Abstract
Activins, members of the transforming growth factor-β family, are pleiotropic growth and differentiation factors. Activin A induces B-cell apoptosis. To identify the genes responsible for activin-induced apoptosis, we performed retrovirus-mediated gene trap screening in a mouse B-cell line. We identified the rasGAP-binding protein Dok-1 (p62) as an essential molecule that links activin receptors with Smad proteins. In B cells overexpressing Dok-1, activin A-induced apoptotic responses were augmented. The expression of bcl-XL was down-regulated by inhibition of the ras/Erk pathway. Activin stimulation triggered association of Dok-1 with Smad3, as well as association of Smad3 with Smad4. Dok-1 also associated with both the type I and type II activin receptors. Dok-1 has been characterized previously as a tyrosine-phosphorylated protein acting downstream of the protein tyrosine kinase pathway: intriguingly, activin signaling did not induce tyrosine phosphorylation of Dok-1. These findings indicate that Dok-1 acts as an adaptor protein that links the activin receptors with the Smads, suggesting a novel function for Dok-1 in activin signaling leading to B-cell apoptosis.
Keywords: activins/apoptosis/Dok-1/p21/Smad
Introduction
Activin, a member of the transforming growth factor-β (TGF-β) superfamily, has numerous biological activities including regulation of secretion of follicle-stimulating hormone from pituitary cells (Ling et al., 1986; Vale et al., 1986) and mesoderm induction in Xenopus embryos (Asashima et al., 1990). In hematopoietic cells, activins have unique functions dependent on the particular cell lineage. In erythroid progenitors, activin stimulates erythrocyte differentiation (Eto et al., 1987). In B-cell lineages, activin potently inhibits cell growth and induces apoptosis (Nishihara et al., 1993).
Activin signals through two types (type I and type II) of membrane-bound receptor complexes on target cells. Both types of receptors belong to the family of serine/threonine kinase receptors. Activin binds directly to the type II receptor, which then recruits the type I receptor to the complex and phosphorylates it, resulting in the activation of the type I receptor and activation of Smads (Miyazono et al., 2001). There are two subtypes of the type II activin receptor, type IIA (ActRIIA) and IIB (ActRIIB), which are encoded by individual genes (Mathews and Vale, 1991; Attisano et al., 1992).
In this study, we identified Dok-1 as an important component of the activin signal mediator using retrovirus-mediated gene trap screening. Dok-1 is characterized as a negative regulator of the ras pathway. In B-lineage cells, Dok-1 is proposed to be a tyrosine-phosphorylated protein acting downstream of a variety of protein tyrosine kinase pathways (Yamanashi et al., 2000).
Signal transduction via transmembrane receptors is regulated by scaffold, anchoring and adaptor proteins (Pawson and Scott, 1997). Recent investigations have revealed that the activities of receptor serine/threonine kinases for the TGF-β superfamily are also regulated by adaptor proteins. For example, the FYVE domain-containing proteins, SARA (Smad anchor for receptor activation) and Hgs/Hrs (hepatic growth factor-regulated tyrosine kinase substrate/hepatic growth factor receptor substrate), are implicated as molecules that transport Smad proteins to receptors to initiate activin/TGF-β signaling (Tsukazaki et al., 1998; Miura et al., 2000). Similarly, PDZ domain-containing proteins are scaffolding proteins that bind both to activin receptors and Smad proteins (Shoji et al., 2000). We report here a novel example in which Dok-1 acts as an adaptor/docking protein linking receptor serine/threonine kinases with Smad proteins in activin signaling.
