Abstract
The Drosophila nucleosome remodeling factor (NURF) is an imitation switch (ISWI)-containing chromatin remodeling complex that can catalyze nucleosome repositioning at promoter regions to regulate access by the transcription machinery. Mononucleosomes reconstituted in vitro by salt dialysis adopt an ensemble of translational positions on DNA templates. NURF induces bi-directional ‘sliding’ of these nucleosomes to a subset of preferred positions. Here we show that mononucleosome sliding catalyzed by NURF bears similarity to nucleosome movement induced by elevated temperature. Moreover, we demonstrate that the GAL4 DNA-binding domain can extend NURF-induced nucleosome movement on a GAL4-E4 promoter, expanding the stretch of histone-free DNA at GAL4 recognition sites. The direction of NURF-induced nucleosome movement can be significantly modulated by asymmetric placement of tandem GAL4 sites relative to the nucleosome core particle. As such, sequence-specific, transcription factor-directed nucleosome sliding is likely to have substantial influence on promoter activation.
Keywords: chromatin/nucleosome/NURF/remodeling/transcription
Introduction
The genome of eukaryotes is packaged into nucleosomes, which limit the access of enzyme complexes that process DNA, including the transcription machinery. This constraint can be relieved in part by the action of ATP-dependent chromatin remodeling complexes. Four families of complexes containing SWI2–SNF2, imitation switch (ISWI), Mi-2 and Ino80 ATPases have been described (Cairns, 1998; Aalfs and Kingston, 2000; Shen et al., 2000; Vignali et al., 2000). The nucleosome remodeling factor (NURF) complex is one of three ISWI-containing, chromatin remodeling factors purified from Drosophila (NURF, ACF and CHRAC) (Tsukiyama et al., 1995; Ito et al., 1997; Varga-Weisz et al., 1997). NURF has a molecular mass of ∼500 kDa and contains three subunits (NURF301, NURF55 and NURF38) in addition to the ISWI ATPase (Tsukiyama et al., 1995; Gdula et al., 1998; Martinez-Balbas et al., 1998; Xiao et al., 2001). NURF is a nucleosome-stimulated ATPase that has been shown to facilitate nucleosome repositioning mediated by sequence-specific DNA-binding factors on the promoters of hsp70 (Tsukiyama et al., 1994; Tsukiyama and Wu, 1995) and GAL4-E4 (Mizuguchi et al., 1997,2001).
Studies using purified ISWI protein or ISWI-containing protein complexes indicate that these enzymes reposition nucleosomes by catalyzing nucleosome sliding, defined as the movement of the histone octamer in cis, without irretrievable displacement from DNA (Corona et al., 1999; Hamiche et al., 1999; Längst et al., 1999). However, the direction of enzyme-catalyzed nucleosome sliding can differ significantly, depending on the individual chromatin remodeling complex. For example, NURF slides reconstituted mononucleosomes from central to distal sites whereas ACF or CHRAC mobilizes nucleosomes in the opposite direction, from distal to central locations (Corona et al., 1999; Hamiche et al., 1999; Längst et al., 1999; Eberharter et al., 2001). These differences appear to be determined by the unique properties of the distinct large subunits, NURF301 and ACF1, of the NURF and ACF–CHRAC complexes, respectively (Ito et al., 1999; Eberharter et al., 2001; Xiao et al., 2001). However, it is unclear how these components cooperate with the ISWI engine to mobilize nucleosomes to different positions. Moreover, the relationship between sequence-dependent nucleosome stability and the outcome of ATP- dependent nucleosome sliding has been largely unexplored.
The relative location of transcription factor-binding sites on a positioned nucleosome can substantially affect site accessibility to proteins. In general, the wrapping of DNA in a nucleosome reduces the affinity of transcription factors for their cognate sites, although in at least one instance, a nucleosome can positively affect factor binding. The liver-enriched transcription factor HNF3 binds more stably to nucleosome core particles than to free DNA (Cirillo and Zaret, 1999). In contrast, glucocorticoid receptor, thyroid hormone receptor and Fos/Jun bind to nucleosomal DNA with slightly lower affinity than binding to free DNA (Li and Wrange, 1993; Wong et al., 1995; Ng et al., 1997). GAL4, c-Myc, heat shock factor (HSF), specificity protein 1 (SP1) and TFIIIA bind to nucleosomal DNA with an affinity at least one order of magnitude lower than their affinities for free DNA (Taylor et al., 1991; Vettese-Dadey et al., 1994; Wechsler et al., 1994; Cirillo and Zaret, 1999). The affinity of nuclear factor 1 and TATA-binding protein (TBP) for nucleosomal DNA is lower by at least two orders of magnitude (Pina et al., 1990; Imbalzano et al., 1994; Blomquist et al., 1999).
ATP-dependent chromatin remodeling complexes can alter the structure of nucleosomes and thereby facilitate the binding of transcription factors (Cote et al., 1994; Imbalzano et al., 1994; Kwon et al., 1994; Cairns et al., 1996; Wang et al., 1996; Burns and Peterson, 1997). In the presence of remodeling enzymes, sequence-specific DNA-binding transcription factors such as GAGA factor, HSF, nuclear factor E2, GAL4 derivatives and nuclear factor κB induce the rearrangement of nucleosome arrays in the vicinity of their DNA-binding sites (Pazin et al., 1994, 1997; Tsukiyama et al., 1994; Wall et al., 1995; Armstrong and Emerson, 1996). In this study, we have investigated the contributions of transcription factor binding to NURF-induced nucleosome sliding in a highly purified system. We show how NURF and the DNA-binding domain (DBD) of GAL4 cooperate in the mobilization of a single nucleosome. Binding of GAL4-DBD in the presence of NURF extends nucleosome sliding to generate a nucleosome-free region at GAL4 recognition sites. Conversely, GAL4-DBD can also create a boundary that prevents nucleosome re-entry, thus constraining the direction of nucleosome sliding. We also compared the thermal induction of nucleosome mobility with NURF-induced sliding, and found that the two processes share interesting similarities.
