Abstract
Budding yeast Mps1p kinase has been implicated in both the duplication of microtubule-organizing centers and the spindle assembly checkpoint. Here we show that hMps1, the human homolog of yeast Mps1p, is a cell cycle-regulated kinase with maximal activity during M phase. hMps1 localizes to kinetochores and its activity and phosphorylation state increase upon activation of the mitotic checkpoint. By antibody microinjection and siRNA, we demonstrate that hMps1 is required for human cells to undergo checkpoint arrest in response to microtubule depolymerization. In contrast, centrosome (re-)duplication as well as cell division occur in the absence of hMps1. We conclude that hMps1 is required for the spindle assembly checkpoint but not for centrosome duplication.
Keywords: cell cycle/centrosome duplication/hMps1/mitosis/mitotic spindle checkpoint
Introduction
The founding member of the Mps1 kinase family, Mps1p of Saccharomyces cerevisiae, has been implicated in the regulation of multiple cell cycle-related processes, including the duplication of the spindle pole body (SPB) (Winey et al., 1991) and the correct segregation of chromosomes during cell division (Weiss and Winey, 1996). A role in SPB duplication has been inferred from the observation that mutations in the MPS1 gene give rise to monopolar spindles (Winey et al., 1991; Schutz and Winey, 1998), but the precise molecular defects in mps1 mutants are not understood. In addition, Mps1p was also found to be involved in the spindle assembly checkpoint (Weiss and Winey, 1996). Whereas wild-type budding yeast arrest in mitosis in response to spindle damage, no such arrest was seen in mps1 mutants (Weiss and Winey, 1996). Conversely, a checkpoint arrest could be triggered by overexpression of Mps1p, and this arrest was dependent on all other checkpoint genes tested (Bub1–3, Mad1–3), suggesting that Mps1p functions upstream in a signaling pathway (Hardwick et al., 1996). Mps1p has also been implicated in the assembly of the spindle apparatus (Jones et al., 1999) and the regulation of sporulation (Straight et al., 2000). At face value, these data suggest that budding yeast Mps1p is a multifunctional enzyme.
Kinases structurally related to Mps1p were described subsequently in Schizosaccharomyces pombe (Mph1p), Arabidopsis thaliana (PPK1), Xenopus laevis (XMps1) and mammals (mMps1/ESK in mouse, hMps1/TTK/PYT in humans) (Douville et al., 1992; Mills et al., 1992; Lindberg et al., 1993; He et al., 1998; Abrieu et al., 2001; Fisk and Winey, 2001). In all these Mps1 family members, the C-terminal catalytic domains show a significant degree of sequence similarity (Fisk and Winey, 2001). Furthermore, both yeast and mammalian Mps1 kinases phosphorylate serine/threonine as well as tyrosine residues, at least in vitro (Mills et al., 1992; Lindberg et al., 1993; Lauze et al., 1995). However, the N-terminal domains show little, if any, sequence conservation. Thus, it clearly will be important to determine to what extent the various functions described for budding yeast Mps1p have been conserved in other organisms.
Fission yeast Mph1 could complement both the spindle checkpoint defect and the SPB duplication defect of a budding yeast mps1 mutant, indicating that it represents a bona fide functional homolog (He et al., 1998). However, although deletion of Mph1 from S.pombe did impair the spindle assembly checkpoint, it did not cause any growth defect in the absence of spindle damage, suggesting that Mph1p is not essential for SPB duplication in fission yeast (He et al., 1998). Initial characterizations have also been reported for putative vertebrate homologs of yeast Mps1p. Mammalian Mps1 family members are expressed in all proliferating cells and tissues (Mills et al., 1992; Hogg et al., 1994), consistent with a function in cell cycle progression. Most recently, amphibian XMps1 was shown to be necessary for the establishment and maintenance of a spindle assembly checkpoint reconstituted in Xenopus egg extracts (Abrieu et al., 2001), whereas mouse mMps1 was reported to regulate centrosome duplication (Fisk and Winey, 2001).
Here, we have undertaken a functional analysis of the putative Mps1 homolog in human cells. This kinase originally was designated as TTK (Mills et al., 1992) or PYT (Lindberg et al., 1993), but recently has been renamed hMps1 (Fisk and Winey, 2001). We show that hMps1 is a cell cycle-regulated kinase, displaying maximal activity during M phase and a conspicuous kinetochore association during early mitosis. By microinjection of highly specific antibodies, as well as the silencing of hMps1 by RNA interference (siRNA), we further demonstrate that hMps1 is required for the establishment and/or maintenance of the spindle assembly checkpoint in human cells. However, contrary to a recent report on murine Mps1 (Fisk and Winey, 2001), our results lead us to conclude that hMps1 is dispensable for centrosome duplication. This conclusion upsets the current assumption that Mps1 kinases have evolutionarily conserved dual functions.
Results
hMps1 is a cell cycle-regulated protein kinase
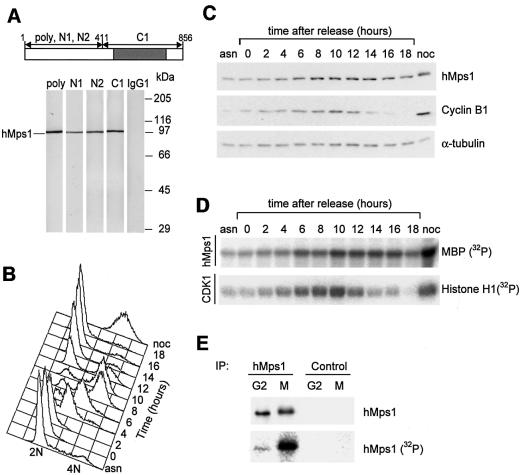
To study the function of hMps1, a polyclonal rabbit antibody and several mouse monoclonal antibodies (mAbs) were generated. As determined by western blotting on appropriate fragments of recombinant hMps1 (data not shown), the three mAbs used in this study (N1, N2 and C1) are directed against the N-terminus (N1 and N2) and the catalytic domain (C1) of hMps1, respectively, as depicted schematically in Figure 1A. Western blot analysis performed on cell extracts from exponentially growing HeLa cells revealed that all anti-hMps1 antibodies were highly specific for hMps1 (Figure 1A). All antibodies also immunoprecipitated hMps1. Interestingly, hMps1 was active in N1 and N2 immunoprecipitates, but not in C1 immunoprecipitates, indicating that this latter antibody inhibits the kinase (data not shown).
