Abstract
NIMA kinases appear to be the least functionally conserved mitotic regulators, being implicated in chromosome condensation in fungi and in spindle function in metazoans. We demonstrate here that the fission yeast NIMA homologue, Fin1p, can induce profound chromosome condensation in the absence of the condensin and topoisomerase II, indicating that Fin1p-induced condensation differs from mitotic condensation. Fin1p expression is transcriptionally and post-translationally cell cycle-regulated, with Fin1p kinase activity maximal from the metaphase–anaphase transition to G1. Fin1p is localized to the spindle pole body and fin1Δ cells are hypersensitive to anti-microtubule drugs, synthetically lethal with a number of spindle mutants and require the spindle checkpoint for viability. Moreover, fin1Δ cells show unusual and extensive elaborations of the nuclear envelope. These data support a role for Fin1p in spindle function and nuclear envelope transactions at or after the metaphase– anaphase transition that may be generally applicable to other NIMA-family members.
Keywords: fin1/mitosis/NIMA/nuclear envelope/spindle
Introduction
The transition from interphase into mitosis is accompanied by a massive re-organization of cellular architecture. The microtubular cytoskeleton is collapsed and reorganized into the mitotic spindle to promote the segregation of chromosomes, which in preparation are substantially condensed to ensure segregation without breakage. In higher eukaryotes, mitosis also sees the breakdown of the nuclear envelope and lamina, whereas fungi undergo a closed mitosis, although there is considerable evidence for structural changes in the nuclear membranes. Notably, the major microtubule organizing centres for the mitotic spindle, the spindle pole bodies (SPBs), move from a perinuclear location to spanning the nuclear membrane for spindle assembly and elongation at the metaphase– anaphase transition and then subsequently exit the membrane to regain their interphase perinuclear position (Ding et al., 1997).
We currently have an understanding of molecular events leading to the onset of and passage through mitosis. Protein phosphorylation plays highly conserved and direct causal roles in many mitotic events. The cyclin-dependent kinase Cdc2p is the key molecule for mitotic entry and acts above an executive of mitotic protein kinases. Its activation leads to the direct regulation of multiple mitotic pathways (Nurse, 1990). Cdc2p activity is ultimately down-regulated by degradation of its cyclin partner through a large multi-subunit ubiquitin ligase known as the anaphase promoting complex (APC), which also controls the degradation of proteins that promote sister chromatid cohesion (Zachariae and Nasmyth, 1999). Other highly conserved mitotic protein kinases are the Polo and Aurora families in which mutations block mitotic progression (Glover et al., 1998). These kinases have been implicated in a number of mitotic events including cyclin B localization (Toyoshima-Morimoto et al., 2001), histone H3 phosphorylation (Hsu et al., 2000), APC activation, spindle formation, sister chromatid separation, mitotic progression and cytokinesis (Glover et al., 1998; Cullen et al., 2000). In addition to its role in nucleocytoplasmic trafficking, the ran GTPase is also a major contributor of spindle formation in mitosis by stabilization of microtubules in the near vicinity of the chromosomes and the regulation of key effectors of spindle formation and movement (Carazo-Salas et al., 2001; Wiese et al., 2001; Wilde et al., 2001). Mutants of fission yeast ran have defects in microtubule stability (Fleig et al., 2000).
Perhaps the most poorly understood family of mitotic kinases is the NIMA family. NIMA was first isolated as conditional cell cycle mutants that failed to enter mitosis in Aspergillus nidulans (Osmani et al., 1987, 1988b). Strains lacking NIMA function activate Cdc2p complexes, but fail to enter mitosis. Inactivation of the APC allows nimA– cells to enter mitosis, with normal chromosome condensation, but aberrations to the spindle and elaborations and unusual defects in nuclear envelope morphology appear (Osmani et al., 1988a; Osmani et al., 1991a,b). NIMA activity is tightly regulated by its degradation and phos phorylation, restricting NIMA activity to mitosis (Osmani et al., 1987; Osmani et al., 1991b). Inappropriate accumulation of the protein causes the initiation of irreversible chromosome condensation, without the induction of other mitotic events (O’Connell et al., 1994; Pu and Osmani, 1995). In normal mitotic progression, a five-protein complex, the condensin, carries out the dramatic condensation of chromosomes at mitosis (Hirano, 2000). It contains two structural maintenance of chromosome (SMC) proteins, associates with chromosomes during mitosis and is directly implicated in the super-coiling of mitotic chromosomes with topoisomerases (Kimura et al., 2001). It was hitherto unknown whether NIMA-induced chromosome condensation, which is also seen in fission yeast, Xenopus oocytes and human cells (O’Connell et al., 1994; Lu and Hunter, 1995), functions through this complex. Nevertheless, these observations led to the hypothesis that NIMA functions downstream of Cdc2p to promote chromosome condensation. The absence of NIMA would then activate a checkpoint-mediated cell cycle arrest in late G2 to prevent other mitotic events.
The closest vertebrate homologue of NIMA is Nek2 (Schultz et al., 1994). Nek2 resides at the centrosome and modulation of Nek2 activity results in defects in the maturation and function of the centrosome in the formation of the mitotic spindle (Fry et al., 1998b). One substrate identified for Nek2 is c-NAP1, which, when phosphorylated, dissociates from centrosomes and they are subsequently separated (Fry et al., 1998a; Mayor et al., 2000). There is no evidence that Nek2 accumulation can induce chromosome condensation and so it remains unknown whether NIMA and Nek2 are indeed homologues or are distinct though related cell cycle regulators.
We have been investigating the function of the closest fission yeast relative of NIMA, Fin1p. Like NIMA, accumulation of Fin1p induces premature and profound compaction of chromosomes and so we have previously proposed that the major role of NIMA-family kinases is to promote chromosome condensation at mitosis (Krien et al., 1998). fin1 is not an essential gene, questioning a role for Fin1p in chromosome condensation and other essential events at mitosis. However, fin1Δ cells are delayed in cell cycle progression and exhibit mitotic abnormalities (Krien et al., 1998). In this report we show that Fin1p is a tightly cell cycle-regulated protein, controlled at both transcriptional and post-translational levels. Fin1p kinase activity is also cell cycle-regulated and does not appear until the metaphase–anaphase transition. This places Fin1p kinase at a point in the cell cycle subsequent to chromosome condensation. We show that the hypercondensation of chromosomes promoted by fin1 overexpression is independent of all known components of the mitotic condensation machinery, suggesting this gain-of- function phenotype is not reflective of normal mitotic chromosome condensation. Fin1p is localized primarily to the SPBs during mitosis. We show that fin1Δ cells are hypersensitive to anti-microtubule drugs, show defects in mitotic spindles, have strong genetic interactions with several regulators of spindle function and that the viability of these cells is dependent on the mitotic spindle checkpoint. Finally, fin1Δ cells, alone or exacerbated by combination with synthetically interacting lethal mutations, have disrupted nuclear envelope structures. We conclude that Fin1p functions at the poles of the spindle during the metaphase–anaphase transition, a time when dramatic re-arrangements of microtubules and nuclear membrane are necessary for successful navigation of mitosis in fungi. These observations, together with the spindle functions of Nek2 and the spindle and nuclear envelope defects of nimA– mutants that enter mitosis when the APC is inactivated, suggest diverse, though partially conserved, roles for this family of mitotic protein kinases.