Results
Nucleosomes adopt multiple positions on the GAL4-E4 promoter
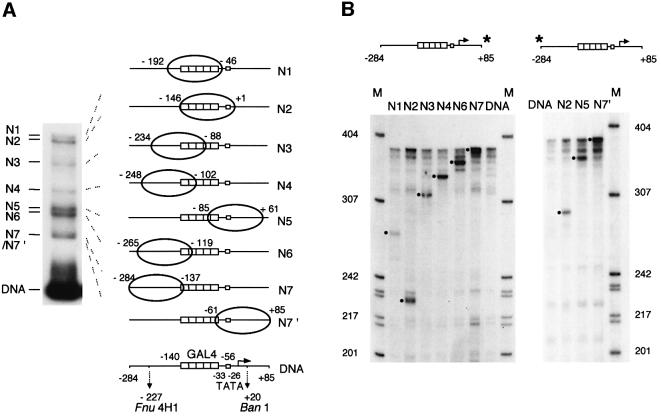
The positioning of a histone octamer with respect to DNA sequence is a function of the intrinsic, sequence-dependent bendability of DNA over the histone octamer (Luger et al., 1997; Travers and Drew, 1997; Richmond and Widom, 2000). We analyzed the distribution of mononucleosomes on a 369 bp DNA fragment containing five tandem GAL4-binding sites upstream of the adenovirus E4 TATA box and minimal core promoter (Pazin et al., 1994; Mizuguchi et al., 1997). Mononucleosomes were reconstituted by salt dialysis using purified histone octamers, and fractionated by native PAGE. The electrophoretic migration of nucleosomes is influenced by the length and spatial orientation of the linker DNA extending from both ends of the core particle—migration is slow for centrally located nucleosomes and increases linearly as nucleosome placement approaches either fragment end (Duband-Goulet et al., 1992; Meersseman et al., 1992). Using this assay, at least seven nucleosome species (N1–N7) could be observed on the 369 bp GAL4-E4 promoter fragment (Figure 1A).
Fig. 1. Reconstituted nucleosomes occupy multiple positions on the GAL4-E4 promoter. (A) Native PAGE of mononucleosomes reconstituted on a 369 bp fragment carrying the GAL4-E4 promoter. Major nucleosome species are indicated as N1–N7′. Nucleosome positions and GAL4-binding sites determined by Exo III footprinting (precise to ± 2 bp) are illustrated on the right. The TATA box and restriction enzyme sites are indicated. (B) Exo III footprinting. Gel bands corresponding to individual N1–N7 nucleosome species (DNA was radiolabeled at either end of GAL4-E4 promoter) were excised, nucleosomes were eluted and digested with Exo III (400 U/ml) for 2 min at 37°C. DNAs were analyzed by electrophoresis in a 6% polyacrylamide gel containing 8 M urea. M indicates pBR322 HpaII fragments as markers. Dots indicate major Exo III pauses. The Exo III footprint of GAL4 (–153 to –44) extends beyond the recognition sites (–140 to –56) (Carey et al., 1989; Carey and Smale, 2000).
We mapped the positions of reconstituted nucleosomes by exonuclease III (Exo III) digestion after elution of each species from the corresponding gel band. The upstream boundary of each nucleosome was determined using the site of a prominent pause in the pattern of Exo III digestion; this pause is absent from the digestion pattern of free DNA (Figure 1B). Downstream boundaries were also determined by Exo III digestion (Figure 1B), or calculated using the location of the upstream nucleosome boundary. Accordingly, N1 and N2 nucleosomes were mapped centrally over the tandem GAL4-binding sites; N3, N4 and N6 nucleosomes were located further upstream; the N5 nucleosome was located over the TATA box and downstream sequences, and N7/N7′, the fastest migrating nucleosomes, mapped to either end of the GAL4-E4 fragment (Figure 1).
NURF catalyzes nucleosome sliding on the GAL4-E4 promoter
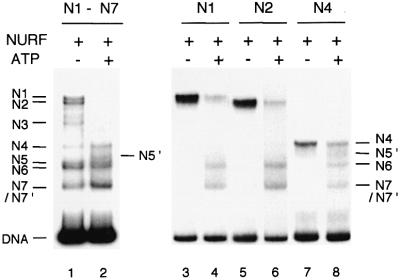
To assess the influence of NURF on the distribution of nucleosomes on the GAL4-E4 fragment, we incubated 369 bp mononucleosomes with NURF and ATP, and analyzed nucleosome positions by native gel electrophoresis. A change in nucleosome distribution was clearly apparent: N1, N2, N3 and N5 nucleosomes were depleted, while N6 and N7/N7′ nucleosomes became more prevalent (Figure 2, lane 2). In addition, the N4 nucleosome was weakly detected, and a novel position located between N4 and N5 (N5′) was sometimes observed. This distribution of nucleosomes did not change after incubation with higher levels of NURF or for a longer period (data not shown), indicating that nucleosome distribution had reached equilibrium.
Fig. 2. NURF mediates nucleosome sliding on the GAL4-E4 promoter. 369 bp GAL4-E4 mononucleosomes (40 nM) were incubated with NURF (0.4 nM) in the absence or presence of ATP for 30 min at 26°C (lanes 1 and 2), followed by native 4.5% PAGE in 0.5× TBE. N1, N2 and N4 nucleosomes were eluted from the gel slice and incubated with NURF and ATP as indicated, before electrophoresis on a second native gel (lanes 3–8).
