Abstract
Dbl-related oncoproteins are guanine nucleotide exchange factors (GEFs) specific for Rho guanosine triphosphatases (GTPases) and invariably possess tandem Dbl (DH) and pleckstrin homology (PH) domains. While it is known that the DH domain is the principal catalytic subunit, recent biochemical data indicate that for some Dbl-family proteins, such as Dbs and Trio, PH domains may cooperate with their associated DH domains in promoting guanine nucleotide exchange of Rho GTPases. In order to gain an understanding of the involvement of these PH domains in guanine nucleotide exchange, we have determined the crystal structure of a DH/PH fragment from Dbs in complex with Cdc42. The complex features the PH domain in a unique conformation distinct from the PH domains in the related structures of Sos1 and Tiam1·Rac1. Consequently, the Dbs PH domain participates with the DH domain in binding Cdc42, primarily through a set of interactions involving switch 2 of the GTPase. Comparative sequence analysis suggests that a subset of Dbl-family proteins will utilize their PH domains similarly to Dbs.
Keywords: Dbs/DH domain/PH domain/Rho GEF/Rho GTPase
Introduction
Rho guanosine triphosphatases (GTPases), such as Cdc42, RhoA and Rac1, comprise a major branch of the Ras superfamily of small GTPases. Like Ras, Rho proteins function as bi-molecular switches by adopting distinct conformational states in response to binding either GDP or GTP. In contrast to GDP-bound Rho, Rho–GTP actively participates in signal transduction networks by interacting with downstream effectors (Van Aelst and D’Souza-Schorey, 1997; Bishop and Hall, 2000). Rho GTPases regulate the organization and structure of the actin cytoskeleton and control the activities of numerous eukaryotic transcription factors (Hall, 1998; Mackay and Hall, 1998). Consequently, Rho GTPases coordinate such diverse cellular processes as adhesion, migration, phagocytosis, cytokinesis, neurite extension and retraction, morphogenesis, polarization, growth, cell cycle progression and proliferation (Van Aelst and D’Souza-Schorey, 1997; Kaibuchi et al., 1999; Chimini and Chavrier, 2000; Evers et al., 2000).
Guanine nucleotide exchange factors (GEFs) convert GTPases to their biologically active state by catalyzing the exchange of bound GDP for GTP. GTPases normally bind guanine nucleotides with high affinity and are typically unstable when nucleotide free. A general reaction scheme for GTPase activation by GEFs involves the formation of an initial, low-affinity GEF·GTPase·GDP ternary complex that rapidly converts to a high-affinity GEF·GTPase binary complex concomitant with expulsion of GDP and Mg2+ (Cherfils and Chardin, 1999). In the absence of exogenous guanine nucleotides, the binary complex is stable. However, the relatively high concentration of GTP in vivo (normally ∼20-fold higher than GDP) favors the binding of GTP followed by dissociation of the GEF and activated GTPase.
The Dbl-family of oncoproteins are Rho-specific GEFs (>50 distinct mammalian family members identified) that contain an ∼300 residue region of sequence homology to Dbl, a transforming protein originally isolated from a diffuse B-cell lymphoma (Eva and Aaronson, 1985; Cerione and Zheng, 1996). This region includes two distinct domains, an ∼200 residue Dbl homology (DH) domain and an ∼100 residue pleckstrin homology (PH) domain which invariably resides immediately C-terminal to the DH domain (Cerione and Zheng, 1996; Whitehead et al., 1997). DH domains interact directly with Rho GTPases to catalyze guanine nucleotide exchange (Ron et al., 1991; Hart et al., 1994; Liu et al., 1998; Aghazadeh et al., 2000), and like other GEFs, DH domains preferentially bind Rho GTPases depleted of nucleotide and Mg2+ (Hart et al., 1994; Glaven et al., 1996). Recent determination of the structure of the DH and PH domains of Tiam1 bound to nucleotide-free Rac1 revealed the mechanism used by Dbl-family GEFs to facilitate guanine nucleotide exchange of Rho GTPases (Worthylake et al., 2000).
Although PH domains are resident in numerous signaling proteins (Lemmon and Ferguson, 1998), the invariant linkage between DH and PH domains strongly suggests a unique role for these PH domains. While DH-associated PH domains can clearly promote the translocation of Dbl-related proteins to plasma membranes (Whitehead et al., 1996, 1999), the PH domains may also participate directly in GTPase binding and regulation of GEF activity. For example, the binding of phosphoinositides to DH-associated PH domains is reported to modulate exchange activity allosterically (Han et al., 1998; Crompton et al., 2000; Russo et al., 2001), although the molecular details of this regulation are currently undefined and subject to speculation (Snyder et al., 2001). Moreover, a fragment of Trio encompassing the DH and PH domains is ∼100 times more efficient at catalyzing guanine nucleotide exchange relative to the isolated DH domain in the absence of phospholipids (Liu et al., 1998).
Dbs (Dbl’s big sister) was originally isolated by its ability to transform NIH 3T3 murine fibroblasts and is ubiquitously expressed (Whitehead et al., 1995). Full-length Dbs encompasses 1149 amino acids and contains, in addition to the DH and PH domains, a putative N-terminal Sec14 domain, two spectrin-like repeats and a C-terminal Src homology 3 domain (SH3) (Figure 1A) (Whitehead et al., 1995; Aravind et al., 1999). However, only the DH and PH domains have been implicated in cellular transformation. Dbl and Dbs are highly similar with ∼50% identity over an 800 residue span and an ∼65% identity within the DH and PH domains (Whitehead et al., 1995). Consistent with their high degree of sequence similarity, Dbl, Dbs and Ost (the Dbs rat ortholog) are GEFs specific for Cdc42 as well as RhoA (Hart et al., 1994; Horii et al., 1994; Whitehead et al., 1999). Also, Dbs and Dbl cause similar transformed foci and activate multiple common pathways including Elk-1, Jun and NF-κB transcription factors and transcriptional stimulation through the cyclin D1 promoter (Westwick et al., 1998; Whitehead et al., 1999). Like Trio, fragments of Dbs or Dbl containing both the DH and PH domains are necessary for optimal GEF activity, further indicating an important functional role for their associated PH domains (Hart et al., 1994; Rossman and Campbell, 2000).
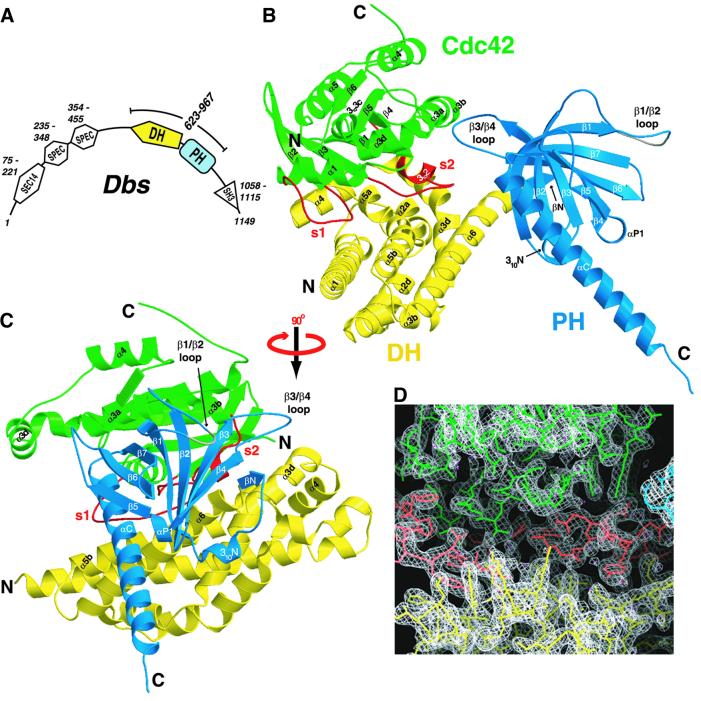
Fig. 1. Structure of the Dbs·Cdc42 complex. (A) A schematic representation of the putative signaling domains and their associated sequence ranges within full-length Dbs (murine, DDBJ/EMBL/GenBank accession No. AAB33461), as predicted by the protein sequence analysis program SMART (Schultz et al., 1998). Also indicated is the fragment of Dbs used for crystallization of the Dbs·Cdc42 complex (residues 623–967). In addition to the DH and PH domains, Dbs also possesses a domain homologous to the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14p (SEC14), two spectrin repeats (SPEC) and Src homology 3 domain (SH3). (B) A ribbon diagram of the Dbs·Cdc42 complex shows that the DH domain (yellow) of Dbs primarily engages the switch regions (red) of Cdc42 (green). The PH domain (blue) also interacts with the GTPase by means of a conformation unique from that of Sos1 and Tiam1·Rac1. The disordered β1/β2 loop within the PH domain is gray. (C) The Dbs·Cdc42 complex is shown rotated 90° about the vertical axis relative to its orientation in (B). (D) The experimental electron density (white) contoured at 1.2σ in the vicinity of the vacant nucleotide binding site. Bound waters are magenta.