Results
Fin1p-mediated premature chromosome condensation is independent of the mitotic chromosome condensation machinery
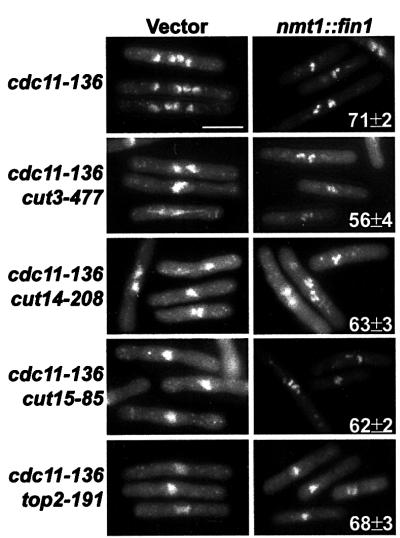
Inappropriate accumulation of Fin1p results in an irreversible and premature hypercondensation of chromosomes (Krien et al., 1998). This phenotype could be induced from any point in the cell cycle. We proposed that by artificially accumulating Fin1p, the cell had bypassed a requirement for Cdc2p for this mitotic event. We investigated whether known factors that control mitotic chromosome condensation were required for this phenotype. Fin1p was overexpressed in a background (cdc11-136) that is unable to perform cytokinesis at 36°C (Le Goff et al., 1999). Inclusion of this mutation allowed us to assay effects of Fin1p overexpression independently from the lethality induced by cell ‘cutting’, in which a septum is laid through unsegregated chromosomes. Temperature-sensitive mutations in condensin subunits (cut3-477 and cut14-208; Saka et al., 1994), importinα (cut15-85; Matsusaka et al., 1998) and topoisomerase II (top2-191; Uemura and Yanagida, 1984), each required to control chromosome condensation at mitosis, were used. In every strain, Fin1p overexpression still resulted in the hypercondensation of chromosomes (Figure 1). Moreover, Fin1p overexpression in these strains and in combinations of these alleles also resulted in hypercondensation in cells previously arrested in S phase (Figure 2). The compacted chromosomes were visible as individual DAPI-stained bodies, with the exception of top2-191 mutants, in which the absence of decantenation resulted in a single compacted mass. Frequently, the nuclear masses were found positioned away from the centre of the cell, which accompanied cell death as described previously with NIMA overexpression (O’Connell et al., 1994). These data show that Fin1p-mediated compaction of the chromosomes is not functionally related to mitotic chromosome condensation and the mechanism by which it occurs remains obscure.
Fig. 1. Fin1p accumulation induces chromosome condensation inde pendently of the condensin or topoisomerase II. Overexpression of Fin1p in the indicated strains was under the control of the nmt1 promoter. Strains were grown in the absence of thiamine for 16 h at 25°C before being shifted to 36°C. The experiment was carried out in the temperature-sensitive septation mutant cdc11-136 to avoid the associated cell lethality with ‘cut’ mutants. Samples for DAPI staining were taken hourly for 6 h, fixed in 3.7% formaldehyde and viewed under UV fluorescence. Hypercondensed chromosomes were observed in all strains tested with fin1 overexpression and the numbers represent the mean ± SD of cells with hypercondensed chromosomes. Vector controls for each strain showed the complete absence of hyper condensed chromosomes in >500 cells.
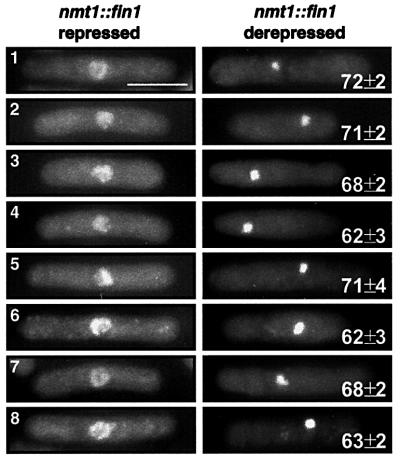
Fig. 2. Fin1p accumulation induces premature chromosome con densation independently of the condensin and topoisomerase II. Overexpression of Fin1p in wild type (1), cut3-477 (2), cut14-208 (3), top2-191 (4), cut3-477 cut14-208 (5), cut3-477 top2-191 (6), cut14-208 top2-19 (7) and cut3-477 cut14-208 top2-191 (8). Strains with fin1 under the control of the nmt1 promoter were grown in the absence and presence of thiamine for 16 h at 25°C, to which hydroxyurea was added to a final concentration of 11 mM and grown for another 4 h at 25°C. Cells were determined to be arrested in early S phase by FACS analysis (data not shown) and then shifted to 36°C for another 4 h with a second addition of hydroxyurea to a final concentration of 11 mM. Samples were taken each hour and fixed in formaldehyde for DAPI staining. Hypercondensed chromosomes were observed in all strains tested with fin1overexpression and the numbers represent the mean ± SD of cells with hypercondensed chromosomes. Vector controls for each strain showed the complete absence of hypercondensed chromosomes in >500 cells.
Cell cycle regulation of Fin1p
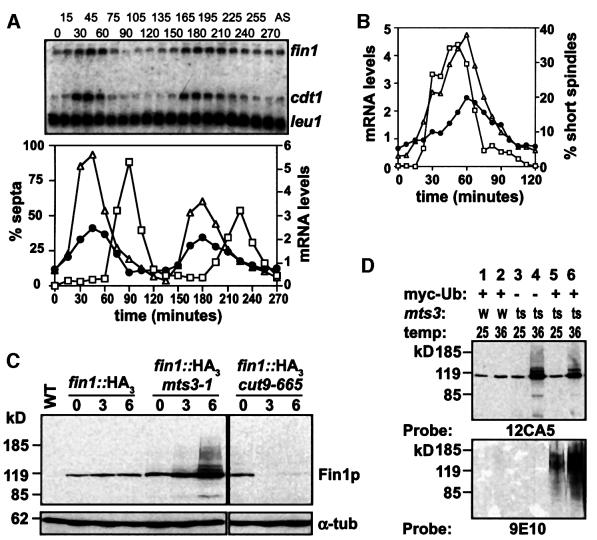
Fin1p levels fluctuated over a three-fold range across the cell cycle, peaking approximately at mitosis (Krien et al., 1998). Northern blot analysis of synchronized cells showed that fin1 mRNA levels fluctuated to approximately the same degree as Fin1p (Figure 3A). The temporal control of fin1 mRNA levels overlapped with cdt1, a target of the Cdc10p/Res1p transcription complex, which is expressed during mitosis (Figure 3; Baum et al., 1998). Consistent with this, fin1 mRNA levels are minimal in cdc10-M17 mutants at restrictive temperatures (data not shown). The reported peak of Fin1p levels was confirmed in these cultures and was at or shortly after the peak in fin1 mRNA (data not shown).
