Abstract
Nijmegen breakage syndrome (NBS) is an autosomal recessive hereditary disease that shares some common defects with ataxia–telangiectasia. The gene product mutated in NBS, named NBS1, is a component of the Mre11 complex that is involved in DNA strand-break repair. To elucidate the physiological roles of NBS1, we disrupted the N-terminal exons of the NBS1 gene in mice. NBS1m/m mice are viable, growth retarded and hypersensitive to ionizing radiation (IR). NBS1m/m mice exhibit multiple lymphoid developmental defects, and rapidly develop thymic lymphoma. In addition, female NBS1m/m mice are sterile due to oogenesis failure. NBS1m/m cells are impaired in cellular responses to IR and defective in cellular proliferation. Most systematic and cellular defects identified in NBS1m/m mice recapitulate those in NBS patients, and are essentially identical to those observed in Atm–/– mice. In contrast to Atm–/– mice, spermatogenesis is normal in NBS1m/m mice, indicating that distinct roles of ATM have differential requirement for NBS1 activity. Thus, NBS1 and ATM have overlapping and distinct functions in animal development and DNA repair.
Keywords: cancer/cell cycle/DNA repair/genetic stability/immunodeficiency
Introduction
Nijmegen breakage syndrome (NBS) is an autosomal recessive genetic disease characterized by multisystemic defects, including microcephaly, growth retardation, immunodeficiency and a high incidence of lymphoid malignancies (van der Burgt et al., 1996). NBS is one of the four genetic instability syndromes including ataxia– telangiectasia (AT), Bloom’s syndrome and Fanconi’s anemia. Like lymphocytes of AT patients, lymphocytes from NBS patients usually harbor chromosomal translocations involving chromosomes 7 and 14, which interrupt the immunoglobulin heavy chain loci and T-cell receptor loci (Kojis et al., 1991; Stumm et al., 2001). NBS cells are also hypersensitive to γ-irradiation and impaired in cellular responses to γ-irradiation, including radioresistant DNA synthesis (RDS) and G2/M checkpoint defects (Sullivan et al., 1997; Ito et al., 1999). The gene mutated in NBS patients, NBS1 (nibrin), has been identified through positional cloning and functional complementation as the p95 component of the Mre11 complex (Carney et al., 1998; Matsuura et al., 1998; Varon et al., 1998). Sequence analysis of NBS1 revealed a fork-head-associated domain and a BRCA C-terminal domain at the N-terminus of NBS1 (Varon et al., 1998). All known NBS1 mutations are within nucleotides 657–1142 and 90% of them contain a deletion (657del5) (Varon et al., 1998).
The NBS cellular defects suggest that NBS1 functions in DNA double-strand break (DSB) repair. Consistent with this notion, soon after the introduction of DSBs, Rad50–Mre11–NBS1 complex forms foci at the sites of DNA DSBs induced by ionizing radiation (IR) or the V(D)J rearrangement process during B- and T-cell development (Carney et al., 1998; Chen et al., 2000). The early association of this complex with damaged DNA suggests that the Mre11 complex might be involved in sensing DNA DSBs and initiating the DNA repair pathway. The importance of the Mre11 complex in cellular responses to DNA strand-break damage is further suggested by the finding that the Mre1l mutation leads to a NBS- and AT-like syndrome in human patients (Stewart et al., 1999). Since Mre11 and Rad50 are cytoplasmic and cannot form foci at the sites of DSBs in the absence of NBS1, one role of NBS1 in DNA DSB repair could involve the recruitment of Mre11/Rad50 into the nucleus (Carney et al., 1998). In addition, while the expression of N-terminal truncated NBS1 can mediate nuclear localization of Mre11 and Rad50, no Mre11 foci can form after DNA strand-break damage (Desai-Mehta et al., 2001). Therefore, NBS1 is required for both nuclear localization of Mre11/Rad50 and foci formation upon DNA strand-break damage. The direct involvement of NBS1 in DNA DSB repair was further suggested by several NBS1-dependent activities, including unwinding of a DNA complex and cleavage of paired hairpin structures (Paull and Gellert, 1999).
The similar defects observed in NBS and AT patients suggest a functional link between ATM and NBS1. In support of this notion, several studies demonstrated that ATM can phosphorylate NBS1 at N-terminal residues and these phosphorylation events are required for NBS1 function in rescuing cellular defects in NBS1 mutant cell lines (Gatei et al., 2000; Lim et al., 2000; Wu et al., 2000; Zhao et al., 2000). In addition, both ATM and NBS1 are required for activation of the Chk2 kinase and the G2/M checkpoint control upon DNA strand-break damage (Dasika et al., 1999; Buscemi et al., 2001). Therefore, these studies suggest that ATM and NBS1 function in a linear pathway in cellular response to DNA strand-break damage. Additionally, these data indicate that ATM is an upstream activator of NBS1.
To analyze the physiological roles of NBS1, the N-terminal exons of NBS1 were disrupted in mice. NBS1 mutant mice recapitulate most of the NBS defects, including growth retardation, impaired lymphoid development, immunodeficiency, oogenesis failure and cancer predisposition. NBS cellular defects are also evident in NBS1 mutant mouse cells. However, unlike Atm–/– mice, no meiosis failure leading to abolished spermatogenesis is detected in NBS1 mutant mice, suggesting that some functions of ATM do not require NBS1. Therefore, our findings provide genetic evidence for a functional interaction between ATM and NBS1. In addition, these findings also reveal the important roles of NBS1 in multiple developmental processes and document the usefulness of NBS1 mutant mice as a mouse model to study the mechanism of pathogenesis in NBS.