In addition to biochemical data verifying the importance of the PH domain in Dbs-catalyzed nucleotide exchange, primary sequence analysis of Tiam1 and Dbs suggests a potentially different interdomain arrangement between the Dbs DH and PH domains. To investigate the possibility that an altered PH domain orientation underlies the Dbs biochemical exchange data, we have determined the crystal structure of the DH/PH fragment from Dbs in complex with Cdc42 at 2.4 Å resolution. Within the complex, the PH domain is observed to interact directly with Cdc42 and to support residues of the DH domain critical for binding switch 2 of Cdc42. The structure and supporting biochemistry illuminate how DH-associated PH domains can act directly in catalyzing the activation of Rho GTPases.
Results
Crystallization and structure determination of the Dbs and Cdc42 heterodimer
The DH and PH portion of murine Dbs (residues 623–967) (Figure 1A) and the placental isoform of human Cdc42 (residues 1–188) were expressed separately in Escherichia coli and purified to homogeneity prior to formation of the Dbs and Cdc42 complex, as described in Materials and methods. Crystallization experiments conducted with the purified Dbs DH/PH·Cdc42 heterodimer yielded well-formed orthorhombic crystals belonging to space group P212121 with two heterodimers in the asymmetric unit. Structure determination utilized seleno-methionine- containing Dbs and a two-wavelength anomalous dispersion data set collected from a single crystal at 100 K. Initial phases were determined using coordinates of 20 selenium atom positions and were improved by solvent flattening, histogram matching and two-fold non-crystallographic symmetry averaging using the program DM (Cowtan, 1994). The resulting high quality electron density map (Figure 1D) was used for model building with the program O (Jones et al., 1991). Model refinement was completed using all data from a single wavelength with |F| > 0 extending to 2.4 Å by iterating cycles of positional and temperature factor refinement and manual intervention. The final model has a crystallographic R-value of 19.7% (free R = 23.9%) using all data from 20–2.4 Å and contains 1032 protein residues (8280 atoms) and 323 water molecules (Table I). Except for subtle differences within the β3/β4 loop of the PH domain, the two complexes within the asymmetric unit are virtually identical and superimpose with a r.m.s.d. (root mean square difference) of 0.91 Å for all 507 common Cα atoms.
Table I. Crystallographic data collection and refinement statistics.
| Data set | Wavelength (Å) | Resolution (shell) (Å) | Observations (total/unique) | Completeness (%) | <I/σI>a | Rsymb (%) |
|---|---|---|---|---|---|---|
| Native λ1 | 0.97924 | 30–2.4(2.49–2.4) | 418 244/49 528 | 90.7 (58.0) | 27.2 (3.2) | 8.9 (43.3) |
| Native λ2 | 0.97858 | 30–2.4 (2.49–2.4) | 402 859/49 536 | 90.7 (57.7) | 26.2 (2.5) | 7.9 (40.2) |
| Y889F | 1.5418 | 50–2.6 (2.69–2.6) | 225 476/43 434 | 99.6 (99.7) | 19.0 (2.9) | 9.1 (58.0) |
| Overall figure of meritc (native) |
before density modification = 0.39 |
after density modification = 0.84 |
||||
| Refinement statistics (λ1) |
Native |
|
Y889F |
|||
| Resolution range (Å) |
20–2.4 |
|
20–2.6 |
|||
| Number of atoms (protein/solvent) | 8280/323 | 8281/115 | ||||
| Number of reflections (work/test) | 46 595/2460 | 41 214/2169 | ||||
| Rworkd (%) | 19.7 | 21.7 | ||||
| Rfreed (%) | 23.9 | 25.9 | ||||
| R.m.s.d. bond distances (Å) | 0.006 | 0.007 | ||||
| R.m.s.d. bond angles (°) | 1.23 | 1.29 | ||||
| Average B-factor all atoms (Å2) | 41.8 | 56.5 | ||||
| Average B-factor DH domain (Å2) | 37.2 | 52.8 | ||||
| Average B-factor PH domain (Å2) | 50.4 | 67.9 | ||||
| Average B-factor Cdc42 (Å2) | 40.8 | 52.5 | ||||
| Average B-factor waters (Å2) | 36.8 | 37.4 | ||||
| Residues in favorable region of Ramachandran plot | 90.6% | 89.4 | ||||
| Residues in allowed region of Ramachandran plot | 9.4% | 10.5 | ||||
| Residues in disallowed region of Ramachandran plot | 0% | 0.1 | ||||
a<I/σI>, mean signal to noise, where I is the integrated intensity of a measured reflection and σI is the estimated error in the measurement.
bRsym = 100 × Σ|I – <I>|/ΣI, where I is the integrated intensity of a measured reflection.
cFigure of merit = <|ΣP(α)eiα/ΣP(α)|>, where α is the structure factor phase and P(α) is the phase probability distribution.
dRcryst = Σ|Fp – Fp(calc)|/ΣFp, where Fp and FP(calc) are the observed and calculated structure factor amplitudes. Rfree is calculated similarly using test set reflections never used during refinement.
Numbers in parentheses pertain to the highest resolution shell.
The more complete heterodimer model includes residues 624–846 and 852–965 of Dbs, and the entire Cdc42 polypeptide. For Dbs, electron density is uninterpretable for terminal residues (623 and 966 through the C-terminal His6 tag) and residues (847–851) within the β1/β2 loop of the PH domain. For illustrative purposes, these loop residues were modeled post-refinement with zero occupancy.
Structural overview
The three-dimensional structure of the Dbs DH/PH·Cdc42 (henceforth Dbs·Cdc42) heterodimer is illustrated in Figure 1B and C. The DH domain (residues 625–817) of Dbs is an elongated helical bundle very similar to other DH domains of known structure (Aghazadeh et al., 1998, 2000; Liu et al., 1998; Soisson et al., 1998; Worthylake et al., 2000). To standardize nomenclature describing this and future DH domain structures, helices within the DH domain of Dbs are named relative to the six major helical axes within DH domains of known structure. There are three highly conserved regions in all DH domains (CRs1–3). CR1 (α1a) and CR3 (α5a–α5b) together with α3d and α6 constitute the major binding surface for Cdc42 (Figures 1B, C, 2A and B). α4 juts out from the body of the helical bundle and is critical to the placement of the subsequent turn and α5a, a region implicated in dictating specificity for binding GTPases (Worthylake et al., 2000; Karnoub et al., 2001). CR2 (α2b–α2d) is on the opposite side of the helical bundle relative to CRs1 and 3 and presumably functions mainly to stabilize the helical bundle. The Dbs·Cdc42 complex does not oligomerize in the crystal structure, in contrast to Tiam1·Rac1, where heterotetramers are observed to form principally through DH domain interactions (Worthylake et al., 2000).
