Abstract
Bridging the gap between the number of gene sequences in databases and the number of gene products that have been functionally characterized in any way is a major challenge for biology. A key characteristic of proteins, which can begin to elucidate their possible functions, is their subcellular location. A number of experimental approaches can reveal the subcellular localization of proteins in mammalian cells. However, genome databases now contain predicted sequences for a large number of potentially novel proteins that have yet to be studied in any way, let alone have their subcellular localization determined. Here we ask whether using bioinformatics tools to analyse the sequence of proteins whose subnuclear localizations have been determined can reveal characteristics or signatures that might allow us to predict localization for novel protein sequences.
Keywords: bioinformatics/nuclear organization/nucleus/protein sequence/protein structure
Related functions in the cytoplasm often take place within the confines of discrete membrane-bounded organelles (e.g. mitochondria, Golgi apparatus, peroxisomes). Despite the absence of such obvious physical compartments, the mammalian nucleus is also thought to be organized into domains associated with different facets of nuclear function. Proteins in common pathways often concentrate together into specific areas of the nucleus. For example, even though proteins involved in pre-mRNA splicing move rapidly around the nucleus, they appear to concentrate into multiple nuclear ‘speckles’ (Phair and Misteli, 2000). Similarly, the events of rDNA processing and ribosome biogenesis predominantly occur within the nucleolus (Lamond and Earnshaw, 1998).
Advances in proteomics and genome sequencing are adding rapidly to our knowledge of the proteins concentrated in subcompartments of the mammalian nucleus. Protein micro-characterization and mass spectrometry have identified many components of the splicing speckles/interchromatin granule clusters (IGCs), nuclear envelope and nucleolus (Neubauer et al., 1998; Mintz, et al., 1999; Dreger et al., 2001; Andersen et al., 2002). Visual screens can reveal the subnuclear location of large numbers of proteins by tagging either the endogenous or ectopically expressed protein with a visual reporter (Rolls et al., 1999; Misawa et al., 2000; Simpson et al., 2000; Sutherland et al., 2001). However, we are far from an understanding of the full complexity of gene products that locate in nuclear compartments.
Here we use a variety of web-based bioinformatics tools to address the question of whether the sequence characteristics of almost 400 human/mouse proteins known to concentrate in different nuclear compartments might allow us to predict potential localization for novel protein sequences in databases.
Characteristics of the primary sequences of nuclear proteins
The most fundamental characteristics of primary protein sequence are size (molecular weight, MW) and isoelectric point (pI). It was recently noted that the pI values of proteins from bacterial and archaeal proteomes have a bimodal distribution (peaks around pI 5 and pI 9). In contrast, the eukaryotic proteomes of Saccharomyces cerevisiae, Drosophila melanogaster and Caenorhabditis elegans appeared to have a tri-modal distribution, with an additional peak at ∼pI 7. This was suggested to reflect different functional requirements of cytoplasmic, membrane-associated and nuclear proteins (Schwartz et al., 2001). Whereas cytoplasmic proteins generally had pI values of ∼5.5, and values for integral membrane proteins clustered at ∼pI 9, values for nuclear proteins appeared to spread across the entire range. It was suggested that it is nuclear proteins that are responsible for the additional peak in pI values seen in eukaryotic proteomes, and that is absent from the proteomes of prokaryotes and archaeans (Schwartz et al., 2001).