Fig. 3. Transcriptional and post-translational cell cycle control of Fin1p levels. (A) A synchronous culture was generated by block and release of the strain cdc25-22; cell cycle progression was monitored by scoring the fraction of septated cells. Cells were harvested at the indicated times after shift to 25°C, total RNA extracted and analysed by northern blotting with the indicated probes. mRNA levels were quantified on a Molecular Dynamics phosphorimager with ImageQuant software. mRNA levels of fin1 (filled circles) and cdt1 (triangles) are expressed as a proportion to the corresponding mRNA in a culture of asynchronously growing cells (AS) and normalized to leu1. The septation index (squares) is also shown. (B) Cells from a cdc25-22 block and release were fixed and stained by indirect immunofluorescence for microtubules to reveal spindle organization as a marker of mitotic progression. mRNA levels of fin1 (filled circles) and cdt1 (triangles) are expressed as a proportion of these in a culture of AS cells. The fraction of cells with short spindles is shown for comparison (squares). (C) Wild type, mts3-1 and cut9-665 strains were derived that contained either the endogenous fin1 gene or a epitope-tagged Fin1p (fin1::HA3) expressed from its own locus. Strains were grown at 25°C, shifted to 36°C and cells were harvested at 0, 3 and 6 h following the shift. Protein extracts were analysed by western blotting with either 12CA5 mAb to detect epitope-tagged fin1p or anti α-tubulin (TAT1) as a loading control. High molecular weight species of Fin1p accumulated when the proteosome was inactivated (mts3-1), but not when the APC was inactivated (cut9-665). (D) fin1::HA3 strains in a wild type background expressing myc-Ubiquitin (myc-Ub; lanes 1 and 2) or an mts3-1 background not expressing (lanes 3 and 4) or expressing myc-Ub (lanes 5 and 6) shifted for 0 and 6 h, respectively. Extracts were prepared, epitope-tagged fin1p purified by Co2+ affinity chromatography and immunoblotted with either 12CA5 mAb to detect epitope-tagged Fin1p or 9E10 mAb to detect purified protein modified by myc-Ub, showing that the higher molecular weight species of Fin1p accumulating after proteosome inactivation were polyubiquitylated.
Deletion of the Fin1p PEST sequence resulted in accumulation of Fin1p and lethal chromatin condensation, suggesting that Fin1p levels must be tightly regulated (Krien et al., 1998). As fin1 mRNA levels mirror Fin1p levels, we hypothesized that Fin1p would be subjected to ubiquitin-dependent proteolysis. We assayed Fin1p levels in cells containing a temperature-sensitive mutation in a core proteosome subunit, Mts3p (Gordon et al., 1996). On shifting to 36°C, Fin1p accumulated, with the appearance of poly-ubiquitylated higher molecular weight Fin1p species (Figure 3C and D). The APC has been suggested to be the E3 for NIMA degradation in A.nidulans (Ye et al., 1998). We tested whether this is also the case in fission yeast by inactivating the APC with a temperature-sensitive mutation in the APC subunit Cut9p (Samejima and Yanagida, 1994). Unlike in Aspergillus, Fin1p protein levels in fission yeast were reduced by APC inactivation, though this may be due to a cell cycle arrest in metaphase, which would promote cell death at a time when fin1 mRNA levels are yet to peak, so we cannot rule out that the APC may play some role in Fin1p proteolysis later in the cell cycle.
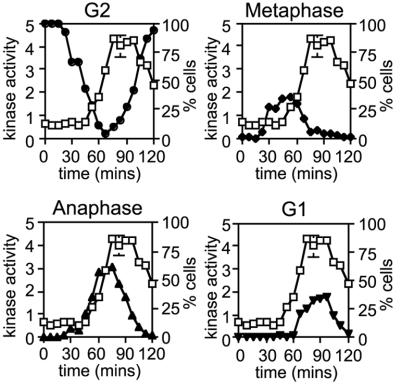
We next assayed Fin1p kinase levels using an assay developed with recombinant Fin1p to provide optimal activity (see Materials and methods). Fin1p was purified from synchronized cells and kinase assays showed that Fin1p activity fluctuated similarly to its mRNA and protein (∼4-fold), though considerable activity was present throughout the cell cycle (Figure 4). When Fin1p activity was compared to microtubule morphology, we found that the activity first rose as the mitotic spindle elongated at the metaphase–anaphase transition and peaked late in mitosis through to early in G1 phase. These data suggest that Fin1p may play a role in late mitosis or in G1 cell cycle progression.
Fig. 4. Fin1p kinase activity appears after the metaphase–anaphase transition. A synchronous culture was generated by block and release of a cdc25-22 strain expressing fin1::HA3. Cells were harvested at the indicated times after shift to 25°C, extracts were prepared, Fin1p immunoprecipitated with 12CA5 mAb and the immunoprecipitates assayed for Fin1p kinase activity, normalized to asynchronous cells. Cells were also fixed and stained by indirect immunofluorescence for microtubules as a marker of cell cycle progression. Fin1p kinase activity (squares) expressed in proportion to that in an AS culture is overlayed with plots of the fraction of cells in G2 with cytoplasmic microtubule arrays (filled circles), in prophase and metaphase with short spindles (filled diamonds), in anaphase and telophase with long spindles (filled upright triangles) and in G1 with post-anaphase arrays (filled inverted triangles). The appearance of Fin1p kinase activity is concomitant with spindle elongation.
Fin1p is localized to the SPB and is required for mitotic spindle function
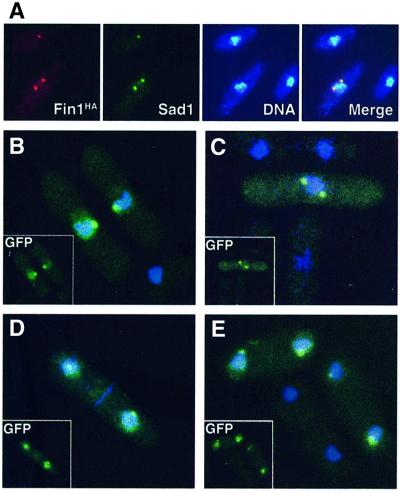
Indirect immunofluorescence using the genomic HA-tagged fin1 allele detected a signal that co-localized with the spindle pole body marker Sad1p (Hagan and Yanagida, 1995) in late G2, mitotic and early G1 cells (Figure 5). In confirmation of these findings, GFP-tagged Fin1p expressed under repressing conditions from the nmt1 promoter, which represents only mild overexpression of Fin1p (Krien et al., 1998), was visible in live cells using deconvolution microscopy. This level of expression has no observable effects on the physiology of the cells. GFP–Fin1p was evident on the nuclei of all cells in late G2 through to early G1 and was concentrated in a single spot on the nuclear periphery of late G2 cells and in two foci in mitotic cells that segregated at mitosis, consistent with the localization to the SPB in fixed cells (Figure 5).