Fig. 1. Cell cycle regulation of hMps1 protein and activity levels. (A) Characterization of hMps1 antibodies. Cell extracts from exponentially growing HeLa cells were separated by SDS–PAGE and probed by western blotting with polyclonal, affinity-purified anti-hMps1 antibody (poly), and with three different anti-hMps1 mAbs (N1, N2 and C1). A non-immune IgG1 was used for control. The migration of hMps1 is shown on the left and molecuar weight markers are indicated on the right. The schematic representation (top) shows the distribution of epitopes recognized by the different mAbs on hMps1, with the catalytic domain marked in gray. (B–D) HeLa cells were released from a thymidine/aphidicolin double block in early S phase, samples were taken for FACS (B), western blot analysis (C) and kinase activity measurements (D) at the time points indicated. For comparison, asynchronously growing (asn) and nocodazole-arrested (noc) cells were analyzed in parallel. (E) hMps1 is phosphorylated specifically upon checkpoint engagement. Cell extracts were prepared from cells labeled in vivo with [32P]inorganic phosphate, and equal amounts of extracts were used for immunoprecipitations with hMps1 antibody or non-immune IgG1. Following SDS–PAGE, recovery of hMps1 protein was monitored by western blotting using mAb N1 (upper panel) and incorporation of 32P into hMps1 visualized by autoradiography (lower panel).
To examine the expression and activity of hMps1 during the cell cycle, HeLa cells were released from a thymidine/aphidicolin double block at the G1/S boundary (Heintz et al., 1983; Sillje et al., 1999). Flow cytometric analysis showed that cells proceeded synchronously through the cell cycle (Figure 1B). hMps1 protein was detectable at all cell cycle stages, but levels clearly increased as cells approached mitosis (Figure 1C), consistent with a previous report (Hogg et al., 1994). hMps1 kinase activity was determined in immunoprecipitates, using myelin basic protein (MBP) as a substrate (Figure 1D). No significant MBP activity was seen in control immunoprecipitates (data not shown). Both hMps1-associated MBP kinase activity and Cdk-associated histone H1 kinase activity peaked upon entry of cells into mitosis (8–10 h after release from the G1/S block). As cells exited mitosis and re-entered G1 phase (12–14 h after release), Cdk1 activity dropped more rapidly than hMps1 activity (Figure 1D). Quantitative analysis showed that hMps1 protein levels increased ∼3-fold as cells entered mitosis, whereas kinase activity increased ∼10-fold (data not shown). This suggests that hMps1 is activated by a post-translational mechanism in every mitosis.
Upon nocodazole-induced spindle assembly checkpoint arrest, hMps1 showed an up to 30-fold increase in activity (Figure 1D, lane noc). Identical results were also obtained when the checkpoint was activated by taxol (data not shown). This indicates that hMps1 is activated maximally regardless of whether the checkpoint was engaged by a lack of microtubule attachment or a loss of tension at kinetochores. Concomitant with checkpoint activation, hMps1 displayed a retarded electrophoretic mobility, suggesting that it might be phosphorylated (Figure 1C, lane noc, and data not shown). To address this issue directly, in vivo labeling with [32P]inorganic phosphate was performed on either interphasic (G2) cells or nocodazole-arrested cells. Immunoprecipitated hMps1 from mitotically arrested cells was clearly phosphorylated, but no significant phosphate incorporation was seen in interphasic hMps1 (Figure 1E, lower panel). Equal recovery of protein was demonstrated by western blotting (Figure 1E, upper panel), and no phosphate incorporation could be detected in control immunoprecipitates (Figure 1E, lanes Control). These results show that both hMps1 protein and activity levels peak during progression through M phase, and that hMps1 is both maximally active and extensively phosphorylated under conditions of spindle checkpoint engagement.
hMps1 localizes to kinetochores during mitosis
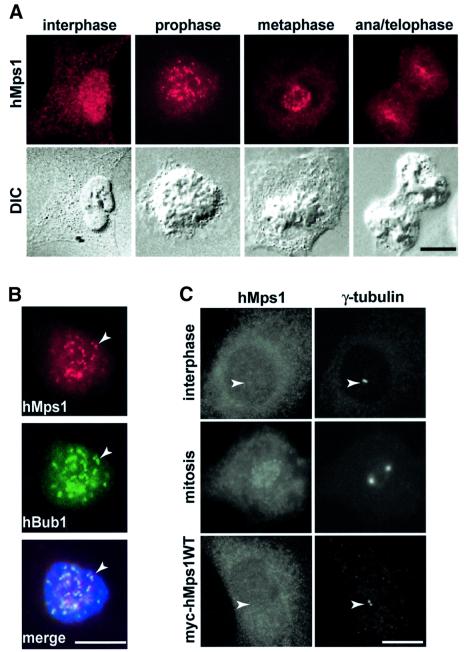
The subcellular localization of hMps1 in exponentially growing U2OS cells was determined by indirect immunofluorescence microscopy (Figure 2). In interphase cells, hMps1 was distributed diffusely throughout the cell. During mitosis, however, staining of a number of bright spots on condensed chromosomes became prominent (Figure 2A). These spots often appeared as closely spaced doublets, reminiscent of kinetochores. Co-localization of hMps1 with hBub1 kinase, a known kinetochore-associated component of the spindle assembly checkpoint (Taylor et al., 1998), confirmed the identity of the hMps1-positive structures with kinetochores (Figure 2B). The kinetochore association of hMps1 was most prominent at early stages of mitosis, from prophase to metaphase, but residual punctate staining could still be seen in telophase cells (Figure 2A). Kinetochore association of hMps1 was not significantly affected by nocodazole treatment, indicating that it does not require either microtubules or spindle checkpoint activation (data not shown). Very similar results were obtained with each of the three anti-hMps1 mAbs in all fixation methods tested.
Fig. 2. Endogenous hMps1 transiently associates with kinetochores in U2OS cells. (A) Exponentially growing U2OS cells were fixed and permeabilized simultaneously with formaldehyde/Triton X-100 and analyzed by indirect immunofluorescence microscopy using anti-hMps1 mAb N1. Corresponding differential interference contrast (DIC) micrographs are shown in the lower panels. Bars = 10 µm. (B) Kinetochores were identified by double staining with anti-hMps1 mAb N1 and rabbit anti-hBub1 antibodies (arrowheads) after fixation as described in (A). DNA was counterstained using DAPI. In the merged panel, anti-hMps1 staining is in red and anti-hBub1 staining in green. Bar = 10 µm. (C) Neither endogenous hMps1 nor ectopically expressed hMps1 localize to centrosomes (arrowheads) in U2OS cells. Interphasic (upper panel) and mitotic (middle panel) U2OS cells were fixed with methanol and analyzed with antibodies against hMps1 (mAb N1; left) and γ-tubulin (right). The bottom panel shows the localization of wild-type, myc-tagged hMps1, after transient expression in U2OS cells and visualization with anti-myc (9E10) antibody (left), again in direct comparison with γ-tubulin localization (right). Bar = 10 µm.
