit ubiquitylation of mPER2
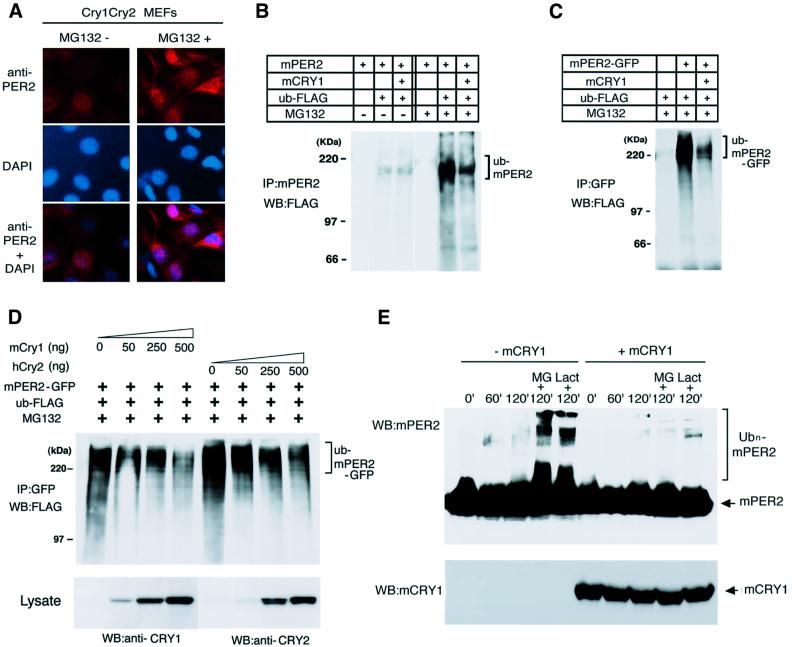
Despite constitutive high levels of mPer2 mRNA (Okamura et al., 1999), mPER2 protein is barely detectable in either nuclei or cytoplasm of SCN cells from mCry1/mCry2 double-mutant mice (Shearman et al., 2000). This observation raises the possibility that mPER2 is not stable in the absence of mCRY1 or mCRY2. Protein stability is frequently controlled by the ubiquitin–proteasome pathway and recently proteasome-mediated proteolysis has been shown to play an important role in the circadian system of fruit flies and plants (Naidoo et al., 1999; Deshaies and Meyerowitz, 2000). Therefore, we first tested the effect of MG132, a specific inhibitor of proteasome-mediated protein degradation, on the stability of endogenous mPER2 in mCry1/mCry2 double-mutant MEFs. Untreated cells show very faint immunocytochemical staining of endogenous mPER2 protein in the nucleus (Figure 6A, left). However, treatment of mCry-deficient MEFs with MG132 resulted in pronounced accumulation of endogenous mPER2 in nucleus and cytoplasm (Figure 6A, right). This suggests that the endogenous mPER2 protein is degraded by the proteasome pathway.
Fig. 6. Ubiqutin–proteasome-mediated degradation of mPER2 is inhibited by mCRY proteins. (A) mCry1/mCry2 double-mutant MEFs were cultured in the absence (left) or presence (right) of MG132. After 5 h of MG132 treatment, the subcellular localization of endogenous mPER2 was determined by immunocytochemistry with anti-mPER2 antibodies (top). Nuclei were stained using the DAPI method (middle). (B) Effect of mCRY1 on the ubiquitylation of non-tagged mPER2 in COS7 cells transiently expressing mPER2 and FLAG-tagged ubiquitin and cultured in the absence or presence of MG132. mPER2 was immunoprecipitated with anti-mPER2 antibodies and precipitates were examined for the presence of FLAG-tagged ubiquitin by western blot analysis using anti-FLAG antibodies. (C) As (B), except that mPER2(full)–GFP and anti-GFP antibodies were used. (D) Dose dependency of mCRY1- and hCRY2-mediated inhibition of mPER2 ubiquitylation. COS7 cells were co-transfected with a constant amount of mPER2(full)–GFP and FLAG–ubiquitin and increasing amounts of either mCry1 or hCry2 expression constructs and cultured in the presence of MG132. mPER2(full)–GFP was immunoprecipitated with anti-GFP antibodies and immunoblots of the precipitate were analyzed for the presence of FLAG-tagged ubiquitin (top) with anti-FLAG antibodies. The amount of expressed mCRY was visualized by immunoblot analysis of the total cell lysate with anti-mCRY1 (TD1119) or anti-mCRY2 (TD2206) antibodies (bottom). (E) Cell-free ubiquitylation of mPER2 in the absence or presence of mCRY1. Total cell lysates of COS7 cells transiently expressing non-tagged mPER2, mCRY1 or FLAG-ubiquitin were mixed as indicated and incubated at room temperature for 0, 60 or 120 min in the absence or presence of MG132 or lactacystin. The presence of ubiquitylated mPER2 in the incubation mixtures was determined by western blot analysis using anti-mPER2 antibodies. The presence of mCRY1 was confirmed using anti-mCRY1 antibodies.
