Abstract
Intermediate filaments (IFs) are key components of the cytoskeleton in higher eukaryotic cells. The elementary IF ‘building block’ is an elongated coiled-coil dimer consisting of four consecutive α-helical segments. The segments 1A and 2B include highly conserved sequences and are critically involved in IF assembly. Based on the crystal structures of three human vimentin fragments at 1.4–2.3 Å resolution (PDB entries 1gk4, 1gk6 and 1gk7), we have established the molecular organization of these two segments. The fragment corresponding to segment 1A forms a single, amphipatic α-helix, which is compatible with a coiled-coil geometry. While this segment might yield a coiled coil within an isolated dimer, monomeric 1A helices are likely to play a role in specific dimer–dimer interactions during IF assembly. The 2B segment reveals a double-stranded coiled coil, which unwinds near residue Phe351 to accommodate a ‘stutter’. A fragment containing the last seven heptads of 2B interferes heavily with IF assembly and also transforms mature vimentin filaments into a new kind of structure. These results provide the first insight into the architecture and functioning of IFs at the atomic level.
Keywords: atomic structure/coiled coil/intermediate filaments/vimentin/X-ray crystallography
Introduction
The cytoskeleton of higher eukaryotic cells contains three distinct types of filaments: microtubules, intermediate filaments (IFs) and microfilaments (Schliwa, 1986; Aebi et al., 1988; Fuchs and Weber, 1994; Herrmann and Aebi, 2000). The integrated network formed by these three filament systems together with various associated proteins is responsible for the mechanical integrity of the cell and is critically involved in such processes as cell division, motility and plasticity. While capable of self-assembly in vitro, the naturally occurring 10 nm wide IFs are dynamic structures that interact with other cytoskeletal components, in particular with motor proteins and plakin-type cross-bridging proteins (Herrmann and Aebi, 2000; Fuchs and Karakesisoglou, 2001). The crystal structures of tubulin and actin, the molecular ‘building blocks’ of microtubules and microfilaments, respectively, have been determined, and atomic models of the respective filaments have been built (Steinmetz et al., 1998; Gigant et al., 2000). However, until now, no atomic resolution structures of any IF component have been determined.
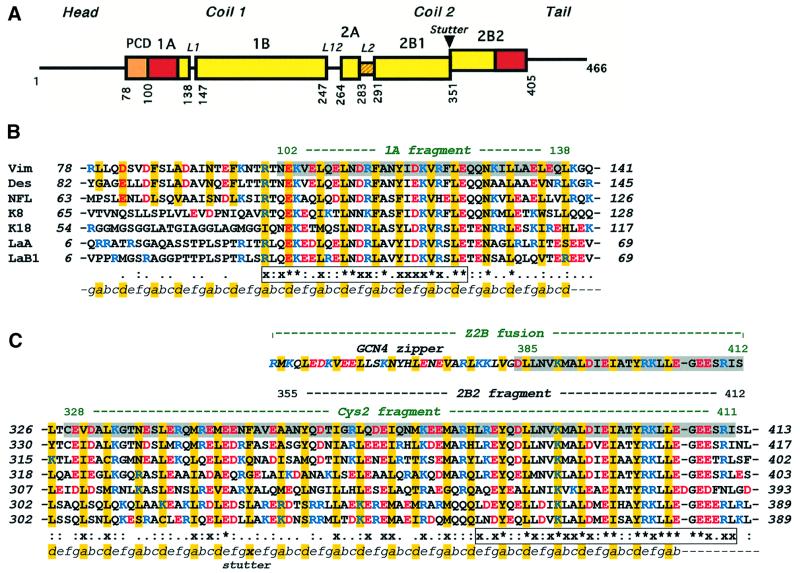
The multigene family of IF proteins includes >60 members, which are grouped into four major sequence homology classes representing cytoplasmic IF proteins and a separate class representing the nuclear lamins (Conway and Parry, 1988; Herrmann and Aebi, 1998a). The amino acid sequences of all IF proteins share a characteristic tripartite structure (Figure 1A) that includes the central highly α-helical ‘rod’ domain flanked by the ‘head’ and ‘tail’ domains at both ends (Geisler and Weber, 1982; Fuchs and Weber, 1994). The rod domain reveals a heptad repeat pattern, which is a signature of a coiled-coil (CC) fold (see, for example, Lupas, 1996). Correspondingly, the elementary ‘building block’ underlying the IF architecture is an elongated, parallel CC dimer. The heptad periodicity within the rod domain is, however, interrupted in several places, resulting in four consecutive α-helical segments 1A, 1B, 2A and 2B that are connected by short linkers L1, L12 and L2. In particular, the rod domain of all vertebrate cytoplasmic IF proteins contains close to 310 residues, and the sizes of the individual α-helical segments are absolutely conserved (Fuchs and Weber, 1994; Parry and Steinert, 1999). In addition, both nuclear lamins and invertebrate IF proteins contain an insertion of six extra heptads in the segment 1B.
Fig. 1. (A) Primary structure of IF proteins. Schematic diagram of human vimentin. Rectangles show α-helical segments, including the pre-coil domain (PCD). (B) Sequence alignment of the 1A segments of human IF proteins including vimentin, desmin, neurofilament L protein, cytokeratins 8 and 18, and nuclear lamins A and B1. (C) Similar alignment of the 2B segments. Vimentin fragments 1A, Cys2, Z2B and 2B2 are highlighted. The heptad repeats are marked as abcdefg, with core positions highlighted with yellow. Basic and acidic residues are shown in blue and red, respectively. The line below the alignment shows the sequence similarity score s of a particular residue in the seven proteins: ‘*’, s = 1.0 (absolutely conserved); ‘x’, 0.75≤s<1.0; ‘:’, 0.5≤s<0.75; ‘.’, 0.25≤s<0.5 (see Materials and methods for details). The two most conserved regions within the 1A segment and in the C-terminal part of the 2B segment, respectively, are shown in boxes.
IF assembly begins with a gradual association of the dimers, governed by several distinct modes of lateral interaction (Steinert et al., 1993; Herrmann and Aebi, 1998a). This process first leads to the formation of tetramers (Herrmann and Aebi, 1999) and ultimately yields so-called unit-length filaments (ULFs). In particular, the vimentin ULFs appear to contain about sixteen ∼46 nm long dimers (Herrmann et al., 1996). Subsequently, the ULFs anneal longitudinally into rather loosely packed filaments. Finally, the extended filaments undergo an internal rearrangement of subunits which manifests itself by a radial compaction of the filament (Herrmann and Aebi, 1998b, 1999). Notably, the mature filament structure includes a short head–tail overlap between consecutive dimers. The resulting IFs appear to have no polarity, as the individual dimers are oriented along its axis in either direction. Finally, the head and tail domains play a critical role in the filament assembly process, despite the fact that they vary considerably in sequence and length among different IF proteins (Heins and Aebi, 1994; Herrmann et al., 1996).