In view of a recent report describing a centrosome association of mMps1 in mouse cells (Fisk and Winey, 2001), the lack of centrosome staining by anti-hMps1 antibodies was unexpected. To examine this issue more carefully, we performed double immunofluorescent labeling with anti-hMps1 and anti-γ-tubulin antibodies, a marker for centrosomes (Fry et al., 1998). None of the three distinct hMps1 antibodies stained either interphasic centrosomes or mitotic spindle poles, no matter what fixation method was used (Figure 2C, and data not shown). Likewise, no detectable centrosome staining was produced by anti-myc antibodies following expression of either wild-type or catalytically inactive (D663A) myc-tagged hMps1 in U2OS cells, although both of these ectopically expressed hMps1 proteins readily localized to kinetochores in mitotic cells (Figure 2C, and data not shown). This latter result indicates that hMps1 localization to mitotic kinetochores is independent of its activity. The observed cell cycle-dependent kinetochore association of hMps1 falls in line with recent studies on XMps1 and mMps1 (Abrieu et al., 2001; Fisk and Winey, 2001). However, contrary to the reported centrosome association of mMps1 (Fisk and Winey, 2001), hMps1 could not be detected on either centrosomes or spindle poles.
Anti-hMps1 antibodies interfere with the spindle assembly checkpoint
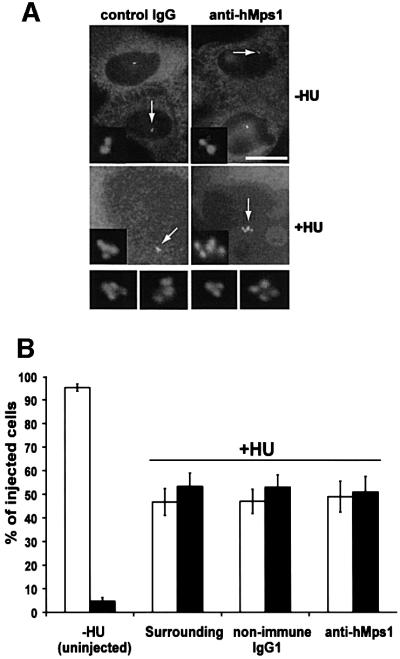
As a first approach to exploring the function of hMps1 in living cells, antibody microinjection experiments were performed. As outlined schematically in Figure 3A, HeLa cells were synchronized at the G1/S boundary, and 3 h after release they were microinjected with either non-immune mouse immunoglobulin (IgG1) or a mixture of anti-hMps1 mAbs (N1, N2 and C1), along with a fluorescent dextran marker. Injections were scattered (one cell per microscopic field), so that any effects of the injected antibodies on subsequent cell divisions could be observed (Lane and Nigg, 1996). Injected cells were cultured for 14 h before they were examined. Alternatively, nocodazole was added 2 h after injection, and the incubation was continued for another 12 h. For quantitative analysis of the fate of injected cells (Figure 3B), these were examined in a live state, using fluorescent dextran to identify the progeny of injected cells. Cells were scored as mitotic or interphasic, depending on whether they displayed a rounded or flattened morphology, respectively. Alternatively, injected cell cultures were fixed and stained for qualitative inspection by fluorescence microscopy (Figure 3C and D).
Fig. 3. hMps1 is an essential component of the spindle assembly checkpoint. (A) A schematic illustration of the experimental protocol used for microinjection experiments. (B) Histograms comparing the morphologies of HeLa cells after cytoplasmic injection of non-immune IgG1 or anti-hMps1 antibodies and subsequent incubation in the absence (left panel) or presence of nocodazole (right panel). Open bars indicate the percentage of injected cells with a flattened, interphasic morphology, whereas black bars indicate the proportion of cells with a rounded morphology (i.e. mitotically arrested cells). Approximately 120–150 injected cells were counted for each experiment. Shown are the averages of three independent experiments, with standard deviations. (C and D) Single, widely spaced HeLa cells were injected with control IgG1 or anti-hMps1 antibodies (as indicated), followed by either a 14 h incubation in the absence of nocodazole (C), or a 12 h incubation with nocodazole (D), before fixation with paraformaldehyde solution. Injected cells were visualized using an anti-mouse IgG secondary antibody and DNA was stained by DAPI. Representative examples of injected HeLa cells are shown. Note that the presence of daughter cells indicates successful cell division. Bars = 10 µm.
The vast majority of microinjected cells divided efficiently, regardless of whether they had been injected with control IgG1 or anti-hMps1 antibodies. More than 95% of all injected cells displayed a flat morphology, indistinguishable from that of surrounding cells (Figure 3B, left panel). Although single cells had been injected with antibodies and fluorescent dextran, virtually all fluorescent cells were present as doublets, indicating that they had divided during the time course of the experiment (Figure 3C). Furthermore, most of the injected cells reached the 4-cell stage by 48 h, showing that they had also been able to proceed through a second round of division (data not shown). Staining of DNA revealed normally shaped nuclei, typical of interphasic cells (Figure 3C). This indicates that antibody-mediated interference with hMps1 did not prevent cell division, although we cannot exclude subtle problems with chromosome segregation.
In response to nocodazole, ∼80% of the cells that had been injected with non-immune IgG1 displayed a rounded morphology and condensed chromatin, indistinguishable from the non-injected surrounding cells (Figure 3B, right panel, and D). This result was expected, since nocodazole triggers the spindle assembly checkpoint and hence causes a prometaphase arrest (Amon, 1999). In stark contrast, the vast majority of the cells that had been injected with anti-hMps1 antibodies showed a flat morphology with decondensed chromatin but aberrantly shaped nuclei (Figure 3B, right panel, and D). Very similar results were observed after separate injection of each of the three anti-hMps1 mAbs, although the percentage of interphasic cells was lower than that observed after combined injection of all three antibodies (data not shown). We conclude that cells injected with anti-hMps1 antibodies were unable to undergo and/or maintain a checkpoint arrest in response to microtubule depolymerization.
Microinjection of anti-hMps1 antibodies does not block centrosome duplication
Recently, murine Mps1 was reported to be required for centrosome duplication (Fisk and Winey, 2001), in line with the purported function of budding yeast Mps1p in SPB duplication (Winey et al., 1991). To explore more directly whether a corresponding function could be attributed to hMps1, U2OS cells were arrested in S phase, the cell cycle stage during which centrosomes duplicate (Hinchcliffe and Sluder, 2001), and injected with either anti-hMps1 antibodies or non-immune IgG1. Cells were then released into fresh medium and, 24–48 h later, centrosome numbers were scored by immunofluorescence microscopy using anti-γ-tubulin antibodies. After 24 h, virtually all injected cells had divided once, and all contained normal numbers of centrosomes, regardless of whether anti-hMps1 mAb or non-immune IgG1 had been injected (Figure 4A, upper panels). Furthermore, 48 h after injection, the vast majority of cells had reached the 4-cell stage and, again, centrosome numbers were normal (data not shown). Although inhibition of centrosome duplication would have been expected to cause monopolar spindle formation and mitotic arrest (Blangy et al., 1995), no such phenotypes were observed.