Fig. 5. Localization of Fin1p kinase to SPB. (A) Indirect immunofluorescence of Fin1HA (C-terminal tag from the endogenous promoter at the fin1 locus) co-localized with the SPB marker Sad1p on the nuclear periphery. (B–E) GFP-fluorescence of N-terminally tagged Fin1p expressed from the nmt1 promoter. Exponentially growing live cells, cultured in the presence of thiamine, were stained with the DNA dye Hoescht 33342. Localization of fin1p to SPB was observed in late G2/early M as assessed by cell length (B), mitosis (C) and G1/S (D and E). GFP signal in the DNA compartment can be seen in all cells.
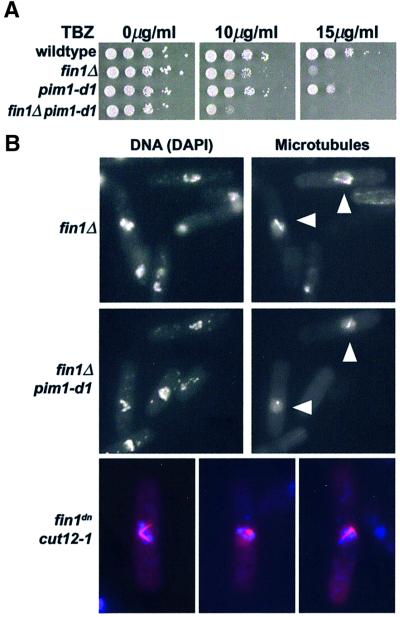
The localization of Fin1p to the spindle poles during mitosis, the time when it is most active as a protein kinase, suggested it may be playing a role in mitotic spindle function. Many mutants defective in spindle assembly and function are hypersensitive to anti-microtubule drugs such as thiabendazole (TBZ). We found fin1Δ cells were also TBZ-hypersensitive compared with wild-type cells (Figure 6). Combining fin1Δ with a temperature-sensitive mutation (pim1-d1) in the nucleotide exchange factor for the ran GTPase (Spi1p; Sazer and Nurse, 1994) resulted in a synthetic arrest at metaphase of mitosis. This contrasts with the G1 cell cycle arrest of pim1-d1 single mutant cells (Krien et al., 1998). Moreover, the pim1-d1fin1Δ double mutant was extremely sensitive to TBZ at the permissive temperature (Figure 6), suggesting both proteins are involved in spindle function, which is consistent with a number of recent findings implicating ran in the assembly of the mitotic spindle (Carazo-Salas et al., 2001; Wilde et al., 2001) and with microtubule stability in fission yeast (Fleig et al., 2000).
Fig. 6. Defects in mitotic spindles in fin1Δ cells (A) Wild type, fin1Δ, pim1-d1 and fin1Δ pim1-d1 strains were spotted onto plates containing the indicated concentrations of TBZ and incubated at 30°C. Note the sensitivity of the fin1Δ and pim1-d1 single mutants to 15 µg/ml TBZ and the hypersensitivity of fin1Δ pim1-d1 to 10 µg/ml. (B) Staining of microtubules showed the aberrant mitoses of fin1Δ cells was accompanied by the presence of abnormal (monopolar) spindles (arrowed), which was enhanced by the presence of the pim1-d1 mutation at 36°C to include all mitotic cells (arrowed). Similarly, fin1dncut12-1 cells at 25°C also arrested as mitotic cells with hypercondensed chromosomes (blue) and aberrant monopolar spindles (red) were evident early in the timecourse following promoter depression (examples shown) though did not persist in later timepoints. Normal bipolar spindles were never observed.
Given these observations, why do fin1Δ cells only show modest levels of mitotic abnormalities? In asynchronously growing cells, aberrant mitoses constitute ∼4% of the cells (Krien et al., 1998) and immunofluorescence showed that these were due to spindle defects (Figure 6). Further, fin1Δ cells spontaneously lost chromosomes at a rate 3.2-fold higher than wild-type cells. Examination of the terminal phenotype of fin1Δpim1-d1 cells at 36°C showed the presence of hypercondensed chromosomes and a complete failure to form a bipolar spindle (Figure 6). We also uncovered a strong genetic interaction with the spindle mutant cut12-1 (Bridge et al., 1998) in that double mutants were inviable at all temperatures. We therefore transformed cut12-1 cells with a dominant negative allele of fin1 (fin1dn) expressed from the nmt1 promoter. This allele had no detectable kinase activity and under repressing conditions, had no effect on wild-type cells, but when derepressed phenocopied fin1Δ. At the permissive temperature of 25°C, cut12-1 cells expressing fin1dn were inviable and terminally arrested in mitosis with condensed chromosomes and aberrant monopolar spindles (Figure 6).
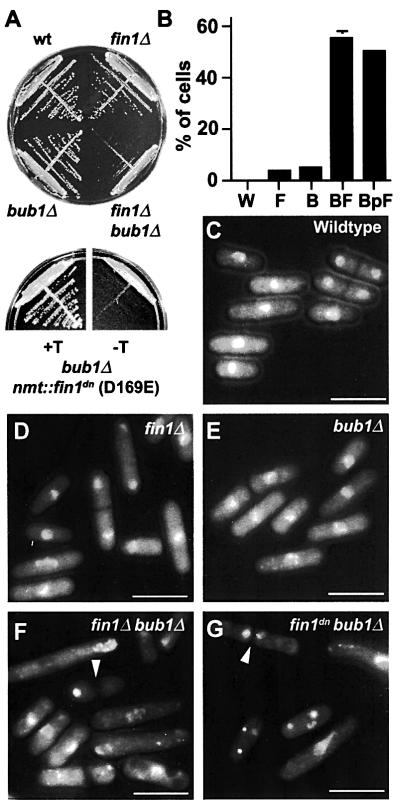
We hypothesized that the mitotic spindle checkpoint may be evoked to sustain mitotic fidelity in the absence of Fin1p. Thus, fin1Δ was crossed into a bub1Δ spindle checkpoint mutant (Bernard et al., 1998). The double fin1Δbub1Δ mutants were viable, though substantially retarded in colony formation and showed extensive chromosome segregation defects (Figure 7A). bub1Δ cells were also transformed with fin1dn expressed from the nmt1 promoter. When fin1dn expression was repressed, bub1Δ cells were viable. Upon derepression of the promoter, cells were inviable and >50% showed mitotic abnormalities and failed to form colonies (Figure 7). Similarly, in a mad2Δ checkpoint mutant (He et al., 1997), 48% of cells showed mitotic defects. The metaphase arrest of fin1dnpim1-d1 cells, due to the inability to form a bipolar spindle was also dependent on bub1 (see Supplementary figure 1). We conclude that the mitotic spindle checkpoint is essential in the absence of fin1 due to a defect in spindle function.