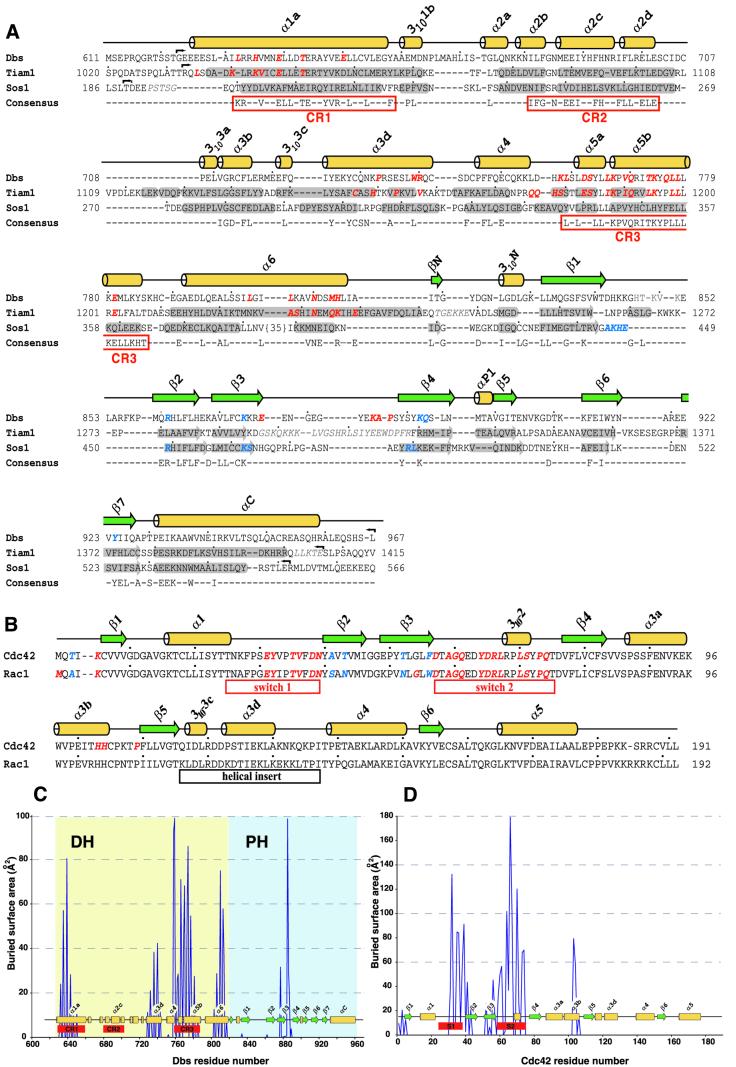
Fig. 2. Sequence alignments of Dbl-family exchange factors and Rho GTPases. (A) Sequences for Dbs, Tiam1 and Sos1 were aligned using Clustal_X v1.8 (Thompson et al., 1997), and manually altered to further align secondary structure elements. The consensus sequence was generated from a Dbl-family alignment of 47 non-redundant sequences and conserved regions (CR) are boxed. Crystallographically determined helices (yellow) and β-strands (green) of Dbs are shown above the alignment, and similar secondary structure elements of Tiam1 and Sos1 are shaded. Red italicized amino acids indicate direct contacts with bound GTPases (Dbs·Cdc42 or Tiam1·Rac1). PH domain residues thought to be involved in binding phospholipids are indicated by blue italics (Koshiba et al., 1997; see Figure 8A). Small arrows indicate construct borders, lightened italicized residues are disordered in the structures, small dots indicate 10-residue spans and numbers indicate amino acid positions within the full-length proteins. DDBJ/EMBL/GenBank accession Nos are: Dbs, AAB33461; Tiam1, Q60610; Sos1, A37488. (B) Sequences for 16 Rho GTPases and Ras were aligned using Clustal_X v1.8 (Thompson et al., 1997) and those for Cdc42 and Rac1 are shown. Residues in Cdc42 and Rac1 that bury >10 Å2 upon complex formation with Dbs or Tiam1, respectively, are indicated in red italics, and nomenclature for secondary structure elements are derived from the structure of Ras (Pai et al., 1990). In blue italics are buried residues that differ between Cdc42 and Rac1 and are probably important for dictating specificity between GTPases and DH domains (Worthylake et al., 2000; Karnoub et al., 2001). Also highlighted (underscore boxes) are the switches that undergo conserved, nucleotide-dependent conformational alteration in GTPases, as well as the 21 amino acid insertion unique to Rho GTPases. The buried accessible surface areas for residues losing >10 Å2 upon complex formation within (C) the Dbs DH/PH domain or (D) Cdc42 were determined and plotted for each residue number.
The interface between Dbs and Cdc42 is extensive and buries nearly 3100 Å2 of accessible surface area (Figure 2C and D). For both the Dbs·Cdc42 and Tiam1·Rac1 structures, the relative orientation of GTPase to the DH domain is nearly identical, and the GTPase switches 1 and 2 (residues 25–39 and 57–75, respectively) are similarly altered. Relative to Cdc42–GDP and excluding the switch regions, the nucleotide binding pocket of Cdc42 bound to Dbs is essentially undisturbed and completely exposed to solvent. There is no discernible electron density for either GDP or Mg2+. Instead, the nucleotide binding pocket contains several ordered water molecules, three of which are distributed across the α- and β-phosphate binding sites.
Similar to other PH domains (Lemmon et al., 1996), the PH domain of Dbs consists of a β-sandwich composed of two anti-parallel β-sheets (strands 1–4 and 5–7) capped at one end by an extended C-terminal α-helix. There are additional secondary structure elements (βN and 310N) N-terminal to the main body of the PH domain. These elements are also found in Tiam1 (contains 310N) and Sos1 (contains both βN and 310N), and are presumably unique to Dbl-family proteins (Koshiba et al., 1997; Zheng et al., 1997; Soisson et al., 1998; Worthylake et al., 2000). Together with hydrophobic residues from β1–β4 (Phe864, His866, Leu871, Tyr889, Tyr891), residues within and between βN and 310N (Ile818, Tyr821, Leu825) form a large hydrophobic cluster supporting hydrogen-bonding interactions between βN and β4 that extends the first anti-parallel β-sheet of the PH domain. Residues Phe864 and Leu871 are highly conserved among DH-associated PH domains, probably due to their key role in stabilizing this region. βN, 310N and β1–β4 of the PH domain also provide the major interface with the DH domain through interactions with the carboxyl portion of α6 (Figure 1C). At the center of this interface lie His814 and Leu815 within α6 of the DH domain, which combine to bury ∼200 Å2 of accessible surface area and make several interactions with the PH domain.
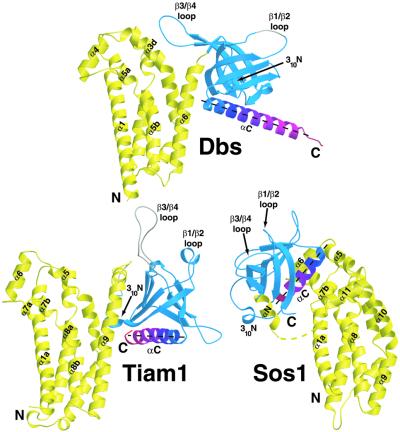
The orientation of the PH domain relative to the DH domain of Dbs is more similar to Tiam1 bound to Rac1 than Sos1, which is radically altered in comparison to both Dbs and Tiam1 (Figure 3) (Soisson et al., 1998; Worthylake et al., 2000). However, the relative orientations of the DH and PH domains of Dbs and Tiam1 differ in two important ways. First, the Dbs PH domain is rotated about α6 by ∼85° towards Cdc42. Secondly, the PH domain of Dbs is translated ∼10 Å toward the N-terminus of α6. Consequently, while there are no direct contacts between the PH domain of Tiam1 and Rac1, the PH domain of Dbs forms several interactions with Cdc42.
Fig. 3. Comparison of the relative orientations between DH and PH domains from Dbs, Tiam1 and Sos1. The DH and PH domains from the structures of Dbs·Cdc42 (top), Tiam1·Rac1 (bottom left) and Sos1 (bottom right) were aligned by least squares superposition of the DH domains conserved regions 1–3. DH domains are colored yellow and PH domains are blue. To aid visualization, the C-terminal helix of each PH domain (αC) is colored from blue to red (N- to C-terminus, respectively) and the αC helical axes are represented by black dashed lines. The yellow dashed line in Sos1 indicates the disordered residues in the DH domain. Whether the PH domains of Tiam1 and Sos1 can adopt Dbs-like conformations to enhance exchange, e.g. at cell membranes or through the actions of accessory proteins, is unclear.