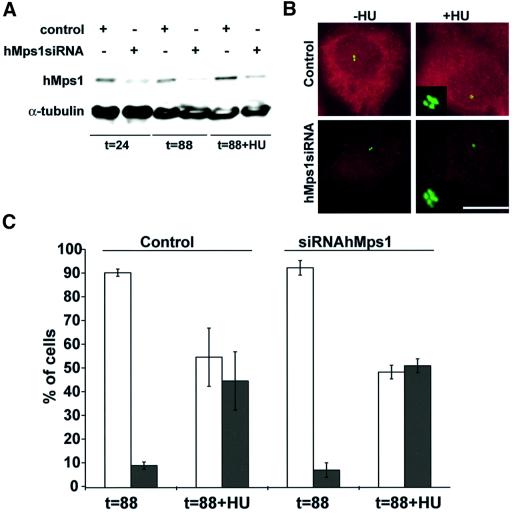
Fig. 4. Microinjection of anti-hMps1 antibodies does not block centrosome duplication. (A) U2OS cells were pre-synchronized for 16 h in 15 mM hydroxyurea (HU) and injected into the cytoplasm with either control IgG1 (left panels) or anti-hMps1 antibodies (right panels). Injections were scattered (i.e. only one cell per microscopic field) for accurate counting. Cells were then incubated for 24 h in the absence of HU (upper panels) or for 64 h in the presence of 15 mM HU (lower panels), before they were methanol fixed and double-stained using an anti-mouse IgG secondary antibody to visualize injected antibodies, and anti-γ-tubulin to visualize centrosomes. Note that cells had divided properly in the absence of HU (upper panels), whereas centrosome reduplication had occurred in the presence of HU-induced S phase arrest, regardless of anti-hMps1 antibody injection. Bar = 10 µm. (B) Quantitative analysis of centrosome reduplication. The histogram indicates the percentages of cells with normal numbers (1–2) of centrosomes (open bars) or extra copies (>2) of centrosomes (black bars). Incubation of U2OS cells with or without HU, as well as the identification of injected cells and centrosome staining, were performed as described in (A). Approximately 80–120 injected cells were counted for each experiment. Shown are the averages of three independent experiments ± SD.
In a complementary experiment, we took advantage of the fact that certain cell types, including U2OS cells, are permissive for multiple rounds of centrosome reduplication under conditions of S phase arrest (Balczon et al., 1995; Matsumoto et al., 1999; Meraldi et al., 1999). U2OS cells were thus treated for 16 h with hydroxyurea (HU), then injected with anti-hMps1 mAbs or non-immune IgG1, and incubated in HU for an additional 64 h. By this time, injected antibodies could still be visualized readily using secondary antibodies (Figure 4A, lower panels). Cells were then stained with anti-γ-tubulin antibodies and centrosomes were counted. [That γ-tubulin staining provides a reliable assay for determining centrosome numbers in such experiments had been demonstrated previously by electron microscopy (Balczon et al., 1995; Meraldi et al., 1999).] As shown in Figure 4B, ∼50% of the uninjected, HU-treated cells contained multiple centrosomes, and the same was true for cells injected with non-immune IgG1. Importantly, the extent of centrosome reduplication was not reduced detectably upon injection of anti-hMps1 mAbs, demonstrating that this process could not be interfered with by antibody-mediated inhibition of hMps1 (Figure 4A and B). Taken together, the above data show that centrosome duplication was unaffected by anti-hMps1 antibodies, although these same antibodies effectively abrogated the spindle assembly checkpoint function of the kinase.
Silencing of hMps1 by siRNA inactivates the spindle assembly checkpoint
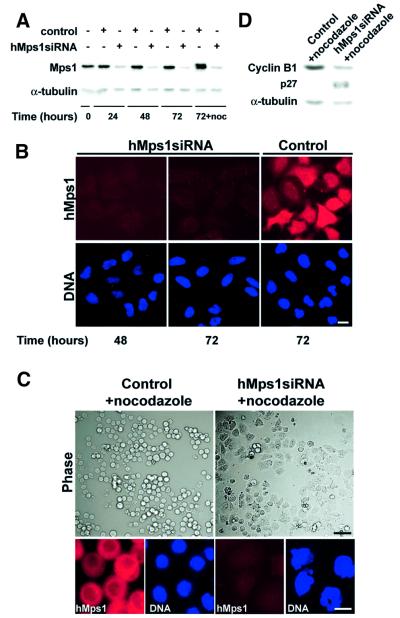
To obtain independent verification of the conclusions reached from the antibody microinjection studies, we turned to gene silencing by small interfering RNA (siRNA) duplexes (Elbashir et al., 2001). HeLa S3 cells were transfected with a 21 nucleotide duplex homologous to the hMps1 sequence and, at different times thereafter, cell extracts were prepared for western blotting (Figure 5A). The level of hMps1 protein was strongly reduced within 24 h of treatment with the hMps1 siRNA duplex, and almost undetectable by 48 h. No reduction in hMps1 levels occurred in control incubations (Figure 5A). Silencing of the hMps1 gene was also confirmed by immunofluorescence microscopy with anti-hMps1 antibodies (Figure 5B). Transfection efficiency in this experiment was ∼90% and, since quantitative analysis of western blots showed a corresponding 90% reduction of the hMps1 protein level in the population, this indicates that hMps1 had been eliminated almost completely in the vast majority of cells. Remarkably, silencing of the hMps1 gene did not interfere in any major way with several rounds of cell division (Figure 5B), in agreement with the microinjection studies described above.
Fig. 5. Silencing of hMps1 by siRNA duplex abolishes the spindle assembly checkpoint. (A) Western blotting shows effective silencing of hMps1 in HeLa S3 cells (transfection efficiency = 90%). Following mock transfection or transfection with hMps1 siRNA duplex, cell extracts were prepared at the time points indicated. Equal amounts of protein were separated by SDS–PAGE and probed by western blotting with anti-hMps1 mAb N1 (upper panel) and anti-α-tubulin antibody as a loading control (lower panel). In the sample marked noc, nocodazole (50 ng/ml) was added at t = 48 h for an additional 24 h. (B) Immunofluorescence microscopy shows effective silencing of hMps1 in HeLaS3 cells. Cells were transfected as described in (A), fixed and permeabilized with paraformaldehyde/Triton X-100, and stained for hMps1 (upper panels) and DNA (lower panels) using anti-hMps1 mAb N1 and DAPI, respectively. Bar = 10 µm. (C) Analysis of HeLa S3 cells after mock transfection (left) or hMps1 silencing by siRNA duplex (right) and subsequent exposure to spindle damage. Nocodazole (50 ng/ml) was added 48 h post-transfection for 24 h before cells were analyzed. Upper panels: phase-contrast pictures. Lower panels: cells stained for hMps1 (left) and DNA (right). Bars = 1 µm (upper panels) and 10 µm (lower panels). (D) Cell extracts, prepared from the samples described in (C), were probed by western blotting with anti-cyclin B1 and anti-p27Kip1 antibodies, respectively. Anti-α-tubulin was used as a loading control.