To examine further the movement of nucleosomes from a single location, we purified N1, N2 and N4 nucleosomes by native gel electrophoresis. After incubation with NURF and ATP, all three individual nucleosome species showed movement primarily to the N6 and N7/N7′ positions, confirming these as the favored locations when nucleosomes are mobilized by NURF (Figure 2, lanes 4, 6 and 8). The results indicate that NURF catalyzes nucleosome movement from central to peripheral positions on the 369 bp GAL4-E4 promoter, leading to partial exposure of GAL4-binding sites (see Figure 1A).
Comparison of NURF- and heat-induced nucleosome sliding
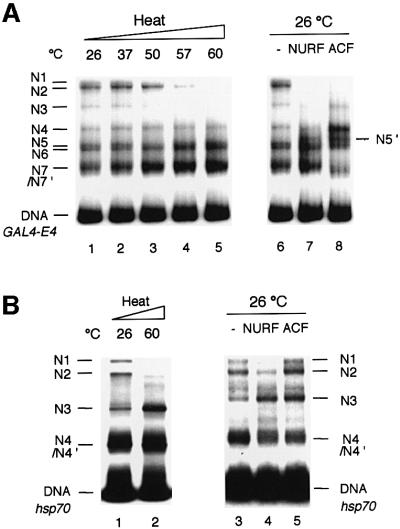
The standard protocol for reconstituting nucleosomes by dialysis of a mixture of core histones and DNA from high salt (Rhodes and Laskey, 1989) apparently traps some nucleosomes in non-equilibrium positions (Drew, 1991; Widom, 1999). Such reconstituted mononucleosomes have been shown to undergo temperature-induced mobility and redistribution (Pennings et al., 1991; Flaus and Richmond, 1998). We examined how heat affects the distribution of mononucleosomes reconstituted on the 369 bp GAL4-E4 promoter. Analysis of nucleosome positions showed that as 369 bp nucleosomes were heated to temperatures up to 60°C before cooling and native gel electrophoresis (at room temperature), a gradual loss of N1, N2 and N3 nucleosomes was observed. Upon heating to 60°C, N5/N6 and N7/N7′ were the preferred nucleosome positions, with N7/N7′ being dominant; the N4 position remained as a weakly detected band (Figure 3A, lanes 1–5). Heat- and NURF-induced relocation of nucleosomes bear interesting similarities, although there are noticeable differences—heating resulted in greater abundance of the N7/N7′ positions and failed to reveal the novel N5′ position.
Fig. 3. Comparison of heat- and NURF-induced nucleosome sliding. Mononucleosomes reconstituted on GAL4-E4 (A) or hsp70 (B) radiolabeled DNAs were incubated for 30 min at the indicated temperatures in TE buffer and 1 mg/ml BSA. Samples were cooled on ice, and analyzed by native PAGE as in Figure 2 legend. Mononucleo somes were incubated with NURF (0.4 nM), [(A) lane 7; (B) lane 4] or ACF (1.6 nM), [(A) lane 8; (B) lane 5] in nucleosome sliding buffer at 26°C. Samples were electrophoresed as in Figure 2 legend.
A similarity between NURF- and heat-induced nucleosome sliding was also observed using the 359 bp hsp70 promoter fragment. Both heat treatment and the action of NURF relocated nucleosomes to the preferred N3 position (Figure 3B, lanes 2 and 4). In contrast, ACF, the ISWI-containing nucleosome assembly and spacing factor (Ito et al., 1997, 1999; Eberharter et al., 2001) mobilized nucleosomes to the N1 and N2 positions as much as the N3 position (Figure 3B, lane 5). On the GAL4-E4 promoter, ACF induced nucleosome movement preferably to N4 more than the N6 and N7 positions (Figure 3A, lane 8).
GAL4-DBD clears nucleosomes from cognate sites
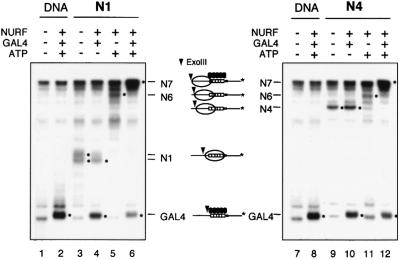
In the absence of other factors, the repositioning of nucleosomes by NURF uncovers some, but not all, of the five GAL4-binding sites on the GAL4-E4 promoter. To evaluate the influence of the GAL4-DBD on nucleosome remodeling, we incubated purified N1 or N4 nucleosomes with NURF and saturating amounts of GAL4-DBD (GAL4 residues 1–147) (Lin et al., 1988; Carey et al., 1990). The resulting complexes of GAL4-DBD and nucleosomes were analyzed by Exo III protection (the 369 bp fragment was 5′ end-labeled at the downstream end). When the N1 nucleosome was mobilized in the presence of both GAL4-DBD and NURF, we observed strong protection from digestion by Exo III at the upstream end of the 369 bp fragment (Figure 4, lane 6). Such enhanced protection from digestion was not detected in the absence of ATP or GAL4-DBD (Figure 4, lanes 4 and 5). Similarly, the N4 nucleosome displayed enhanced protection from Exo III digestion at the upstream fragment end (Figure 4, lanes 10–12). The results indicate that nucleosomes are relocated to the upstream end (or the downstream end; see below) of the DNA fragment.
Fig. 4. GAL4-DBD and NURF-induced extension of nucleosome sliding—Exo III footprinting analysis. Mononucleosomes were reconstituted on the end-labeled 369 bp GAL4-E4 promoter. Gel-purified nucleosomes (0.025 nM) and carrier DNA (20 µg/ml), N1 (lanes 1–6), N4 (lanes 7 and 8) were incubated with NURF (0.2 nM) and GAL4-DBD (0.3 nM) in the presence or absence of ATP as indicated. Reaction mixtures were digested by Exo III (400 U/ml) and analyzed on an 8 M urea–6% polyacrylamide gel. Dots indicate major Exo III pauses. Diagrams show the position of nucleosomes (open circles), GAL4 (filled ovals) and Exo III pauses (arrowheads).