Fig. 7. fin1Δ requires the spindle checkpoint for viability. (A) Growth of the indicated strains for 4 days at 30°C (upper plate) and bub1Δ cells harbouring a plasmid expressing fin1dn (D135E; see Materials and methods) from the nmt1 promoter under repressing (+T) or derepressing (–T) conditions. (B) Percentage of exponentially growing cells showing mitotic defects. Strains are wild type (W), fin1Δ (F), bub1Δ (B), fin1Δbub1Δ (BF) and bub1Δ fin1dn (BpF). (C–G) DAPI-stained cells showing nuclear phenotypes for wild-type cells (C), fin1Δ (D), bub1Δ (E), fin1Δbub1Δ (F), bub1Δ fin1dn (G). Examples of some of the cells with mitotic defects are arrowed in (F) and (G). Bar = 10 µm.
We also made double mutants between fin1Δ and a large collection of mitotic mutants. We have previously shown a synthetic interaction with top2-191, a temperature-sensitive allele of type II topoisomerase (Krien et al., 1998). The only other strong genetic interactions we uncovered were with the cohesin subunit gene, rad21 and with cut11 (see below). Double mutants between fin1Δ and the temperature-sensitive allele rad21-K1 (Tatebayashi et al., 1998) were synthetically lethal at all temperatures. Double mutants between fin1Δ and the weaker rad21 allele, rad21-45 (Birkenbihl and Subramani, 1992), showed 33% of cells arrested with aberrant mitoses (data not shown). These data are consistent with defective sister chromatid cohesion and spindle tension exacerbating the spindle defects of fin1Δ cells. All of these observations are consistent with a role for Fin1p in mitotic spindle function.
Evidence for a role for Fin1p at the nuclear envelope
Another important gene in the formation of the spindle is cut11 (West et al., 1998). Cut11 encodes a transmembrane protein that is embedded in the nuclear envelope, where it co-localizes with nuclear pore complexes in interphase and with the SPBs at mitosis and may anchor these complexes into the nuclear envelope (West et al., 1998).
fin1Δ was crossed to each of the six existing temperature sensitive alleles of cut11, the progeny were analysed by tetrad dissection and no double mutants were obtained at the permissive temperature of 25°C in multiple asci per cross. Similarly strong interactions have been described between these cut11 alleles and cut12-1.
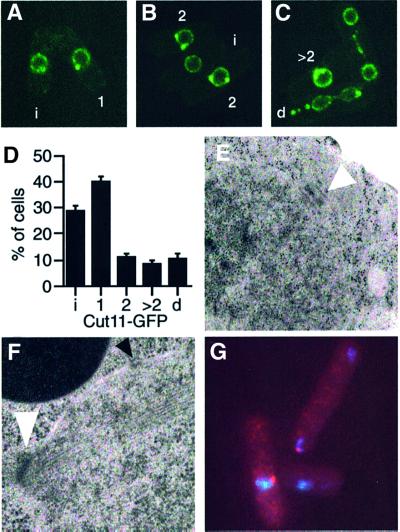
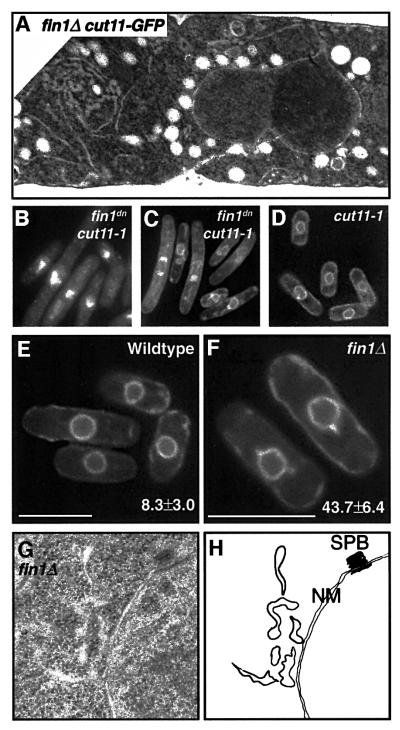
To more closely examine the terminal phenotype of the loss of Fin1p function in cut11ts mutants, these strains were transformed with nmt1-driven fin1dn. Only cut11-1 and cut11-3 could tolerate the repressed levels of expression of this construct (see Supplementary figure 2). Both cut11 alleles overexpressing dominant negative Fin1p exhibited a terminal phenotype of elongated cells with aberrant DNA morphologies. This is in stark contrast to cut11 single mutants that show a paradigm mitotic arrest resulting from the inability to anchor the SPB into the nuclear membrane (West et al., 1998). Given the SPB localization of Fin1p, we next assessed whether the cell cycle-regulated localization of Cut11p was perturbed in fin1Δ cells. To this end, fin1Δ was crossed into a background expressing GFP-tagged Cut11p integrated at the cut11 locus. The GFP–Cut11p allele appears not to be fully functional; exhibiting synthetic phenotypes in the presence of a temperature sensitive allele of the fission yeast ran homologue, spi1 (Fleig et al., 2000). Similarly, we found synthetic interactions between fin1Δ and cut11::GFP supporting this. In wild-type cells, GFP– Cut11p is found at the SPBs of ∼10% of cells, representing those in mitosis. In contrast, fin1Δ cells show bright GFP–Cut11p foci on the nuclear periphery in 72% of cells. In addition to either one or two foci of GFP–Cut11p, nearly half these cells showed aberrant patterns of GFP– Cut11p localization including multiple foci and elaborations of the nuclear envelope (Figure 8). Furthermore, examination of the terminal phenotype of cut11-1fin1dn cells using a GFP-tagged membrane protein revealed extensive architectural abnormalities with the nuclear envelope (Figure 9). Electron microscopy of fin1Δ cut11:: GFP confirmed the unusual nuclear envelope structures in these cells, suggesting the lethal synthetic interaction between fin1Δ and cut11 might relate to mis-regulation of the nuclear envelope rather than SPB anchoring during mitosis (Figure 9). The SPBs in fin1Δ and fin1ΔGFP–cut11 cells was assessed by light and electron microscopy and found to be normal (Figure 8; Krien et al., 1998).
Fig. 8. Synthetic interactions between fin1and cut11. (A–C) Localiz ation of Cut11p-GFP in live fin1Δ cells. Cut11p–GFP is localized to one or more nuclear foci in 72% of cells (c.f. 10% for wild-type cells). Shown are examples of morphologically normal localization to the nuclear envelope (i), one spot on the nucleus (1) and two nuclear foci (2). In addition, X% of cells show abnormal Cut11p–GFP localization of >two foci (>2) or foci displaced from the nucleus (d). (D) Quantification of Cut11p-GFP localization in wild-type and fin1Δ cells. Similar distribution of Cut11p–GFP is seen is fin1dn cells (not shown). (E) Transmission electron micrograph (TEM) showing normal SPB in fin1Δcut11–GFP cells. (F) TEM of a fin1Δ cell in mitosis. Normal spindle, SPB (white arrow) and nuclear pore complex (black arrow) morphology and position. (G) Normal Sad1 staining (red) of SPBs in fin1Δ cells.