Results
Targeted disruption of NBS1 gene in mice
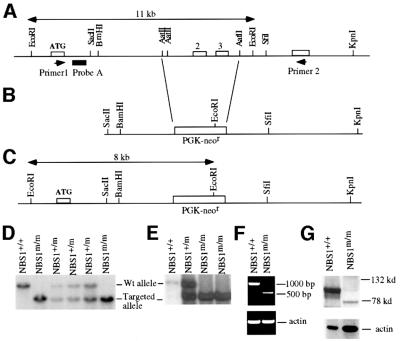
To disrupt the NBS1 gene in mice, a targeting vector was constructed to replace exons 2 and 3 (the exon containing the initiation codon ATG is designated exon 1) of the mouse NBS1 gene. These two exons encode codons 12–194 of NBS1 (Figure 1A and B). The targeting vector was electroporated into mouse embryonic stem (ES) cells, which were subsequently selected with G418. Homologous recombinants were screened by Southern blot analysis of genomic DNA derived from G418-resistant ES cell clones with EcoRI digestion and hybridization to probe A, giving an 11 kb germline band and an 8 kb mutant band (Figure 1A, C and D). Heterozygous NBS1 mutant ES cells (NBS1+/m) were injected into mouse blastocysts to generate chimeric mice, which transmitted the mutation into the germline. Breeding of NBS1+/m mice gave rise to homozygous mutant (NBS1m/m) mice (Figure 1D). To generate NBS1m/m ES cells, NBS1+/m ES cells were cultured under the selection of increasing concentrations of G418 as described previously (Xu and Baltimore, 1996). The NBS1m/m ES cells were subcloned and confirmed by Southern blotting (Figure 1E).
Fig. 1. Targeted disruption of the NBS1 gene in mice. (A) Germline configuration of the mouse NBS1 locus. Open boxes represent NBS1 exons. Primers 1 and 2 for RT–PCR analysis are indicated by arrowheads. The lengths of the germline EcoRI restriction fragment and probe A are indicated. (B) Targeting vector to replace exons 2 and 3 with the PGK-neor gene. (C) Targeted NBS1 allele. The size of the mutant EcoRI restriction fragment is indicated. Southern blot analysis of the genomic DNA derived from the tails of the offspring of NBS1+/m intercross (D) or NBS1m/m ES cells (E). The EcoRI restriction fragment derived from the wild-type and mutant allele is indicated. The genotypes are indicated at the top. (F) RT–PCR analysis of the NBS1 mRNA in wild-type and NBS1m/m MEFs. The size of the DNA marker is indicated on the right and the genotypes at the top. (G) Western blot analysis of NBS1 protein expression in wild-type and NBS1m/m MEFs. Mouse NBS1 was identified by a polyclonal antibody specific for the C-terminus of mouse NBS1 (Chen et al., 2000). The size of the molecular weight marker is indicated on the right and genotypes at the top.
To confirm that exons 2 and 3 of the wild-type NBS1 gene were disrupted in NBS1m/m mice, RT–PCR was employed to examine the expression of NBS1 mRNA in NBS1+/+ and NBS1m/m mouse embryonic fibroblasts (MEFs) as described previously (Xu and Baltimore, 1996). Primers 1 and 2 used for PCR reaction are located 5′ of exon 2 and 3′ of exon 3, respectively (Figure 1A). While no PCR product of the wild-type NBS1 mRNA was detected in NBS1m/m cells, a lower level PCR product of a truncated NBS1 mRNA was present in NBS1m/m cells (Figure 1F). Sequence analysis of this truncated NBS1 mRNA confirmed the deletion of exons 2 and 3 from the mRNA, and indicated the trans-splicing of exon 1 to exon 4 of the NBS1 pre-mRNA, transcribed from the disrupted NBS1 allele. Splicing of exon 1 to exon 4 introduces a frameshift mutation. To further test whether truncated NBS1 proteins were present in NBS1m/m cells, protein extracts derived from NBS1m/m and control NBS1+/+ MEFs were analyzed by western blotting with a polyclonal antibody specific for the extreme C-terminus of mouse NBS1 (Chen et al., 2000). While abundant wild-type NBS1 was detected at ∼95 kDa in the protein extracts from NBS1+/+ cells, no full-length NBS1 protein can be detected in NBS1m/m cells (Figure 1G). However, a truncated NBS1 polypeptide at ∼75 kDa was detected at a much lower level in NBS1m/m cells, suggesting an internal initiation of translation within the truncated NBS1 mRNA, as has been observed in cells derived from NBS patients (Maser et al., 2001).
Radiosensitivity of NBS1m/m cells and mice
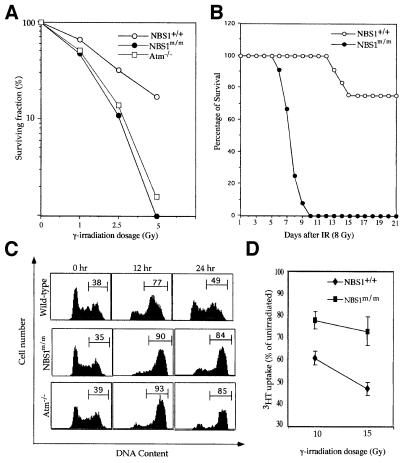
One of the hallmarks of NBS is hypersensitivity to γ-irradiation (van der Burgt et al., 1996). Therefore, we examined the sensitivity of NBS1m/m and control NBS1+/+ ES cells to γ-irradiation using a clonal survival assay, as described previously (Xu and Baltimore, 1996). Similar to Atm–/– ES cells, NBS1m/m ES cells are hypersensitive to γ-irradiation (Figure 2A). We also examined the sensitivity of NBS1m/m and NBS1+/+ mice to γ-irradiation by exposing them to 8 Gy of γ-irradiation. Consistent with the cellular hypersensitivity to γ-irradiation, irradiated NBS1m/m mice all died by 10 days after irradiation, while most NBS1+/+ mice were viable for at least 2 months after irradiation (Figure 2B). Similarly to Atm–/– mice, the hypersensitivity of NBS1m/m mice to γ-irradiation is due to acute radiation-induced toxicity to the intestinal tract (data not shown; Barlow et al., 1996).
Fig. 2. Radiosensitivity, G2 arrest and RDS after γ-irradiation. (A) Clonogenic survival of wild-type, NBS1m/m and Atm–/– ES cells after exposure to graded doses of γ-irradiation. Consistent data were obtained from three independent experiments. (B) Sensitivity of wild-type and NBS1m/m mice to γ-irradiation. Twelve pairs of NBS1m/m and control NBS1+/+ mice were exposed to 8 Gy of γ-irradiation and survival after irradiation was monitored. (C) G2 accumulation in NBS1+/+, NBS1m/m and Atm–/– ES cells after γ-irradiation. ES cells were exposed to 5 Gy of γ-irradiation and cell cycle profile analyzed by staining with propidium iodide. The percentages of cells residing in G2/M phases are indicated. Consistent data were obtained from three independent experiments. (D) RDS in NBS1m/m MEFs after γ-irradiation. The mean value from three independent experiments is presented with error bars.