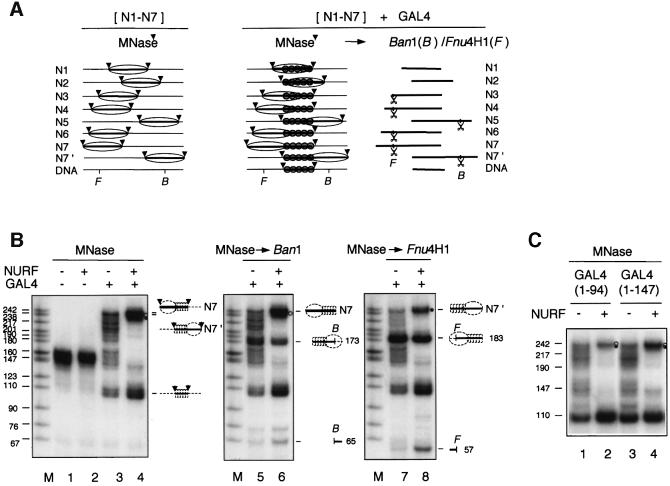
To confirm these findings, we analyzed the positioning of nucleosomes by restriction enzyme analysis after micrococcal nuclease (MNase) digestion and DNA purification (Dong et al., 1990; Studitsky et al., 1994; Davey et al., 1998; Studitsky, 1999). Nucleosomes reconstituted with uniformly labeled, 369 bp GAL4-E4 DNA were purified by glycerol gradient centrifugation to remove free DNA and dinucleosome particles. Analysis of the size of core particle DNA upon MNase digestion showed the expected ∼150 bp DNA (Figure 5A, left panel and B, lane 1). When GAL4-DBD was incubated with 369 bp nucleosomes, MNase digestion and DNA purification revealed fragments of ∼100 bp and from ∼145 to 240 bp (Figure 5B, lane 3). We attribute the ∼100 and ∼145 bp fragments to protection from cleavage by tandemly bound GAL4-DBDs and the nucleosome core particle, respectively, and the fragment series between ∼145 and 240 bp to combined protection by N1–N7 positioned core particles and GAL4-DBDs (Figure 5A, middle panel). When NURF and ATP were included in the incubation of GAL4-DBD and the N1–N7 nucleosome population, the MNase digestion pattern strikingly resolved into a major DNA species of ∼240 bp (Figure 5B, lane 4 and illustration), which is attributed to combined protection by tandemly bound GAL4-DBDs and a nucleosome relocated to either end of the 369 bp fragment. In such experiments, we also observed a faint, ∼145 bp fragment, and increased abundance of the ∼100 bp fragment. This increase may result from MNase cleavage at a more accessible junction between the GAL4-DBDs and the relocated nucleosome core particle, or reflect some loss of histone octamers during nucleosome repositioning.
Fig. 5. GAL4-DBD and NURF-induced extension of nucleosome sliding—MNase and restriction enzyme analysis. Nucleosomes were reconstituted on 32P uniformly labeled 369 bp GAL4-E4 fragment and incubated with GAL4 and/or NURF prior to MNase digestion. To map boundaries of MNase protection, DNA was purified and digested with BanI (B) or Fnu4H1 (F). (A) Schematic illustration showing mononucleosomes and GAL4-DBD binding on 369 bp GAL4-E4 promoter. Arrowheads mark the boundaries of MNase protection. (B) Mononucleosome fractions (0.7 nM) purified by glycerol gradient centrifugation were incubated with NURF (0.2 nM) and/or GAL4 (30 nM monomer) for 30 min at 26°C, followed by MNase (25 U/ml) digestion (lanes 1–4). DNA was purified and further digested with BanI (1300 U/ml, lanes 5 and 6) or Fnu4H1 (333 U/ml, lanes 7 and 8) and analyzed by 8% PAGE in TBE and autoradiography. Dots (filled and unfilled) indicate fragments protected by complexes of GAL4-DBD and N7 or N7′ nucleosomes, respectively. Binding of GAL4-DBD to free DNA or nucleosomes generates 109 and 145–240 bp MNase resistant fragments, respectively. In the absence of GAL4-DBD, MNase digestion of N1–N7 nucleosomes yields 145–155 bp fragments from core particles. (C) Mononucleosomes were incubated with GAL4(1–94) and GAL4(1–147) (20 nM monomer) in the presence or absence of NURF, and analyzed by MNase digestion and DNA gel electrophoresis. MNase cleavage at sites flanking the tandemly bound GAL4-DBDs appears to be more efficient for GAL4(1–94).
We confirmed the positional assignments of the relocated nucleosomes by restriction enzyme analysis, using a BanI site at downstream position +20 and a Fnu4H1 site at –227 upstream of the GAL4-E4 promoter (Figure 5A, right). BanI digested a fraction (33%) of the 240 bp DNA, generating 173 and 65 bp fragments. This fraction represents nucleosomes that moved to the downstream end (N7′) of the GAL4-E4 fragment (Figure 5B, lane 6 and illustration). Fnu4H1 digested 68% of the 240 bp DNA, generating fragments of 183 and 57 bp; this fraction represents nucleosomes that moved to the upstream end (N7) (Figure 5B, lane 8 and illustration). Collectively, our results indicate that the combined actions of GAL4-DBD and NURF mobilize nucleosomes to either one or the other end of the 369 bp fragment, away from the tandem GAL4-binding sites.
We also studied the effects on nucleosome positioning of GAL4(1–94), which lacks the cryptic activation region of GAL4(1–147) (Lin et al., 1988; Carey et al., 1989, 1990; Workman et al., 1991). GAL4(1–94) also directs NURF-induced movement of nucleosomes to either end of the 369 bp fragment (Figure 5C, lanes 2 and 4). The results indicate that the minimal DNA binding and dimerization domains of GAL4 are sufficient to direct nucleosome repositioning.