Since ubiquitylation is an important step in targeting proteins for degradation by the proteasome (Pickart, 2000), we examined whether mPER2 is subject to ubiquitylation and if so, whether mCRY1 would affect ubiquitylation of mPER2. Hence, we expressed non-tagged mPER2 and FLAG-tagged ubiquitin in COS7 cells in the absence or presence of the proteasome inhibitor MG132 and precipitated mPER2 with an antibody raised against the latter protein. Immunoblot analysis of the immunoprecipitate with anti-FLAG antibodies revealed the presence of ubiquitylated mPER2 in cells that had not been exposed to MG132 (Figure 6B). Co-expression of mCRY1 did not dramatically alter the amount of ubiquitylated mPER2 (Figure 6B). However, treatment of cells with MG132 caused a strong increase in the amount of ubiquitylated mPER2, probably due to the block of the proteasome-mediated proteolysis step. Interestingly, under this condition, co-expression of mCRY1 and mPER2 in the presence of MG132 results in a clear reduction of the level of ubiquitylated mPER2. A similar result was obtained when GFP-tagged (Figure 6C) or HA-tagged versions of mPER2 (data not shown) were used. As shown in Figure 6D, the amount of ubiquitylated mPER2 in the presence of MG132 was inversely proportional to the amount of mCRY1 present in the cell. Importantly, hCRY2 can also suppress the ubiquitylation of mPER2 in the COS7 cell system in a dose-dependent manner. Since the proteasomal degradation machinery is blocked in the presence of MG132, the reduced levels of ubiquitylated mPER2 can only be explained by assuming that mCRY1 directly or indirectly interferes with ubiquitylation of mPER2. In conclusion, the above findings suggest that mCRY proteins inhibit ubiquitylation of mPER2, thereby likely preventing rapid degradation of mPER2 by the proteasome pathway.
To confirm the ubiquitylation of mPER2 and its inhibition by mCRY1, we analyzed the ubiquitylation of exogenously expressed mPER2 in a cell-free system. To this end, we mixed cell lysates from COS7 cells transiently transfected with either mPER2 or FLAG-tagged ubiquitin and incubated this mixture in the presence or absence of a cell lysate from mCRY1 transfected COS7 cells and/or the proteasome inhibitors MG132 and lactastatin (Figure 6E). In the presence of a proteasome inhibitor, but in the absence of mCRY1, western blot analysis of the reaction mixtures revealed the presence of ubiquitylated mPER2, which showed that the cell-free ubiquitylation assay was effective. However, when mCRY1 protein was present during the 2 h incubation period, the amount of ubiquitylated mPER2 was significantly reduced, which suggests that mCRY1 negatively affects ubiquitylation of mPER2. When proteasome inhibitors were omitted from the reaction mixture, ubiquitylated mPER2 was hardly detectable, independent of whether or not mCRY1 protein was present during the reaction. This indicates that ubiquitylated mPER2 is degraded by the proteasome system in vitro. Taken together, these in vivo and in vitro results show that mCRY1 directly inhibits the ubiquitylation of mPER2.
mPER2 inhibits ubiquitylation of mCRY proteins
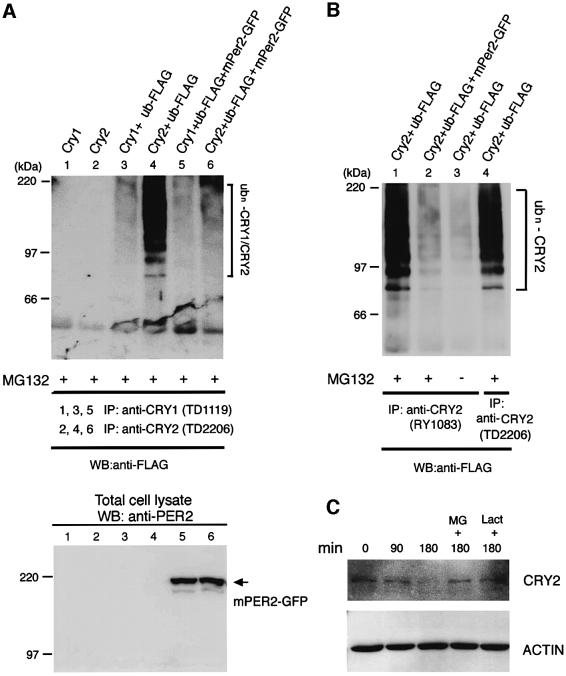
Since the levels of mCRY1 protein have been reported to be severely affected in mPer2 knockout mice (Bae et al., 2001), we first examined the effect of mPER2 on ubiquitylation of mCRY proteins in transiently transfected COS7 cells (Figure 7A). When mCRY1 was co-expressed with FLAG-tagged ubiquitin in the presence of MG132, immunoprecipitated mCRY1 was weakly ubiquitylated. In contrast, when a similar experiment was performed with hCRY2, pronounced ubiquitylation of the protein was observed. Next, we investigated the effects of co-expression of mPER2(full)–GFP on the ubiquitylation of CRY proteins (Figure 7A). Despite the low level of ubiquitylated mCRY1 protein, co-expression with mPER2 caused a small but significant decrease in ubiquitylation of mCRY1. Moreover, the presence of mPER2(full)–GFP dramatically reduced the ubiquitylation of hCRY2. The ubiquitylation of hCRY2 and the inhibitory effect of mPER2 thereon were confirmed using another anti-CRY2 antibody (Figure 7B). When cells were grown in the absence of proteasome inhibitors, the amount of ubiquitylated hCRY2 was reduced, likely as a result of degradation by the proteasome. These data indicate that mPER2 inhibits the ubiquitylation of mCRY proteins and ubiquitylated hCRY2 can be degraded by the proteasome.