G2 accumulation and radioresistant DNA synthesis in NBS1m/m cells after γ-irradiation
Cells derived from AT and NBS patients accumulate at G2 upon γ-irradiation (Beamish and Lavin, 1994; Hong et al., 1994). Therefore, we examined NBS1m/m ES cells for G2 accumulation because wild-type ES cells undergo typical G2/M arrest following γ-irradiation (Hirao et al., 2000). While the cell cycle profile of wild-type ES cells returned largely to normal 24 h post γ-irradiation, most NBS1m/m and Atm–/– ES cells remained in G2/M phase 24 h post γ-irradiation (Figure 2C). Thus, as observed in AT and NBS cells, both NBS1m/m and Atm–/– ES cells undergo prolonged G2 accumulation after γ-irradiation. Another hallmark of NBS cellular defects is RDS. Therefore, we tested NBS1+/+ and NBS1m/m MEFs for RDS. When compared with NBS1+/+ MEFs, NBS1m/m MEFs exhibit increased RDS after 10 and 15 Gy of γ-irradiation (Figure 2D). Together, these findings indicate that NBS1m/m mouse cells recapitulate the cellular defects characteristic of NBS upon γ-irradiation.
Growth retardation in NBS1m/m mice
Genotyping of the offspring from the intercrossing of heterozygous mutant mice showed a ratio of NBS1+/+: NBS1+/m:NBS1m/m of 66:156:68 (1:2.2:1), close to the expected 1:2:1 Mendelian ratio. Therefore, NBS1m/m mice were viable and matured into adulthood. However, they were growth retarded at all development stages with a body weight 77 ± 5% (mean value from 10 sets of 6- to 9-week-old NBS1m/m and NBS1+/+ mice) of that of sex-matched NBS1+/+ littermates. The weight of the whole brain of NBS1m/m mice was similar to that of control NBS1+/+ mice and histological examination revealed no microencephaly in NBS1m/m mice. In addition, histological analysis of the brains of NBS1m/m and NBS1+/+ mice identified no apparent abnormalities in the development of the central nervous system (data not shown).
Impaired cellular proliferation in NBS1m/m MEFs
Fibroblasts derived from Atm–/– mice are defective in their cellular proliferation (Barlow et al., 1996; Xu and Baltimore, 1996). Therefore, we analyzed the cellular proliferation of NBS1m/m and NBS1+/+ MEFs. NBS1m/m MEFs of passage 2–5 proliferated slower than control NBS1+/+ MEFs with a lower saturation density (Figure 3A; data not shown). In addition, NBS1m/m MEFs underwent premature senescence at high passages (Figure 3B). p21 is a universal inhibitor of cyclin-dependent kinases and can inhibit G1/S transition as well as promote senescence (Serrano and Blasco, 2001). A high basal level of p21 was detected in Atm–/– MEFs and is the primary cause of the impaired cellular proliferation as well as premature senescence observed in Atm–/– MEFs (Xu and Baltimore, 1996; Xu et al., 1998). Therefore, we analyzed the protein level of p21 in NBS1m/m and NBS1+/+ MEFs of various passage numbers by western blotting. While p21 protein levels were similar in wt and NBS1m/m MEFs at very early passage, similar to that in Atm–/– MEFs, a significantly higher basal level of p21 was detected in NBS1m/m MEFs of passage 3 and higher (Figure 3C). Since p53 transactivates the expression of p21 in MEFs, we analyzed the basal protein level of p53 in NBS1m/m and NBS1+/+ MEFs of different passage numbers by western blotting. p53 was not detectable in either NBS1m/m or NBS1+/+ MEFs, indicating that the basal protein level of p53 was not significantly increased in NBS1m/m MEFs (data not shown). However, it is possible that the activity of p53 is induced in NBS1m/m MEFs due to the accumulation of unrepaired DNA damage. Since a high level of p21 inhibits G1/S phase transition, we analyzed the efficiency of mutant MEFs to enter S phase after serum stimulation. NBS1m/m and NBS1+/+ MEFs of passage 3 were synchronized at G0 by serum starvation, and subsequently released into the medium containing 10% fetal calf serum. The percentage of cells in S phase was determined 12, 24 and 36 h post-serum stimulation. A consistently lower percentage of NBS1m/m MEFs had entered S phase at all time points analyzed when compared with NBS1+/+ MEFs (Figure 3D). Therefore, NBS1m/m MEFs are impaired in G1/S transition during cellular proliferation.
Fig. 3. Cellular proliferation and p21 protein level in NBS1+/+ and NBS1m/m MEFs. The proliferation curve of MEFs at passage 4 (A) and passage 6 (B) is presented. The cell number represents the average of three plates counted at each time point. (C) The p21 protein level in proliferating NBS1+/+ and NBS1m/m MEFs. The genotypes and passage numbers are indicated at the top and p21/actin indicated on the right. (D) S-phase entry after serum stimulation of NBS1+/+ and NBS1m/m MEFs synchronized at G0. The S-phase cells (BrdU+) were revealed by anti-BrdU antibody and DNA content by propidium iodide staining. Boxes representing G0/G1, S and G2/M phases and percentage of cells in S phase are indicated. Consistent data were obtained from two independent experiments.