Relative placement of GAL4 modulates direction of nucleosome movement
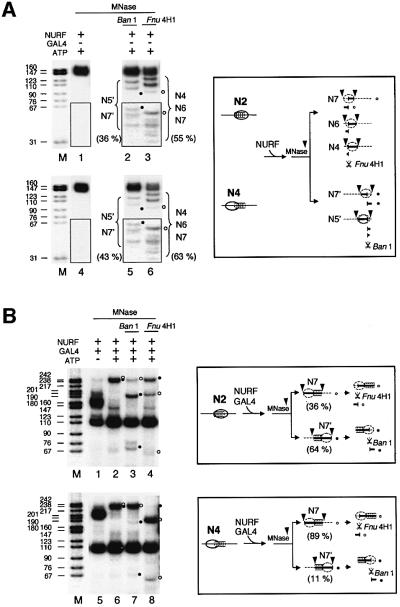
To explore further how placement of GAL4 sites in relation to the nucleosome core particle might direct nucleosome movements, we analyzed the repositioning of purified nucleosomes in which the GAL4 sites are located asymmetrically. In the N2 nucleosome, GAL4 sites are placed inside the core particle, bordering the upstream edge, whereas the N4 nucleosome has GAL4 sites placed in the linker and overlapping the downstream edge of the core particle (Figure 6). Control experiments were performed in which purified N2 or N4 nucleosomes were treated with NURF in the absence of GAL4-DBD, followed by MNase digestion, DNA purification and BanI or Fnu4H1 cleavage to map nucleosome positions. The repositioning of nucleosomes by NURF (alone) shows a moderate preference of N2 or N4 nucleosomes for upstream locations. The N2 species moves upstream to N7, N6, N4 positions (55% Fnu4H1-sensitive; ∼13% N7), or moves downstream to N7′, N5′ positions (36% BanI-sensitive; ∼6% N7′). The N4 species also moves upstream to N7, N6, N4 positions (63% Fnu4H1-sensitive; ∼15% N7), or downstream to N7′, N5′ positions (43% BanI-sensitive; ∼3% N7′) (Figure 6A).
Fig. 6. Placement of GAL4 sites modulates direction of nucleosome sliding. (A) Gel-purified 369 bp GAL4-E4 mononucleosomes N2 (upper panel) and N4 (lower panel) were incubated with NURF in the presence of ATP and digested with MNase; DNA was purified, further digested with BanI and Fnu4H1, and analyzed by PAGE and auto radiography as in the legend to Figure 5. The lower part of the gel (indicated by the box) was exposed to film ∼3 times longer than the upper part and contrast enhanced to visually reveal small DNA fragments. Quantitation of radioactive band intensity was performed by phosphoimager analysis. The percentage restriction enzyme cleavage was calculated by subtraction of the residual ∼150 bp nucleosome core particle DNA from an equivalent, uncleaved sample (lanes 1 and 4). Restriction fragments derived from N7 and N7′ nucleosomes are indicated by unfilled and filled dots, respectively. The 81 bp fragment derived from BanI cleavage of N7′ core particle DNA is not visible at this exposure; note that the N7 and N7′ percentage values given in the text are approximate. (B) Gel-purified 369 bp GAL4-E4 mononucleo somes N2 (upper panel) and N4 (lower panel) were incubated with NURF and GAL4-DBD, in the presence or absence of ATP, and analyzed as above. Dots (unfilled and filled) indicate fragments corresponding to, or derived from, N7 and N7′ nucleosomes, respectively. The distribution of N7 and N7′ nucleosomes was calculated by phosphoimager analysis of radioactive fragments, and averaged from three sets of experiments.
The inclusion of GAL4-DBD in the reaction significantly alters the outcome of NURF-induced nucleosome mobility. As indicated by the presence of the 240 bp fragment protected from MNase digestion, the combination of NURF and GAL4-DBD drives the N2 nucleosome to one or the other end of the 369 bp fragment (Figure 6B, lane 2). Digestion of the 240 bp fragment with BanI (Figure 6B, lane 3) and Fnu4H1 (Figure 6B, lane 4), indicates that N7 (36%; Fnu4H1-sensitive) and N7′ (64%; BanI-sensitive) are the only major nucleosome positions adopted. The N4 nucleosome shows highly preferential movement to the upstream N7 nucleosome position. Eighty-nine percent of N4 nucleosomes were driven to the N7 (Fnu4H1-sensitive) position, while 11% moved to the N7′ (BanI-sensitive) position (Figure 6B, lanes 7 and 8). Hence, the relative placement of the GAL4-binding sites on the nucleosome core particle can have a significant influence on the direction of nucleosome movement. Given the bi-directional nature of NURF-induced nucleosome mobility (Hamiche et al., 1999), the results also imply that a DNA-binding protein can impose a substantial barrier to re-entry of nucleosomes that were moved away from its cognate sites.
Discussion
Recent studies have demonstrated that several ATP-dependent chromatin remodeling complexes have the ability to generate nucleosome movement in cis on a specific DNA fragment (Hamiche et al., 1999; Längst et al., 1999; Whitehouse et al., 1999; Brehm et al., 2000; Guschin et al., 2000). For the ISWI complex NURF, sliding of mononucleosomes has been so far demonstrated on the hsp70 promoter and the sea urchin 5S RNA gene (Hamiche et al., 1999). In this report, we extended the analysis of nucleosome sliding by NURF to the GAL4-E4 model promoter, and characterized the impact of the GAL4 DNA-binding motif on the outcome of nucleosome remodeling. We also compared features of NURF-induced nucleosome sliding and nucleosome mobility induced by heat treatment.