The sequence conservation across different IF proteins is especially pronounced within two regions that are located at the opposite ends of the CC rod (Figure 1B and C). The first conserved region spans 26 residues corresponding to about two-thirds of the α-helical segment 1A, and involves eight residues which are absolutely conserved within the sequence comparison presented in Figure 1. The second conserved region is situated at the very end of the 2B segment. This region spans 32 residues, of which 13 are absolutely conserved, and harbors the so-called IF ‘consensus’ motif YRKLLEGEE (Herrmann et al., 2000). Chemical cross-linking studies have revealed that both of these highly conserved regions are critically involved in various dimer–dimer interactions within the mature filament (Steinert et al., 1993; Wu et al., 2000). The significance of the two conserved regions is supported further by the fact that they harbor a number of phenotypically pronounced point mutations (Parry and Steinert, 1999). These include, in particular, hereditary defects in keratins which were linked to various skin blistering diseases in humans, such as epider molysis bullosa simplex (for reviews see Fuchs and Coulombe, 1992; Fuchs and Weber, 1994; Parry and Steinert, 1999).
In addition, the 2B segment contains a discontinuity in the heptad repeat pattern, a so-called ‘stutter’, which is equivalent to an insertion of four extra residues at the end of a heptad (Brown et al., 1996; Lupas, 1996). The stutter appears to be an obligatory feature in all IF proteins (Weber and Geisler, 1985), and its position at the end of the eighth full heptad of 2B is absolutely conserved despite a high sequence variability in this region (Figure 1C).
The major obstacle preventing high-resolution X-ray studies of IFs has always been the difficulty in growing suitable three-dimensional crystals. The breakthrough came after we suggested that, instead of crystallizing the full-length IF protein, one should use multiple overlapping fragments thereof. Following this ‘divide-and-conquer’ strategy, we recently have obtained X-ray quality crystals from six fragments of human vimentin (Herrmann et al., 2000; Strelkov et al., 2001). Here we present the atomic structures of three such fragments as well as accompanying biochemical experiments on IF assembly. Taken together, these data reveal the molecular architecture and function of the highly conserved α-helical segments 1A and 2B of the IF dimer.
Results
Crystallographic structure determination
We have obtained crystal structures of three fragments of human vimentin, named 1A, Z2B and Cys2, respectively (Figure 1B and C). The design and crystallization of these fragments were described previously (Strelkov et al., 2001). The 1A fragment contains residues 102–138 corresponding to the first α-helical segment of the vimentin rod. Z2B is a chimeric peptide which includes vimentin residues 385–412, i.e. the highly conserved region of the 2B segment, fused at the N-terminus to the 31 residue leucine zipper domain from the yeast transcription activator GCN4 (O’Shea et al., 1991). This fusion was designed to provide for formation of a proper double-stranded CC within the relatively short vimentin sequence (Herrmann et al., 2000). Finally, the Cys2 fragment (residues 328–411) includes the major part of the α-helical segment 2B starting with residue Cys328. It therefore contains the vimentin part of the Z2B chimera within itself.
X-ray diffraction data for all three vimentin fragments were collected at resolutions between 1.4 and 2.3 Å (Table I). The 1A and Z2B crystals contain a monomeric α-helix and a double-stranded CC, respectively, per asymmetric unit (Figures 2A and 3A). Both structures were phased by molecular replacement (MR) using models derived from the GCN4 leucine zipper. While MR is generally known to be difficult for ‘rod-like’ molecules (Turkenburg and Dodson, 1996), the case of the 1A fragment was complicated further by the fact that an α-helical structure repeats itself after a screw rotation about the helix axis. Since a CC possesses a similar property, the MR searches for the Z2B construct were also rather elaborate despite the presence of the exact GCN4 leucine zipper sequence within the molecule. The diffraction data for the Cys2 fragment could only be phased by multiple isomorphous replacement with anomalous scattering (MIRAS) using three heavy atom derivatives. The Cys2 crystals contain three independent, ∼120 Å long CC dimers per asymmetric unit, denoted AB, CD and EF, respectively. The dimers are aligned approximately along the longest unit cell axis c (Figure 3B). All three crystal structures were refined to good crystallographic R-factors while securing quality stereochemical parameters (Table I).
Table I. Crystallographic data.
| Fragment name | 1A | Z2B | Cys2 |
|---|---|---|---|
| Vimentin residues included | 102–138 | 385–412 | 328–411 |
| No. of residues per chaina |
39 |
59 |
84 |
| Diffraction data |
|
|
|
| Space group | P6222 | P3121 | I222 |
| Cell constants a × b × c (Å) | 56.8 × 56.8 × 58.4 | 98.8 × 98.8 × 36.5 | 76.6 × 84.3 × 240.8 |
| Resolution limitsb,c (Å) | 50.0–1.4 (1.45–1.40) | 35.0–1.9 (1.97–1.90) | 35.0–2.3 (2.33–2.30) |
| No. of independent reflections | 11 440 (1122) | 15 422 (1604) | 34 937 (1159) |
| Redundancy | 10.3 (5.5) | 4.0 (3.7) | 4.5 (4.0) |
| Completeness (%) | 99.7 (99.6) | 99.2 (99.4) | 99.5 (99.7) |
| <I/σ> | 17.0 (1.9) | 14.6 (3.3) | 13.5 (1.8) |
|
Rsymd |
0.037 (0.560) |
0.063 (0.372) |
0.058 (0.405) |
| Refined model |
|
|
|
| Protein chains/asymmetric unit | 1 | 2 | 6 |
| Ordered vimentin residues in each chain | A: 102–138 | A: 385–409 | A: 328–406 |
| B: 385–406 | B: 328–406 | ||
| C: 337–406 | |||
| D: 330–407 | |||
| E: 337–406 | |||
| F: 333–406 | |||
| No. of solvent molecules | 35 | 194 | 455 |
| Total no. of non-H atoms | 375 | 1066 | 4170 |
| Average model B-factor | 23.5 | 31.5 | 53.8 |
| Rworke | 0.197 | 0.199 | 0.242 |
| Rfreee,f | 0.216 (461) | 0.227 (778) | 0.262 (1095) |
| R.m.s.d. bondsg (Å) | 0.020 | 0.017 | 0.008 |
| R.m.s.d. anglesg (°) | 2.0 | 1.7 | 1.1 |
aBesides the authentic vimentin residues, the 1A fragment contains two extra residues, GlySer, at the N-terminus (see Strelkov et al., 2001). The Z2B chimera includes the GCN4 leucine zipper.
bData in parentheses are for the highest resolution shell.
cCys2 crystals exhibited anisotropic diffraction, which extended up to 1.9 Å resolution in the c* direction and to 2.3 Å resolution in the a* and b* directions.
eR = ∑|Fobs – Fcalc|/∑Fobs.
fIn parentheses is the number of randomly selected reflections that were excluded from refinement and used to calculate the ‘free’ R-factor (Brünger and Nilges, 1993).
gR.m.s.ds from the Engh and Huber standard parameters.