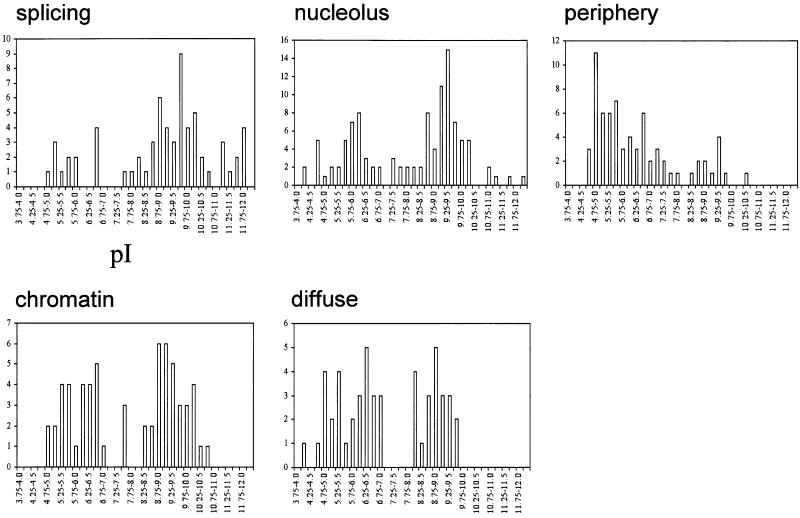
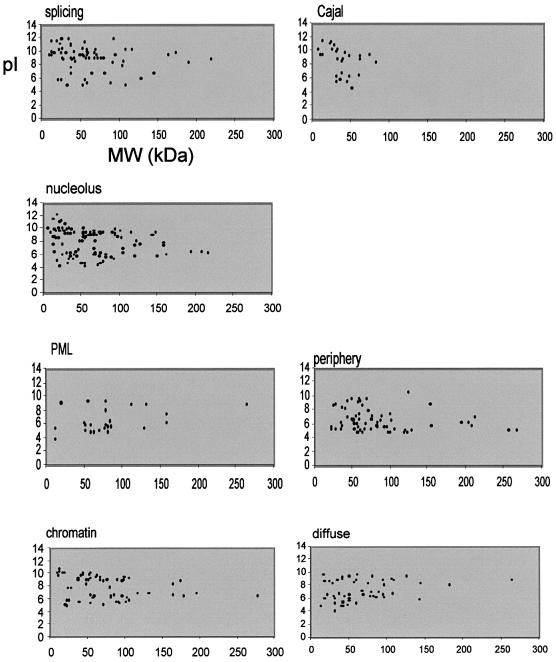
Do all subcompartments of the nucleus contain proteins with a similarly broad range of pI values? The ProtParam tool (http://www.expasy.ch/tools/protparam.html) was used to determine the predicted MW and pI for ∼400 proteins of known subnuclear localization and that are stored in a Nuclear Protein Database (http://www.hgu.mrc.ac.uk/NPD/) (Sutherland et al., 2001). Significant differences were found between both the average and the distribution of pI values for proteins assigned to different nuclear compartments (Table I; Figure 1). The compartment with, on average, the most basic proteins is the splicing speckles (Table I). pI values for these proteins peak between 9.5 and 9.75, and 28% of them have pI >10 (Figures 1 and 2). Proteins in Cajal bodies, nuclear domains that are thought to be involved in snRNP maturation and hence are functionally related to the splicing compartment (Sleeman and Lamond, 1999), also tend to be basic (Table I; Figure 2). Indeed, over half of the nuclear proteins in our survey with pI >10.5 are reported as concentrating in splicing speckles and 25% of them concentrate in Cajal bodies. The basic nature of the proteins in the splicing speckles might simply reflect their functional interactions with (m)RNA. However, this does not seem to be a pre-requisite for RNA binding or interacting proteins, since proteins in the other nuclear compartment dealing extensively with RNA processing (the nucleolus) do not necessarily share this characteristic. Nucleolar proteins have a very broad range of pI values in comparison with proteins in other nuclear compartments (Figure 1).
Table I. Size and pIs of proteins in subnuclear compartments.
| Compartment | MW (kDa) |
pI |
No. of known proteins | ||
|---|---|---|---|---|---|
| Mean | Median | Mean | Median | ||
| Splicing speckles | 62.8 | 53.7 | 9.0 | 9.5 | 65 |
| Cajal bodies | 38.5 | 34.9 | 8.6 | 9.2 | 28 |
| Nucleolus | 68.5 | 57.7 | 8.0 | 8.7 | 109 |
| PML/ND10 bodies | 84.4 | 77.7 | 6.4 | 5.8 | 24 |
| Nuclear periphery | 88.4 | 68.6 | 6.4 | 5.9 | 70 |
| Chromatin associated | 78.6 | 61.5 | 7.8 | 8.4 | 62 |
| Diffuse | 67.7 | 51.2 | 7.0 | 6.5 | 50 |
The physical and chemical parameters of proteins, which have been reported in the literature to be concentrated in nuclear compartments, were determined with the ProtParam tool (http://www.expasy.ch/tools/protparam.html).