Fig. 7. mPER2 proteins inhibit mCRY1 and hCRY2 ubiquitylation. (A) Effect of mPER2 on the ubiquitylation of CRY proteins in COS7 cells transiently expressing mouse CRY1, hCRY2, mPER2(full)–GFP and/or FLAG-tagged ubiquitin (as indicated) and cultured in the presence of MG132. CRY proteins were immunoprecipitated with anti-CRY1 (TD1119) or anti-CRY2 (TD2206) antibodies and precipitates were examined by western blot analysis using an anti-FLAG (top) antibody. Expression of mPER2–GFP was confirmed by western blot analysis of total cell lysates using anti-mPER2 antibodies (bottom). (B) As (A), except that another antiserum against CRY2 (RY1083) was used for immunoprecipitation and one cell lysate was prepared from cells grown in the absence of MG132. Note the low levels of ubiquitylated mPER2 in the latter sample. (C) Cell-free degradation of transiently expressed hCRY2. COS7 cell lysates were incubated at room temperature for the indicated time in the absence or presence of MG132 (50 µM) or lactacystin (50 µM). hCRY2 was visualized by western blot analysis using anti-CRY2 antibodies. Anti-actine antibodies were used as a loading control.
Finally, we studied the proteasome-dependent degradation of transiently expressed hCRY2 using a cell-free degradation assay (Figure 7C). When the hCRY2-expressing COS7 cell lysate was incubated with ubiquitin-expressing COS7 cell lysate at room temperature in the absence of proteasome inhibitors, a time-dependent disappearance of hCRY2 was observed, which was virtually complete after 3 h. When the assay was performed in the presence of MG132 or lactacystin, the level of hCRY2 remained unaltered, indicating that, similar to mPER2, hCRY2 is degraded by the proteasome degradation machinery.
Discussion
The timing of nuclear accumulation of clock proteins, constitutes an important step in the transcription– translation feedback loop driving the circadian core oscillator and is believed to be controlled by nuclear localization mechanisms and protein turnover (Young and Kay, 2001). In Drosophila, nuclear entry of the PER protein requires complex formation with TIMELESS (TIM) (Young, 1998). In mammals, nuclear import of mPER2 has been suggested to involve the mCRY proteins, as in cellular transfection experiments, nuclear accumulation of exogenous mPER2 is facilitated by co-expression of mCRY proteins (Kume et al., 1999). Yet, cells in the piriform cortex (Shearman et al., 2000) from mCry-deficient mice show nuclear mPER2, indicating that nuclear translocation of mPER2 can occur in the absence of mCRY proteins. To understand the mechanism underlying the nuclear accumulation of the mPER2 protein and solve the role of mCRY proteins therein, we have studied the subcellular localization of GFP-tagged full-length and truncated mPER2 proteins under various conditions in transiently transfected COS7 cells as well as mCry1/mCry2-deficient MEFs.
We have identified domains in the mPER2 protein involved in subcellular localization of the mPER2 protein. The primary structure of mPER2 has been reported to include a putative bipartite NLS sequence in the middle portion of the protein as well as PAS domains and a putative CLD region (Shearman et al., 1997; Takumi et al., 1998). Comparison of the subcellular localization patterns of the various truncated mPER2–GFP fusion proteins revealed that residues 596–881 (which include the reported NLS region) are required for nuclear translocation of mPER2. This observation fits with a recent deletion mutant study of the rat PER2 protein, identifying a functional NLS at residues 778–794 (Miyazaki et al., 2001). Interestingly, despite the absence of the NLS, mPER2(1–381)–GFP, mPER2(1–460)–GFP and mPER2 (882–1257)–GFP accumulate considerably in the nucleus when cells are cultured in the presence of LMB, indicating that these truncated mPER2 proteins have an alternative way for nuclear entry. Although we do not exclude the presence of unidentified NLS sequences in the N- and C-terminal regions of mPER2, the most likely interpretation is that mPER2(1–460)–GFP and mPER2(882– 1257)–GFP can translocate into the nucleus via association with other proteins. One such protein could be mPER3, which we have previously shown to interact with and promote nuclear translocation of mPER1 and mPER2 in cells under serum-shock conditions (Yagita et al., 2000). It is not known whether, and to what extent, co-transport might contribute to nuclear localization of mPER2 under physiological conditions.
Our localization studies with truncated mPER2 proteins indicated that the N- and C-terminal regions must contain domains that inhibit nuclear accumulation of mPER2. Analysis of the primary amino acid structure of mouse and human PER2 revealed the presence of three conserved leucine-rich nuclear export signal domains, NES1 (LIR TLKELKV, residues 109–118), NES2 (LQLNLLQL, residues 983–990) and NES3 (LTEQIHRLLM, residues 460–469), potentially allowing mPER2 to leave the nucleus again via the CRM1/Exportin1 nuclear export machinery. Substitution of hydrophobic amino acid residues in NES1 of full-length mPER2–GFP resulted in an increased percentage of cells expressing the protein exclusively in the nucleus. A less pronounced effect was observed when NES2 or NES3 were mutagenized. This suggests that all NES sequences in mPER2 are functional and that NES1 might be more effective than NES2 and NES3. Neither mutations in only one NES nor inactivation of two out of three NES sequences (data not shown) could completely abolish cytoplasmic localization of mPER2. This suggests that the presence of only one functional NES is sufficient to allow nuclear export. Indeed, only simultaneous inactivation of all three NES sequences caused a near complete loss of cells solely showing cytoplasmic mPER2 and a strong increase in the percentage of cells with nuclear mPER2 only. The active role of the NES domains is further underscored by our observation that treatment of transfected COS7 cells with LMB, an inhibitor of CRM1/Exportin1-mediated nuclear export, resulted in almost exclusive nuclear accumulation of pre-existing mPER2, thereby providing direct evidence that mPER2 can leave the nucleus via the nuclear export system. Furthermore, in contrast to Drosophila PER (Saez and Young, 1996), our data argue against a function of the putative CLD in cytoplasmic accumulation of mPER2. In conclusion, cytoplasmic localization of exogenously expressed mPER2 protein in cultured mammalian cells is controlled by NES sequences in mPER2 itself.