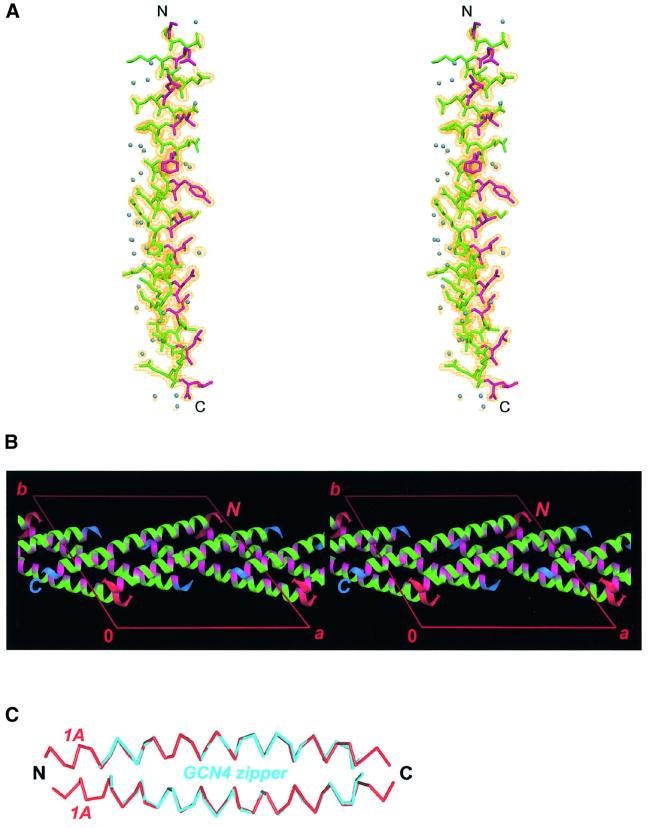
Fig. 2. Crystal structure of the vimentin fragment 1A. (A) Stereo view of the atomic model and electron density map with coefficients 2Fobs – Fcalc contoured at 1.2σ. Residues in the a and d positions of the putative heptad repeat are shown in magenta. Solvent molecules are shown as blue spheres. (B) The crystal packing arrangement of 1A shown in stereo. The a and d positions are highlighted with magenta. The N- and C-termini of the helices are marked in red and blue, respectively. (C) Modeling of a parallel coiled-coil by docking two 1A helices (red) while using the GCN4 zipper structure (cyan) as a ruler.
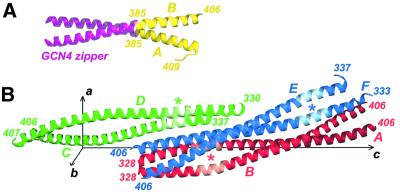
Fig. 3. Crystal structures of the Z2B and Cys2 fragments. (A) Ribbon diagram of the Z2B structure. The GCN4 leucine zipper and the authentic vimentin residues are shown in magenta and yellow, respectively. (B) Ribbon diagrams of the three symmetry-independent Cys2 dimers AB (red), CD (green) and EF (blue) in the unit cell. The position of the stutter in each dimer is marked with an asterisk.
Atomic structure of the 1A segment
We have found previously that the recombinant vimentin fragment 1A is monomeric in solution, while its circular dichroism spectra reveal ∼30% α-helical and 70% random structure (Strelkov et al., 2001). Most strikingly, the 1A crystals also contain a single polypeptide chain per asymmetric unit (Figure 2A). Furthermore, all of its 39 residues are strictly α-helical (φ approximately –65°, ψ approximately –45°). As a consequence of the pronounced heptad repeat pattern, most apolar side chains locate approximately on one side of the relatively short α-helix. Interestingly, the helix is slightly bent (radius of curvature r = 84 Å) in such a way that the hydrophobic patch locates on the concave side of the curvature. Furthermore, the 1A crystal packing arrangement reveals a ‘layered’ pattern (Figure 2B). Each layer is formed by alternating antiparallel 1A helices located in a plane perpendicular to the crystallographic 6-fold axis. The residues in the a and d positions are situated on one side of the layer, rendering this side substantially more hydrophobic than the other. Moreover, the orientations of consecutive layers alternate between ‘up’ and ‘down’. As a result, distinct ‘bilayers’ are observed, with a hydrophobic interface between the two layers within a bilayer. The crossing angle between the axes of helices in the two layers is 45°. Notably, about three-quarters of all ordered solvent molecules are located on the hydrophilic side of the α-helix (see Figure 2A) and thus between the adjacent bilayers.
The crystal packing arrangement of the 1A fragment does not reveal CCs of any kind (parallel or antiparallel, aligned in register or staggered). However, the bending of the 1A helix results in a conformation which is fully compatible with a double-stranded parallel CC geometry. This can be shown by fitting two 1A helices as rigid bodies onto either chain of the GCN4 leucine zipper, with heptad repeats in phase (Figure 2C). After some adjustment of the side chain conformation in the a and d positions, such an artificial dimer would have a perfect CC structure. Furthermore, the crystal structure of the 1A fragment does not reveal any intrahelical salt bridges. However, within the constructed dimer, an interhelical salt bridge between Lys120 (heptad position g) and Glu125 (position e′ of the following heptad) is feasible. Last but not least, these data suggest that, generally speaking, the curvature of an α-helix within a CC is not a consequence of the hydrophobic core formation, as may be thought. Instead, it appears to be an intrinsic feature of an α-helix with most apolar side chains localized on one side. However, this hypothesis requires further experimental confirmation.
Atomic structure of the 2B segment
The crystal structures of the Z2B and Cys2 fragments document that the 2B segment of vimentin forms a continuous double-stranded CC (Figure 3A and B). Importantly, both structures independently indicate that this segment terminates with residue Glu405. While the latter is the last residue in an α-helical conformation, Leu404 in an a position is the last residue involved in the hydrophobic seam between the two helices. The residues past Gly406 appear to be disordered in most of the cases. An important exception is one chain of the Z2B construct which reveals that the residues Glu407, Glu408 and Ser409 fold back onto the CC away from its axis (Figure 3A, see also Figure 4B). At the same time, a least-squares superposition of the three crystallographically independent Cys2 dimers and the Z2B structure reveals some variability in the overall geometry of the CC (Figure 4A) and also considerable differences in the side chain conformation of certain residues (Figure 4B). This points to the inherent conformational flexibility of the CC structure. In addition, as documented in Table I, it turns out that only one of the three Cys2 dimers, AB, includes an ordered CC starting with the N-terminal Cys328 residues of both chains. The two other dimers, CD and EF, reveal a proper CC geometry starting only with residue Leu340 in a d position, while the preceding residues are partially disordered. In particular, in the electron density maps, both chains C and E are only traceable starting with residue Asn337, and their residues 337–339 are clearly folding away from the CC axis (Figures 3B and 4A). While the differences between the three dimers are due to a different crystallographic environment, these data suggest that the CC between residues 328 and 340 is relatively labile.
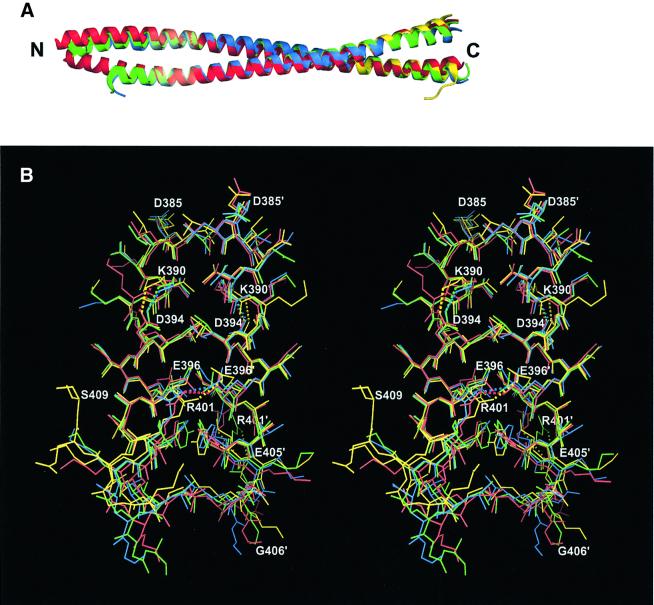
Fig. 4. Superposition of the Cys2 and Z2B structures. (A) Ribbon diagrams of the three Cys2 dimers (AB, red; CD, green; and EF, blue) and the Z2B dimer (yellow). (B) The C-terminal part of the 2B segment shown in stereo (coloring as above). Salt bridges are shown with dotted lines.