As a result of their variable orientations, the DH and PH domains of Dbs interact to bury considerably less solvent-accessible surface area relative to Tiam1 (∼886 versus 1430 Å2, respectively). For example, in Tiam1·Rac1, 310N inserts into a hydrophobic cleft underneath α9 (analogous to α6 in Dbs) of the DH domain to stabilize the DH/PH interface. In Dbs, however, rotation of the PH domain pulls 310N away from the DH domain and consequently 310N contributes appreciably less buried surface area to the DH/PH domain interface (23 versus 187 Å2 in Tiam1).
The interface between the PH domain of Dbs and Cdc42
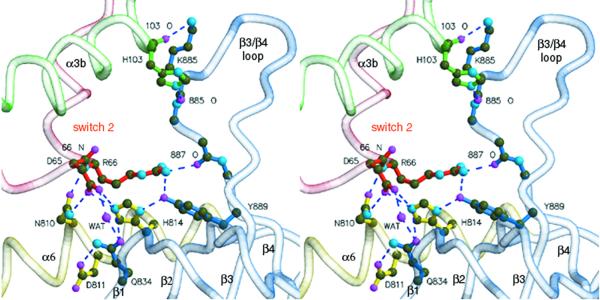
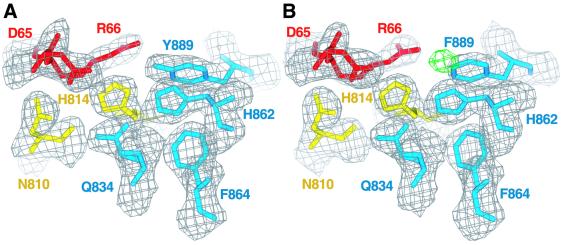
In contrast to Tiam1·Rac1, the PH domain of Dbs rotates around α6 of the DH domain and directly contacts Cdc42 (Figure 4). Supported by α6 of the DH domain, portions of β1, β4 and the β3/β4 loop of the PH domain contact switch 2 (Asp65, Arg66) and α3b (His103) of Cdc42. The side chain of Asp65 directly hydrogen bonds to the side chains of Asn810 and His814 within α6 of the DH domain. The equivalent of Asn810 is nearly invariant in all DH domains and the analogous residue within Tiam1 (Asn1232) essentially duplicates the interactions seen in Dbs·Cdc42. However, His814 is unique to Dbs and hydrogen bonds Tyr889 within β4 of the PH domain. In addition, Gln834 (within β1) simultaneously interacts with Asp65 of switch 2 via a water-mediated hydrogen bond and Asp811 within the DH domain.
Fig. 4. Interactions between the PH domain of Dbs and Cdc42. Stereo view of the PH domain (blue) participating with α6 of the DH domain (yellow) to bind switch 2 (red) and α3b (green) of Cdc42. Dashed lines indicate hydrogen bonds (<3.3Å).
In the Tiam1·Rac1 structure, Arg66 from Rac1 ion pairs with Glu1239, a nearly invariant residue located within α6 of DH domains. For Dbs, the equivalent of Glu1239 is replaced with Ala817 and thus a similar interaction cannot occur with Cdc42 (Figure 2A). In Dbs·Cdc42, Arg66 occupies an alternate rotamer conformation and its guanidinium group hydrogen bonds with the carbonyl oxygen of Pro887 (within the β3/β4 loop), as well as the hydroxyl of Tyr889 of the PH domain. Thus, Tyr889 is intimately involved in binding Cdc42 by stabilizing His814 of the DH domain, as well as through direct interaction with Arg66 of switch 2.
In addition, the β3/β4 loop of the PH domain of Dbs interacts with several residues within the α3b of Cdc42. Chief among these interactions are a pair of hydrogen bonds between His103 (within α3b of Cdc42) and Lys885 (within the β3/β4 loop of the PH domain). The GEFs, RCC1 (Renault et al., 2001), Sos (Boriack-Sjodin et al., 1998) and EF-Ts (Kawashima et al., 1996; Wang et al., 1997), also engage portions of switch 2 and α3 upon binding their respective GTPases, indicating a common ‘footprint’ of interaction (Day et al., 1998).
Biochemical mapping of functional interactions
Typically, mutations within PH domains associated with DH domains have been designed either to eliminate phosphoinositide binding or disrupt the global PH domain fold (Chen et al., 1997; Freshney et al., 1997; Han et al., 1998; Sterpetti et al., 1999). For example, mutation of the highly conserved tryptophan (W937L) within the C-terminal α-helix of the PH domain of Dbs is generally assumed to disrupt global structure and results in loss of cellular transformation promoted by Dbs (Whitehead et al., 1995). The structure of Dbs·Cdc42 now provides a framework for a more directed probing of PH domain functions within the context of exchange activity. Consequently, we have designed substitutions and deletions to disrupt specific interactions involving the PH domain and have assessed their functional effects on nucleotide exchange (Figure 5; Tables II and III).
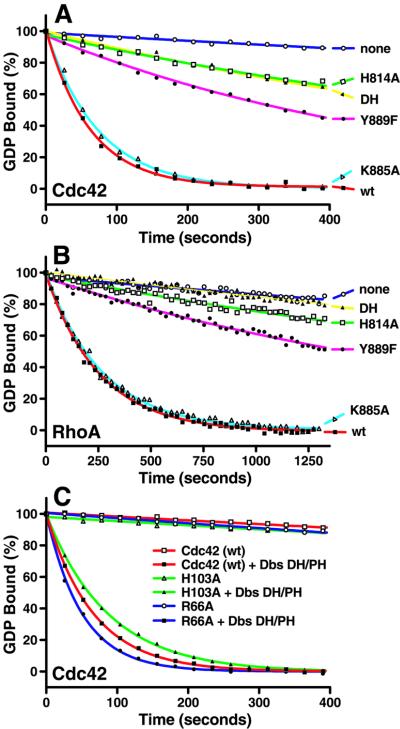
Fig. 5. Biochemical analysis of PH domain-mediated interactions with Cdc42. Substitutions within the DH/PH fragment of Dbs (K885A, Y889F, H814A) remove specific interactions with Cdc42 and differentially affect exchange activity on (A) Cdc42 or (B) RhoA. For comparison, also shown is the wild-type (wt) DH/PH fragment, as well as the isolated DH domain (DH) and no exchange factor (none). (C) Substitutions in Cdc42 (R66A, H103A) at sites in contact with the Dbs PH domain exhibit minor effects on Dbs-stimulated guanine nucleotide exchange compared with wild-type Cdc42.
Table II. Rate constants of guanine nucleotide exchange reactions catalyzed by wild-type and mutant Dbs proteins on Cdc42 and RhoA.
| Dbs | Cdc42 |
RhoA |
||
|---|---|---|---|---|
| kobs (s–1 × 10–3) | Fold stimulation | kobs (s–1 × 10–3) | Fold stimulation | |
| None | 0.28 ± 0.03 | – | 0.16 ± 0.01 | – |
| DH/PH (wt) | 15.49 ± 0.70 | 57 ± 5 | 4.11 ± 0.02 | 26 ± 1 |
| H814A | 1.22 ± 0.33 | 4 ± 1 | 0.25 ± 0.01 | 2 ± 1 |
| Y889F | 1.98 ± 0.17 | 7 ± 1 | 0.43 ± 0.01 | 3 ± 1 |
| K885A | 13.95 ± 0.47 | 49 ± 6 | 3.65 ± 0.02 | 23 ± 1 |
| DH (wt) | 1.42 ± 0.39 | 5 ± 1 | 0.17 ± 0.00 | 1 ± 0 |
The rates (kobs) of guanine nucleotide exchange for 1 µM wild-type Cdc42 (left) or RhoA (right) stimulated by 0.2 µM various Dbs proteins were determined by fitting the data from Figure 5A and B as single exponential decays. The fold stimulation for each Dbs protein reflects the ratio of the kobs measured for the GEF-containing reaction to the unstimulated reaction containing no GEF (none).