The stability of nucleosomes is influenced by the intrinsic, sequence-dependent bendability of the double helix (Luger et al., 1997; Travers and Drew, 1997; Richmond and Widom, 2000). We observed at least six different translational positions of the nucleosome reconstituted by salt dialysis on the 369 bp GAL4-E4 promoter fragment. Upon heating, the N1, N2 and N3 nucleosomes are induced to slide, generating a distribution that bears some similarity to nucleosomes mobilized by the ATP-dependent activity of NURF. These findings suggest that the mechanism of NURF action has a component that may mimic the thermal alterations of nucleosomes that lead to nucleosome mobility.
NURF-induced nucleosome movement on the GAL4-E4 promoter renders GAL4 sites only partially free of nucleosomes. The distribution of nucleosome positions can be further perturbed by introduction of GAL4-DBD into the purified remodeling system. In accordance with previous observations, we found that binding of GAL4-DBD to nucleosomal DNA in the absence of remodeling enzymes does not lead to the repositioning of nucleosomes (Pazin et al., 1994; Owen-Hughes et al., 1996; Mizuguchi et al., 1997). However, the combined activities of GAL4-DBD and NURF proteins result in extended sliding of nucleosomes to clear the GAL4-binding sites, leaving an expanded stretch of nucleosome-free DNA. We suggest that a similar mechanism of extended nucleosome sliding could account for the documented repositioning of nucleosomes by GAGA factor, HSF and NURF on the hsp70 and hsp26 promoters (Tsukiyama et al., 1994; Tsukiyama and Wu, 1995; Wall et al., 1995). Such generation of chromatin accessibility is likely to be required to facilitate other steps in promoter activation, e.g. binding of TFIID and recruitment of RNA polymerase II. In this context, it is of interest that a recent report demonstrates contributions to nucleosome sliding as well by the TATA-binding protein and other proteins that bend promoter DNA (Lomvardas and Thanos, 2001).
The relative placement of GAL4 sites on the nucleosome can also have a strong influence on the direction of nucleosome movement. When tandem GAL4 sites are placed asymmetrically on the nucleosome core particle (e.g. overlapping a linker and one edge of the core particle), GAL4-DBD binding in the presence of NURF activity mobilizes nucleosomes preferentially in the direction of the other, unbound linker DNA. It has been previously shown that GAL4-DBD binds with greater affinity to free DNA than to nucleosomes (Taylor et al., 1991), and proceeds cooperatively from one end of the core particle until all sites are occupied (Vettese-Dadey et al., 1994). Hence, GAL4-DBD binding initiating in the linker DNA and spreading into the core particle should constrain the direction of nucleosome sliding by providing a dominant boundary against back-sliding due to the bi-directional nature of NURF-induced nucleosome mobility (Hamiche et al., 1999). Taken together, these findings indicate that a DNA-binding factor should be continuously required for the maintenance of NURF-induced nucleosome positioning, a conclusion consistent with the results of an earlier study using a crude chromatin assembly system containing ISWI complexes (Pazin et al., 1997).
Under our assay conditions, binding of GAL4-DBD extended NURF-induced sliding largely in cis. However, it has been reported that binding of transcription factors such as GAL4 or glucocorticoid receptor to nucleosomal DNA stimulates SWI–SNF-induced displacement in trans (Owen-Hughes and Workman, 1996; Owen-Hughes et al., 1996; Ostlund Farrants et al., 1997). These findings underscore differences in the remodeling mechanism between NURF and other ATP-dependent chromatin remodeling complexes. NURF-induced nucleosome sliding may be functionally effective only when the intrinsic DNA structure of specific promoter sequences allows nucleosome movement away from elements for recognition by the transcription machinery. In situations where promoter sequences dictate inherently stable nucleosome positions, or where immobile neighbors block NURF-induced nucleosome sliding, it may be necessary to bring on additional remodeling proteins like SWI–SNF with capacity for enhanced sliding or nucleosome eviction. Our present studies should provide a stimulus to additional investigations on the relative nucleosome sliding strengths of the family of ATP-dependent chromatin remodeling complexes.
Materials and methods
Preparation of DNA fragments
A 369 bp DNA fragment containing five GAL4-binding sites and the TATA box from the adenovirus E4 promoter was generated by PCR from pGIE-0 (Pazin et al., 1994) with the primers 5′-GCGAATTCAGATCTCCAGATGCTACACAATTAG-3′ and 5′-GCGAATTCTCGAGCTTACCAGTAAAAAAGAAAACCTA-3′ using Pfu DNA polymerase (Stratagene). DNA fragments for Exo III footprinting were prepared by labeling one of the primers at the 5′-end with [γ-32P]ATP and performing PCR with the other primer unlabeled. For MNase analysis, the DNA fragment was uniformly labeled by PCR synthesis using [α-32P]dNTPs. The double-stranded PCR products were gel-purified and concentrated by ethanol precipitation. The radiolabeled 359 bp hsp70 DNA fragment was prepared according to Hamiche et al. (1999).
Nucleosome reconstitution
Nucleosomes were assembled from a 32P-labeled 369 bp GAL4-E4 DNA fragment and Drosophila histone octamers by the salt jump method as described previously (Hamiche et al., 1999). Two micrograms of carrier plasmid DNA and 50–200 ng of 32P-labeled linear DNA fragment were mixed with 0.8–2.4 µg of core histones in 2 M NaCl, 10 mM Tris–HCl pH 7.5, 0.1 mg/ml bovine serum albumin (BSA), in a total volume of 10 µl. This mixture was diluted to 0.5 M NaCl, and then dialyzed against TE buffer (10 mM Tris pH 7.5, 1 mM EDTA). Reconstituted nucleosomes were analyzed on a 4.5% polyacrylamide gel (acrylamide:bisacrylamide, 29:1) containing 0.5× TBE or TE buffer. Nucleosome species were excised and eluted from the gel in buffer containing carrier DNA (50 µg/ml) as described previously (Hamiche et al., 1999). For the experiments in Figures 5 and 6, mononucleosomes were separated from excess free DNA and dinucleosomes by glycerol gradient sedimentation. Glycerol gradient (5–20%) in TE was centrifuged in an SW40 Ti rotor at 36 000 r.p.m. for 16 h at 4°C. Mononucleosome peak fractions were directly used in reactions with NURF and GAL4-DBD, followed by MNase digestion.