Importantly, the Z2B and Cys2 structures reveal a network of intra- and interhelical salt bridges within the highly conserved C-terminal region of the 2B segment (Figure 4B). The first, intrahelical i to i + 4 type salt bridge, links residues Lys390 and Asp394 located in heptad positions a and e, respectively. This salt bridge is present in all eight symmetry-independent polypeptide chains of the two crystal structures. The second, g–e′ type interhelical salt bridge, is formed between the residues Glu396 and Arg401. This salt bridge was found only in three out of the eight cases. In the remaining five cases, the side chains of Glu396 and Arg401, while being visible in the electron density maps, are separated by distances that exclude effective ionic interactions (>4.0 Å). However, in two cases, the Arg401 residue forms an intrahelical i to i + 4 type salt bridge with Glu405 instead (Figure 4B).
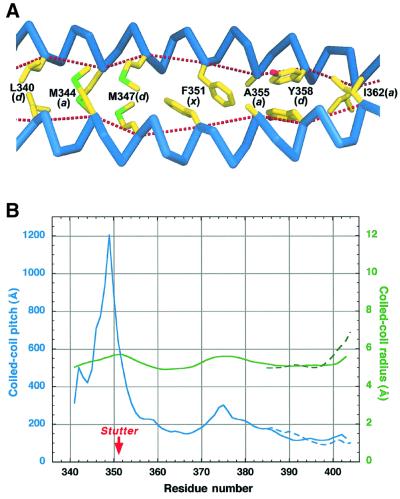
The Cys2 structure documents that the stutter occurring near the vimentin residue 351 can be tolerated without destroying the CC geometry (Figure 5A). Indeed, the two α-helices run continuously through the stutter site, and the main chain hydrogen bonding pattern is fully preserved as well. Interestingly, the two Phe351 rings are arranged asymmetrically with respect to the CC axis, and locate one after the other in the hydrophobic core. This arrangement appears to be possible due to the presence of the small Ala355 residue in the next core position (a). These phenylalanine and alanine residues are highly conserved in vimentins and desmins of vertebrates (Herrmann and Aebi, 1999), but not in more distant IF proteins (Figure 1C).
Fig. 5. Effect of the coiled-coil stutter within the 2B segment. (A) Hydrophobic core organization near the stutter of the Cys2 dimer. The letters in parentheses after the residue number indicate the heptad position. The dotted lines connect the Cα atoms of the consecutive core residues. (B) Coiled-coil radius (green) and pitch (blue) as a function of residue number. The data for the Cys2 structure (averages over three independent dimers) and Z2B structure are shown with solid and dashed lines, respectively.
Geometrical parameters of a CC are often estimated globally by fitting an idealized CC structure to the experimental coordinates (O’Shea et al., 1991; Tao et al., 1997). We have developed a new algorithm to analyze the CC geometry in considerably more detail by directly determining each of these parameters locally as a function of residue number (see Materials and methods). Figure 5B shows the variability of the two principal parameters, namely the superhelix radius and pitch, within the 2B segment. Starting with residue 355, i.e. downstream of the stutter, the 2B segment reveals a fairly even CC geometry with a radius of 5.22 ± 0.24 Å and a pitch of 160.5 ± 43.0 Å (average values over the three Cys2 dimers). For comparison, Tao et al. (1997) have found an average radius of 5.0 Å and an average pitch of 128.5 Å for several typical double-stranded CCs. The stutter, however, results in a sharp local increase of the CC pitch (Figure 5B), which corresponds to the two α-helices becoming nearly parallel (cf. Figure 3B). In addition, there is a slight increase of the CC radius, while both the α-helical radius and pitch remain essentially constant (data not shown). It is therefore the local unwinding of the CC that chiefly compensates for the stutter. This observation is in accordance with earlier theoretical calculations (Brown et al., 1996). Averaged over the three Cys2 dimers, the unwinding corresponds to a 28.3° ‘delay’ of the CC phase, compared with a regular CC with a pitch of 161 Å for the region between residues 341 and 354 (cf. Figure 5B).
Influence of the 2B2 fragment on IF assembly
In addition to the X-ray crystallographic studies, we have examined the influence of a further vimentin construct, 2B2, incorporating residues 355–412, on IF assembly (Figure 1C). Like the Z2B and Cys2 fragments, this construct includes the highly conserved sequence at the C-terminal end of the rod and forms CC dimers in solution (Strelkov et al., 2001). We have evaluated, first, the co-assembly of wild-type human vimentin with the 2B2 fragment and, secondly, the effect of this fragment on the already assembled recombinant human vimentin IFs.
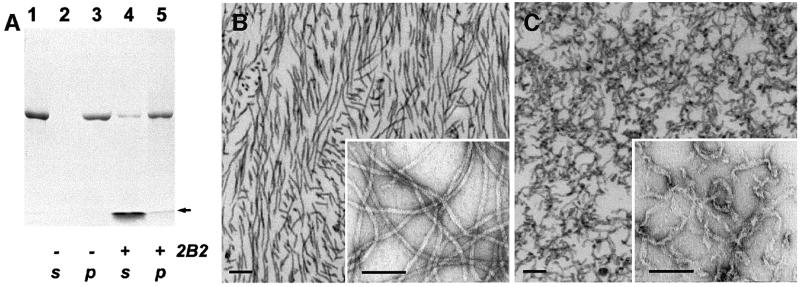
In the absence of 2B2, addition of the ‘filament buffer’ to a solution of vimentin tetramers (see Materials and methods) resulted in the formation of bona fide IFs (Figure 6B). Upon centrifugation, the filaments went entirely into the pelleted fraction (Figure 6A, lanes 2 and 3). When the filament assembly was initiated in the presence of a 10-fold excess of 2B2, most of the wild-type vimentin was again found in the pellet, with only a trace amount remaining in the soluble fraction (Figure 6A, lanes 4 and 5). Remarkably, some of the 2B2 fragment also went into the pellet. Thus some kind of assembly does occur also in the presence of 2B2. However, electron microscopy of the pellets after the co-assembly already at a 1:1 molar ratio revealed flat, short and partially unraveled fibrillar structures (not shown), suggesting that IF elongation was substantially inhibited. Most strikingly, similar aberrant structures could be observed already within several minutes after the 2B2 fragment was added to pre-assembled vimentin IFs, as nearly all IFs were severed and transformed into short, ribbon-like fibers (Figure 6C). Taken together, these results indicate that the 2B2 fragment readily interferes with both vimentin tetramers and mature IFs.