Table III. Rate constants of Dbs-catalyzed guanine nucleotide exchange of mutant Cdc42 proteins.
| 2 µM Cdc42 | Intrinsic rate | + 0.2 µM Dbs | Fold stimulation |
|---|---|---|---|
| R66A | 0.34 ± 0.01 | 19.69 ± 0.13 | 57 ± 2 |
| H103A | 0.27 ± 0.06 | 11.19 ± 0.35 | 41 ± 1 |
His814 within the DH domain is a keystone residue interacting with both switch 2 (Asp65) of Cdc42 and β4 (Tyr889) of the PH domain. Not surprisingly, H814A severely reduces Dbs-facilitated nucleotide exchange within both Cdc42 and RhoA (4- and 2-fold stimulation over intrinsic rates, respectively, versus 57- and 26-fold for wild type, respectively) (Figure 5A and B; Table II). Similarly, the associated substitution Y889F is also extremely detrimental to catalyzed exchange on both GTPases and these effects are not due to the global disruption of the GEF (see next section). The reduced exchange activity is significant for both H814A and Y889F, as either mutation in Dbs results in the complete loss of the transforming activity of GEF in vivo (K.Rossman, L.Cheng, I.Whitehead and J.Sondek, in preparation).
Alanine substitutions of either K885 (49- and 23-fold over intrinsic rates for Cdc42 and RhoA, respectively) or the proximal His103 (41-fold over intrinsic) (Figure 5C; Table III) have minimal effects on catalyzed exchange, suggesting that the relatively disordered β3/β4 loop contributes little to functional engagement of GTPases under these solution conditions.
Arg66 of switch 2 buries more solvent-accessible surface area than any other residue within Cdc42 upon complex formation, and hydrogen bonds with both Pro887 and Tyr889 of the PH domain. Yet surprisingly, R66A fails to diminish the rate of Dbs-catalyzed exchange on Cdc42.
As for some other Dbl-family members (Hart et al., 1994; Liu et al., 1998), the isolated DH domain of Dbs has greatly reduced nucleotide exchange activity relative to the DH/PH fragment (Figure 5A and B; Table II). In particular, the isolated DH domain of Dbs is inactive on RhoA while retaining limited activity toward Cdc42 (5-fold over intrinsic). This result suggests that the Dbs PH domain makes additional interactions with RhoA, not observed within Dbs·Cdc42, which favorably influence exchange.
Dbs(Y889F) in complex with Cdc42
The formal possibly that Y889F disrupts the overall fold of Dbs, leading to decreased exchange, is ruled out by the crystallographic structure determination of the complex of Dbs(Y889F) bound to Cdc42 (Figure 6). Except for a slight rotation of the PH domain about α6 (∼3°), consistent with loss of the Y889 to H814 hydrogen bond, the structure is essentially identical to the original complex with a r.m.s.d. of 0.76 Å for all common atoms. Given that R66A in Cdc42 does not impair guanine nucleotide exchange catalyzed by Dbs (Figure 5C), Tyr889 presumably contributes to exchange via interactions mediated through His814 of the DH domain. Two roles for Tyr889 can be envisioned: (i) restricting the conformation of His814 in unbound Dbs; and (ii) stabilizing the electronic polarization of the imidazole group of His814. Both characteristics would favor interaction of His814 of the DH domain with Asp65 of switch 2. This arrangement between the side chains of Tyr889 of the PH domain, His814 of the DH domain and Asp65 of Cdc42 is reminiscent of catalytic triads found within serine proteases. However, for Dbs and Cdc42, the triad of residues favors intermolecular association and consequent nucleotide exchange. A similar spatial arrangement of residues has been suggested to stabilize the fold of the WD repeats of Gβγ (Garcia-Higuera et al., 1996; Sondek et al., 1996).
Fig. 6. Comparison of native and Dbs(Y889F)·Cdc42 structures. (A) 2Fo – Fc density contoured at 1.2σ on the final coordinates of the native complex and (B) Dbs(Y889F)·Cdc42 in the vicinity of Tyr889. For illustrative purposes, the wild-type coordinates including Tyr889 were refined using the Dbs(Y889F)·Cdc42 data. Fo – Fc difference density (green) contoured at –6.0σ surrounds the position of the inappropriate tyrosine hydroxyl oxygen atom.
Discussion
Dbs as a model for PH domain-assisted exchange
Here we present structural and biochemical evidence demonstrating a critical role for the PH domain of Dbs in assisting effective guanine nucleotide exchange. The Dbs·Cdc42 crystal structure highlights previously unappreciated interactions between the PH domain of Dbs and switch 2 and α3b of Cdc42. Most importantly, Tyr889 within the PH domain is essential for productive exchange, and presumably is required to stabilize the structural and electronic conformation of His814 within the DH domain for high-affinity interaction with Asp65 of Cdc42. Similarly, Gln834 interacts indirectly with Asp65 through a water-mediated hydrogen bond. Interestingly, even though His103 and Arg66 of Cdc42 interact directly with the PH domain, and Arg66 buries considerable solvent-accessible surface area upon complex formation, neither residue appears critical for exchange in our in vitro conditions.
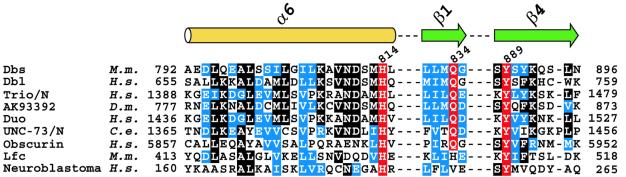
Primary sequence conservation indicates that a subset of Dbl-family members is likely to use their PH domains to support nucleotide exchange (Figure 7). Residues analogous to His814, Gln834 and Tyr889 of Dbs are highly conserved among this group, suggesting that their PH domains adopt a Dbs-like interface with their associated DH domains and cognate Rho GTPases. These observations may explain the requirements for the PH domains of Trio and Dbl for efficient exchange of Rac1 and Cdc42, respectively (Hart et al., 1994; Liu et al., 1998). Further structural and biochemical analyses are required to test these predictions.
Fig. 7. Sequence alignment of Dbs-like Dbl-family members. A multiple sequence alignment of non-redundant Dbl-family members was used to identify sequences containing identity to residues in Dbs important for PH domain enhancement of nucleotide exchange. Only the regions corresponding to α6 of the DH domain and β1 and β4 of the PH domain of Dbs are shown. Identical residues are shown boxed in black; similar conserved residues are boxed blue. Highlighted in red are the highly conserved residues corresponding to His814, Gln834 and Tyr889 of Dbs. Trio/N and Unc-73/N refer to the N-terminal DH and PH domains within these proteins. M.m., Mus musculus; H.s., Homo sapiens; D.m., Drosophila melanogaster; C.e., Caenorhabditis elegans.
Orientation of Dbs on cell membranes
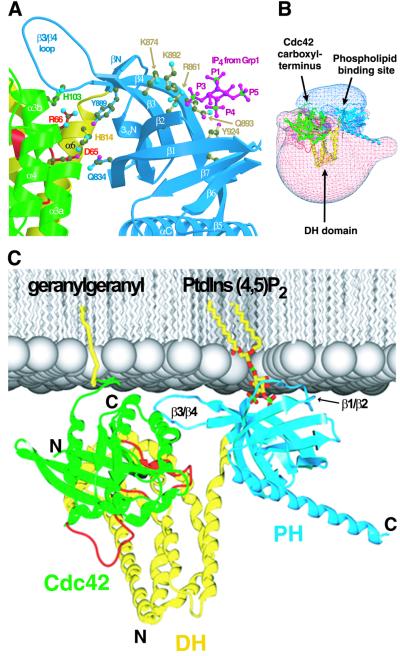
The PH domain of Dbs promiscuously binds phosphoinositides (Snyder et al., 2001) and is required in larger fragments of Dbs for proper membrane localization, cellular signaling and transformation (Whitehead et al., 1995, 1999; Westwick et al., 1998). To identify the putative phospholipid binding site within Dbs, the Dbs PH domain was compared with structures of other PH domains bound with inositol phosphates (Figure 8A) (Ferguson et al., 2000; Lietzke et al., 2000). Residues that appear necessary for binding to lipid head groups include Arg861 (at the base of β2), Lys874 (within β3), Lys892 and Gln893 (within β4), and Tyr924 (within β7). Similar to other PH domains, residues within the β1/β2 loop are also expected to be required for binding phospholipids; however, this region is disordered within Dbs·Cdc42.