In principle, the presence of functional NLS and NES sequences in the mPER2 allows the protein to continuously shuttle between the cytoplasm and nucleus. Indeed, using a heterokaryon assay, in which NIH 3T3 cells containing nuclear GFP-tagged mPER2 were fused with COS7 cells and subsequent redistribution of fluorescence was determined after release of a nuclear export block, we have shown that pre-existing mPER2 can move from one nucleus to the other. This is the first demonstration of a clock gene shuttling between the cytoplasm and the nucleus. The potential relevance of mPER2 nuclear– cytoplasmic shuttling for clock functioning is discussed in more detail below.
mPER2 has been reported previously to physically associate with mCRY proteins (Griffin et al., 1999; Kume et al., 1999). We have shown that the mCRY1-binding domain resides in the C-terminal region of mPER2 (between residues 917 and 1257), most likely in the last 100 residues of mPER2, as has been shown for rat PER2 (Miyazaki et al., 2001). However, nuclear translocation of mPER2 does not per se require mCRY proteins since (i) GFP-tagged full-length and truncated mPER2 are found in nuclei of mCRY-deficient mCry1/mCry2 mutant MEFs and (ii) deletion of the mCRY-binding domain of mPER2 [as in mPER2(1–916)–GFP] does not abolish nuclear localization of the protein. Although co-expression with mCRY proteins did not significantly alter the subcellular distribution of truncated mPER2 proteins in mCry1/mCry2 double-mutant MEFs, mCRY1 could still considerably enhance nuclear localization of mPER2(full)–GFP. This indicates that although mPER2 can translocate in the nucleus in an mCRY-independent manner, mCRY1 proteins promote nuclear accumulation of mPER2 via another mechanism.
How can mCRY proteins affect the mPER2 levels in the nucleus? mPER2 is not detected in the nucleus and cytoplasm of SCN neurons of mCry1/mCry2 double-mutant mice (Shearman et al., 2000), despite high levels of the corresponding transcript in these tissues (Okamura et al., 1999). This opens the possibility that synthesized mPER2 is degraded in the absence of mCRY. Indeed, endogenous mPER2 can be visualized in mCry1/mCry2 double-mutant MEFs after inactivation of the proteasomal degradation pathway with MG132, showing that the mPER2 protein is synthesized but rapidly degraded in these cells. Degradation of proteins via the proteasome pathway requires that target proteins are first marked for degradation by covalent attachment of ubiquitin moieties (Ciechanover et al., 2000; Weissman, 2001). Here, we have shown that transiently transfected COS7 cells express low levels of ubiquitylated mPER2 protein, which strongly increased when the proteasomal degradation machinery was blocked. Taken together, these data show that in overexpression studies as well as under physiological conditions, mPER2 is degraded via the ubiquitin– proteasome pathway.
Importantly, mPER2 ubiquitylation is reduced by mCRY1 as well as hCRY2 in a dose-dependent manner. This suggests that CRY proteins stabilize mPER2 by preventing ubiquitylation and subsequent degradation of the latter. A concept of mCRY-mediated protection of mPER2 against ubiquitylation and degradation very much resembles the mechanism of TIM-mediated PER stabilization in Drosophila. The fly PER protein is rapidly degraded upon phosphorylation by DOUBLE-TIME (Price et al., 1998). However, physical interaction with TIM prevents the degradation of PER and, as a consequence, promotes accumulation of the protein in the nucleus (Kloss et al., 2001; Young and Kay, 2001). In accordance with the above concept, the absence of nuclear mPER2 in SCN neurons of mCry1/mCry2-deficient mice (Shearman et al., 2000) can now be explained in terms of enhanced degradation, rather than impaired nuclear import of the protein. In some tissues of mCry1/mCry2-deficient mice, such as neurons in the piriform cortex, mPER2 is constitutively present in the nucleus (Shearman et al., 2000). It might be that in these cells nuclear export, ubiquitylation and/or proteasome-mediated protein degradation is less efficient.