Fig. 1. Frequency histograms showing the pI values, estimated using the ProtParam tool (http://www.expasy.ch/tools/protparam.html) (at 0.25 unit intervals) of nuclear proteins that have been reported to concentrate in different nuclear compartments (splicing speckles, nucleolus, nuclear periphery, and chromatin-associated). A selection of proteins that appear to be diffusely localized through the nucleoplasm are included for comparison.
Fig. 2. Scatter plots of pI versus molecular weight (kDa), estimated using the ProtParam tool (http://www.expasy.ch/tools/protparam.html), for proteins reported to concentrate in nuclear compartments. For considerations of scale, only proteins with molecular weights of <300 kDa are shown here, but these large proteins are included in the analyses summarized in Table I.
At the other end of the spectrum, proteins concentrated at the nuclear periphery, including many integral nuclear membrane proteins, are generally acidic (Table I; Figure 1). This contrasts strongly with the basic pI values seen among general integral membrane proteins in the cell (Schwartz et al., 2001). Forty percent of the nuclear proteins with pI <5 are proteins of the nuclear periphery, although none of these has a pI <4.5. Promyelocytic leukemia bodies (PML/ND10 bodies) also contain on average rather acidic proteins (Table I; Figure 2). The biological significance of this is unclear.
Chromatin-associated proteins within the mammalian nucleus have pI values clustering into two broad peaks of between 5.25–6.75 and 8.75–9.25 (Figures 1 and 2). The overall distribution of pI values for these proteins is narrow, with few proteins having pI values >10 or <5. The only chromatin proteins with pI >10 are small histone-like or HMG-like proteins. We only considered proteins to be chromatin associated if they had been demonstrated visually to co-localize in nuclei with DAPI counterstain, with domains of constitutive heterochromatin or facultative heterochromatin (inactive X and PcG domains), or to be associated with mitotic chromosomes. Hence we did not include most transcription factors in this class of proteins, for they are classified under proteins with a diffuse nucleoplasmic distribution. We also did not include core histone proteins in our analysis. As a comparison we sampled 50 proteins that have been reported to have a diffuse localization in the nucleus. These are probably the most abundant class of nuclear proteins, but their detailed subcellular localization is often poorly described in the literature. The pI characteristics of these proteins in our sample seem quite similar to those of chromatin-associated proteins, although >10% of the diffuse proteins analysed had a pI <5 and there were none with pI >9.75 (Figure 1).
There is no evidence in our survey for large numbers of proteins in discrete nuclear compartments with pI ∼7 that could account for the peak at this value in the analysis of Schwartz et al. (2001). Although some nuclear compartments contain a significant proportion of proteins with pI values ∼7, there is a distinct lack of proteins in most nuclear compartments with pIs close to the pH of the nucleus (pH ∼7.4) (Jackson et al., 1988) (Figure 1). Proteins with pI ∼7.4 might tend to coalesce and precipitate at physiological pH since proteins are generally the least soluble near their pIs.
The proteins associated with the nuclear periphery and PML bodies are the largest on average (mean MW, 88 and 84 kDa, respectively; Table I; Figure 2). In contrast, proteins from the major compartments involved in RNA metabolism (nucleolus and slicing speckles) tend to be smaller (69 and 63 kDa, respectively), and proteins in Cajal bodies are the smallest of all (mean MW, 38.5 kDa; Table I). Interestingly, despite their similar pI characteristics, chromatin-associated proteins tend to be larger than the diffusely localized proteins (mean MW, 79 and 68 kDa, respectively; Table I).