Defective lymphoid development and immunodeficiency in NBS1m/m mice
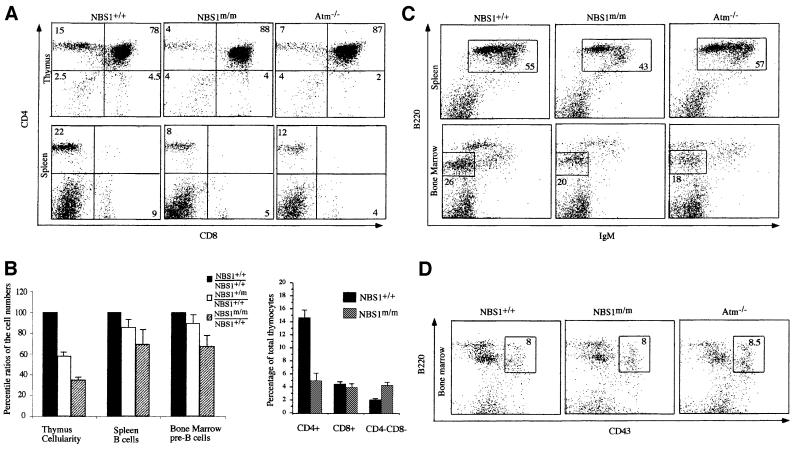
NBS patients are immunodeficient (van der Burgt et al., 1996). Therefore, we examined the lymphoid development in 5- to 12-week-old NBS1m/m and control NBS1+/+ mice. Similar to that in Atm–/– mice, the thymus cellularity of NBS1m/m mice was greatly reduced and ∼30% of that in NBS1+/+ mice (Figure 4B). Thymocyte development in mice can be broadly divided into three distinct stages by surface expression of CD4 and CD8 markers (Kisielow and von Boehmer, 1995). CD4–CD8– double-negative (DN) thymocytes represent T-cell precursors, which develop into CD4+CD8+ double-positive (DP) immature thymocytes. DP thymocytes are subsequently positively selected into CD4+ or CD8+ single-positive (SP) mature thymocytes or alternatively undergo apoptosis. Flow cytometric analysis for CD4 and CD8 markers showed that the percentage of CD4+ SP mature thymocytes is more dramatically reduced in NBS1m/m mice, leading to a >5-fold reduction in the absolute number of CD4+ thymocytes in NBS1m/m mice (Figure 4A and B). In this context, the absolute numbers of CD4+ thymocytes in 6- to 8-week-old NBS1+/+ and NBS1m/m mice are 17 ± 2 × 106 and 3.1 ± 0.8 × 106, respectively. Consistent with this finding, mature T cells were similarly reduced in the peripheral lymphoid organs of NBS1m/m mice (Figure 4A). The reduction of thymus cellularity in NBS1m/m mice is not due to fewer T-cell precursors because the percentage of DN precursor cells was increased in the NBS1m/m thymus, leading to similar absolute numbers of DN precursors in the NBS1m/m and control NBS1+/+ mice (Figure 4A and B). In this context, the absolute numbers of DN thymocytes in 6- to 8-week-old NBS1+/+ and NBS1m/m mice are 3.5 ± 0.28 × 106 and 3.2 ± 0.38 × 106, respectively.
Fig. 4. Lymphocyte development in NBS1+/+ and NBS1m/m mice. (A) Flow cytometric analysis of cells derived from thymus and spleen of NBS1+/+ and NBS1m/m mice for CD4 and CD8 surface markers. The percentage of total cells in the lymphoid gate is indicated. (B) Statistical analysis of the ratios of thymocytes, bone marrow pre-B and spleen mature B cells in NBS1m/m and NBS1+/m mice versus those in NBS1+/+ mice. Data were obtained from four sets of mice. The mean value is presented with error bars. Flow cytometric analysis of cells derived from spleen and bone marrow for B220/IgM (C) and B220/CD43 (D) surface markers. The percentage of total cells residing in the lymphoid gate is indicated.
B-cell development in the bone marrow can be broadly divided into three stages based on surface expression of B220, CD43 and IgM (Hardy and Hayakawa, 2001). B220+CD43+ B-lineage precursors in bone marrow develop into B220+IgM–CD43– pre-B cells, which develop into B220+IgM+ mature B cells. Similarly to Atm–/– mice, the numbers of B220+IgM– pre-B cells were reduced in NBS1m/m mice when compared with their control NBS1+/+ mice (Xu et al., 1996; Figure 4B and C). The reduction of pre-B cells in NBS1m/m mice is not due to fewer B-cell precursors since the number of B220+CD43+ pro-B cells is similar in NBS1m/m and control NBS1+/+ mice (Figure 4D). However, unlike Atm–/– mice, mature B cells in the peripheral lymphoid organs of NBS1m/m mice were also reduced compared with those in NBS1+/+ mice (Figure 4B and C).
We also observed an intermediate reduction of total thymocyte number in NBS1+/m mice when compared with NBS1+/+ and NBS1m/m mice (Figure 4B). However, the numbers of B and pre-B cells in NBS1+/m mice are comparable to those in NBS1+/+ mice.
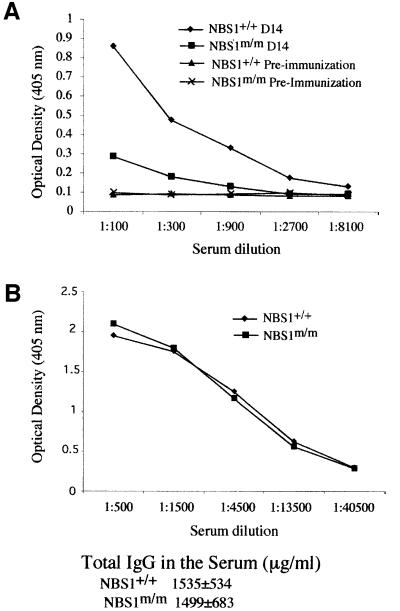
Clonal expansion of antigen-specific lymphocytes is the hallmark of adaptive immune responses. Therefore, reduced lymphocyte numbers in mice might not lead to severe immunodeficiency. To determine the T-dependent immune responses in NBS1m/m mice, we tested the T-dependent antibody responses to heptan conjugated to ovalbumin (NP5–OVA) in NBS1m/m and control NBS1+/+ mice. Consistent with the immunodeficiency observed in NBS patients, NBS1m/m mice were severely defective in the T-dependent antibody response (Figure 5). The lack of antibody responses in NBS1m/m mice could be due to impaired T-cell help or intrinsic defects in class switching in NBS1m/m B cells. Therefore, we determined the total IgG levels in the serum of 6- to 8-week-old NBS1m/m and control NBS1+/+ mice because intrinsic defects in class switching in B cells will lead to reduced serum IgG levels (Durandy and Honjo, 2001). Our analysis indicated similar levels of total IgG in the serum of NBS1m/m and control NBS1+/+ mice, suggesting that there are no gross defects in class switching in NBS1m/m B cells.
Fig. 5. T-dependent immune responses in NBS1+/+ and NBS1m/m mice. (A) Serial dilutions of serum (pre-immunization and 14 days after immunization) were analyzed for NP-specific IgG. Consistent data were obtained from four sets of immunized NBS1+/+ and NBS1m/m mice. (B) Serial dilutions of serum derived from NBS1+/+ and NBS1m/m mice were analyzed for total IgG. The mean value of the absolute amount of serum IgG in four pairs of 6- to 8-week-old NBS1+/+ and NBS1m/m mice is presented with the SD.