Fig. 6. Effect of the 2B2 fragment on IF assembly. (A) Denaturing gel electrophoresis (SDS–PAGE) illustrating the assembly of human recombinant vimentin alone (lanes 2 and 3) and in the presence of the 2B2 fragment (lanes 4 and 5). The 2B2 fragment was added to the vimentin sample (lane 1) at a 10-fold molar excess, and then filament assembly in the test and reference (i.e. without the fragment addition) samples was performed as described in Materials and methods. The samples subsequently were centrifuged in a Beckman Airfuge for 30 min at 10 p.s.i. yielding the supernatant (lanes 2 and 4) and pelleted (lanes 3 and 5) fractions. The arrow indicates the location of the gel front. (B) Negatively stained EM images of vimentin IFs assembled in vitro. The samples were prepared by ultrathin sectioning (main figure) or on grids (inset). Scale bars are 100 nm. (C) Similarly assembled IFs, which subsequently were incubated with a 10-fold molar excess of the 2B2 fragment for 1 h at 37°C.
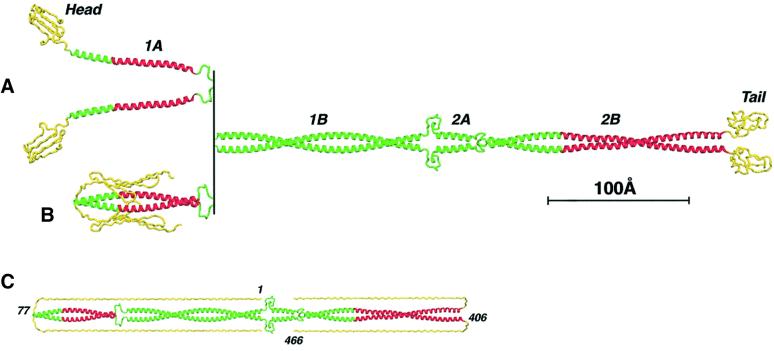
Model of the IF dimer
Taken together, the crystal structures of the 1A, Z2B and Cys2 fragments describe the molecular organization of the α-helical segments 1A and 2B. In addition, preliminary studies of another fragment corresponding to the 1B segment point to a double-stranded CC (A.Lustig and S.V.Strelkov, unpublished). Based on these data accompanied by homologous modeling, we have assembled a three-dimensional model of the vimentin dimer (Figure 7A and B). This model, in particular, illustrates the ‘open’ and ‘closed’ conformations of the 1A segments within the whole molecule (see Discussion for details). Interestingly, a triple-stranded CC of bacteriophage T4 fibritin (Tao et al., 1997) consists of 13 consecutive segments connected by short non-α-helical loops and thus closely resembles the vimentin dimer. In addition, North et al. (1994) have proposed earlier that the conformation of the linker L12 may involve a 180° shift in the CC phase.
Fig. 7. Atomic model of the IF dimer. (A) ‘Open’ conformation of the 1A segments. Regions corresponding to the three described crystal structures are shown in red. (B) ‘Closed’ conformation of the 1A segments. (C) Modeling of the fully extended conformations of the head and tail domains (yellow).
Currently, no high-resolution structural data on the head and tail domains are available. However, secondary structure prediction for the head suggests ∼25% β-structure and 75% random coil, which points to its high conformational flexibility. In addition, folding of the head domain in vimentins and keratins may be based on the stacking of its six aromatic residues, mainly tyrosines (Parry and Steinert, 1995; Herrmann and Aebi, 1998a). Furthermore, it appears likely that, at least under some conditions, the head folds back onto the rod domain (see Parry and Steinert, 1999; see also Discussion). Similarly, the tail domains may also fold back onto the CC, as supported by the observed conformation of residues 406–409 in the Z2B structure. In the extreme case of fully extended conformations (which are unlikely to occur), the N-terminus of the backfolded head domain would reach up to the linker L12, while the tail domain would span the whole length of the 2B segment (Figure 7C).
Discussion
While compatible with a coiled-coil geometry, monomeric 1A helices are likely to play a critical role during IF assembly
Segment 1A of vimentin reveals a distinct heptad repeat pattern, especially in its C-terminal part where nearly all a and d positions are occupied by the preferred residues leucine and valine (Figure 1B). Computer sequence analysis of this segment suggests both high α-helicity and high probability for CC formation. Based on these arguments, the 1A segment previously was believed to form a parallel double-stranded α-helical CC (Parry and Steinert, 1999).
Contrary to these expectations, we have found that the vimentin fragment corresponding to segment 1A of the rod forms a monomeric α-helix in both solution and crystals. This rather surprising result suggests that the two 1A helices may also stay separate in the full-length structure (Figure 7A). In this context, it should be recalled that the head domain is indispensable for IF assembly; indeed, ‘headless’ IF proteins fail to assemble beyond the elementary dimer under standard conditions (Hatzfeld and Burba, 1994; Herrmann et al., 1996). Moreover, the two head domains of an IF dimer are believed to make specific interactions with distinct sites within the rod domain of an adjacent dimer. Such sites may involve the segments 2A, 2B and possibly 1B, depending on the particular mode of the dimer–dimer alignment (see Geisler et al., 1992; Steinert et al., 1993; Herrmann and Aebi, 1999). Furthermore, such specific interactions are likely to be driven by electrostatic attraction, as the head domains of vimentin and other IF proteins are very basic (Parry and Steinert, 1999), whereas each of the four rod segments and also the tail domain are acidic (in particular, segment 1A of human vimentin has a pI of 4.8). Finally, the linker L1 between the 1A and 1B segments is predicted to be highly flexible, based on its amino acid composition (Smith et al., 2002).
We believe that the two monomeric 1A helices may act as mobile ‘connectors’ between the head domains and the remainder of the dimer. Such a device might be necessary to allow the head domains to move into positions required for an effective interaction with the rods of adjacent dimers within the complete IF structure. Interestingly, a vimentin mutant with an insertion of three residues, AlaAlaLeu, into the linker L1, which leads to an uninterrupted heptad periodicity throughout segments 1A and 1B, does not assemble beyond the ULFs (Herrmann et al., 1999). This implies that the formation of a continuous CC within the 1A and 1B segments is preventing the proper longitudinal annealing of the ULFs into extended filaments. On the other hand, the distinctly amphipatic 1A helices should have a strong tendency for intermolecular interactions. In the crystal structure, this tendency is satisfied by the contacts between symmetry-related molecules (Figure 2B). In the complete IFs, however, one would expect the interaction of the 1A helices with the heads or, possibly, distinct regions of the rod domains of adjacent dimers.
Importantly, we have found previously that the vimentin fragment HCC which includes residues 1–138, i.e. the full head and the 1A segment, is predominantly dimeric in solution (Strelkov et al., 2001). We therefore conclude that the dimerization of 1A is facilitated by a cross-wise interaction between the backfolded head domain of one chain and the 1A segment of the other chain, as shown in Figure 7B. Furthermore, the slightly bent conformation of the 1A helix observed in the crystals is fully compatible with a CC structure (Figure 2C). Taken together, these data suggest that, at least within an isolated dimer, the two 1A helices may form a proper CC. At the same time, we believe that this CC should be relatively labile, and its ability to ‘unzip’ is likely to play an essential role during IF assembly and dynamics.
The specific functions of the 2B segment during filament assembly are based on its three-dimensional structure
From the primary sequence, the 2B segment of vimentin was predicted to start with residue Tyr291 and to end with residue Ser412 (Conway and Parry, 1988; Parry and Steinert, 1999). In contrast, our crystallographic data reveal that the CC segment 2B ends with residue Glu405 (Figure 4). The termination of the CC is apparently mediated by the repulsion between the acidic clusters Glu405–Gly406–Glu407–Glu408, so that the two chains fold away from the dimer axis. It should be noted that this acidic cluster corresponds to the second half of the nearly absolutely conserved YRKLLEGEE motif (Herrmann et al., 2000). In addition, based on computer modeling, the eight-residue linker L2 appears to have a defined conformation (North et al., 1994). Correspondingly, the 2.5 heptad repeat 2A segment and the 16 heptad repeat 2B segment are likely to form a common, relatively rigid rod-like structure.