Fig. 9. Perturbations at the nuclear envelope. (A) TEM of fin1Δ cut11::GFP, demonstrating elaborations of the nuclear membrane and irregular nuclear membrane morphology. A wild-type control is shown in Supplementary figure 4. (B) DAPI staining of cut11-1 cells expressing nmt::fin1dn at 25°C. (C) GFP-marked membranes in cut11-1 cells expressing nmt::fin1dn at 25°C. Note that cells are elongated (delayed in interphase) and there are elaborations to the nuclear envelope. (D) Wild-type nuclear envelopes in cut11-1 cells at 25°C transformed with vector only. (E–F) Visualization of membranes using a GFP-marker in wild type (E) and fin1Δ (F) cells. Note the membranous protrusions (numbers are mean ± SD) emanating from the nucleus in 43.7% of fin1Δ cells. Minor protrusions were seen in 8.3% of wild type cells. Bars = 10 µm. (G) TEM of fin1Δ showing elaborations of nuclear membranes, with normal interphase SPB. (H) Map of membranous structures in (G). SPB, spindle pole body; NM, nuclear membrane.
Examination of fin1Δ cells by electron microscopy and using GFP-marked nuclear membranes (Ding et al., 2000) showed the existence of abnormal membranes protruding from the nucleus (Figure 9). These data, together with the synthetic lethality with cut11 alleles suggest a role for Fin1p and Cut11p in regulating the morphology of the nuclear envelope. As Fin1p is located at the spindle pole body, it may exert this effect within this proximity, which undergoes substantial changes with the movement of the spindle pole bodies into and especially out of the nuclear envelope when Fin1p is active at mitotic exit. The partitioning of a single nuclear envelope into two during anaphase also places considerable stress on the nuclear membrane. These observations are also reminiscent of nimA– mutant in A.nidulans when in combination with APC mutations (S.A.Osmani et al., 1988a).
Discussion
Waves of phosphorylation drive entry into and passage through mitosis. In addition to Cdc2p, the conservation of protein kinase families Polo and Aurora are well established, regulating such events as spindle formation, spindle–chromosome association, induction of anaphase and cytokinesis. The evolutionary conservation in function and regulation of mitotic kinases highlight the ancient origin of the G2/M transition. Contrary to these mitotic regulators, the NIMA family of protein kinases are less conserved. At least one function of NIMA at mitotic entry in A.nidulans appears to be in promoting chromosome condensation (De Souza et al., 2000). The closest metazoan homologue, Nek2, is conversely implicated in the centrosome cycle (Fry et al., 1998a,b, 2000; Mayor et al., 2000) and is not able to induce chromosome condensation (M.O’Connell, unpublished data) and so the relationship between Nek2 and NIMA is not clear. Unlike NIMA, Fin1p is not required for entry into mitosis in fission yeast nor is it required for the effective condensation of chromosomes in early mitosis. Here we have demonstrated the up-regulation of Fin1p activity in mid-mitosis and presented evidence of an important role for Fin1p in spindle function and in nuclear envelope dynamics during mitosis and its exit thereafter.
Fin1p is not a regulator of mitotic entry
Our evidence suggests that Fin1p does not function in early mitosis to promote chromosome condensation. First, we demonstrated the independence of Fin1p-mediated chromosome compaction from any of the known effectors of mitotic chromosome condensation. Furthermore, double mutant analysis with fin1Δ and conditional alleles of cut3, cut14 and cut15 failed to yield any synthetic interactions supporting the independence of Fin1p and mitotic chromosome condensation (data not shown). We therefore conclude that the compaction observed upon overexpression of Fin1p is unlikely to be relevant to mitotic chromosome condensation. It is difficult to speculate on the nature of NIMA/Fin1p-induced chromosome compaction. NIMA has been proposed to be a S10 histone H3 kinase in Aspergillus (De Souza et al., 2000), though we found no evidence for Fin1p playing a similar role in fission yeast (see Supplementary figure 3); however, it is possible only a small sub-population is affected.
We demonstrated here that the Fin1p activity is cell cycle-regulated, first appearing at the metaphase–anaphase transition and maintained until the cells exit mitosis into G1. This is coincidental with localization of the protein to the SPB, suggesting a role for Fin1p during mitosis rather than at the G2/M transition.
Fin1 and spindle function
We have presented several lines of evidence demonstrating a role for Fin1p in spindle function: (i) fin1Δ requires an intact spindle checkpoint for viability, (ii) fin1Δ is hypersensitive to the microtubule poison TBZ, (iii) fin1Δ shows mitotic spindle abnormalities and loses chromosomes at an elevated rate, (iv) we have established a synthetic interaction between cut12-1 and fin1Δ and Cut12p is localized to the SPB throughout the cell cycle and is required for the formation of a bipolar spindle, (v) fin1Δ cells are synthetically lethal with alleles of the cohesin subunit gene rad21 and (vi) we demonstrated that the metaphase arrest in the fin1Δ pim1-d1 double mutant at restrictive temperatures is mediated by the spindle checkpoint, due to an inability of these cells to make a bipolar spindle. These data suggest the necessity of Fin1p for a fully functional bipolar spindle. Why then is fin1 not an essential gene? Other compensatory mechanisms, e.g. the spindle checkpoint and/or a heightened requirement for the ran system may be employed to ensure bipolar spindle formation in cells lacking Fin1p function.
Nuclear envelope regulation
Anaphase poses an additional challenge for fungi: the division of the nuclear envelope into two equal daughters. fin1Δ cells showed nuclear envelope elaborations and these were greatly exacerbated when fin1Δ was in combination with loss-of-function alleles in cut11. Cut11p functions in the insertion of the SPB into the nuclear envelope during mitosis, and associates with NPCs during interphase. Loss-of-function mutants in cut11 result in a mitotic arrest preceding lethal septation due to an inability to form a functional spindle. In contrast, the fin1– cut11ts double mutants appear to arrest in interphase and exaggerated elaborations of the nuclear envelope similar to numerous NPC mutants in budding yeast (Marelli et al., 2001 and references therein). This interaction between fin1Δ and cut11– proposes a further role for Cut11p in nuclear envelope architecture in cooperation with Fin1p. Cut11p contains two NIMA consensus phosphorylation sequences (FRKS208, FRKT475) and we investigated whether Fin1p could phosphorylate Cut11p but were unable to show direct phosphorylation of Cut11p by Fin1p in vitro (data not shown). Given that Cut11p contains seven transmembrane domains, the bacterially expressed products may not fold in a manner appropriate for substrate presentation. However, we would speculate that Fin1p might regulate, either directly or indirectly, Cut11p and nuclear envelope dynamics during mitosis. Given the difference in severity of phenotypes between fin1Δ and cut11ts fin1Δ, we favour the existence of other factors cooperating with Fin1p to regulate nuclear envelope dynamics via Cut11p.