Fig. 8. The structure of Dbs·Cdc42 suggests a model for membrane engagement. (A) The structures of the Grp1 and Dapp1 PH domains bound to Ins(1,3,4,5)P4 (RCSB accession Nos 1FGY and 1FAO, respectively) were superimposed upon the Dbs·Cdc42 crystal structure coordinates to identify residues within the Dbs PH domain likely to participate in phospholipid binding. Shown are the Ins(1,3,4,5)P4 (magenta and green) from the Grp1 structure and a subset of Dbs residues (Arg861, Lys874, Lys892, Gln893 and Tyr924) (light brown) at positions common to Dapp1 or Grp1 that are utilized for inositol phosphate recognition. (B) The electrostatic potential (contoured from –2 kT to +2 kT) for the Dbs·Cdc42 complex. Regions covered by blue and red mesh indicate positive and negative electrostatic potential, respectively. (C) The probable orientation of Dbs·Cdc42 (colored as in Figure 1) at negatively charged membranes (silver) as suggested by the highly polarized electrostatic potential, and known sites of membrane engagement by Cdc42 (geranylgeranyl) and PH domains [phosphatidylinositol (4,5) bisphosphate].
Dbl-family proteins may engage Rho GTPases at lipid membranes, and the structure of Dbs·Cdc42 provides an understanding of how this may occur. In particular, the putative phospholipid binding site (Figure 8A), the dramatically bipolar electrostatic potential of the complex (Figure 8B) and the site of geranylgeranylation required to tether Cdc42 to membranes (Ghomashchi et al., 1995; Johnson, 1999) will strictly limit orientations of the Dbs·Cdc42 complex relative to cell membranes (Figure 8C). This mode of membrane engagement is probably important for proper function in vivo and conserved for other Dbl-family proteins. For example, the locations of the C-terminus of Rac1 and the loops of the PH domain of Tiam1 thought to bind phospholipids are consistent with the mode of binding shown in Figure 8C.
The interface between Cdc42 and the Dbs PH domain is juxtaposed to the lipid binding site, suggesting the potential for allosteric regulation of guanine nucleotide exchange through lipid binding to the PH domain (Figure 8A). Unfortunately, we have been unable to detect any modulation of exchange activity for several Dbl-family proteins, including Dbs, by various phospholipids in vitro (Snyder et al., 2001). Allosteric regulation of Dbs-catalyzed nucleotide exchange by phospholipids could occur under more physiological conditions, i.e. at cellular membranes containing lipid-modified GTPases. However, no direct evidence has been reported for such allosterism operating within any Dbl-family member in vivo. Nevertheless, it is reasonable to expect coordinated activation of Rho GTPases to rely on the timely co-localization of GEF, GTPase and relevant phospholipid. The molecular details regarding phospholipid regulation of Dbl-family proteins await future scrutiny.
In conclusion, the invariant association of PH domains with DH domains strongly suggests a critical, interactive role for both domains in activating Rho GTPases. The Dbs·Cdc42 structure, accompanying mutational analysis and previous cell-based functional studies demonstrate that the PH domain of Dbs will not only serve to direct subcellular localization, but will also participate in binding Rho GTPases to facilitate guanine nucleotide exchange. Finally, comparative sequence analysis suggests that a subset of Dbl-family proteins will operate similarly.
Materials and methods
Protein expression and purification
PCR-amplified murine Dbs (residues 623–967) spanning the DH and PH domains was ligated into pET-28a (Novagen) between NcoI and XhoI for expression in E.coli. Seleno-methionine (SeMet)-incorporated Dbs DH/PH domain, containing an additional N-terminal methionine, an unintentional E940A substitution and a Glu(His6) C-terminal tag were prepared by expressing pET-28/Dbs in the methionine auxotrophic E.coli strain B834(DE3). Cell cultures were grown at 37°C in l-SeMet-enriched media (Doublie, 1997) with kanamycin (50 µg/ml), and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 25°C. Cell pellets were resuspended in 50 mM NaH2PO4 pH 8.0, 300 mM NaCl (buffer A) and 5 mM imidazole, then lysed using a French press (Aminco) operating at 15 000 p.s.i. Lysates were clarified by centrifugation at 40 000 g for 25 min at 4°C. Clarified supernatant was loaded onto a nickel-charged metal chelating column (Pharmacia) equilibrated in buffer A containing 5 mM imidazole. Protein was then washed with buffer A containing 20 mM imidazole and eluted with 300 mM imidazole. Protein was next loaded onto an S-200 size exclusion column (Pharmacia) equilibrated in 50 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol (DTT) and 5% glycerol (buffer B). Fractions containing purified Dbs DH/PH domain were pooled, concentrated and stored at –80°C.
A pET21 (Novagen) bacterial expression plasmid encoding a C-terminal truncation mutant of human placental Cdc42 (residues 1–188, C188S) was constructed and transformed into the E.coli strain BL21(DE3). Cell cultures were grown in LB/ampicillin (50 µg/ml) at 37°C before inducing protein expression with 1 mM IPTG at 30°C. Cell pellets were resuspended in 10 mM Tris pH 8.0, 1 mM MgCl2, 1 mM DTT, 10 µM GDP and 5% glycerol (buffer C), lysed, then clarified by centrifugation at 40 000 g for 25 min at 4°C. Lysate was then loaded onto a Fast Flow Q (Pharmacia) column equilibrated in buffer C and eluted with a linear gradient of 0–500 mM NaCl. Cdc42 was next injected onto an S-200 size exclusion column (Pharmacia) equilibrated in buffer C containing 150 mM NaCl. Cdc42 was exchanged into buffer C, passed over a Source Q (Pharmacia) column equilibrated in buffer C and eluted with a linear gradient of 0–300 mM NaCl.
To prepare Dbs bound to Cdc42, SeMet-incorporated Dbs DH/PH was incubated with a 2-fold molar excess of Cdc42 and buffer exchanged into buffer D containing 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 5 mM DTT and 5% glycerol. The protein solution was loaded onto a Superdex 75 (Pharmacia) size-exclusion column equilibrated in buffer D. Two resolved peaks were eluted from the column, with the leading peak composed exclusively of the Dbs·Cdc42 complex and a trailing peak containing unbound Cdc42. Fractions containing the purified SeMet Dbs·Cdc42 complex were pooled, buffer exchanged into a buffer containing 10 mM Tris pH 8.0, 25 mM NaCl, 2 mM DTT and 1 mM EDTA, and stored at –80°C.
Crystallization of the Dbs·Cdc42 complex
SeMet-substituted Dbs·Cdc42 was crystallized using vapor diffusion. Drops were formed by combining equal volumes of protein solution (∼14 mg/ml in 10 mM Tris pH 8.0, 25 mM NaCl, 2 mM DTT and 1 mM EDTA) and reservoir solution [50 mM Tris pH 7.0, 12% PEG 8K (Fluka), 650 mM sodium formate, 2 mM DTT], then equilibrated against 1 ml of reservoir solution at 18°C. Crystals typically appeared after 1 day and grew to final dimensions of 0.2 × 0.4 × 0.5 mm after 1 week. Preparation of SeMet Dbs·Cdc42 crystals for data collection at 100 K involved a stepwise adjustment of a drop containing crystals from 0 to 16% in glycerol using 2% increments, followed by an overnight incubation against a well solution containing 50 mM Tris pH 7.0, 12% PEG 8000, 650 mM sodium formate, 2 mM DTT and 25% glycerol. Cryoprotected crystals were suspended in a rayon loop (Hampton Research) and snap frozen by immersion into liquid nitrogen. Crystals belong to space group P212121 with unit cell parameters a = 68.2 Å, b = 92.6 Å, c = 232.2 Å.