Amino acid composition
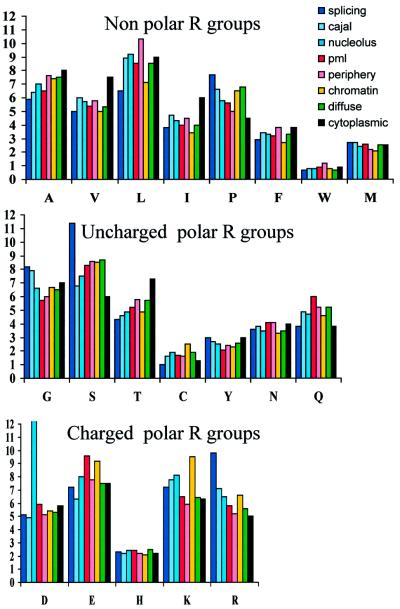
Variation in pIs amongst proteins in different nuclear compartments must reflect their differing amino acid compositions. Amino acid compositions generally characteristic of nuclear proteins have previously been noted. Ponting (2001) suggests that nuclear proteins often have unusually high occurrences of lysine (Lys) and arginine (Arg) residues, perhaps in part due to the presence of these amino acids in nuclear localization signals (NLSs). However, Figure 3 shows that this is not generally the case, e.g. the prevalence of Lys and Arg in proteins concentrated in PML bodies or diffusely distributed across the nucleoplasm is almost identical to that found in cytoplasmic proteins. Schwartz et al. (2001) suggest that serine (Ser) and proline (Pro) are unusually prevalent in nuclear proteins compared with cytoplasmic proteins. This does appear to be the case (Figure 3) and in addition, proteins in all the nuclear compartments are depleted for isoleucine, valine and threonine compared with cytoplasmic proteins.
Fig. 3. Average amino acid frequencies for proteins reported to concentrate in nuclear compartments. Amino acids are grouped into those with non-polar, and those with uncharged and charged polar R groups. The values for cytoplasmic proteins, taken from Schwartz et al. (2001), are shown (black) for comparison.
As well as these general differences in amino acid composition between nuclear and cytoplasmic proteins, our survey reveals that some residues are particularly common, or depleted, in proteins concentrated in specific subnuclear domains (Figure 3). Most striking is the prevalence of Ser and Arg in the proteins that concentrate in splicing speckles. This is due to the presence of RS domains in many splicing proteins (Birney et al., 1993), and also accounts for the very basic nature of the proteins in this compartment (14/18 of the splicing proteins with pI >10 contain an RS domain). Pro residues also seem to be relatively abundant in splicing proteins, in contrast to the general depletion of other non-polar amino acid residues, especially leucine (Leu). This suggests that large parts of these proteins may be unstructured.
Leu is prevalent in general cellular integral membrane proteins (Schwartz et al., 2001). Figure 3 shows that it is also rather abundant in the proteins concentrated at the nuclear periphery, but this includes many proteins of the nuclear pore complex and the lamins, not just the integral nuclear membrane proteins. The low pI values of proteins at the nuclear periphery appears to be due to a depletion of basic residues, rather than an abundance of glutamic acid (Glu) or aspartic acid residues. In contrast, the acidic nature of PML body-associated proteins can be attributed to the abundance of Glu (Figure 3).
The other striking feature apparent in Figure 3 is the prevalence of Lys residues in chromatin-associated proteins. That this does not result in high average pI values for the proteins in this group is due to the equal abundance of Glu residues in these proteins. It is not clear why it is Lys, rather than Arg, that is so abundant in chromatin-associated proteins. One possibility is that modification, e.g. by acetylation, of lysine residues in many chromatin proteins (not just core histones) is a widespread mechanism in the chromatin-mediated control of gene expression (Roth et al., 2001). In contrast, the diffusely localized nuclear proteins do not have an abundance of charged residues (Figure 3).
The PSORT tool (http://psort.nibb.ac.jp/form2.html) tries to predict whether a protein will be located in the nucleus or cytoplasm using a heuristic that nuclear proteins are generally rich in basic residues (Lys + Arg >20%) (Reinhardt and Hubbard, 1998). It correctly predicts a nuclear location for most of the chromosome-associated proteins (89%) and the proteins localized to splicing speckles (82%), as might be expected from the prevalence of Lys and Arg in these proteins (Figure 3). However, it is poor at predicting localization for proteins at the nuclear periphery (47%) and in Cajal bodies (65%) (see below). The results using PSORT to correctly predict nuclear localization, for proteins in subnuclear compartments, using the k-nearest neighbour method (Nakai and Horton, 1999) are similar (84% prediction for chromatin-associated proteins, 40% prediction for proteins at the nuclear periphery).