We have provided evidence that co-expression of mPER2 (as well as HA-mPER1 and HA-mPER3; data not shown) dramatically reduced the ubiquitylation of hCRY2 at least. Thus, both proteins apparently mutually protect each other against ubiquitylation and, as a consequence, degradation. Interestingly, such a mechanism can enhance robustness of protein oscillations and, in principle, would even allow rhythmic expression of proteins originating from constitutively expressed genes. In fact, cycling of mCry2 mRNA in the mouse SCN and cultured fibroblasts is very much blunted or even absent (Okamura et al., 1999; Vitaterna et al., 1999; Yagita et al., 2001), which is in marked contrast to the robust oscillation of mCRY2 immunoreactivity in SCN cells (Kume et al., 1999). As mCRY2 and mPER2 oscillate with nearly the same phase (Kume et al., 1999), the robust oscillation of mCRY2 localized in the nucleus could originate from rhythmic protection against ubiquitylation/degradation by cyclically available mPER2. The ability of interacting clock protein partners to mutually protect each other from degradation not only allows amplification of the effect of oscillating transcription, but can also synchronize oscillation of protein partners from genes that oscillate with a (small) phase difference.
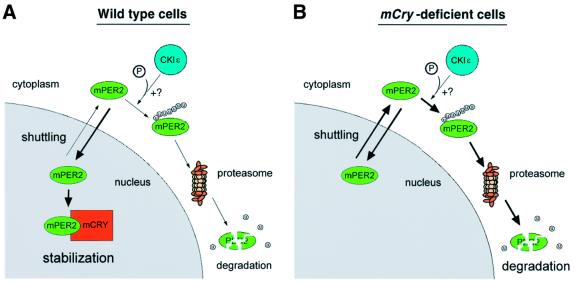
In conclusion, we have shown that the mPER2 protein can shuttle between the cytoplasm and the nucleus and that mCRY proteins promote nuclear accumulation of mPER2 by preventing ubiquitylation and subsequent proteasomal degradation of the latter. A regulation mechanism using shuttling and ubiquitylation has also been described for the Mdm2-mediated degradation of the p53 tumor suppressor protein (Boyd et al., 2000; Geyer et al., 2000; Zhang and Xiong, 2001). Recently, it has been shown that the c-Abl protein can prevent ubiquitylation and nuclear export of p53 by Mdm2 and that c-abl null cells fail to accumulate p53 following exposure to DNA damage (Vogt Sionov et al., 2001). It is tempting to speculate that the mCRY-mediated stabilization and nuclear accumulation of mPER2 proceeds via a similar mechanism. Therefore, we propose a working model (Figure 8) in which the mPER2 protein shuttles between cytoplasm and nucleus. In the cytoplasm, mPER2 can get ubiquitylated, ultimately leading to its degradation via the proteasome pathway. To date, it is not clear whether ubiquitylation of cytoplasmic mPER2 occurs on free protein, depends on physical interaction with other proteins or requires modification of the protein. It is interesting to note that human PER1 and PER2 proteins can associate with CKIε and δ and that phosphorylation reduces the stability of the mPER1 and mPER2 proteins (Keesler et al., 2000; Lowrey et al., 2000; Camacho et al., 2001). Thus, phosphorylation might facilitate ubiquitylation of mPER2 proteins, thereby retarding the increase in mPER2 levels, but experimental evidence is lacking so far. Once sufficient amounts of mCRY protein are synthesized and translocated into the nucleus, mCRY will bind to the C-terminal region of mPER2, thereby preventing or retarding nuclear export of mPER2 again via the nuclear export system (possibly via masking of NES2) and, as a consequence, slowing down multi-ubiquitylation of mPER2 in the cytoplasm. This model, in which mCRY acts at the level of nuclear export rather than import of mPER2, explains very well the apparent discrepancy between the finding that mCRY proteins on the one hand are not required for nuclear translocation (this study) and on the other hand can bind to mPER2 and promote its nuclear accumulation (Kume et al., 1999; this study). Interestingly, mCRY1 only associates with the C-terminal region of mPER2. This leaves NES1 and NES3 in the N-terminal and central region of mPER2 as a potential targets for other proteins that, in a way similar to mCRY proteins, may promote nuclear accumulation of mPER2. In a previous study, we have shown that a serum-shock promotes mPER2–mPER3 complex formation which enhances nuclear accumulation of mPER2 (Yagita et al., 2000). It would be interesting to investigate whether mPER3 could interact with the N-terminal region of mPER2 and thereby preventing ubiquitylation and degradation of mPER2 protein synthesized after non-photic stimuli. Moreover, induction of mPER2 protein in the SCN after a clock resetting light-pulse during the subjective night is quite subtle, despite the fact that mPer2 mRNA is strongly induced (Field et al., 2000). This may be due to degradation of newly synthesized mPER2, which can not be trapped by nuclear mCRYs in light-responsive SCN cells.
Fig. 8. Working model for mCRY-mediated nuclear accumulation of mPER2. Under normal conditions (A), newly synthesized mPER2 protein is translocated into the nucleus. This process most likely involves the NLS sequence, but we cannot exclude the possibility that other proteins (but not mCRY) can co-import mPER2 into the nucleus. Once in the nucleus, mPER2 is transported back to the cytoplasm via the CRM1/Exportin1 nuclear export system. The protein keeps on shuttling between nucleus and cytoplasm until (i) mPER2 is ubiquitylated and subsequently degraded by the proteasome system or (ii) the C-terminal region of nuclear mPER2 binds to mCRY1 or mCRY2, preventing nuclear export and allowing mPER2 to perform its function. As such, nuclear–cytoplasmic shuttling, coupled to cytoplasmic and nuclear degradation of mPER2 by the ubiquitin–proteasome system contributes to creating a phase delay between mPer2 mRNA and mPER2 protein peaks, characteristic for the transcription–translation-based circadian feedback loop. It is possible that, analogous to the situation of mPER1, CKIε-mediated phosphorylation of mPER2 further promotes its degradation. In the absence of mCRY proteins (B), mPER2 still shuttles between nucleus and cytoplasm but can not be retained in the nucleus, leading to enhanced degradation of the protein by the ubiquitin–proteasome machinery. Although not indicated in the figure, the concentration of mPER2 and CRY proteins may be controlled by ubiquitylation and the following degradation. mPER2, green; mCRY1, orange; mCRY2, blue, CKIε.