Stutters, along with stammers, skips and other heptad repeat discontinuities, are found in numerous two- and three-stranded CCs, and were shown to be functionally significant in many cases (Brown et al., 1996; Lupas, 1996). The structure of the vimentin Cys2 fragment provides the first example of a stutter within a double-stranded CC analyzed in atomic detail. It reveals several unexpected features, most remarkably the special hydrophobic core organization at the stutter site (Figure 5A). Importantly, we show that the stutter results in a local unwinding of the CC by ∼28°. This means that the difference of the CC phase angles at the opposite ends of the 2B segment is smaller than it would be with a continuous heptad repeat. As pointed out earlier (Brown et al., 1996), the presence of the stutter therefore has a ‘global’ effect on the dimer structure. In particular, the stutter contributes to the relative azimuthal orientation of the functionally relevant groups in the N- and C-terminal parts of the vimentin rod, respectively, and distortion of this optimal orientation may adversely affect the correct intermolecular interactions during the annealing step. Indeed, this was confirmed experimentally by studying a mutant vimentin which has an insertion of three residues, SerAlaThr, at the stutter site and therefore possesses a continuous heptad repeat. This mutant was found to assemble into apparently normal ULFs, but these failed to anneal into stable filaments (Herrmann et al., 1999). Hence we conclude that the stutter within the 2B segment is not only absolutely conserved among all IF proteins, but it also has an important biological function.
Intra- and interhelical salt bridges were shown to be essential for CC formation in a number of proteins. In particular, the salt bridges appear to contribute in a crucial way to the folding initiation (‘trigger’) sites of CCs (Spek et al., 1998; Burkhard et al., 2000, 2001). In cytokeratins, two such sites were localized within the C-terminal parts of segments 1B and 2B, respectively (Wu et al., 2000). The second trigger site lies roughly in the middle of the highly conserved region of the 2B segment (Figure 1C). In vimentin, the corresponding sequence includes a potential i to i + 4 type intrahelical salt bridge between residues Lys390 and Asp394 and a potential g–e′ type interhelical salt bridge between Glu396 and Arg401 (Figure 4B). The intrahelical salt bridge is clearly observable in all eight helices present within the Z2B and Cys2 crystal structures. The interhelical salt bridge, however, is only present in three out of eight potentially possible cases, which may point to the relative weakness of this interaction.
Notably, mutation of the Glu396 residue to aspartate in vimentin has been demonstrated to seriously influence IF assembly in vitro, as abnormal, loose ‘ribbon-like’ filaments were observed (McCormick et al., 1993). Moreover, after the introduction of the Glu396Asp mutant into human vimentin-free cultured cells, only dot-like aggregates and no filaments were detected. To explain these data, we note that the Cα atoms of Glu396 and Arg401 in the two vimentin chains are separated by a distance of 10.5 Å. Correspondingly, an effective ionic interaction requires a rather ‘stretched’ configuration of either side chain (Figure 4B). Moreover, modeling suggests that, after the glutamate is replaced by an even shorter aspartate side chain, the salt bridge is still plausible stereochemically. However, this would require a full extension of both side chains, which is entropically unfavorable.
Last, but not least, we have found that the presence of the 2B2 fragment during the assembly of vimentin IFs drastically alters ULF formation as well as their longitudinal annealing and compaction into mature filaments. Even more strikingly, the 2B2 fragment is highly potent in interacting with pre-assembled IFs, destroying the normal IF architecture. This implies that the highly conserved C-terminal region of the rod is interfering crucially with the proper dimer–dimer interactions within IFs. These data are in excellent agreement with earlier results of Hatzfeld and Weber (1992). These authors found that a synthetic 20-residue peptide corresponding to the highly conserved region of segment 2B interfered with assembly, and disassembled pre-formed keratin IFs. Similarly to the 2B2 fragment, the fragment corresponding to the highly conserved 1A segment was also shown to interfere strongly with the assembly of vimentin IFs, while giving rise to a different phenotype (Goldman et al., 1996).
Outlook
All IF proteins share a common structural organization of the dimer, which is the elementary ‘building block’ of the filament. This implies that the crystallographic detail obtained for vimentin may serve as a paradigm for the molecular organization of IFs in general. Furthermore, our results provide a solid structural basis for the interpretation of the vast amount of data on the IF structure and assembly obtained by various other methods, therefore opening up a perspective of understanding the architecture and functioning of bona fide IFs in atomic detail. In particular, this concerns the IF proteins such as cytokeratins, desmin and glial fibrillary acidic protein (GFAP), mutations in which were linked to disease phenotypes (Fuchs and Coulombe, 1992; Fuchs and Weber, 1994), as well as the nuclear lamins (Cohen et al., 2001).
In addition, we suggest that the application of the X-ray crystallographic technique is not limited to the elementary IF dimer. First, future crystallographic studies will involve homo- and heterotypic complexes formed by various IF fragments (Strelkov et al., 2001). This should provide atomic resolution detail of the particular dimer–dimer interactions and thus drastically extend the knowledge obtained from the cross-linking experiments. Secondly, we recently have discovered several mutations in full-length vimentin that restrict the IF assembly to the stage of the ULFs (H.Herrmann, T.Wedig, P.M.Steinert and U.Aebi, in preparation). Crystallization and structure determination of such complex macromolecular assemblies (molecular mass of ∼1.7 MDa) are no longer beyond the scope of the current methodology, as demonstrated recently for ribosomal subunits (see for example Wimberly et al., 2000). Finally, the crystallographic data will be complemented substantially by cryo-electron microscopy-based tomography of the ULFs as well as mature filaments, which is likely to provide further structural insight into the successive dimer association events underlying IF assembly.
Materials and methods
Sequence alignment and analysis
The amino acid sequences of α-helical segments 1A and 2B from seven human IF proteins, including vimentin (SWISS-PROT database accession code P08670), desmin (P17661), neurofilament L protein (P07196), cytokeratins 8 (P05787) and 18 (P05783), nuclear lamins A (P02545) and B1 (P20700) were aligned manually based on the heptad repeat. For each position of the alignment, the similarity score s was calculated as s = (Ncons + w × Nsim – 1)/(N – 1), where Ncons is the occurrence of the consensus (i.e. present in the largest number of sequences) residue, Nsim is the occurrence of ‘conservative’ substitutions (V↔L↔I↔M, K↔R, E↔D, Y↔F, N↔Q, S↔T and A↔G) with respect to the consensus residue, N is the number of sequences compared and w = 0.7 – weighting factor. Secondary structure prediction from the primary sequence was carried out with the program PREDATOR (http://mips.gsf.de/mips/staff/frishman/). The probability of CC formation was estimated using the program COILS (http://www.ch.embnet.org/software/COILS_form. html).