Protein motifs and domains
Most proteins must be specifically imported into the nucleus, by interaction of NLSs with importin β (Nakielny and Dreyfuss, 1999). The PSORT tool (http://psort.nibb.ac.jp/form2.html) detects NLSs in 80–86% of proteins localizing to the nucleolus, splicing speckles, chromatin or PML bodies. However, only 62% of proteins localized to Cajal bodies and 41% of proteins concentrated at the nuclear periphery contain an NLS, suggesting that many of the proteins in these compartments enter the nucleus in other ways, e.g. via the endoplasmic reticulum, or complexed to other proteins. We assessed the incidence of other conserved domains, and motifs amongst nuclear proteins using the SMART tool (http://smart.embl- heidelberg.de). This showed that motifs and domains are often shared amongst proteins co-localized within the same subnuclear compartment. Conversely, some generally abundant motifs/domains are lacking from the proteins concentrated in some areas of the nucleus.
Proteins reported to concentrate in the nucleolus appear to have simple domain architecture with rarely more than one type of recognizable conserved motif or domain. Two motifs were especially common. The first motif common amongst the nucleolar proteins surveyed here is that characteristic of DEAD (or DExD/H) box putative RNA helicases or RNPases (Table II) (Tanner and Linder, 2001). The DEAD box helicase motif is rarely partnered with other recognizable motifs except, in two cases, with a helicase and RNase D C-terminal (HRDC) domain, that has a putative role in nucleic acid binding. Notably, all three human proteins in SwissProt known to contain an HRDC domain are located in the nucleous (BLM, WRN and PMScl-100). The DEAD-box helicase is also the most common motif identified among nucleolar proteins identified in a gene-trap screen and proteins identified in purified nucleoli (Sutherland et al., 2001; Andersen et al., 2002). The other most common motif recurring in nucleolar proteins is the WD40 repeat, an abundant motif in the human proteome probably involved in protein– protein interactions (Table II).
Table II. The most abundant domains and motifs in proteins from different nuclear compartments.
| Compartment | Most common motif amongst known proteins | Abundance in human proteome |
|---|---|---|
| Splicing speckles | RRM 25/65 (38%) | 7th |
| RS 21/65 (32%) | not known | |
| Nucleolus | DExD/H box helicase 6/109 (6%) | 38th |
| WD40 6/109 (6%) | 8th | |
| PML bodies | bromodomain 3/24 (13%) | 168th |
| Sand 3/24 (13%) | 405th | |
| Nuclear periphery | transmembrane 18/70 (26%) | not known |
| FG repeats 11/70 (16%) | not known | |
| Chromatin | chromodomain 10/62 (16%) | 184th |
| bromodomain 7/62 (11%) | 168th | |
| AT hook 7/62 (11%) | not known | |
| PHD finger 6/62 (10%) | 75th | |
| C2H2 zinc finger 6/62 (10%) | 2nd | |
| Diffuse | C2H2 zinc finger 6/50 (12%) | 2nd |
| HLH 6/50 (12%) | 37th |
The most frequent motifs or domains present in the sequences of proteins, which have been reported in the literature in nuclear subcompartments, were identified using the SMART tool (http://smart.embl-heidelberg.de/). The proportions (%) of the localized proteins containing these sequences were compared with the frequency with which the same motifs have been detected in the human genome sequence (http://www.ensembl.org/IPtop500.html).
Compared with nucleolar proteins, proteins concentrated in splicing speckles have a more complex architecture, with half of the proteins containing two or more recognizable conserved motifs or domains. The domain that occurs most commonly amongst proteins in the splicing speckles is the RNA recognition motif RRM (25/65 proteins) (Table II). This is an abundant motif in the human proteome (http://www.ensembl.org/IPtop500. html). Despite the fact that the nucleolus is also involved in (ribosomal) RNA processing, RRM domains are not that abundant amongst published nucleolar proteins (3/97), but instead a more diverse array of other RNA-binding motifs (e.g. KH and RGG domains) appears to be utilized by these proteins. Half of the splicing proteins with RRM(s) also contain an RS domain. RS domain-containing proteins are abundant amongst splicing proteins (21/65) (Mintz et al., 1999; Sutherland et al., 2001). Despite the functional relationship between Cajal bodies and splicing proteins, only one of the proteins located in this body contains an RRM and no RS domain-containing proteins are found concentrated there. The proteins in common between splicing speckles and Cajal bodies are the Sm proteins.