Data collection and structure determination
A two-wavelength anomalous dispersion data set was collected using a single frozen crystal at beamline 9-2 at the Stanford Synchrotron Radiation Laboratory. Data were collected at energies corresponding to the minimum of dispersive differences (λ1 = 12 687.14 eV), and the maximum of anomalous differences (λ2 = 12 695.69 eV). Data were integrated and scaled using DENZO and Scalepack (Otwinowski, 1991). Initially, 16 selenium atoms were located using λ2 anomalous differences and the Patterson peak-searching program SHELXS (Sheldrick, 1986). Phases were calculated using the anomalous differences in both wavelengths and the dispersive differences between λ2 and λ1 using MLPHARE (1991) from the CCP4 suite of programs (CCP4, 1994). Resulting phases and λ2 anomalous differences were used to locate the remaining four selenium atoms prior to phase improvement with solvent flattening, histogram matching and non-crystallographic symmetry (NCS) averaging using DM (Cowtan, 1994).
Model building and refinement
The interactive graphics program O (Jones et al., 1991) and electron density maps calculated using λ1 amplitudes and DM (solvent-flattened, histogram-matched) phases were used for model building. Automated structure refinement utilized CNS (Brünger et al., 1998) with bulk solvent correction. Initial rounds of refinement employed NCS restraints defined for regions of the model in similar packing environments. In later stages of refinement, the NCS restraints were removed. The final model contains 8456 atoms (8280 protein atoms, 323 solvent atoms) with Rcryst = 19.7% (Rfree = 23.9%), and has 90.6% of all residues in the core region of the Ramachandran plot with no residues in disallowed regions. The mean temperature factor for all atoms is 41.8 Å2 (the Wilson temperature factor for data in the range 2.90 Å > d > 2.46 Å = 37.5 Å2).
Structure determination of Dbs(Y889F)·Cdc42
Crystals of Dbs(Y889F)·Cdc42 were grown and cryoprotected as described for the native complex. Data were collected in-house on a single crystal using a Rigaku RUH3R generator and an R-Axis IV++ area detector, and reduced as described for wild type. Crystals belong to the space group P212121 with unit cell parameters a = 67.32, b = 88.19, c = 232.78. For structural determination, the native coordinates truncated to the Cβ atom at position 889 were used as a starting model. The model was refined using alternating cycles of positional and temperature factor refinement.
Guanine nucleotide exchange assays
C-terminal His6-tagged, wild-type Dbs DH domain (residues 623–832) and DH/PH domain (residues 623–967) expression constructs were prepared as above by isolating the appropriate cDNA fragments by PCR amplification from a mouse brain cDNA library. Mutations were introduced into wild-type Dbs DH/PH domain (H814A, K885A, Y889F) and Cdc42 (R66A and H103A) using the Quikchange site-directed mutagenesis kit (Stratagene) as per the manufacturer’s instructions. cDNA sequences of all protein expression constructs were verified by automated sequencing. The protein mass of Cdc42(R66A) was determined on a Bruker Reflex III MALDI-TOF mass spectrometer to further verify the mutation. Dbs DH/PH domain and Cdc42 proteins were prepared from BL21(DE3) cultures grown in LB media and purified as described above. The Dbs DH domain protein was prepared similar to the DH/PH domain protein. Full-length RhoA was expressed and purified similar to Cdc42.
Fluorescence spectroscopic analysis of N-methylanthraniloyl (mant)-GTP incorporation into bacterially purified Cdc42 and RhoA was carried out using a Perkin-Elmer LS 50B spectrometer at 25°C. Exchange reaction assay mixtures containing 20 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT and 100 µM mant-GTP (Biomol), and 1 µM either Cdc42 or RhoA protein were prepared and allowed to equilibrate with continuous stirring. After equilibration, the Dbs DH or DH/PH domain proteins were added at 200 nM, and the rates of nucleotide loading (kobs) of Rho GTPases were determined by monitoring the decrease in Cdc42 or RhoA tryptophan fluorescence (λex = 295 nm, λem = 335 nm) in response to binding mant-GTP (Klebe et al., 1993, 1995; Leonard et al., 1994). The rates (kobs) of guanine nucleotide exchange were determined by fitting the data as single exponential decays utilizing the program GraphPad Prizm. Data were normalized to wild-type curves to yield percent GDP released.
Figures 1B, C, 3, 4 and 8A were generated using MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Murphy, 1994). Figures 6, 8B and C were generated using SPOCK (Christopher, 1998).
Atomic coordinates have been deposited at the Research Collaboratory for Structural Biology (RCSB) under codes 1KZ7 and 1KZG.
Acknowledgments
Acknowledgements
We are grateful to M.Pham and S.Gershburg for technical assistance. We also thank the staff at SSRL and C.P.Hill, M.Mathews and L.Betts for help with data collection. K.L.R. is a recipient of a 2001 Lineberger Graduate Fellow Award; D.K.W. is supported by an American Cancer Society Postdoctoral Fellowship PF-00-163-01-GMC; D.P.S. is a Year 2000 Scholar of The EJLB Foundation (Montréal, Canada) and a recipient of the Burroughs-Wellcome Fund New Investigator Award in the Pharmacological Sciences; J.S. acknowledges support by National Institutes of Health grant GM62299 and the Pew Charitable Trusts.
References
- Aghazadeh B., Zhu,K., Kubiseski,T.J., Liu,G.A., Pawson,T., Zheng,Y. and Rosen,M.K. (1998) Structure and mutagenesis of the Dbl homology domain. Nature Struct. Biol., 5, 1098–1107. [DOI] [PubMed] [Google Scholar]
- Aghazadeh B., Lowry,W.E., Huang,X.Y. and Rosen,M.K. (2000) Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell, 102, 625–633. [DOI] [PubMed] [Google Scholar]
- Aravind L., Neuwald,A.F. and Ponting,C.P. (1999) Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr. Biol., 9, R195–R197. [DOI] [PubMed] [Google Scholar]
- Bishop A.L. and Hall,A. (2000) Rho GTPases and their effector proteins. Biochem. J., 348, 241–255. [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin P.A., Margarit,S.M., Bar-Sagi,D. and Kuriyan,J. (1998) The structural basis of the activation of Ras by Sos. Nature, 394, 337–343. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cerione R.A. and Zheng,Y. (1996) The Dbl family of oncogenes. Curr. Opin. Cell Biol., 8, 216–222. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Corbalan-Garcia,S. and Bar-Sagi,D. (1997) The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J., 16, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J. and Chardin,P. (1999) GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci., 24, 306–311. [DOI] [PubMed] [Google Scholar]
- Chimini G. and Chavrier,P. (2000) Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nature Cell Biol., 2, E191–E196. [DOI] [PubMed] [Google Scholar]
- Christopher J.A. (1998) SPOCK: the Structural Properties Observation and Calculation Kit. The Center for Macromolecular Design, Texas A&M University, College Station, TX.