One of the key features of the self-sustaining circadian feedback loop is the phase delay of several hours between mRNA and protein peaks for genes involved in the circadian core oscillator (Dunlap, 1999; Young, 2000). A mechanism of nuclear–cytoplasmic shuttling and ubiquitin–proteasome-dependent degradation obviously adds another level at which this delay can be modulated. Examination of the predicted amino acid sequence of other clock proteins revealed the presence of three putative NES domains in mPER1 (residues 138–149, 489–498 and 981–988) and mPER3 (residuess 54–63, 399–408 and 913–920), all with conservation of the critical hydrophobic amino acid residues (Fukuda et al., 1997). In addition, human BMAL1 also contains two potential NES sequences (residues 105–114 and 124–134). It remains to be determined whether the mechanism of nuclear– cytoplasmic shuttling in conjunction with the ubiquitin– proteasome system is restricted to the mPER2 protein or whether it extends to other clock proteins. Similarly, it is not known whether shuttling of mPER2 (and perhaps other clock proteins) in combination with ubiquitin– proteasome-mediated degradation is clock regulated or whether it is a general mechanism involved in protein stabilization. Given the parallel with the p53/Mdm2 system we favor the latter option. Ubiquitylation and proteasomal degradation of clock proteins by itself is not a new finding. Light-dependent proteasomal degradation of TIMELESS has been described in Drosophila (Naidoo et al., 1999). However, to our knowledge, this is the first example of the (light-independent) involvement of the ubiquitin–proteasome pathway in the core mechanism of the circadian oscillator.
Materials and methods
Plasmids
Enhanced GFP fusion constructs were made from pBSIImPer2, containing the complete mPer2 cDNA cloned into the NotI site of pBlueScript II (Takumi et al., 1998). For mPER2(full)–GFP, a PCR fragment containing the complete coding sequence, in which the stop codon was replaced by a BamHI site, was cloned into NheI–BamHI digested pd2EGFP-N1 (Clontech). The sequence of the insert was verified for the absence of PCR-based errors. For mPER2(1–916)–GFP and mPER2(1–460)–GFP, pBSIImPer2 was digested with SpeI–StuI and SpeI–SacI, respectively and ligated into NheI–SmaI and NheI–SacI sites, respectively, on pd2EGFP-N1. For mPER2(596–1257)-GFP and mPER2(882–1257)-GFP, pBSIImPer2 was digested with KpnI–SalI and EcoRI–SalI, respectively and ligated into pEGFP–C2 (Clontech). HA-mPER2 was made via a PCR-based method as described previously (Yagita et al., 2000). For full-length mPER2 without any tags, a NotI–SalI cDNA fragment, spanning the complete coding sequence, was cloned into NotI–SalI digested pTRE2 (Clontech). For HA-mCRY1, a HA tag was fused to the 5′ end of full-length mCry1 cDNA and cloned into pcDNA3–Hyg (Invitrogen). For mCRY1 and hCRY2 expression vectors, full-length cDNAs were cloned into pcDNA3 (Invitrogen) as described previously (Yagita et al., 2000). The FLAG-tagged ubiqutin expression construct was kindly provided by Dr Kazuhiro Iwai (Kyoto University, Kyoto, Japan).
Site-directed mutagenesis
Amino acid substitutions in the mPER2 NES domains of mPER2(full)-GFP were generated using a QuickChange site-directed mutagenesis kit (Stratagene). Mutagenesis primers for the NES1 mutant were 5′-GATAAGGACCGCGAAGGAGGCGAAGGTCCACCTC-3′ and its complementary strand, for the NES2 mutant 5′-CTACAAGCTAACGCGG CTCAGGCAGAGGAGGCGCC-3′ and its complementary strand and for NES3 5′-CTCACAGAACAAGCCCACCGGGCACTGATGCAGCC-3′ and its complementary strand. The presence of the expected base pair substitutions was confirmed by DNA sequencing.
Cell culture
Wild-type and mCry1mCry2 double knockout MEFs, COS7, HeLa and NIH 3T3 cells were cultured and transfected (using LipofectAMIN Plus Reagent; Gibco BRL) as previously described (Yagita et al., 2000). Nuclear export of proteins was studied by culturing cells for 2 h in the presence of 10 ng/ml LMB (Sigma), an inhibitor of the CRM1/Exportin1 nuclear export system. In some experiments, de novo protein synthesis was blocked 6 h prior to the start of LMB treatment by addition of CHX (Sigma) to the culture medium at a final concentration of 50 µg/ml. For ubiquitylation studies, the proteasomal protein degradation machinery was inhibited by culturing cells in the presence of 25 µM MG132 (Calbiochem). Changes in the subcellular distribution of GFP-tagged proteins were statistically analyzed using the Chi square test.