Many of the proteins concentrated at the nuclear periphery are quite distinctive. There is an abundance of predicted transmembrane spanning domains (18/70 proteins), and proteins that contain the FG repeats characteristic of many nuclear pore complex proteins (11/70) (Radu et al., 1995) (Table II).
Amongst the proteins reported to concentrate within PML bodies, two domains, Sand and bromo, are the most abundant (3/24 proteins). The two PML proteins that contain both Sand and bromodomains also have a PHD finger. PHD fingers and bromodomains are commonly found together in chromatin-associated proteins (see below). The function of the Sand domain is unknown, but it may be a DNA-binding domain (Kumar et al., 2001). PML bodies are targets of viral infection, and it has also been suggested that they are involved in transcriptional regulation, and they do associate with specific regions of the human genome (Shiels et al., 2001). The domain architecture of PML-associated proteins strongly supports a role for them in the chromatin-mediated control of gene expression. Intriguingly, the AIRE protein (mutated in APECED syndrome) contains a Sand domain in combination with two PHD fingers, and it localizes to nuclear bodies that appear to be very similar to, but are distinct from, PML bodies (Bjorses et al., 1999). This suggests that there may be another nuclear compartment related to PML bodies in appearance, protein composition and possibly function. It is not clear why PML-associated proteins are acidic or so large.
Chromatin-associated proteins contain a diversity of motifs/domains. Half of them have two or more (up to a maximum of five) different conserved motifs or domains each, and many of these are considered to be protein– protein interaction motifs. This may reflect the multiplicity of interactions that occur between chromatin-associated proteins and other nuclear proteins, e.g. transcriptional machinery and replication apparatus. Hence, chromatin-associated proteins may act as landing pads for many other proteins or protein complexes. The single most abundant domain identified amongst known chromatin proteins is the chromodomain (10/63) (Table II). In some chromatin-associated proteins this domain may bind to methylated lysine residues in histones (Bannister et al., 2001). Ten chromatin-associated proteins have zinc fingers (in six of these cases they are of the C2H2 type). Seven of the chromatin-associated proteins contain a bromodomain, a domain that may interact with acetylated lysine residues (Dhalluin et al., 1999). In PML-associated proteins, bromodomains are often found associated with PHD fingers, and indeed all except one of the chromatin-associated bromodomain-containing proteins also contain at least one PHD finger. PHD fingers may be protein– protein interaction domains specialized for use in chromatin (Aasland et al., 1995). In one of the chromatin-associated proteins, KAP-1, the PHD finger and the bromodomain have been shown to form a cooperative unit that recruits a chromatin-remodelling complex (Schultz et al., 2001). This arrangement of PHD finger immediately N-terminal to the bromodomain is conserved in all six of the chromatin-associated proteins that carry both domains, although these motifs can be at either the N- or C-terminus of the proteins. Interestingly in HRX, where three of the four PHD fingers are just N-terminal to the bromodomain, many of the leukemia-associated breakpoints that occur in this protein disrupt this organization (Rowley, 1998). Lastly, AT hooks, a conserved DNA-binding motif that preferentially binds to the minor groove of AT-rich DNA, are present in seven of the chromatin-associated proteins (Table II).
In contrast to the chromatin-associated proteins, proteins diffusely localized in the nucleoplasm have fewer conserved motifs/domains per protein (a third have two or more motifs/domains). The most common domains are the C2H2 zinc finger (a DNA binding domain that is the second most abundant motif/domain in the human proteome), and the helix–loop–helix (HLH) dimerization domain (Table II).