Purification of GAL4 transcription factors
GAL4(1–147) is described in Mizuguchi et al. (1997). GAL4-DBD(1–94) was cloned into the NcoI and BamHI sites of the pET 28a expression vector. GAL4-DBD proteins were expressed in Escherichia coli and purified with heparin–Sepharose CL6B (Amersham Pharmacia Biotech) and GAL4-binding affinity resin as described previously (Mizuguchi et al., 1997). The amount of each GAL4 protein was normalized based on DNA-binding activity.
Exo III protection
Native gel bands corresponding to each nucleosome species (radiolabeled at either end of GAL4-E4 promoter) were excised from 4.5% poly acrylamide gel containing TE as described previously (Hamiche et al., 1999). Nucleosomes were eluted and digested with Exo III (400 U/ml) for 2 min at 37°C, followed by electrophoresis in a 6% polyacrylamide gel containing 8 M urea.
Nucleosome remodeling
NURF was purified to the final glycerol gradient step from 0 to 12 h Drosophila embryos as described previously (Tsukiyama and Wu, 1995; Sandaltzopoulos et al., 1999) except that Mono-Q and Mono-S columns (Amersham Pharmacia Biotech) were substituted for Q–Sepharose and phosphocellulose resin. Mononucleosomes reconstituted on GAL4-E4 promoter fragment (40 nM), or each nucleosome species eluted from the gel, were incubated with NURF (0.2–0.4 nM) or purified recombinant ACF (0.8–1.6 nM; a kind gift from J.Kadonaga’s laboratory) in sliding buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 3 mM MgCl2, 1 mM β-mercaptoethanol, 1 mg/ml BSA) containing 1 mM ATP for 30 min at 26°C. Nucleosome sliding was analyzed by 4.5% PAGE in 0.5× TBE.
Micrococcal nuclease and restriction enzyme digestion
Nucleosomes were digested with MNase (25 U/ml) in the presence of 20 µg/ml sheared sperm salmon DNA and 1 mM CaCl2 for 30 min on ice and 3 min at 37°C as recommended by Davey et al. (1998). The digestions were stopped with the addition of 100 µl of stop solution (20 mM EDTA, 0.2 M NaCl, 1% SDS, 0.25 mg/ml glycogen, 0.2 mg/ml proteinase K) and incubated for 30 min at 37°C. MNase digests were purified by phenol–chloroform extraction and ethanol precipitation. Purified DNAs were further digested with BanI (1300 U/ml) or Fnu4H1 (333 U/ml) at 37°C for 30 min, analyzed by TBE–8% PAGE at 250 V for 2 h and visualized by autoradiography.
Acknowledgments
Acknowledgements
We thank Tim Richmond and Karolin Luger for helpful discussions, Jim Kadonaga and Dmitry Fyodorov for a kind gift of purified ACF, Gaku Mizuguchi for helping initial purification of NURF and GAL4 proteins, Hua Xiao for assisting construction of GAL4(1–94) and members of our laboratory for helpful suggestions. This work was supported by the Intramural Research Program of the National Cancer Institute.
References
- Alfas J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- Armstrong J.A. and Emerson,B.M. (1996) NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vitro. Mol. Cell. Biol., 16, 5634–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist P., Belikov,S. and Wrange,O. (1999) Increased nuclear factor 1 binding to its nucleosomal site mediated by sequence-dependent DNA structure. Nucleic Acids Res., 27, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Langst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns L.G. and Peterson,C.L. (1997) The yeast SWI–SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell. Biol., 17, 4811–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R. (1998) Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci., 23, 20–25. [DOI] [PubMed] [Google Scholar]
- Cairns B.R. et al. (1996) RSC, an essential, abundant chromatin-remodeling complex. Cell, 87, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Carey M. and Smale,S.T. (2000) Transcriptional Regulation in Eukaryotes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Carey M., Kakidani,H., Leatherwood,J., Mostashari,F. and Ptashne,M. (1989) An amino-terminal fragment of GAL4 binds DNA as a dimer. J. Mol. Biol., 209, 423–432. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin,Y.S., Green,M.R. and Ptashne,M. (1990) A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature, 345, 361–364. [DOI] [PubMed] [Google Scholar]
- Cirillo L.A. and Zaret,K.S. (1999) An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell, 4, 961–969. [DOI] [PubMed] [Google Scholar]
- Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Cote J., Quinn,J., Workman,J.L. and Peterson,C.L. (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science, 265, 53–60. [DOI] [PubMed] [Google Scholar]
- Davey C., Pennings,S. and Allan,J. (1998) Reconstitution and analysis of nucleosome positioning. In Gould,H. (ed.), Chromatin: A Practical Approach. Oxford University Press, Oxford, UK, pp. 153–172. [Google Scholar]
- Dong F., Hansen,J.C. and van Holde,K.E. (1990) DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc. Natl Acad. Sci. USA, 87, 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H.R. (1991) Can one measure the free energy of binding of the histone octamer to different DNA sequences by salt-dependent reconstitution? J. Mol. Biol., 219, 391–392. [DOI] [PubMed] [Google Scholar]
- Duband-Goulet I., Carot,V., Ulyanov,A.V., Douc-Rasy,S. and Prunell,A. (1992) Chromatin reconstitution on small DNA rings. IV. DNA supercoiling and nucleosome sequence preference. J. Mol. Biol., 224, 981–1001. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Ferrari,S., Langst,G., Straub,T., Imhof,A., Varga-Weisz,P., Wilm,M. and Becker,P.B. (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J., 20, 3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A. and Richmond,T.J. (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol., 275, 427–441. [DOI] [PubMed] [Google Scholar]
- Gdula D.A., Sandaltzopoulos,R., Tsukiyama,T., Ossipow,V. and Wu,C. (1998) Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev., 12, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Imbalzano A.N., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Imbalzano,A.N., Khavari,P.A., Kingston,R.E. and Green,M.R. (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature, 370, 477–481. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Li Q. and Wrange,O. (1993) Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev., 7, 2471–2482. [DOI] [PubMed] [Google Scholar]