To confirm that inhibition of hMps1 abrogates the spindle assembly checkpoint, cells were transfected with hMps1 siRNA duplex for 48 h, before nocodazole was added for an additional 24 h. Mock-transfected cells responded to nocodazole by arresting with a rounded morphology and condensed chromosomes, reflecting the activation of the spindle assembly checkpoint (Figure 5C, left panels). In contrast, cells in which hMps1 had been silenced by siRNA were unable to sustain such an arrest. Instead, most of these cells flattened out and reformed readily discernible interphase nuclei (Figure 5C, upper right panel). When examined by DNA staining, most of the nuclei showed highly aberrant, multilobular shapes (Figure 5C, lower right panel), indistinguishable from those seen after injection of anti-hMps1 mAbs prior to challenging cells with nocodazole (see above). To confirm that cells had exited mitosis and proceeded into the next G1 (as opposed to returning to G2), cell extracts were analyzed by western blotting for cyclin B1, a protein known to be degraded upon mitotic exit (Clute and Pines, 1999), and for the Cdk inhibitor p27Kip1, a marker for G1 phase (Hengst and Reed, 1996). As expected for a spindle checkpoint arrest, nocodazole-treated, mock-transfected cells showed high levels of cyclin B1 but no detectable p27Kip1 (Figure 5D). In contrast, after silencing of hMps1, nocodazole-treated populations showed reduced levels of cyclin B1 and increased levels of p27Kip1 (Figure 5D), indicating that most cells had progressed from M phase into the next G1 phase. These siRNA data thus confirm our conclusion that hMps1 is essential for a spindle assembly checkpoint-induced mitotic arrest in human cells.
Silencing of hMps1 by siRNA does not inhibit centrosome duplication
To explore further a possible role for hMps1 in centrosome duplication, we asked whether silencing of hMps1 by siRNA would interfere with the ability of cells to re-duplicate centrosomes. To this end, U2OS cells were transfected with the hMps1 RNAi duplex, and the extent of silencing was assessed by western blotting (Figure 6A) and immunofluorescence microscopy (Figure 6B). By 24 h after transfection, the level of hMps1 was reduced by ∼70% (with a corresponding transfection efficiency of 70% in this particular experiment), and remained low for the entire duration of the experiment (88 h). Yet, virtually all transfected cells continued to divide and form apparently normal bipolar spindles (not shown), and no evidence was obtained for disruption of the centrosome duplication cycle (Figure 6B, left panel, and C). Furthermore, centrosome reduplication occurred to the same extent in HU-treated cells, whether or not hMps1 had been silenced (Figure 6B, right panels, and C). Thus, both siRNA and antibody microinjection studies concur to indicate that hMps1 is not required for centrosome duplication in human cells.
Fig. 6. Silencing of hMps1 by siRNA duplex does not interfere with centrosome (re-)duplication. (A) Western blotting demonstrates effective silencing of hMps1 in U2OS cells (transfection efficiency = 70%). Cells were transfected with GL-2 control or hMps1 siRNA duplexes, and extracts were prepared at the times indicated. Equal amounts of protein were resolved by SDS–PAGE and probed by western blotting with anti-hMps1 mAb N1 (upper panel) and anti-α-tubulin as a loading control (lower panel). In the sample marked t = 88 h + HU, hydroxyurea (15 mM) was added at t = 24 h for an additional 64 h. (B) Immunofluorescence microscopy shows effective silencing of hMps1 in U2OS cells. Cells were transfected as described in (A), fixed with methanol and stained for hMps1 (red) and γ-tubulin (green). Bar = 10 µm. (C) Histogram comparing the efficiency of centrosome reduplication in the samples described above. Open bars indicate the percentage of cells with normal numbers (1–2) of centrosomes, whereas black bars denote cells with multiple centrosomes (≥3). Only cells displaying low levels of hMps1 staining were counted. Shown are the averages of three independent experiments (counting ∼200 cells each) ± SD.
Overexpression of hMps1 does not influence centrosome duplication
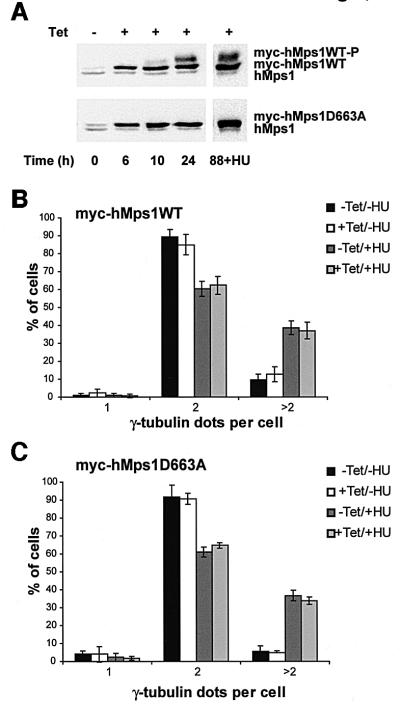
The above conclusion is in stark contrast to a recent report attributing a key role to Mps1 kinases in centrosome duplication (Fisk and Winey, 2001). Considering that Fisk and Winey had used primarily overexpression of wild-type and catalytically inactive mMps1 as a means to modulate the centrosome duplication cycle, one could argue that our discordant conclusion stems from the use of different experimental approaches. Thus, as a final test, we have constructed U2OS cell lines allowing the expression of both myc-tagged wild-type and catalytically inactive hMps1 under tetracycline control. Both proteins were induced ∼6-fold over endogenous hMps1 in response to tetracycline addition, and the activity of the wild-type kinase was demonstrated by the appearance of a band with retarded electrophoretic mobility, almost certainly reflecting autophosphorylation (Figure 7A). No deregulation of the centrosome duplication cycle could be detected upon propagation of cells overexpressing either wild-type (Figure 7B) or catalytically inactive hMps1 (Figure 7C). Likewise, centrosome reduplication in the presence of HU was unaffected (Figure 7B and C). These results also confirm our conclusion that hMps1 does not regulate centrosome duplication in human cells.