Although we are far from a full understanding of the complexity of nuclear organization, it is clear that there are distinct characteristics often shared by proteins with similar subnuclear localization. It seems likely that, in some cases, it will be possible to combine information on motif/domain architecture, amino acid composition, size and pI, to predict new protein constituents of nuclear compartments from genome databases, and possibly to predict the biological pathways in which they function (Eisenhaber and Bork, 1998). This hypothesis should now be tested.
Acknowledgments
Acknowledgements
We thank Paul McLaughlin (University of Edinburgh) for useful discussions on protein architecture and Colin Semple for reading the manuscript. Most of all, we thank Rachel Farrall for constructing the Nuclear Protein Database. H.S. is supported by a fellowship from the AICR. W.A.B. is a Centennial Fellow of the James S.McDonnell Foundation.
References
- Aasland R., Gibson,J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K.L., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorses P., Pelto-Huikko,M., Kaukonen,J., Aaltonen,J., Peltonen,L. and Ulmanen,I. (1999) Localization of the APECED protein in distinct nuclear structures. Hum. Mol. Genet., 8, 259–266. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Dreger M., Bengtsson,L., Schöneberg,T., Otto,H. and Hucho,F. (2001) Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl Acad. Sci. USA, 98, 11943–11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber F. and Bork,P. (1998) Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol., 8, 169–170. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Yuan,J. and Cook,P.R. (1988) A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J. Cell Sci., 90, 365–378. [DOI] [PubMed] [Google Scholar]
- Kumar P.G., Laloraya,M., Wang,C.Y., Ruan,Q.G., Davoodi-Semiromi,A., Kao,K.J. and She,J.X. (2001) The Autoimmune Regulator (AIRE) is a DNA-binding protein. J. Biol. Chem., 276, 41357–41364. [DOI] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Mintz P.J., Patterson,S.D., Neuwald,A.F., Spahr,C.S. and Spector,D.L. (1999) Purification and biochemical characterization of interchromatin granule clusters. EMBO J., 18, 4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa K., Nosaka,T., Morita,S., Kaneko,A., Nakahat,T., Asano,S. and Kitamura,T. (2000) A method to identify cDNAs based on localization of green fluorescent protein fusion products. Proc. Natl Acad. Sci. USA, 97, 3062–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their sub-cellular localization. Trends Biochem. Sci., 24, 34–35. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Phair R.D. and Mistelli,T. (2000) High mobility of proteins in the mammalian nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. (2001) Issues in predicting protein function from sequence. Brief. Bioinform., 2, 19–29. [DOI] [PubMed] [Google Scholar]
- Radu A., Moore,M.S. and Blobel,G. (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell, 81, 215–222. [DOI] [PubMed] [Google Scholar]
- Reinhardt A. and Hubbard,T. (1998) Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res., 26, 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Stein,P.A., Taylor,S.S., Ha,E., McKeon,F. and Rapoport,T.A. (1999) A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell Biol., 146, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.T., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Rowley J.D. (1998) The critical role of chromosome translocations in human leukemias. Annu. Rev. Genet., 32, 495–519. [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Friedman,J.R. and Rauscher,F.J.,III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev., 15, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R., Ting,C.S. and King,J. (2001) Whole proteome values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res., 11, 703–709. [DOI] [PubMed] [Google Scholar]
- Shiels C., Islam,S.A., Vatcheva,R., Sasieni,P., Sternberg,M.J.E., Freemont,P.S. and Sheer,D. (2001) PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci., 114, 3705–3716. [DOI] [PubMed] [Google Scholar]
- Simpson J.C., Wellenreuther,R., Poustka,A., Pepperkok,R. and Wiemann,S. (2000) Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO rep., 1, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999) Newly assembled snRNPs associate with coiled bodies before splicing speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Sutherland H.G.E., Mumford,G.K., Newton,K., Ford,L.V., Farral,R., Dellaire,G., Cáceres,J.F. and Bickmore,W.A. (2001) Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet., 10, 1995–2011. [DOI] [PubMed] [Google Scholar]
- Tanner N.K. and Linder,P. (2001) DexD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell, 8, 251–262. [DOI] [PubMed] [Google Scholar]