Heterokaryon nuclear–cytoplasmic shuttling assay
Heterokaryon assays were performed as described (Borer et al., 1989; Wu et al., 1999). Briefly, NIH 3T3 cells were transfected with a plasmid encoding mPER2(1–916)–GFP. Seventeen hours after transfection, LMB (10 ng/ml) was added to the culture medium. Five hours later, NIH 3T3 cells were trypsinized, mixed with an equal amount of LMB-treated (non-transfected) HeLa cells and cultured for 3 h on glass cover slips in the presence of LMB (10 ng/ml) and CHX (50 µg/ml). Thirty minutes before cell fusion, the CHX concentration was increased to 100 µg/ml. Next, cells were washed with phosphate-buffered saline (PBS), fused by treatment with 50% PEG-4000 for 2 min and washed again with PBS. Finally, cells were cultured in fresh medium containing 100 µg/ml of CHX and in the presence or absence of 10 ng/ml LMB. Four hours later, the cells were fixed in 4% paraformaldehyde, immunostained by anti-hERCC1-specific antibody to recognize HeLa cells and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; nacalai tesque, Kyoto, Japan) to visualize the nuclei. Samples were analyzed with a Zeiss Axiovert microscope.
Immunofluorescence and immunoprecipitation
Immunocytochemistry was performed on cells fixed with 4% paraformaldehyde in PBS. To detect HA-mCry1 in transfected COS7 cells, we used a high affinity anti-HA monoclonal antibody (1:1000; Roche) and Cy3-conjugated anti-rat IgG antibody (1:800; Jackson Immuno Research Laboratories) as primary antibody and secondary antibody, respectively. Endogenous mPER2 in mCry1/mCry2 MEFs was detected using a rabbit mPER2 (affinity purified, 1:500; ADI) antibodies followed by a Cy3-conjugated anti-rabbit IgG antibody (1:800; Jackson Immuno Research Laboratories). We used DAPI (nacalai tesque; Kyoto, Japan) for nuclear staining.
For (co)immunoprecipitation studies, transfected cells were harvested in 300 µl of immunoprecipitation (IP) buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40) and incubated on ice for 15 min. After centrifugation, lysates were incubated with anti-mPER2 (ADI), anti-GFP (Clontech), anti-mCRY1 and anti-mCRY2 (TD1119 and TD2206 respectively; kindly provided by Dr T.Todo, Kyoto University, Kyoto, Japan) or anti-mCRY2 antibodies (RY1083) in the presence of protein A agarose (Roche) for 2 h at 4°C with rotation. Samples were washed three times with IP buffer, boiled at 100°C for 3 min, separated by SDS–PAGE on a 6.5% gel and transferred to a PVDF membrane. Next, immunoblot analysis was performed using the various antibodies at a 1:2000 dilution (as indicated in the figures), except for anti-FLAG M2 antibodies (monoclonal; Sigma), which were used at a 1:500 dilution. As secondary antibodies, we used horseradish peroxidase-conjugated anti-rabbit IgG (Amersham), anti-mouse IgG (Amersham) and anti-rat IgG (Santa Cruz) at a 1:2000 dilution. Chemiluminescence was performed using Renaissance western blot reagent plus (NEN).
Cell-free ubiquitylation and degradation assay
For the cell-free ubiquitylation or degradation assay, COS7 cells independently expressing either mPER2, ubiquitin-FLAG or mCRYs were harvested in ubiquitylation reaction buffer (5 mM MgCl2, 10 mM ATP, 50 mM HEPES pH 7.5, 40 mM KCl, 1% NP-40, 100 µM PMSF). Total cell lysates were mixed in various combinations (as indicated in Figures 6 and 7), divided over five tubes and incubated at room temperature for the indicated time in the absence or presence of 50 µM MG132 (Sigma) or 50 µM lactacystin (Calbiochem). After the incubation, lysates were subjected to western blot analysis.
Acknowledgments
Acknowledgements
The authors wish to thank Dr Kazuhiro Iwai (Kyoto University) for donating a FLAG-tagged ubiquitin expression construct, Dr Takeshi Todo (Kyoto University) for providing anti-CRY1 and anti-CRY2 antibodies and Dr Wim Vermeulen (Erasmus University) for providing the anti-human ERCC1 antibody. This work was supported in part by grants from the Special Coordination Funds and the Grant-in-Aid for the Scientific Research on Priority Areas of the Ministry of Education, Culture, Sports, Science and Technology of Japan and a SPINOZA premium from the Netherlands Organization for Scientific Research (NWO).