- Cowtan K.D. (1994) DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF–EACBM Newsl. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Crompton A.M., Foley,L.H., Wood,A., Roscoe,W., Stokoe,D., McCormick,F., Symons,M. and Bollag,G. (2000) Regulation of Tiam1 nucleotide exchange activity by pleckstrin domain binding ligands. J. Biol. Chem., 275, 25751–25759. [DOI] [PubMed] [Google Scholar]
- Day G.J., Mosteller,R.D. and Broek,D. (1998) Distinct subclasses of small GTPases interact with guanine nucleotide exchange factors in a similar manner. Mol. Cell. Biol., 18, 7444–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Eva A. and Aaronson,S.A. (1985) Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature, 316, 273–275. [DOI] [PubMed] [Google Scholar]
- Evers E.E., Zondag,G.C., Malliri,A., Price,L.S., ten Klooster,J.P., van der Kammen,R.A. and Collard,J.G. (2000) Rho family proteins in cell adhesion and cell migration. Eur. J. Cancer, 36, 1269–1274. [DOI] [PubMed] [Google Scholar]
- Ferguson K.M., Kavran,J.M., Sankaran,V.G., Fournier,E., Isakoff,S.J., Skolnik,E.Y. and Lemmon,M.A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell, 6, 373–384. [DOI] [PubMed] [Google Scholar]
- Freshney N.W., Goonesekera,S.D. and Feig,L.A. (1997) Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett., 407, 111–116. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I., Fenoglio,J., Li,Y., Lewis,C., Panchenko,M.P., Reiner,O., Smith,T.F. and Neer,E.J. (1996) Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein β subunit. Biochemistry, 35, 13985–13994. [DOI] [PubMed] [Google Scholar]
- Ghomashchi F., Zhang,X., Liu,L. and Gelb,M.H. (1995) Binding of prenylated and polybasic peptides to membranes: affinities and intervesicle exchange. Biochemistry, 34, 11910–11918. [DOI] [PubMed] [Google Scholar]
- Glaven J.A., Whitehead,I.P., Nomanbhoy,T., Kay,R. and Cerione,R.A. (1996) Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J. Biol. Chem., 271, 27374–27381. [DOI] [PubMed] [Google Scholar]
- Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science, 279, 509–514. [DOI] [PubMed] [Google Scholar]
- Han J. et al. (1998) Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science, 279, 558–560. [DOI] [PubMed] [Google Scholar]
- Hart M.J., Eva,A., Zangrilli,D., Aaronson,S.A., Evans,T., Cerione,R.A. and Zheng,Y. (1994) Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J. Biol. Chem., 269, 62–65. [PubMed] [Google Scholar]
- Horii Y., Beeler,J.F., Sakaguchi,K., Tachibana,M. and Miki,T. (1994) A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J., 13, 4776–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.I. (1999) Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev., 63, 54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K., Kuroda,S. and Amano,M. (1999) Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem., 68, 459–486. [DOI] [PubMed] [Google Scholar]
- Karnoub A.E., Worthylake,D.K., Rossman,K.L., Pruitt,W.M., Campbell,S.L., Sondek,J. and Der,C.J. (2001) Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nature Struct. Biol., 8, 1037–1041. [DOI] [PubMed] [Google Scholar]
- Kawashima T., Berthet-Colominas,C., Wulff,M., Cusack,S. and Leberman,R. (1996) The structure of the Escherichia coli EF-Tu·EF-Ts complex at 2.5 Å resolution. Nature, 379, 511–518. [DOI] [PubMed] [Google Scholar]
- Klebe C., Nishimoto,T. and Wittinghofer,F. (1993) Functional expression in Escherichia coli of the mitotic regulator proteins p24ran and p45rcc1 and fluorescence measurements of their interaction. Biochemistry, 32, 11923–11928. [DOI] [PubMed] [Google Scholar]
- Klebe C., Bischoff,F.R., Ponstingl,H. and Wittinghofer,A. (1995) Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry, 34, 639–647. [DOI] [PubMed] [Google Scholar]
- Koshiba S., Kigawa,T., Kim,J.H., Shirouzu,M., Bowtell,D. and Yokoyama,S. (1997) The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1). J. Mol. Biol., 269, 579–591. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lemmon M.A. and Ferguson,K.M. (1998) Pleckstrin homology domains. Curr. Top. Microbiol. Immunol., 228, 39–74. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., Ferguson,K.M. and Schlessinger,J. (1996) PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell, 85, 621–624. [DOI] [PubMed] [Google Scholar]
- Leonard D.A., Evans,T., Hart,M., Cerione,R.A. and Manor,D. (1994) Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using fluorescence spectroscopy. Biochemistry, 33, 12323–12328. [DOI] [PubMed] [Google Scholar]
- Lietzke S.E., Bose,S., Cronin,T., Klarlund,J., Chawla,A., Czech,M.P. and Lambright,D.G. (2000) Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell, 6, 385–394. [DOI] [PubMed] [Google Scholar]
- Liu X. et al. (1998) NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell, 95, 269–277. [DOI] [PubMed] [Google Scholar]
- Mackay D.J. and Hall,A. (1998) Rho GTPases. J. Biol. Chem., 273, 20685–20688. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (ed.) (1991) Maximum Likelihood Refinement of Heavy Atom Parameters. Daresbury Laboratory, Warrington, UK.
- Pai E.F., Krengel,U., Petsko,G.A., Goody,R.S., Kabsch,W. and Wittinghofer,A. (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J., 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Kuhlmann,J., Henkel,A. and Wittinghofer,A. (2001) Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell, 105, 245–255. [DOI] [PubMed] [Google Scholar]
- Ron D., Zannini,M., Lewis,M., Wickner,R.B., Hunt,L.T., Graziani,G., Tronick,S.R., Aaronson,S.A. and Eva,A. (1991) A region of Proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24 and the human breakpoint cluster gene, bcr. New Biol., 3, 372–379. [PubMed] [Google Scholar]
- Rossman K.L. and Campbell,S.L. (2000) Bacterial expressed DH and DH/PH domains. Methods Enzymol., 325, 25–38. [DOI] [PubMed] [Google Scholar]
- Russo C., Gao,Y., Mancini,P., Vanni,C., Porotto,M., Falasca,M., Torrisi,M.R., Zheng,Y. and Eva,A. (2001) Modulation of oncogenic DBL activity by phosphoinositol phosphate binding to pleckstrin homology domain. J. Biol. Chem., 276, 19524–19531. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G.M. (1986) SHELXS86—Program for Crystal Structural Solution. University of Göttingen, Göttingen, Germany.
- Snyder J.T., Rossman,K.L., Baumeister,M.A., Pruitt,W.M., Siderovski,D.P., Der,C.J., Lemmon,M.A. and Sondek,J. (2001) Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J. Biol. Chem., 276, 45868–45875. [DOI] [PubMed] [Google Scholar]
- Soisson S.M., Nimnual,A.S., Uy,M., Bar-Sagi,D. and Kuriyan,J. (1998) Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell, 95, 259–268. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm,A., Lambright,D.G., Hamm,H.E. and Sigler,P.B. (1996) Crystal structure of a G-protein βγ dimer at 2.1 Å resolution. Nature, 379, 369–374. [DOI] [PubMed] [Google Scholar]
- Sterpetti P., Hack,A.A., Bashar,M.P., Park,B., Cheng,S.D., Knoll,J.H., Urano,T., Feig,L.A. and Toksoz,D. (1999) Activation of the Lbc Rho exchange factor proto-oncogene by truncation of an extended C terminus that regulates transformation and targeting. Mol. Cell. Biol., 19, 1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L. and D’Souza-Schorey,C. (1997) Rho GTPases and signaling networks. Genes Dev., 11, 2295–2322. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang,Y., Meyering-Voss,M., Sprinzl,M. and Sigler,P.B. (1997) Crystal structure of the EF-Tu·EF-Ts complex from Thermus thermophilus. Nature Struct. Biol., 4, 650–656. [DOI] [PubMed] [Google Scholar]
- Westwick J.K., Lee,R.J., Lambert,Q.T., Symons,M., Pestell,R.G., Der,C.J. and Whitehead,I.P. (1998) Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J. Biol. Chem., 273, 16739–16747. [DOI] [PubMed] [Google Scholar]
- Whitehead I.P., Kirk,H. and Kay,R. (1995) Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene, 10, 713–721. [PubMed] [Google Scholar]
- Whitehead I.P., Khosravi-Far,R., Kirk,H., Trigo-Gonzalez,G., Der,C.J. and Kay,R. (1996) Expression cloning of lsc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J. Biol. Chem., 271, 18643–18650. [DOI] [PubMed] [Google Scholar]
- Whitehead I.P., Campbell,S., Rossman,K.L. and Der,C.J. (1997) Dbl family proteins. Biochim. Biophys. Acta, 1332, F1–F23. [DOI] [PubMed] [Google Scholar]
- Whitehead I.P. et al. (1999) Dependence of dbl and dbs transformation on MEK and NF-κB activation. Mol. Cell. Biol., 19, 7759–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake D.K., Rossman,K.L. and Sondek,J. (2000) Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature, 408, 682–688. [DOI] [PubMed] [Google Scholar]
- Zheng J., Chen,R.H., Corblan-Garcia,S., Cahill,S.M., Bar-Sagi,D. and Cowburn,D. (1997) The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains. J. Biol. Chem., 272, 30340–30344. [DOI] [PubMed] [Google Scholar]