Crystallization and X-ray data collection
Recombinant vimentin fragments 1A, Z2B and Cys2 were prepared and crystallized as described (Strelkov et al., 2001). Oscillation images were collected from flash-cooled crystals at 110 K using synchrotron radiation with a wavelength of 0.9–1 Å and an image plate detector at EMBL/DESY, Hamburg, and at the BM1 beamline at ESRF, Grenoble. The obtained data were integrated and scaled with the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). Subsequent crystallographic processing was carried out with the CCP4 program suite (CCP4, 1994), unless noted otherwise.
Crystallographic structure determination
To obtain initial phasing of the 1A structure, molecular replacement was employed. Search models were derived from a single α-helix of the GCN4 leucine zipper (Protein Data Bank accession code 2zta) with all long side chains reduced to alanine. During the first stage of the MR procedure using the program MOLREP (Vagin and Teplyakov, 1997), helices of various lengths were tried consecutively. For each model, rotation function was calculated with data between 10 and 2.8 Å resolution, whereby the optimal integration radius was selected automatically by the MOLREP algorithm. The top 20 maxima of the rotation function were then subjected to a translational search with packing restraints, followed by rigid-body fitting. Comparison of the top-scoring solutions for different models revealed one common solution that, while yielding only a marginally better correlation coefficient compared with other solutions, was present for several of the longer search models (GCN4 residues 3–31, 3–29, 3–27 and 3–26). Furthermore, this common solution was only detectable in the space group P6222 and not in the enantiomorphic group P6422. For the second stage of the MR procedure, one copy of the model including residues 3–31 was fixed at the found position and the translation search was repeated for the second copy, this time without any packing restraints. This search revealed a clear solution that partially overlapped with the fixed molecule, so that the two molecules formed a continuous α-helix with a relative register shift of 13 residues. The model combining the two solutions was subjected to several cycles of refinement with the program REFMAC 5 (Murshudov et al., 1997) and manual rebuilding in the program O (Jones et al., 1991). At the later stages, solvent molecules as well as one sulfate ion were added, and individual anisotropic temperature factors were refined using all data up to 1.4 Å resolution.
Phasing of the Z2B structure was obtained by MR using the program AMoRe (Navaza, 1994) with the complete GCN4 leucine zipper (dimeric CC with 31 residues per chain with full side chains) as a search model. The first 40 maxima of the rotation function (integration radius 17 Å, resolution limits 10–2.4 Å) were submitted to a translational search that yielded a distinct top-scoring solution in space group P3121 which was the correct enantiomorph. Once this solution was fixed, two additional solutions could be found. These solutions were partially overlapping with the initial solution, with register shifts of +7 and –7 residues, respectively, yielding a single, continuous CC dimer. After initial refinement of this model, the correct sequence register could be identified. Surprisingly, none of the three obtained MR solutions corresponded to the actual position of the GCN4 leucine zipper within the Z2B chimera, but rather were related to this position by +7, +14 and +21 register shifts, respectively. At the final stages of refinement, residues 406–409 of chain A, which deviate from the CC geometry, could be traced.
Three isomorphous derivatives of the Cys2 crystals were obtained by soaking in solutions of Sm acetate (0.3 mM, 2 h), K2PtCl4 (0.7 mM, 12 h) and Na p-chloromercuriobenzoate (5 mM, 5 h). All three derivatives diffracted X-rays to at least 3.0 Å, yielding Rdiff = Σ|Fderiv – Fnat|/ΣFnat of 0.155, 0.166 and 0.168, respectively, versus the native data. Relatively low concentrations of Sm acetate and short soaking times were necessary to preserve the diffraction quality of the crystals. At the same time, isomorphous Patterson syntheses for this derivative were readily interpretable, revealing a single major binding site. After initial refinement in SHARP (de la Fortelle and Bricogne, 1997), cross-phasing with this site allowed the location of three heavy atom-binding sites in either of the remaining two derivatives. After solvent flattening with the program DM (Cowtan and Main, 1998), the mean figure of merit of the refined MIRAS phases was 0.57. The resulting electron density maps revealed three symmetry-independent Cys2 dimers which were built independently using standard methods (Jones et al., 1991). The structure was refined in CNS (Brünger et al., 1998).
Structure analysis and three-dimensional modeling
Local geometrical parameters of the CC (Crick, 1953) were determined using the program TWISTER (Strelkov and Burkhard, 2002). This program uses an original algorithm that is based on analyzing short (four residues per chain) consecutive fragments of the CC. Superpositions of protein structures were carried out with LSQKAB using the Cα coordinates. The program DINO (http://www.dino3d.org) was used for molecular graphics.
The atomic model of the vimentin dimer was based on the 1A and Cys2 structures complemented by homologous modeling. The CC segments 1B and 2A were modeled by a standard geometry with a pitch of 140 Å and a radius of 5.0 Å. The linkers L1 and L2 were both based on the eight-residue loop L10 of bacteriophage T4 fibritin (PDB code 1aa0), while the linker L12 was based on the 12-residue loop L11 of fibritin. Head and tail domains were modeled qualitatively by polypeptides of appropriate length.
The crystal structures of fragments 1A, Z2B and Cys2 as well as experimentally determined structure factors were deposited in the Protein Data Bank (http://www.rcsb.org) under the accession codes 1gk7, 1gk6 and 1gk4, respectively.
IF assembly assays
For in vitro assembly assays, a 0.2 mg/ml solution of recombinant human vimentin in 5 mM Tris–HCl pH 8.4 was used. Filament formation was initiated by the addition of a 1/9 vol. of 200 mM Tris–HCl pH 7.0 and 1.6 M NaCl (concentrated ‘filament buffer’; see also Herrmann et al., 1996). Prior to further analysis, the samples were incubated at 37°C for 1 h. Electron microscopy was performed as described (Herrmann et al., 1996, 2000).
Acknowledgments
Acknowledgements
We are most grateful to Professor David D.A.Parry for stimulating discussions and to our colleagues at Biozentrum Basel for practical help. We thank Christine Grund for assistance with electron microscopic analysis. We furthermore thank Professor Werner W.Franke for continuous interest and support. This work was supported by the ‘Förderprogramm der Gemeinsamen Forschungskommission der Medizinischen Fakultät Heidelberg’ (Project No. 158/96 to H.H.), a grant from the Swiss National Science Foundation (to U.A. and P.B.), as well as by Canton Basel-Stadt and the M.E.Müller Foundation of Switzerland. The synchrotron data collection was made possible through the European Molecular Biology Laboratory Outstation (Hamburg) and the European Synchrotron Radiation Facility (Grenoble).