Thymic lymphomas in NBS1m/m mice
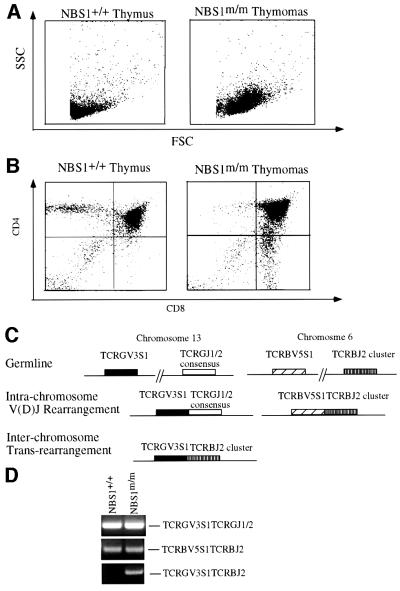
NBS patients suffer from a high incidence of lymphoid tumorigenesis (van der Burgt et al., 1996). NBS1m/m mice are also prone to thymic lymphomas. Among 32 3- to 8-month-old NBS1m/m mice monitored for tumorigenesis, five died of thymic lymphoma. Thymic lymphomas were detected in another four mutant mice when the surviving 3- to 8-month-old NBS1m/m mice were killed and examined. On the contrary, no tumors were detected in the NBS1+/+ littermate control. While the youngest NBS1m/m mouse that died of thymic lymphomas so far is 7 weeks old, the onset of tumorigenesis in NBS1m/m mice is significantly slower than that in Atm–/– mice, which mostly die of thymic lymphomas by 4 months of age (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996). Flow cytometric analysis of the tumor cells indicates that they are mostly of CD4+CD8+ immature thymocyte origin and composed of large sized lymphoblasts (Figure 6A and B). Histological examination of the thymic tumor indicated that the blasting tumor cells completely disrupted the cortical-medullar structure of the normal thymus (data not shown).
Fig. 6. Analysis of thymic lymphomas in NBS1m/m mice. (A and B) Cells derived from a thymic lymphoma of a 5-month-old NBS1m/m mouse as well as from a healthy wild-type littermate were analyzed by flow cytometry. The plot of FSC versus SSC (A) indicates large-sized lymphoblasts in the lymphomas. The plot of CD4 and CD8 markers (B) indicates that most lymphoma cells are of CD4+CD8+ immature thymocyte origin. (C) Schematic diagram of intra- and inter-chromosomal rearrangements in TCRβ and γ loci as described by Lista et al. (1997). TCRGV3S1TCRBJ2 rearrangement represents the formation of a t(6, 13) chromosomal translocation. (D) PCR analysis of the intra- and inter-chromosomal rearrangements. The PCR conditions were essentially the same as those described in Lista et al. (1997), and 100 ng of genomic DNA derived from thymocytes of NBS1+/+ and NBS1m/m mice were used for PCR amplification. The PCR products from the three rearrangements are indicated on the right and the genotypes at the top.
NBS is a genetic instability syndrome and NBS lymphocytes usually harbor chromosomal translocation involving the T-cell receptor locus (Kojis et al., 1991; Stumm et al., 2001). In an attempt to understand the mechanism of the high incidence of thymic tumors in NBS1m/m mice, we employed a PCR assay designed by Lista et al. (1997) to identify chromosomal translocations involving TCRβ and TCRγ loci in thymocytes derived from 4- to 6-week-old pre-tumor NBS1m/m mice and control NBS1+/+ mice (Figure 6C; Lista et al., 1997). The intra-locus V(D)J recombination at the analyzed TCRβ and TCRγ loci was similar in NBS1m/m and NBS1+/+ thymocytes, indicating that there are no gross defects in V(D)J joining in NBS1m/m thymocytes (Figure 6D). However, the frequency of interchromosomal translocation between Tcrβ and Tcrγ loci is greatly increased in NBS1m/m thymocytes (Figure 6D).
Germ cell development in NBS1m/m mice
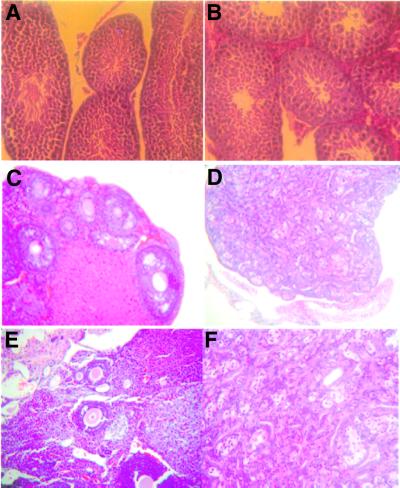
Since spermatogenesis is blocked at the early stages of development due to meiosis failure in Atm–/– mice (Barlow et al., 1996; Xu et al., 1996), we examined spermatogenesis in NBS1m/m and control NBS1+/+ mice. Spermatocytes of all developmental stages were identified in the seminiferous tubules of NBS1m/m and NBS1+/+ mice, and no apparent degeneration was detectable (Figure 7A and B). Consistent with these findings, NBS1m/m male mice were fertile. Since impaired oogenesis has been observed in NBS patients, we examined the ovaries derived from 8-week-old NBS1m/m and NBS1+/+ mice. While ovaries derived from the wild-type mice have primary oocytes and follicles of various developmental stages (Figure 7C and E), ovaries derived from NBS1m/m females were highly degenerated and completely lacked any oocytes and follicles (Figure 7D and F). Therefore, oogenesis is abolished in NBS1m/m females.
Fig. 7. Germ cell development in NBS1+/+ and NBS1m/m mice. Spermatogenesis in wild-type (A) and NBS1m/m (B) male mice. Histological sections of seminiferous tubules of 3-month-old wild-type and NBS1m/m mice were stained with hematoxylin–eosin. Hematoxylin–eosin-stained sections of ovaries from 6-week-old NBS1+/+ (C and E) and NBS1m/m (D and F) female mice. (C and D) Gross ovarian morphology at a low magnification. (E and F) Follicles of various developmental stages at a higher magnification.