References
- Albrecht U., Sun,Z.S., Eichele,G. and Lee,C.C. (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell, 91, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Bae K., Jin,X., Maywood,E.S., Hastings,M.H., Reppert,S.M. and Weaver,D.R. (2001) Differntial function of mPer1, mPer2 and mPer3 in the SCN circadian clock. Neuron, 30, 525–536. [DOI] [PubMed] [Google Scholar]
- Borer R.A., Lehner,C.F., Eppenberger,H.M. and Nigg,E.A. (1989) Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell, 56, 379–390. [DOI] [PubMed] [Google Scholar]
- Boyd S.D., Tsai,K.Y. and Jacks,T. (2000) A intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nature Cell Biol., 2, 563–568. [DOI] [PubMed] [Google Scholar]
- Camacho F. et al. (2001) Human casein kinase I δ phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett., 489, 159–165. [DOI] [PubMed] [Google Scholar]
- Cermakian N. and Sassone-Corsi,P. (2000) Multilevel regulation of the circadian clock. Nature Rev. Mol. Cell. Biol., 1, 59–67. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian,A. and Schwartz,A.L. (2000) The ubiquitin-mediated proteolytic pathway: biological regulation via destruction. BioEssays, 22, 442–451. [DOI] [PubMed] [Google Scholar]
- Dunlap J. (1999) Molecular bases for circadian biological clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. and Meyerowitz,E. (2000) COP1 patrols the night beat. Nature Cell Biol., 2, E102–E104. [DOI] [PubMed] [Google Scholar]
- Field M.D., Maywood,E.S., O’Brien,J.A., Weaver,D.R., Reppert,S.M. and Hastings,M.H. (2000) Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron, 25, 437–447. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Staknis,D., Nguyen,H.B., Davis,F.C., Wilsbacher,L.D., King,D.P., Tahahashi,J.S. and Weitz,C.J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science, 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- Geyer R.K., Yu,Z.K. and Maki,C.G. (2000) The MDM2 RING-finger domain is required to promote p53 nuclear export. Nature Cell Biol., 2, 569–573. [DOI] [PubMed] [Google Scholar]
- Griffin E.A. Jr, Staknis,D. and Weitz,C.J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science, 286, 768–771. [DOI] [PubMed] [Google Scholar]
- Keesler G.A., Camacho,F., Guo,Y., Virshup,D., Mondadori,C. and Yao,Z. (2000) Phosphorylation and destabilization of human period 1 clock protein by human casein kinase I ε. Neuroreport, 11, 951–955. [DOI] [PubMed] [Google Scholar]
- Klein D.C., Moore,R.Y. and Reppert,S.M. (1991) Suprachiasmatic Nucleus. The Mind’s Clock. Oxford University Press, New York, NY.
- Kloss B., Rothenfluh,A., Young,M.W. and Saez,L. (2001) Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD and TIMELESS in the Drosophila clock. Neuron, 30, 699–706. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka,M.J., Sriram,S., Shearman,L.P., Weaver,D.R., Jin,X., Maywood,E.S., Hastings,M.H. and Reppert,S.M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura,K., Antoch,M.P., Yamazaki,S., Zemenides,P.D., Ralph,M.R., Menaker,M. and Takahashi,J.S. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science, 288, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Mesaki,M. and Ishida,N. (2001) Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol. Cell. Biol., 21, 6651–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N., Song,W., Hunter-Ensor,M. and Sehgal,A. (1999) A role for the proteasome in the light response of the Timeless clock protein. Science, 285, 1737–1741. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Okamura H., Miyake,S., Sumi,Y., Yamaguchi,S., Yasui,A., Muijtjens,M., Hoeijmarkers,J.H.J. and van der Horst,G.T.J. (1999) Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science, 286, 2531–2534. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. (2000) Ubiquitin in chains. Trends Biochem. Sci., 25, 544–548. [DOI] [PubMed] [Google Scholar]
- Price J.L., Blau,J., Rothenfluh,A., Abodeely,M., Kloss,B. and Young,M.W. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. and Weaver,D.R. (2001) Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol., 63, 647–676. [DOI] [PubMed] [Google Scholar]
- Saez L. and Young,M.W. (1996) Regulation of nuclear entry of the Drosophila clock proteins Period and Timeless. Neuron, 17, 911–920. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Zylka,M.J., Weaver,D.R., Kolakowski,L.F.J. and Reppert,S.M. (1997) Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron, 19, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Shearman L.P. et al. (2000) Interacting molecular loops in the mammalian circadian clock. Science, 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Takumi T. et al. (1998) A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells, 3, 167–176. [DOI] [PubMed] [Google Scholar]
- van der Horst G.T.J. et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature, 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H. et al. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA, 96, 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt Sionov R., Coen,S., Goldberg,Z., Berger,M., Bercovich,B., Ben-Neriah,Y., Ciechanover,A. and Haupt,Y. (2001) cAbl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol. Cell. Biol., 21, 5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitilation. Nature Rev. Mol. Cell. Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Wu J., Zhou,L., Tonissen,K., Tee,R. and Artzt,K. (1999) The Quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem., 274, 29202–29210. [DOI] [PubMed] [Google Scholar]
- Yagita K., Yamaguchi,S., Tamanini,F., van der Horst,G.T.J., Hoeijmakers,J.H.J., Yasui,A., Loros,J.J., Dunlap,J.C. and Okamura,H. (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev., 14, 1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Yagita K., Tamanini,F., van der Horst,G.T.J and Okamura,H. (2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science, 292, 278–281. [DOI] [PubMed] [Google Scholar]
- Young M.W. (1998) The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu. Rev. Biochem., 67, 135–152. [DOI] [PubMed] [Google Scholar]
- Young M.W. (2000) Life’s 24-hour clock: molecular control of circadian rhythms in animal cells. Trends Biochem. Sci., 25, 601–606. [DOI] [PubMed] [Google Scholar]
- Young M.W. and Kay,S.T. (2001) Time zones: a comparative genetics of circadian clocks. Nature Rev. Genet., 2, 702–715. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Xiong,Y. (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science, 292, 1910–1915. [DOI] [PubMed] [Google Scholar]
- Zheng B. et al. (2001) Nonredundant roles of mPer1 and mPer2 genes in the mammalian circadian clock. Cell, 105, 683–694. [DOI] [PubMed] [Google Scholar]