References
- Aebi U., Haner,M., Troncoso,J., Eichner,R. and Engel,A. (1988) Unifying principles in intermediate filament structure and assembly. Protoplasma, 145, 73–81. [Google Scholar]
- Brown J.H., Cohen,C. and Parry,D.A. (1996) Heptad breaks in α-helical coiled coils: stutters and stammers. Proteins, 26, 134–145. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. and Nilges,M. (1993) Computational challenges for macromolecular structure determination by X-ray crystallography and solution NMR-spectroscopy. Q. Rev. Biophys., 26, 49–125. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Kammerer,R.A., Steinmetz,M.O., Bourenkov,G.P. and Aebi,U. (2000) The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of inter- and intra-helical salt-bridges. Structure, 8, 223–230. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld,J. and Strelkov,S.V. (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol., 11, 82–88. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cohen M., Lee,K.K., Wilson,K.L. and Gruenbaum,Y. (2001) Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci., 26, 41–47. [DOI] [PubMed] [Google Scholar]
- Conway J.F. and Parry,D.A.D. (1988) Intermediate filament structure. 3. Analysis of sequence homologies. Int. J. Biol. Macromol., 10, 79–98. [Google Scholar]
- Cowtan K. and Main,P. (1998) Miscellaneous algorithms for density modification. Acta Crystallogr. D, 54, 487–493. [DOI] [PubMed] [Google Scholar]
- Crick F.H.C. (1953) The packing of α-helices: simple coiled coils. Acta Crystallogr., 6, 689–697. [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–493. [DOI] [PubMed] [Google Scholar]
- Fuchs E. and Coulombe,P.A. (1992) Of mice and men: genetic skin diseases of keratin. Cell, 69, 899–902. [DOI] [PubMed] [Google Scholar]
- Fuchs E. and Karakesisoglou,I. (2001) Bridging cytoskeletal inter sections. Genes Dev., 15, 1–14. [DOI] [PubMed] [Google Scholar]
- Fuchs E. and Weber,K. (1994) Intermediate filaments: structure, dynamics, function and disease. Annu. Rev. Biochem., 63, 345–382. [DOI] [PubMed] [Google Scholar]
- Geisler N. and Weber,K. (1982) The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J., 1, 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Schunemann,J. and Weber,K. (1992) Chemical cross-linking indicates a staggered and antiparallel protofilament of desmin intermediate filaments and characterizes one higher-level complex between protofilaments. Eur. J. Biochem., 206, 841–852. [DOI] [PubMed] [Google Scholar]
- Gigant B., Curmi,P.A., Martin-Barbey,C., Charbaut,E., Lachkar,S., Lebeau,L., Siavoshian,S., Sobel,A. and Knossow,M. (2000) The 4 Å X-ray structure of a tubulin:stathmin-like domain complex. Cell, 102, 809–816. [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Khuon,S., Chou,Y.H., Opal,P. and Steinert,P.M. (1996) The function of intermediate filaments in cell shape and cytoskeletal integrity. J. Cell Biol., 134, 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M. and Burba,M. (1994) Function of type I and type II keratin head domains: their role in dimer, tetramer and filament formation. J. Cell Sci., 107, 1959–1972. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. and Weber,K. (1992) A synthetic peptide representing the consensus sequence motif at the carboxy-terminal end of the rod domain inhibits intermediate filament assembly and disassembles preformed filaments. J. Cell Biol., 116, 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins S. and Aebi,U. (1994) Making heads and tails of intermediate filament assembly, dynamics and networks. Curr. Opin. Cell Biol., 6, 25–33. [DOI] [PubMed] [Google Scholar]
- Herrmann H. and Aebi,U. (1998a) Structure, assembly and dynamics of intermediate filaments. Subcell. Biochem., 31, 319–362. [PubMed] [Google Scholar]
- Herrmann H. and Aebi,U. (1998b) Intermediate filament assembly: fibrillogenesis is driven by decisive dimer–dimer interactions. Curr. Opin. Struct. Biol., 8, 177–185. [DOI] [PubMed] [Google Scholar]
- Herrmann H. and Aebi,U. (1999) Intermediate filament assembly: temperature sensitivity and polymorphism. Cell Mol. Life Sci., 55, 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H. and Aebi,U. (2000) Intermediate filaments and their associates: multi-talented structural elements specifying cyto architecture and cytodynamics. Curr. Opin. Cell Biol., 12, 79–90. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Haner,M., Brettel,M., Muller,S.A., Goldie,K.N., Fedtke,B., Lustig,A., Franke,W.W. and Aebi,U. (1996) Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J. Mol. Biol., 264, 933–953. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Haner,M., Brettel,M., Ku,N.O. and Aebi,U. (1999) Characterization of distinct early assembly units of different intermediate filament proteins. J. Mol. Biol., 286, 1403–1420. [DOI] [PubMed] [Google Scholar]
- Herrmann H. et al. (2000) The intermediate filament protein consensus motif of helix 2B: its atomic structure and contribution to assembly. J. Mol. Biol., 298, 817–832. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- McCormick M.B., Kouklis,P., Syder,A. and Fuchs,E. (1993) The roles of the rod end and the tail in vimentin IF assembly and IF network formation. J. Cell Biol., 122, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- North A.C., Steinert,P.M. and Parry,D.A. (1994) Coiled-coil stutter and link segments in keratin and other intermediate filament molecules: a computer modeling study. Proteins, 20, 174–184. [DOI] [PubMed] [Google Scholar]
- O’Shea E.K., Klemm,J.D., Kim,P.S. and Alber,T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science, 254, 539–544. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 277, 307–325. [DOI] [PubMed] [Google Scholar]
- Parry D.A.D. and Steinert,P.M. (1995) Intermediate Filament Structure. Springer Verlag, Heidelberg, Germany.
- Parry D.A.D. and Steinert,P.M. (1999) Intermediate filaments: molecular architecture, assembly, dynamics and polymorphism. Q. Rev. Biophys., 32, 99–187. [DOI] [PubMed] [Google Scholar]
- Schliwa M. (1986) The Cytoskeleton. Cell Biology Monographs, Vol. 13. Springer-Verlag, Vienna, Austria.
- Smith T.A., Strelkov,S.V., Burkhard,P., Aebi,U. and Parry,D.A.D. (2002) Sequence comparisons of intermediate filament chains: evidence of a unique functional/structural role for coiled-coil segment 1A and linker L1. J. Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Spek E.J., Bui,A.H., Lu,M. and Kallenbach,N.R. (1998) Surface salt bridges stabilize the GCN4 leucine zipper. Protein Sci., 7, 2431–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P.M., Marekov,L.N. and Parry,D.A. (1993) Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. J. Biol. Chem., 268, 24916–24925. [PubMed] [Google Scholar]
- Steinmetz M.O., Hoenger,A., Tittmann,P., Fuchs,K.H., Gross,H. and Aebi,U. (1998) An atomic model of crystalline actin tubes: combining electron microscopy with X-ray crystallography. J. Mol. Biol., 278, 703–711. [DOI] [PubMed] [Google Scholar]
- Strelkov S.V. and Burkhard,P. (2002) Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Strelkov S.V., Herrmann,H., Geisler,N., Lustig,A., Ivaninskii,S., Zimbelmann,R., Burkhard,P. and Aebi,U. (2001) Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J. Mol. Biol., 306, 773–781. [DOI] [PubMed] [Google Scholar]
- Tao Y., Strelkov,S.V., Mesyanzhinov,V.V. and Rossmann,M.G. (1997) Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure, 5, 789–798. [DOI] [PubMed] [Google Scholar]
- Turkenburg J.P. and Dodson,E.J. (1996) Modern developments in molecular replacement. Curr. Opin. Struct. Biol., 6, 604–610. [DOI] [PubMed] [Google Scholar]
- Vagin A. and Teplyakov,A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr., 30, 1022–1025. [Google Scholar]
- Weber K. and Geisler,N. (1985) Intermediate filaments: structural conservation and divergence. Ann. NY Acad. Sci., 455, 126–143. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M., Jr, Morgan-Warren, R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Wu K.C., Bryan,J.T., Morasso,M.I., Jang,S.I., Lee,J.H., Yang,J.M., Marekov,L.N., Parry,D.A. and Steinert,P.M. (2000) Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol. Biol. Cell, 11, 3539–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]