Discussion
By disrupting the N-terminal exons of the NBS1 gene in mice, we have created a NBS mouse model that recapitulates most of the NBS defects, including growth retardation, impaired lymphocyte development, immunodeficiency, abolished oogenesis and a high incidence of lymphoid tumors. In addition, impaired cellular responses to γ-irradiation characteristic of NBS are also evident in the NBS1 mutant cells. Therefore, the NBS1 mutant mice represent a mouse model to study the mechanisms of pathogenesis in NBS patients. However, one published study of NBS1 disruption in mice indicated that disruption of NBS1 leads to early embryonic lethality (Zhu et al., 2001). To account for this discrepancy, we examined the targeting strategy in the two studies. Our targeting vector is designed to disrupt exons 2 and 3 of the mouse NBS1 gene. The targeting vector in the previous study disrupted intron 1 and exon 1, containing the initiation ATG of the NBS1 transcript (Zhu et al., 2001). While it cannot be ruled out that the targeting vector in the earlier experiment might simultaneously disrupt an overlapping gene whose product is essential for embryonic survival, it is more likely that the discrepancy in the two studies is due to the different regions of the NBS1 gene that were targeted. While not examined in the previous study, the disruption of intron 1 of the NBS1 gene might interrupt the promoter of the NBS1 gene and thus abolish any NBS1 transcription from the targeted allele. However, in our study, the targeted allele can still express an N-terminal truncated NBS1, although at a greatly reduced level. It is possible that the NBS1 function retained by the truncated NBS1 protein could prevent the embryonic lethality. In this context, the NBS1 mutant mice described here share the same characteristics with NBS patients, since the NBS1 mutant allele in NBS patients also produces an N-terminal truncated NBS1 protein of ∼70 kDa (Maser et al., 2001).
Several defects that we observed in NBS1 mutant mice and cells strongly support an essential role for NBS1 in the repair of DNA DSBs. One such observation is the defective lymphocyte development observed in NBS1 mutant mice. Lymphocytes mature by expressing B- or T-cell receptor on the surface through somatic V(D)J rearrangement, which is a physiological process of DNA DSB repair (Weaver and Alt, 1997). The potential roles of NBS1 in V(D)J recombination have been suggested by the finding that NBS1 foci can be detected at V(D)J recombination-induced DNA DSBs (Chen et al., 2000). Our analysis of NBS1 mutant mice indicates that both immature B and T cells are significantly reduced. This suggests that V(D)J recombination might be impaired in NBS1 mutant lymphocytes since productive V(D)J recombination at the immunoglobulin heavy chain and TCRβ loci is required for the significant cellular expansion of immature lymphocytes (Sleckman et al., 1996). In addition, the more dramatic reduction of CD4+ SP thymocytes suggests impaired V(D)J recombination since productive V(D)J rearrangement at the TCRα loci is required for the transition from the DP stage to the CD4 SP stage (Ellmeier et al., 1999). However, PCR analysis of the rearrangement of a number of V, D, J gene segments at TCRβ and γ loci showed no gross defects in the joining of the V, D, J gene segments. Therefore, it is possible that the efficiency of productive V(D)J recombination is reduced in NBS1m/m thymocytes. In this context, increased chromosomal translocations involving TCR loci in NBS1m/m thymocytes could contribute to the low efficiency of productive V(D)J recombination. The increase in chromosomal translocations observed in NBS1m/m thymocytes also recapitulates the genetic instability observed in NBS lymphocytes and can contribute to tumorigenesis. Defective cellular proliferation might also contribute to the immune defects observed in NBS1 mutant mice, since it has been shown that similar T-cell developmental defects in Atm–/– mice are partially due to impaired thymocyte expansion (Chao et al., 2000).
The similarity between the many phenotypes of NBS1 and Atm mutant mice indicates that some ATM functions in the cellular responses to DNA strand-break damage are mediated through NBS1-dependent pathways. Both ATM and NBS1 are required for activation of Chk2 after DNA strand-break damage (Matsuoka et al., 1998; Ahn et al., 2000; Melchionna et al., 2000; Buscemi et al., 2001). In addition, activation of Chk2 by ATM after DNA damage is dependent on a functional NBS1 (Buscemi et al., 2001). Since activation of Chk2 is required for both S phase and G2/M checkpoints, the functions of ATM in S and G2/M checkpoints are probably mediated by NBS1. Together, these findings indicate that ATM and the Mre11 complex function in the same pathways in response to DNA strand-break damage. In addition, similar cellular proliferative defects observed in Atm–/– and NBS1 mutant cells also suggest that Atm function in cellular proliferation depends on NBS1 activity.
Several findings also suggest that Atm and NBS1 have distinct functions in animal development. Meiosis failure leading to blocked spermatogenesis in Atm–/– mice is not observed in NBS1 mutant mice. While it is possible that the N-terminal truncated NBS1 could provide sufficient wild-type NBS1 activity to allow normal spermatogenesis, it is also possible that the essential role of ATM in meiosis is not dependent on NBS1 since ATM activates a number of other DNA repair pathways upon DNA strand-break damage. In this context, Brca1, which is also activated by ATM through phosphorylation after DNA strand-break damage, is required for homologous recombination and thus might mediate ATM-dependent functions during meiosis (Cortez et al., 1999; Moynahan et al., 1999; Li et al., 2000). But in any case, it is apparent from our studies that various ATM functions have a differential requirement for NBS1 activity. Our findings also suggest that NBS1 has functions independent of ATM. In this context, there is a significant reduction of mature B cells in NBS1 mutant mice, but not in Atm–/– mice. In addition, while Atm-null mutant mice are viable, NBS1-null mice are embryonic lethal, further supporting the notion that NBS1 has essential functions independent of Atm during animal development (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Zhu et al., 2001). In conclusion, Atm and NBS1 function in overlapping as well as distinct pathways in regulating cellular responses to DNA damage and animal development.
Materials and methods
Construction of NBS1 mutant ES cells and mice
The N-terminal mouse NBS1 cDNA fragment was used as a probe to isolate NBS1 mouse genomic DNA from a mouse 129 genomic DNA phage library (Stategene) as described previously (Xu and Baltimore, 1996). The mouse NBS1 genomic DNA was cloned into pBluescript and exon–intron structure characterized by restriction digestion, Southern blot analysis and DNA sequencing. The targeting vector was constructed by replacing exons 2 and 3 of the mouse NBS1 gene with the PGK-neor gene (Figure 1A and B). Targeting vector was linearized by SalI digestion and electroporated into ES cells. ES cell transfectants were selected with G418 (0.3 mg/ml) and homologous recombination events were screened by Southern blotting with EcoRI digestion and hybridization to probe A (Figure 1A and C). To generate homozygous mutant ES cells, heterozygous mutant (NBS1+/m) ES cells were selected with increasing concentrations of G418 as described previously (Xu and Baltimore, 1996). NBS1+/m ES were also microinjected into C57BL/6 blastocysts to generate chimeric mice, which transmitted the mutation into mouse germline. NBS1+/m mice were intercrossed to generate NBS1m/m mice.