Fig. 7. Overexpression of wild-type myc-hMps1 (myc-hMps1WT) and myc-hMps1D663A does not affect centrosome reduplication. (A) Tetracycline-inducible expression of myc-hMps1WT and myc-hMps1D663A. Stably transfected U2OS-TRex cells were induced with tetracycline (1 µg/ml), and levels of hMps1 determined by immunoblotting. (B) Myc-hMps1WT does not enhance centrosome (re-)duplication. After an initial 24 h induction of myc-hMps1WT by tetracycline (1 µg/ml) in the absence of HU, myc-hMps1WT expression was continued for 64 h in the presence or absence of 15 mM HU, as indicated. Cells were then fixed with methanol and stained for myc-hMps1 and γ-tubulin, and the numbers of centrosomes counted. The histogram shows the results of three independent experiments ± SD (at least 200 cells were counted in each experiment). (C) Myc-hMps1D663A does not block centrosome (re-)duplication. Induction of myc-hMps1D663A and analysis of cells were performed as described in (B).
Discussion
In this study, we have examined the expression, localization and function of hMps1, the putative human homolog of Mps1p of S.cerevisiae. All our data concur to demonstrate that hMps1 is essential for the establishment and/or maintenance of the spindle assembly checkpoint, but dispensable for centrosome duplication. This conclusion contradicts the proposed dual role of Mps1 family members in vertebrate organisms.
Expression and localization of hMps1
hMps1 protein levels and kinase activity were detectable at all stages of the cell cycle, but they were maximal during passage through mitosis. Furthermore, hMps1 showed a particularly high activity in nocodazole-treated cells, indicating that it is activated upon engagement of the spindle assembly checkpoint. Concomitantly, hMps1 displayed a retarded electrophoretic mobility due to phosphorylation. Whether this reflects the activity of hMps1 itself or that of an upstream kinase remains to be determined.
By immunofluorescence microscopy, hMps1 was localized to both the nucleus and cytoplasm of interphasic cells. During mitosis, however, pronounced labeling of kinetochores could be seen, confirming and extending recent results with mMps1 (Fisk and Winey, 2001) and XMps1 (Abrieu et al., 2001). This labeling was particularly strong at early stages of mitosis, but was still detectable after silencing of the spindle assembly checkpoint and anaphase onset. Kinetochore association of hMps1 did not depend on either kinase activity or the presence of microtubules. Very similar results were obtained when analyzing the localization of myc-tagged, ectopically expressed hMps1. Recently, a centrosome association was reported for murine Mps1 (Fisk and Winey, 2001), but no such localization could be seen for hMps1 in this study. While we are confident of the specificity of the anti-hMps1 antibodies used here, the commercial antibody used by Fisk and Winey (2001) recognized several bands by western blotting in our hands (data not shown). Although we did see weak centrosome staining in rare cells when expressing a green fluorescent protein (GFP)-tagged version of hMps1 (L.Arnaud and E.A.Nigg, unpublished results), we are wary that this is an artifact. In fact, myc-tagged exogenous hMps1 was never detected at centrosomes.
hMps1 is an essential component of the spindle assembly checkpoint in human cells
By both antibody microinjection and gene silencing, we demonstrate that hMps1 is required for establishing and/or maintaining the spindle assembly checkpoint. This checkpoint was originally defined through genetic analyses in budding yeast (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996), and several relevant genes were identified, notably the kinase Bub1p and its binding partner Bub3p, BubR1/Mad3, the Mad1p–Mad2p complex and the Mps1p kinase (Amon, 1999; Wassmann and Benezra, 2001). Subsequent studies on homologous proteins in higher eukaryotes have revealed that all these checkpoint components localize to kinetochores during early mitosis (Wassmann and Benezra, 2001). They are thus thought to cooperate in a signaling pathway that prevents anaphase onset until all kinetochores have undergone correct bipolar attachment. In the absence of appropriate microtubule–kinetochore interactions, an inhibitory signal is relayed to the anaphase-promoting complex/cyclosome (APC/C), a ubiquitin ligase controlling the proteolytic degradation of anaphase onset inhibitors, mitotic cyclins and other proteins (Morgan, 1999; Peters, 1999). The function of Mps1 kinases in this pathway remains unclear, but the available evidence suggests that it operates upstream of all known Mad and Bub gene products (Hardwick et al., 1996; Abrieu et al., 2001).
We have shown here that hMps1 is absolutely required for spindle checkpoint arrest in response to microtubule depolymerization by nocodazole. However, in the absence of a deliberate experimental insult to the spindle apparatus, the antibody-mediated inhibition of hMps1 function did not interfere detectably with mitotic progression. Similarly, the almost complete elimination of hMps1 by siRNA did not prevent cells from undergoing several rounds of division, although it remains possible that subtle defects in chromosome segregation did occur. In striking contrast, antibody-mediated interference with Mad2 severely affected mitotic progression (Gorbsky et al., 1998), and the elimination, by gene targeting, of either murine Bub3 or Mad2 resulted in embryonic lethality, apparently due to extensive chromosome mis-segregation (Dobles et al., 2000; Kalitsis et al., 2000). In future studies, it will be interesting to measure the fidelity of chromosome segregation in mammalian cells from which Mps1 has been eliminated by gene targeting.
Are vertebrate Mps1 kinases required for centrosome duplication?
Extensive studies on budding yeast Mps1p have led to the notion that this kinase functions in at least two ostensibly distinct processes, namely SPB duplication and the spindle assembly checkpoint (Winey et al., 1991; Weiss and Winey, 1996). The present study establishes that the human Mps1 homolog is essential for the spindle assembly checkpoint in mammalian cells in vivo, in excellent agreement with recent data showing that Xenopus Mps1 is required for a similar checkpoint reconstituted in Xenopus egg extracts (Abrieu et al., 2001). However, in contradiction to a recent report implicating murine Mps1 in the control of centrosome duplication (Fisk and Winey, 2001), we argue here that human Mps1 is not required for centrosome duplication. Of course, it is possible that centrosome duplication requires only trace amounts of Mps1 activity, or that individual mammalian species and cell types differ with regard to their requirements for centrosome duplication. Alternatively, a second human Mps1 isoform, devoted to a centrosomal function, could have gone undetected in spite of the virtually complete elucidation of the genome. We cannot formally exclude these explanations, but consider them unlikely. Instead, we propose that mammalian Mps1 kinases are not required for centrosome duplication and that the primary function of these kinases relates to the spindle checkpoint in all eukaryotes.
Materials and methods
Plasmid constructions
An expressed sequence tag (EST) coding for hMps1 was obtained from the Image consortium (ID 511705). It was completely sequenced and found to code for a near full-length hMps1 (residues 3–856). [The sequence of this EST cDNA was identical to the one reported for TTK (Mills et al., 1992), except for the fact that the expected Gln420 was absent. The reason for this discrepancy presently is unclear, but several ESTs containing or lacking the corresponding codon have been reported.] PCR was performed to add the two missing residues (ATG, Met and GAA, Glu) to the 5′ end of the available cDNA. To prepare catalytically inactive hMps1 (hMps1D663A), codon 663 (GAT, Asp) was mutated to GCT (Ala). N-terminally myc-tagged hMps1 was prepared as an in-frame fusion in pBluescript-myc (Schmidt-Zachmann and Nigg, 1993). For construction of mammalian expression plasmids, myc-hMps1 was excised and subcloned into pRcCMV (Invitrogen). All PCR fragments and mutations were checked by sequencing.