NIMA family kinases
Our observations presented here suggest Fin1p may play a number of roles during mitotic progression. Similar multi-tasking has been described for the Polo and Aurora family of mitotic kinases. Clearly, a common theme between Fin1p, NIMA and Nek2 is localization to the poles of the spindle and a function in ensuring bipolar attachment and chromosome segregation. The fungal kinases appear to have an additional role at the nuclear envelope, a feature enforced by the necessity of a closed mitosis. Identific ation of substrates for NIMA kinases in various systems, like those for other mitotic kinases, will be very fruitful in determining details of molecular function.
Materials and methods
Fission yeast methods
All strains are derivatives of 972h– and 975h+. Standard genetic methods where used for strain propagation and construction (Moreno et al., 1991). Methods for transformation, synchronization of cultures, microscopy, protein expression and detection have been described (O’Connell et al., 1994, 1997). Chromosome loss experiments were performed as described (Verkade et al., 1999).
Microscopy
Immunofluorescence images were viewed on a Zeiss Axioscop and captured with a Diagnostic Instruments Spot2 camera. For GFP imaging, live cells were immobilized in 0.5% low gelling temperature agarose and viewed immediately. Alternatively, images were collected with an intelligent imagine innovations’ deconvolution microscope captured with a Cooke SensiCam CCD camera. Images of GFP–Fin1p were deconvolved with the nearest neighbour algorithm. Electron microscopy was performed on cells that were high pressure frozen, freeze-substituted in acetone with osmium and uranyl acetate and embedded in epon as described (Ding et al., 1993).
DNA manipulations
To construct an integrated C-terminally epitope-tagged version of Fin1p, a BamHI site was inserted between codons 717 and 718 of the genomic sequence by site directed mutagenesis. A BglII fragment encoding a His6–HA3 tag was inserted and integrated at the fin1 locus. Human myc-tagged ubiquitin cDNA was inserted into pART1 for constitutive expression from the adh1 promoter. For bacterial expression of glutathione S-transferase (GST)–Fin1p, fin1 cDNA was subcloned into pGEX4T-1. To construct a dominant negative mutant of fin1, aspartic acid 169 was mutated to glutamic acid (D169E). The cDNA was cloned onto the full-strength nmt1 promoter. This allele has no detectable protein kinase activity and is dominant negative mimicking fin1Δ by all genetic assays (Krien et al., 1998).
Bacterial expression and glutathione affinity purification
GST–Fin1p was expressed in Escherichia coli BL21, extracted in phosphate-buffered saline, 2% Triton X-100, 1 mM dithiothreitol (DTT) and 1 mM PMSF. The extract was cleared by centrifugation at 13 000 g for 15 min at 4°C and incubated with GSH–Sepharose for 30 min at 4°C. The matrix was washed three times with the extraction buffer and three times with dilution buffer: 50% glycerol (MB grade), 50 mM Tris–HCl pH 8.0 and 1 mM DTT. GST or GST–fin1p was eluted from the matrix by incubation in dilution buffer plus 20 mM reduced glutathione for 15 min at 4°C. The eluate was dialysed against dilution buffer (three times) for 24 h at 4°C, quantified by densitometry against standards in silver stained gels and stored at –20°C.
Fin1p kinase assays
Purified GST–Fin1p or immunoprecipitated Fin1p, was incubated for 30 min at 30°C in kinase buffer containing: 25% glycerol (MB grade), 2% Triton X-100, 50 mM Tris–HCl pH 9.5, 10 mM MgCl2, 1 mM DTT, 100 mM ATP, 300 mCi/ml [γ-32P]ATP and 0.5 mg/ml of a peptide MARFRRSRRMIAKKK, with the phosphorylated serine underlined. Reaction products were spotted onto P81 paper, washed three times in 75 mM phosphoric acid, dried and counted with scintillant. For enzyme kinetics, EnzFit software (EAG) was used. Under the conditions of the assay, Fin1p kinase activity was linear over time with increasing enzyme concentration. Fin1p exhibits a strong preference for basic reaction conditions with maximal activity evident at pH 9.5. A requirement for divalent cations in the reaction was fulfilled by MgCl2 at an optimal concentration of 5 mM. MnCl2 could substitute for MgCl2 only at a concentration of 0.625 mM; at higher concentrations it was sharply inhibitory to Fin1p kinase activity. Kinase activity was also significantly inhibited by NaCl concentrations >6.25 mM. The temperature dependency of Fin1p kinase demonstrated that under these assay conditions activity was maximal in the physiological range of Schizosaccharomyces pombe. Analysis of the kinetics of Fin1p kinase activity derived Km values for ATP of 169.1 ± 26.8 mM, for the peptide substrate of 97.8 ± 20.5 mM and a Vmax of 596 ± 78 nmol/min/mg.
Purification of tagged-Fin1p from cellular extracts
For kinase assays and western blots, frozen cellular pellets were disrupted with glass beads in: 25% glycerol (MB grade), 2% Triton X-100, 50 mM Tris–HCl pH 8.0, 1 mM DTT, 0.5 mM NaVO4, 2 mg/ml leupeptin, 2 pg/ml aprotinin, 0.2 mg/pepstatin and 1 mM PMSF. The extract was cleared by centrifugation at 13 000 g for 10 min at 4°C. Tagged Fin1p was immunoprecipitated from 0.5 to 2 mg extract by incubation on ice with 2 µg 12CA5 for 1 h and immobilized by incubation with Protein A–Sepharose for 30 min. The matrix was washed three times with extraction buffer and three times with kinase buffer lacking ATP. For experiments assaying proteolysis and polyubiquitylation of Fin1p, denatured extracts (8 M urea, 100 mM Na2HPO4, 10 mM Tris–HCl pH 8.0) were made and Co2+ affinity purification of Fin1p complexes carried out as previously described (O’Connell et al., 1997).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs Mitsuhiro Yanagida, Iain Hagan and Alison Pidoux for S.pombe strains and reagents, Dr Keith Gull for TAT-1 antibody, Dr Rick Pearson and Mr Colin House for help with kinase assays and protein expression and Drs Doris Germain, Andrew Cuddihy and Ms Susan Harvey for critical reading of the manuscript. We thank Mary Morphew for Fin1 electron microscopy. This work was supported by ARC Grant No. A09804303 to M.O’C. and NIH grant GM-33787 to J.R.M., who is a Research Professor of the American Cancer Society. M.J.E.K. is a recipient of an Australian Postgraduate Award. M.O’C. is a scholar of the Leukemia and Lymphoma Society.