- Lin Y.S., Carey,M.F., Ptashne,M. and Green,M.R. (1988) GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell, 54, 659–664. [DOI] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos,D. (2001) Nucleosome sliding via TBP DNA binding in vivo. Cell, 106, 685–696. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Tsukiyama,T., Gdula,D. and Wu,C. (1998) Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl Acad. Sci. USA, 95, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meersseman G., Pennings,S. and Bradbury,E.M. (1992) Mobile nucleosomes–a general behavior. EMBO J., 11, 2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Tsukiyama,T., Wisniewski,J. and Wu,C. (1997) Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell, 1, 141–150. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Vassilev,A., Tsukiyama,T., Nakatani,Y. and Wu,C. (2001) ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J. Biol. Chem., 276, 14773–14783. [DOI] [PubMed] [Google Scholar]
- Ng K.W., Ridgway,P., Cohen,D.R. and Tremethick,D.J. (1997) The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J., 16, 2072–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund Farrants A.K., Blomquist,P., Kwon,H. and Wrange,O. (1997) Glucocorticoid receptor–glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol., 17, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T. and Workman,J.L. (1996) Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J., 15, 4702–4712. [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T., Utley,R.T., Cote,J., Peterson,C.L. and Workman,J.L. (1996) Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science, 273, 513–516. [DOI] [PubMed] [Google Scholar]
- Pazin M.J., Kamakaka,R.T. and Kadonaga,J.T. (1994) ATP-dependent nucleosome reconfiguration and transcriptional activation from pre assembled chromatin templates. Science, 266, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Pazin M.J., Bhargava,P., Geiduschek,E.P. and Kadonaga,J.T. (1997) Nucleosome mobility and the maintenance of nucleosome positioning. Science, 276, 809–812. [DOI] [PubMed] [Google Scholar]
- Pennings S., Meersseman,G. and Bradbury,E.M. (1991) Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol., 220, 101–110. [DOI] [PubMed] [Google Scholar]
- Pina B., Bruggemeier,U. and Beato,M. (1990) Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell, 60, 719–731. [DOI] [PubMed] [Google Scholar]
- Rhodes D. and Laskey,R.A. (1989) Assembly of nucleosomes and chromatin in vitro. Methods Enzymol., 170, 575–585. [DOI] [PubMed] [Google Scholar]
- Richmond T. and Widom,J. (2000) In Elgin,S.C. and Workman,J.L. (eds), Chromatin Structure and Gene Expression. Oxford University Press, Oxford, UK. [Google Scholar]
- Sandaltzopoulos R., Ossipow,V., Gdula,D.A., Tsukiyama,T. and Wu,C. (1999) Purification of Drosophila nucleosome remodeling factor. Methods Enzymol., 304, 757–765. [DOI] [PubMed] [Google Scholar]
- Shen X., Mizuguchi,G., Hamiche,A. and Wu,C. (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature, 406, 541–544. [DOI] [PubMed] [Google Scholar]
- Studitsky V.M. (1999) Preparation and analysis of positioned nucleosomes. Methods Mol. Biol., 119, 17–26. [DOI] [PubMed] [Google Scholar]
- Studitsky V.M., Clark,D.J. and Felsenfeld,G. (1994) A histone octamer can step around a transcribing polymerase without leaving the template. Cell, 76, 371–382. [DOI] [PubMed] [Google Scholar]
- Taylor I.C., Workman,J.L., Schuetz,T.J. and Kingston,R.E. (1991) Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev., 5, 1285–1298. [DOI] [PubMed] [Google Scholar]
- Travers A. and Drew,H. (1997) DNA recognition and nucleosome organization. Biopolymers, 44, 423–433. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu,C. (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Becker,P.B. and Wu,C. (1994) ATP-dependent nucleo some disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature, 367, 525–532. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel,C., Tamkun,J. and Wu,C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor Cell, 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M., Walter,P., Chen,H., Juan,L.J. and Workman,J.L. (1994) Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol. Cell. Biol., 14, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Hassan,A.H., Neely,K.E. and Workman,J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall G., Varga-Weisz,P.D., Sandaltzopoulos,R. and Becker,P.B. (1995) Chromatin remodeling by GAGA factor and heat shock factor at the hypersensitive Drosophila hsp26 promoter in vitro. EMBO J., 14, 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (1996) Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J., 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wechsler D.S., Papoulas,O., Dang,C.V. and Kingston,R.E. (1994) Differential binding of c-Myc and Max to nucleosomal DNA. Mol. Cell. Biol., 14, 4097–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I., Flaus,A., Cairns,B.R., White,M.F., Workman,J.L. and Owen-Hughes,T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Widom J. (1999) Equilibrium and dynamic nucleosome stability. Methods Mol. Biol., 119, 61–77. [DOI] [PubMed] [Google Scholar]
- Wong J., Shi,Y.B. and Wolffe,A.P. (1995) A role for nucleosome assembly in both silencing and activation of the Xenopus TR βA gene by the thyroid hormone receptor. Genes Dev., 9, 2696–2711. [DOI] [PubMed] [Google Scholar]
- Workman J.L., Taylor,I.C. and Kingston,R.E. (1991) Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell, 64, 533–544. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos,R., Wang,H.M., Hamiche,A., Ranallo,R., Lee,K.M., Fu,D. and Wu,C. (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell, 8, 1–20. [DOI] [PubMed] [Google Scholar]