Antibody production
To generate polyclonal antibodies against hMps1, a fragment corresponding to residues 3–516 was expressed in Escherichia coli using the QIAexpress System (Qiagen) and used for immunizing rabbits (Elevage Scientifique des Dombes, Chatillon sur Chalaronne, France). Antibodies were affinity purified on His6-hMps1D663A that had been purified from insect cells and immobilized on Ni-NTA–agarose (Qiagen). For the production of mAbs, Balb/c mice were immunized by repeated subcutaneous injections of 100–150 µg of His6-hMps1D663A, using either Freund’s or aluminium hydroxide as an adjuvant. Spleen cells were fused with PAIB3Ag81 mouse myeloma cells. Supernatant screening was performed by indirect enzyme-linked immunosorbent assays (ELISA) using His6-hMps1D663A as an antigen, by dot blot assays and by western blotting on total HeLa cell extracts. mAb isotyping revealed that N1, N2 and C1 are all IgG1.
Antibody microinjection
Antibody microinjection experiments were performed as described previously (Lane and Nigg, 1996) using either a 1:1:1 mixture of anti-hMps1 N1, N2 and C1 mAbs or a non-immune IgG1 (Sigma-Aldrich) for control. Antibodies were prepared from tissue culture supernatants by affinity purification on protein G–Sepharose™ (Amersham Pharmacia Biotech), washed with phosphate-buffered saline (PBS) and concentrated with an Ultrafree-0.5 centrifugal filter (Amicon bioseparation; Millipore). They were injected at 1 mg/ml. To allow live cell imaging, fluorescein isothiocyanate (FITC)–dextran (Mr >2 MDa) (Sigma-Aldrich) was co-injected at a final concentration of 0.5 mg/ml.
Cell culture and transfections
HeLa and U2OS cells were cultured at 37°C in a 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (Gibco-BRL), supplemented with 10% heat-inactivated fetal calf serum (FCS; Gibco-BRL) and penicillin– streptomycin (100 IU/ml and 100 mg/ml, respectively). To induce spindle checkpoint arrest, HeLa cells were treated with nocodazole (500 ng/ml) or taxol (10 µM) for 16 h before cell extracts were prepared for western blot analysis and kinase assays. U20S cells were transiently transfected using the calcium phosphate method as described by Meraldi et al. (1999). To generate tetracycline-inducible cell lines expressing myc-tagged wild-type and D663A mutant hMps1, U2OS-TRex™ cells (Invitrogen) were transfected using FuGENE™ 6 transfection reagent (Roche). Stably transfected lines were established by selection with 200 µg/ml Zeocin™ (Invitrogen) and 50 µg/ml hygromycin (Merck).
For labeling with [32P]inorganic phosphate, HeLa cells were synchronized at the G1/S boundary as described above. To obtain labeled G2 extracts, cells were released for 6 h into fresh medium, followed by a 4 h and 20 min incubation in DMEM, supplemented with 0.25 mCi/ml of [32P]orthophosphate (Amersham). Extracts from checkpoint-arrested cells were prepared after a release from the G1/S boundary for 10 h and an arrest in nocodazole (500 ng/ml) for 4 h. Rounded cells were then collected by shake-off and resuspended for 4 h in [32P]orthophosphate labeling medium, washed and lysed as described below.
Cell extracts, immunoprecipitation and immunoblotting
HeLa cells were washed once in ice-cold PBS, 1 mM phenylmethylsulfonyl fluoride (PMSF) and resuspended in HeLa lysis buffer (50 mM Tris–HCl pH 8.0, 1% NP-40, 150 mM NaCl) containing 30 µg/ml RNase A, 30 µg/ml DNase, protease inhibitors (1 mM PMSF, 1 µg/ml leupeptin, 10 µg/ml pepstatin A, 10 µg/ml aprotinin, 10 µg/ml bestatin) and phosphatase inhibitors (20 mM β-glycerophosphate, 5 mM NaF, 100 µM Na3VO4). After 30 min on ice, cells were collected by scraping and centrifuged at 14 000 r.p.m. for 15 min at 4°C. Protein concentrations were determined using the Dc protein assay (Bio-Rad). For immunoblotting, equal amounts of protein were resolved by SDS–PAGE and electrophoretically transferred to nitrocellulose membranes (Schleicher & Schuell). Membranes were stained with Ponceau S, blocked in 5% non-fat dry milk in PBS for 1 h at room temperature, and incubated with either polyclonal, affinity-purified primary hMps1 antibody (1.7 µg/ml), N1 mAb (undiluted culture supernatant), polyclonal anti-p27Kip1 antibody (Santa Cruz Biotechnology, 1:1000) or monoclonal anti-α-tubulin (Sigma-Aldrich, 1:4000) for 2 h at room temperature (or overnight at 4°C), followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies or goat anti-mouse antibodies, respectively (Amersham, 1:3000) for 1 h at room temperature. Bound HRP-conjugated antibodies were visualized with the enhanced chemiluminescence (ECL) detection system.
For immunoprecipitations, equal amounts of HeLa cell extracts were pre-cleared using protein G–Sepharose™ for 1 h at 4°C. They were then incubated for 3 h with protein G–Sepharose™ carrying bound mAb N1 on a rotating wheel, spun down and washed three times in wash buffer I (1 M NaCl, 1% NP-40, 50 mM Tris–HCl pH 8.0), three times in wash buffer II (200 mM NaCl, 0.1% NP-40, 50 mM Tris–HCl pH 8.0) and three times in wash buffer III (150 mM NaCl, 0.01% NP-40, 50 mM Tris–HCl pH 8.0).
In vitro kinase assays
Following immunoprecipitation, hMps1 immune complexes were washed three times in hMps1 kinase buffer [50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.5 mM dithiothreitol (DTT), 10 mM β-glycerophosphate, 100 µM Na3VO4]. Kinase reactions were carried out for 30 min at 30°C in hMps1 kinase buffer supplemented with 10 µM ATP, 2 µCi of [γ32P]ATP (Amersham) and 0.5 mg/ml of MBP as a substrate. Reactions were stopped by addition of gel sample buffer and heating at 95°C for 5 min. Proteins were then resolved by SDS–PAGE and 32P incorporation was visualized by autoradiography. Cdk–cyclin complexes were collected using p9suc1 beads (Maridor et al., 1993), and histone H1 kinase activity was assayed as described previously (Golsteyn et al., 1995).