References
- Baum B., Nishitani,H., Yanow,S. and Nurse,P. (1998) Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J., 17, 5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Hardwick,K. and Javerzat,J.P. (1998) Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol., 143, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R.P. and Subramani,S. (1992) Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res., 20, 6605–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J., Morphew,M., Bartlett,R. and Hagan,I.M. (1998) The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev., 12, 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Gruss,O.J., Mattaj,I.W. and Karsenti,E. (2001) Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nature Cell Biol., 3, 228–234. [DOI] [PubMed] [Google Scholar]
- Cullen C.F., May,K.M., Hagan,I.M., Glover,D.M. and Ohkura,H. (2000) A new genetic method for isolating functionally interacting genes: high plo1+-dependent mutants and their suppressors define genes in mitotic and septation pathways in fission yeast. Genetics, 155, 1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSouza C.P., Osmani,A.H., Wu,L.P., Spotts,J.L. and Osmani,S.A. (2000) Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell, 102, 293–302. [DOI] [PubMed] [Google Scholar]
- Ding D.-Q., Tomita,Y., Yamamoto,A., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (2000) Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells, 5, 169–190. [DOI] [PubMed] [Google Scholar]
- Ding R., McDonald,K.L. and McIntosh,J.R. (1993) Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol., 120, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., West,R.R., Morphew,D.M., Oakley,B.R. and McIntosh,J.R. (1997) The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell., 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U., Salus,S.S., Karig,I. and Sazer,S. (2000) The fission yeast ran GTPase is required for microtubule integrity. J. Cell Biol., 151, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Mayor,T., Meraldi,P., Stierhof,Y.D., Tanaka,K. and Nigg,E.A. (1998a) C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol., 141, 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi,P. and Nigg,E.A. (1998b) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J., 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Descombes,P., Twomey,C., Bacchieri,R. and Nigg,E.A. (2000) The NIMA-related kinase X-Nek2B is required for efficient assembly of the zygotic centrosome in Xenopus laevis. J. Cell Sci., 113, 1973–1984. [DOI] [PubMed] [Google Scholar]
- Glover D.M., Hagan,I.M. and Tavares,A.A. (1998) Polo-like kinases: a team that plays throughout mitosis. Genes Dev., 12, 3777–3787. [DOI] [PubMed] [Google Scholar]
- Gordon C., McGurk,G., Wallace,M. and Hastie,N.D. (1996) A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J. Biol. Chem., 271, 5704–5711. [DOI] [PubMed] [Google Scholar]
- Hagan I. and Yanagida,M. (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol., 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2000) Chromosome cohesion, condensation and separation. Annu. Rev. Biochem., 69, 115–144. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y. et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell, 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Kimura K., Cuvier,O. and Hirano,T. (2001) Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem., 276, 5417–5420. [DOI] [PubMed] [Google Scholar]
- Krien M.J.E., Bugg,S.J., Palatsides,M., Ausoline,G., Morimyo,M. and O’Connell,M.J. (1998) A NIMA homologue promotes chromatin condensation in fission yeast. J. Cell Sci., 111, 967–976. [DOI] [PubMed] [Google Scholar]
- LeGoff X., Utzig,S. and Simanis,V. (1999) Controlling septation in fission yeast: finding the middle and timing it right. Curr. Genet., 35, 571–584. [DOI] [PubMed] [Google Scholar]
- Lu K.P. and Hunter,Y. (1995) Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell, 81, 413–424. [DOI] [PubMed] [Google Scholar]
- Marelli M., Lusk,C.P., Chan,H., Aitchison,J.D. and Wozniak,R.W. (2001) A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol., 153, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T., Imamoto,N., Yoneda,Y. and Yanagida,M. (1998) Mutations in fission yeast Cut15, an importin α homolog, lead to mitotic progression without chromosome condensation. Curr. Biol., 8, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Mayor T., Stierhof,Y.D., Tanaka,K., Fry,A.M. and Nigg,E.A. (2000) The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol., 151, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 715–723. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1990) Universal control mechanism regulating onset of M-phase. Nature, 344, 503–508. [DOI] [PubMed] [Google Scholar]
- O’Connell M.J., Norbury,C. and Nurse,P. (1994) Premature chromatin condensation upon accumulation of NIMA. EMBO J., 13, 4926–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.J., Raleigh,J.M., Verkade,H.D. and Nurse,P. (1997) Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J., 16, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A.H., McGuire,S.L. and Osmani,S.A. (1991a) Parallel activation of the nimA and p34cdc2 cell cycle-regulated protein-kinases is required to initiate mitosis in Aspergillus nidulans. Cell, 67, 283–291. [DOI] [PubMed] [Google Scholar]
- Osmani A.H., O’Donnell,K., Pu,R.T. and Osmani,S.A. (1991b) Activation of the nimA protein kinase plays a unique role during mitosis that cannot be bypassed in the absence of the bimE checkpoint. EMBO J., 10, 2669–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S.A., May,G.S. and Morris,N.R. (1987) Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans.J. Cell Biol., 104, 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S.A., Engle,D., Doonan,J. and Morris,N.R. (1988a) Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell, 52, 241–251. [DOI] [PubMed] [Google Scholar]
- Osmani S.A., Pu,R.T. and Morris,N.R. (1988b) Mitotic induction and maintenance by over-expression of a G2-specific gene that encodes a potential protein kinase. Cell, 53, 237–244. [DOI] [PubMed] [Google Scholar]
- Pu R.T. and Osmani,S.A. (1995) Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J., 14, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Sutani,T., Yamashita,Y., Saitoh,S., Takeuchi,M., Nakaseko,Y. and Yanagida,M. (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J., 13, 4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I. and Yanagida,M. (1994) Bypassing anaphase by fission yeast cut9 mutation: requirement of cut9+ to initiate anaphase. J. Cell Biol., 127, 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S. and Nurse,P. (1994) A fission yeast rcc1-related protein is required for the mitosis to interphase transition. EMBO J., 13, 606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.J., Fry,A.M., Sutterlin,C., Ried,T. and Nigg,E.A. (1994) Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ., 5, 625–635. [PubMed] [Google Scholar]
- Tatebayashi K., Kato,J. and Ikeda,H. (1998) Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics, 148, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F., Taniguchi,E., Shinya,N., Iwamatsu,A. and Nishida,E. (2001) Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature, 410, 215–220. [DOI] [PubMed] [Google Scholar]
- Uemura T. and Yanagida,M. (1984) Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J., 3, 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade H.M., Bugg,S.J., Lindsay,H.D., Carr,A.M. and O’Connell,M.J. (1999) Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell, 10, 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Vaisberg,E.V., Ding,R., Nurse,P. and McIntosh,J.R. (1998) cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell, 9, 2839–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Wilde,A., Moore,M.S., Adam,S.A., Merdes,A. and Zheng,Y. (2001) Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science, 291, 653–656. [DOI] [PubMed] [Google Scholar]
- Wilde A., Lizarraga,S.B., Zhang,L., Wiese,C., Gliksman,N.R., Walczak,C.E. and Zheng,Y. (2001) Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nature Cell Biol., 3, 221–227. [DOI] [PubMed] [Google Scholar]
- Ye X.S., Fincher,R.R., Tang,A., Osmani,A.H. and Osmani,S.A. (1998) Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA. Mol. Biol. Cell, 9, 3019–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]