Analysis of NBS1 mRNA in MEFs by RT–PCR
To analyze the expression of NBS1 transcript in NBS1m/m cells, total RNA was isolated from NBS1+/+ and NBS1m/m MEFs using TRI Reagent (Molecular Research Center, Inc.) and reversely transcribed into cDNA as described previously (Xu and Baltimore, 1996). cDNA was amplified by PCR with primers 1 and 2, which are located upstream and downstream of the targeted exons (Figure 1A). The sequences of primers are as follows: primer 1, 5′-ATGTGGAAGCTGCTCCCGGCCGCC-3′; primer 2, 5′-CATATTCACTGGAGCACTTGGGAT-3′.
Culture of primary cells
ES cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal calf serum (FCS), 1× glutamine, penicillin/streptomycin, non-essential amino acids, 100 µM β-mercaptoethanol and leukemia inhibitory factor.
MEFs were derived from day 14 NBS1+/+ and NBS1m/m embryos as described previously (Xu and Baltimore, 1996). MEFs were cultured in DMEM supplemented with 10% FCS, 1× glutamine, penicillin/streptomycin and 50 µM β-mercaptoethanol.
Radiation treatment of cells and mice
To test the clonogenic survival of NBS1+/+ and NBS1m/m ES cells after γ-irradiation, ES cells were plated on the feeder layer in a series of 6-well plates at a density of 1 × 103/well. Twenty-four hours after plating, ES cells were exposed to graded dosages of γ-irradiation. The irradiated ES cells were maintained with medium change every 2 days. After 8 days of culture, the surviving ES cell colonies in each well were counted and compared with untreated controls. To determine the sensitivity of NBS1+/+ and NBS1m/m mice to γ-irradiation, 12 pairs of NBS1+/+ and NBS1m/m littermates were irradiated with a single dose of 8 Gy and their survival was monitored by daily inspection.
Radioresistant DNA synthesis
NBS1+/+ and NBS1m/m MEFs were plated in 24-well tissue culture plates at 15 000 cells/well. The next day, medium was replaced and cells irradiated at 0, 10 and 15 Gy. Ten microcuries of [3H]thymidine were added to each well and cells incubated for 4 h before [3H]thymidine incorporation was determined.
Protein analysis and antibodies
Western blot analysis was performed as described previously (Xu and Baltimore, 1996). Rabbit polyclonal anti-p21 antibody was purchased from Santa Cruz Biotech., Inc. Rabbit polyclonal anti-mouse NBS1 antibody recognizes the extreme C-terminus of mouse NBS1 and was developed in Dr Andre Nussenzweig’s laboratory (Chen et al., 2000).
Flow cytometric analysis
Single-cell suspensions were prepared from thymus, spleen, bone marrow and thymic lymphomas. Half a million cells were stained simultaneously with phycoerythrin-conjugated (CD4, B220) and fluorescein isothiocyanate (FITC)-conjugated (CD8) or biotinylated (IgM, CD43) antibodies. Biotinylated antibodies were revealed with FITC-conjugated streptavidin. All antibodies were obtained from PharMingen. Stained cells were analyzed with a FACscan (Becton-Dickinson) using CellQuest Software.
Analysis of T-dependent immune response
Two- to 3-month-old NBS1+/+ and NBS1m/m mice were injected with alum-precipitated NP5–OVA (100 µg per mouse) and sera collected before injection and at day 14 after immunization. IgG specific for NP was detected with a sandwiched enzyme-linked immunosorbent assay (ELISA) assay as described, using NP3–BSA as the capturing antigen (Xu et al., 1996). ELISA to detect total serum IgG levels was performed as described previously (Xu et al., 1996).
Histological analysis
Harvested tissues were fixed in buffered formalin, dehydrated, and embedded in paraffin. Section were cut and stained with hematoxylin– eosin. The histological image was examined with a light microscope and captured with a CCD camera using Photoshop.
Acknowledgments
Acknowledgements
We thank Dr Andre Nussenzweig for anti-mouse NBS1 antibody and Dr Cornelis Murre for critically reading this manuscript. This work was supported by a NIH grant (CA77563) to Y.X.