Immunofluorescence microscopy
Growth of cells on coverslips and immunostaining procedures, using either paraformaldehyde/Triton X-100 or methanol/acetone fixation and permeabilization, were performed as described (Fry et al., 1998). In some experiments, cells were fixed and permeabilized simultaneously in 20 mM PIPES pH 6.8, 4% formaldehyde, 0.2% Triton X-100, 10 mM EGTA, 1 mM MgCl2 for 15 min at room temperature. All fixed cells were incubated for 30 min at room temperature in blocking solution (PBS, 3% bovine serum albumin). Primary antibodies were anti-hMps1 mAbs (undiluted culture supernatant), 9E10 anti-myc mAb (undiluted culture supernatant), crude anti-hBub1 serum (1:500; kind gift of Silvia Martin-Lluesma) and anti-γ-tubulin IgG (2 µg/ml; Fry et al., 1998). All antibody incubations were carried out for 1 h at room temperature in a humidified chamber, followed by three washes in PBS. Primary antibodies were detected with Texas Red-conjugated, FITC-conjugated or AlexaFluor488-conjugated secondary antibodies. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; 2 µg/ml). Immuno fluorescence microscopy was performed using a Zeiss Axioplan II microscope and 10× or 40× and 63× oil immersion objectives, respectively. Photographs were taken using a Micromax (Princeton Instruments) CCD camera and Metaview (Universal Imaging) software. Images were processed with Adobe Photoshop (Adobe Systems, Mountain View, CA).
siRNA experiments
The siRNA sequence used for silencing of hMps1 corresponds to the coding region 133–155 (relative to the start codon). As a control, a duplex (GL-2) targeting luciferase was used (Elbashir et al., 2001). The 21 nucleotide RNA duplexes were purchased from Dharmacon Research, Inc. Annealing of the siRNAs and transfections using Oligofectamine (Life Technologies) were performed as described (Elbashir et al., 2001).
Acknowledgments
Acknowledgements
We thank Silvia Martin-Lluesma for a generous gift of anti-hBub1 serum, Thibault Mayor for 32P in vivo labeled cell extracts, Alicja Baskaya and Elena Nigg for excellent technical assistance in the production of monoclonal hMps1 antibodies, and K.Weber and J.Harborth (MPI, Göttingen) for help with siRNA technology. We also thank all members of the Barr and Nigg laboratories for helpful discussions.
References
- Abrieu A., Magnaghi-Jaulin,L., Kahana,J.A., Peter,M., Castro,A., Vigneron,S., Lorca,T., Cleveland,D.W. and Labbe,J.C. (2001) Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell, 106, 83–93. [DOI] [PubMed] [Google Scholar]
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Balczon R., Bao,L., Zimmer,W.E., Brown,K., Zinkowski,R.P. and Brinkley,B.R. (1995) Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol., 130, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A., Lane,H.A., d’Herin,P., Harper,M., Kress,M. and Nigg,E.A. (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell, 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Clute P. and Pines,J. (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell Biol., 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Dobles M., Liberal,V., Scott,M.L., Benezra,R. and Sorger,P.K. (2000) Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell, 101, 635–645. [DOI] [PubMed] [Google Scholar]
- Douville E.M., Afar,D.E., Howell,B.W., Letwin,K., Tannock,L., Ben David,Y., Pawson,T. and Bell,J.C. (1992) Multiple cDNAs encoding the esk kinase predict transmembrane and intracellular enzyme isoforms. Mol. Cell. Biol., 12, 2681–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Fisk H.A. and Winey,M. (2001) The mouse Mps1p-like kinase regulates centrosome duplication. Cell, 106, 95–104. [DOI] [PubMed] [Google Scholar]
- Fry A.M., Meraldi,P. and Nigg,E.A. (1998) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J., 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn R.M., Mundt,K.E., Fry,A.M. and Nigg,E.A. (1995) Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol., 129, 1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen,R.H. and Murray,A.W. (1998) Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol., 141, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss,E., Luca,F.C., Winey,M. and Murray,A.W. (1996) Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science, 273, 953–956. [DOI] [PubMed] [Google Scholar]
- He X., Jones,M.H., Winey,M. and Sazer,S. (1998) mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S.pombe. J. Cell Sci., 111, 1635–1647. [DOI] [PubMed] [Google Scholar]
- Heintz N., Sive,H.L. and Roeder,R.G. (1983) Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol., 3, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst L. and Reed,S.I. (1996) Translational control of p27Kip1 accumulation during the cell cycle. Science, 271, 1861–1864. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H. and Sluder,G. (2001) ‘It takes two to tango’: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev., 15, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Hogg D., Guidos,C., Bailey,D., Amendola,A., Groves,T., Davidson,J., Schmandt,R. and Mills,G. (1994) Cell cycle dependent regulation of the protein kinase TTK. Oncogene, 9, 89–96. [PubMed] [Google Scholar]
- Hoyt M.A., Totis,L. and Roberts,B.T. (1991) S.cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Bachant,J.B., Castillo,A.R., Giddings,T.H.,Jr and Winey,M. (1999) Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell, 10, 2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Earle,E., Fowler,K.J. and Choo,K.H. (2000) Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev., 14, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H.A. and Nigg,E.A. (1996) Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol., 135, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauze E., Stoelcker,B., Luca,F.C., Weiss,E., Schutz,A.R. and Winey,M. (1995) Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J., 14, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Lindberg R.A., Fischer,W.H. and Hunter,T. (1993) Characterization of a human protein threonine kinase isolated by screening an expression library with antibodies to phosphotyrosine. Oncogene, 8, 351–359. [PubMed] [Google Scholar]
- Maridor G., Gallant,P., Golsteyn,R. and Nigg,E.A. (1993) Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J. Cell Sci., 106, 535–544. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Hayashi,K. and Nishida,E. (1999) Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol., 9, 429–432. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas,J., Fry,A.M., Bartek,J. and Nigg,E.A. (1999) Centro some duplication in mammalian somatic cells requires E2F and Cdk2–cyclin A. Nature Cell Biol., 1, 88–93. [DOI] [PubMed] [Google Scholar]
- Mills G.B. et al. (1992) Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J. Biol. Chem., 267, 16000–16006. [PubMed] [Google Scholar]
- Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nature Cell Biol., 1, 47–53. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (1999) Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res., 248, 339–349. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M.S. and Nigg,E.A. (1993) Protein localization to the nucleolus: a search for targeting domains in nucleolin. J. Cell Sci., 105, 799–806. [DOI] [PubMed] [Google Scholar]
- Schutz A.R. and Winey,M. (1998) New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol. Biol. Cell, 9, 759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje H.H., Takahashi,K., Tanaka,K., Van Houwe,G. and Nigg,E.A. (1999) Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J., 18, 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight P.D., Giddings,T.H.,Jr and Winey,M. (2000) Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell, 11, 3525–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (2001) Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev., 11, 83–90. [DOI] [PubMed] [Google Scholar]
- Weiss E. and Winey,M. (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol., 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch,L., Baum,P. and Byers,B. (1991) MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol., 114, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]