References
- Ahn J.Y., Schwarz,J.K., Piwnica-Worms,H. and Canman,C.E. (2000) Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res., 60, 5934–5936. [PubMed] [Google Scholar]
- Barlow C. et al. (1996) Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell, 86, 159–171. [DOI] [PubMed] [Google Scholar]
- Beamish H. and Lavin,M.F. (1994) Radiosensitivity in ataxia-telangiectasia: anomalies in radiation-induced cell cycle delay. Int. J. Radiat. Biol., 65, 175–184. [DOI] [PubMed] [Google Scholar]
- Buscemi G. et al. (2001) Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol., 21, 5214–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney J.P., Maser,R.S., Olivares,H., Davis,E.M., Le Beau,M., Yates,J.R.,III, Hays,L., Morgan,W.F. and Petrini,J.H. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell, 93, 477–486. [DOI] [PubMed] [Google Scholar]
- Chao C., Yang,E.M. and Xu,Y. (2000) Rescue of defective T cell development and function in Atm–/– mice by a functional TCR αβ transgene. J. Immunol., 164, 345–349. [DOI] [PubMed] [Google Scholar]
- Chen H.T. et al. (2000) Response to RAG-mediated VDJ cleavage by NBS1 and γ-H2AX. Science, 290, 1962–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Wang,Y., Qin,J. and Elledge,S.J. (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science, 286, 1162–1166. [DOI] [PubMed] [Google Scholar]
- Dasika G.K., Lin,S.C., Zhao,S., Sung,P., Tomkinson,A. and Lee,E.Y. (1999) DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene, 18, 7883–7899. [DOI] [PubMed] [Google Scholar]
- Desai-Mehta A., Cerosaletti,K.M. and Concannon,P. (2001) Distinct functional domains of nibrin mediate Mre11 binding, focus formation and nuclear localization. Mol. Cell. Biol., 21, 2184–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy A. and Honjo,T. (2001) Human genetic defects in class-switch recombination (hyper-IgM syndromes). Curr. Opin. Immunol., 13, 543–548. [DOI] [PubMed] [Google Scholar]
- Ellmeier W., Sawada,S. and Littman,D.R. (1999) The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol., 17, 523–554. [DOI] [PubMed] [Google Scholar]
- Elson A., Wang,Y., Daugherty,C.J., Morton,C.C., Zhou,F., Campos-Torres,J. and Leder,P. (1996) Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl Acad. Sci. USA, 93, 13084–13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei M. et al. (2000) ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nature Genet., 25, 115–119. [DOI] [PubMed] [Google Scholar]
- Hardy R.R. and Hayakawa,K. (2001) B cell development pathways. Annu. Rev. Immunol., 19, 595–621. [DOI] [PubMed] [Google Scholar]
- Hirao A., Kong,Y.Y., Matsuoka,S., Wakeham,A., Ruland,J., Yoshida,H., Liu,D., Elledge,S.J. and Mak,T.W. (2000) DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science, 287, 1824–1827. [DOI] [PubMed] [Google Scholar]
- Hong J.H., Gatti,R.A., Huo,Y.K., Chiang,C.S. and McBride,W.H. (1994) G2/M-phase arrest and release in ataxia telangiectasia and normal cells after exposure to ionizing radiation. Radiat. Res., 140, 17–23. [PubMed] [Google Scholar]
- Ito A., Tauchi,H., Kobayashi,J., Morishima,K., Nakamura,A., Hirokawa,Y., Matsuura,S., Ito,K. and Komatsu,K. (1999) Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem. Biophys. Res. Commun., 265, 716–721. [DOI] [PubMed] [Google Scholar]
- Kisielow P. and von Boehmer,H. (1995) Development and selection of T cells: facts and puzzles. Adv. Immunol., 58, 187–209. [DOI] [PubMed] [Google Scholar]
- Kojis T.L., Gatti,R.A. and Sparkes,R.S. (1991) The cytogenetics of ataxia telangiectasia. Cancer Genet. Cytogenet., 56, 143–156. [DOI] [PubMed] [Google Scholar]
- Li S., Ting,N.S., Zheng,L., Chen,P.L., Ziv,Y., Shiloh,Y., Lee,E.Y. and Lee,W.H. (2000) Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature, 406, 210–215. [DOI] [PubMed] [Google Scholar]
- Lim D.S., Kim,S.T., Xu,B., Maser,R.S., Lin,J., Petrini,J.H. and Kastan,M.B. (2000) ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature, 404, 613–617. [DOI] [PubMed] [Google Scholar]
- Lista F., Bertness,V., Guidos,C.J., Danska,J.S. and Kirsch,I.R. (1997) The absolute number of trans-rearrangements between the TCRG and TCRB loci is predictive of lymphoma risk: a severe combined immune deficiency (SCID) murine model. Cancer Res., 57, 4408–4413. [PubMed] [Google Scholar]
- Maser R.S., Zinkel,R. and Petrini,J.H. (2001) An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nature Genet., 27, 417–421. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Huang,M. and Elledge,S.J. (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science, 282, 1893–1897. [DOI] [PubMed] [Google Scholar]
- Matsuura S. et al. (1998) Positional cloning of the gene for Nijmegen breakage syndrome. Nature Genet., 19, 179–181. [DOI] [PubMed] [Google Scholar]
- Melchionna R., Chen,X.B., Blasina,A. and McGowan,C.H. (2000) Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nature Cell Biol., 2, 762–765. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu,J.W., Koller,B.H. and Jasin,M. (1999) Brca1 controls homology-directed DNA repair. Mol. Cell, 4, 511–518. [DOI] [PubMed] [Google Scholar]
- Paull T.T. and Gellert,M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev., 13, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M. and Blasco,M.A. (2001) Putting the stress on senescence. Curr. Opin. Cell Biol., 13, 748–753. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman,J.R. and Alt,F.W. (1996) Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol., 14, 459–481. [DOI] [PubMed] [Google Scholar]
- Stewart G.S. et al. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell, 99, 577–587. [DOI] [PubMed] [Google Scholar]
- Stumm M., Neubauer,S., Keindorff,S., Wegner,R.D., Wieacker,P. and Sauer,R. (2001) High frequency of spontaneous translocations revealed by FISH in cells from patients with the cancer-prone syndromes ataxia telangiectasia and Nijmegen breakage syndrome. Cytogenet. Cell Genet., 92, 186–191. [DOI] [PubMed] [Google Scholar]
- Sullivan K.E., Veksler,E., Lederman,H. and Lees-Miller,S.P. (1997) Cell cycle checkpoints and DNA repair in Nijmegen breakage syndrome. Clin. Immunol. Immunopathol., 82, 43–48. [DOI] [PubMed] [Google Scholar]
- van der Burgt I., Chrzanowska,K.H., Smeets,D. and Weemaes,C. (1996) Nijmegen breakage syndrome. J. Med. Genet., 33, 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R. et al. (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell, 93, 467–476. [DOI] [PubMed] [Google Scholar]
- Weaver D.T. and Alt,F.W. (1997) V(D)J recombination. From RAGs to stitches. Nature, 388, 428–429. [DOI] [PubMed] [Google Scholar]
- Wu X. et al. (2000) ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature, 405, 477–482. [DOI] [PubMed] [Google Scholar]
- Xu Y. and Baltimore,D. (1996) Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev., 10, 2401–2410. [DOI] [PubMed] [Google Scholar]
- Xu Y., Ashley,T., Brainerd,E.E., Bronson,R.T., Meyn,M.S. and Baltimore,D. (1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects and thymic lymphoma. Genes Dev., 10, 2411–2422. [DOI] [PubMed] [Google Scholar]
- Xu Y., Yang,E.M., Brugarolas,J., Jacks,T. and Baltimore,D. (1998) Involvement of p53 and p21 in cellular defects and tumorigenesis in atm–/– mice. Mol. Cell. Biol., 18, 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. et al. (2000) Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature, 405, 473–477. [DOI] [PubMed] [Google Scholar]
- Zhu J., Petersen,S., Tessarollo,L. and Nussenzweig,A. (2001) Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol., 11, 105–109. [DOI] [PubMed] [Google Scholar]