Results
Isolation of gene trap B-cell lines resistant to activin A-induced apoptosis
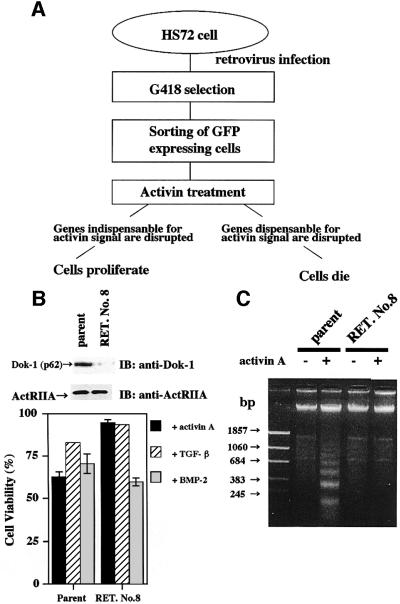
To understand the mechanism of activin A-induced apoptosis in B cells, we performed gene trap screening by using a retroviral removal exon trap (RET) vector (Ishida and Leder, 1999). In this study, mouse B-cell hybridoma HS72 cells were used, since apoptosis of this cell line is induced easily by activin A (Nishihara et al., 1993). As shown in Figure 1A, we reasoned that these cells can proliferate only when genes indispensable for activin signaling are disrupted by the gene trap. When HS72 cells were infected by the RET retrovirus, ∼10% of cells were found to be positive for green fluorescent protein (GFP) expression by fluorescence-activated cell sorting (FACS) analysis (data not shown). The infected cells were selected by culturing in media containing G418 for 1 week (Koseki et al., 1995). Then, strongly GFP-positive cells that expressed the trapped genes efficiently were sorted by cell sorter. The sorted cells were incubated with activin A for 2 weeks, and those proliferating in the presence of activin A were isolated. One of the proliferating cell lines, named RET. No. 8, was found to have the RET vector integrated in the first intron of the Dok-1 gene of mouse chromosome 6. As a result, Dok-1 was disrupted at amino acid position 269. The resulting truncated Dok-1 was composed of the pleckstrin homology (PH) and phosphotyrosine-binding (PTB) domains, but lacked the C- terminal tail harboring multiple potential tyrosine phosphorylation sites. As shown in Figure 1B and C, growth inhibition and DNA fragmentation were not observed in RET. No. 8 cells even in the presence of activin A, indicating that apoptosis of RET. No. 8 cells was no longer induced by activin. We also tested the effects of TGF-β and bone morphogenetic protein 2 (BMP-2) on RET. No. 8 cells. As reported, the extent of TGF-β-induced apoptosis of parental HS72 cells was weaker than that of activin A- or BMP-2-induced apoptosis. The responses of RET. No. 8 cells to TGF-β and BMP-2 were indistinguishable from those of parental cells (Figure 1B). Importantly, this finding strongly suggests that Dok-1 is a specific and essential mediator of activin-induced apoptosis. Our finding that Dok-1 is essential for growth inhibition of B cells is consistent with an analysis of Dok-1 knockout mice showing the role of Dok-1 in negative regulation of B-cell proliferation (Yamanashi et al., 2000).
Fig. 1. Retrovirus-mediated gene trap screening identifies Dok-1 as a mediator of activin A-induced apoptosis. (A) Outline of gene trapping screening. HS72 cells were infected with RET retrovirus, selected by G418, then GFP-positive clones were selected by cell sorting. Sorted cells were stimulated with 25 ng/ml of activin A for 2 weeks. If genes indispensable for activin signaling are disrupted, cells continue to proliferate, whereas if genes dispensable for activin signaling are disrupted, cells will die. (B) Comparison of cell viability of parental and RET. No. 8 cells. Cells were treated with activin A (25 ng/ml), TGF-β (5 ng/ml) or BMP-2 (25 ng/ml) for 24 h, and cell viability was determined by MTT assay (lower panel). The values are the mean ± SE of quadruplicate determinations. Expression of Dok-1 or ActRIIA in parental and RET. No. 8 cells was shown by immunoblotting (upper panel). Total proteins (40 µg) were separated by 7.5% SDS–PAGE, transferred to a PVDF membrane and probed with the anti-Dok-1 monoclonal antibody or the anti-ActRIIA antibody. (C) DNA fragmentation analysis. Cells were treated with 25 ng/ml of activin A for 24 h. Genomic DNAs were extracted and electrophoresed in 1.5% agarose gel containing ethidium bromide and visualized; 1.5 μg of DNA was analyzed in each lane.
Establishment of B-cell lines stably expressing Dok-1 and characterization of activin A-induced apoptotic responses
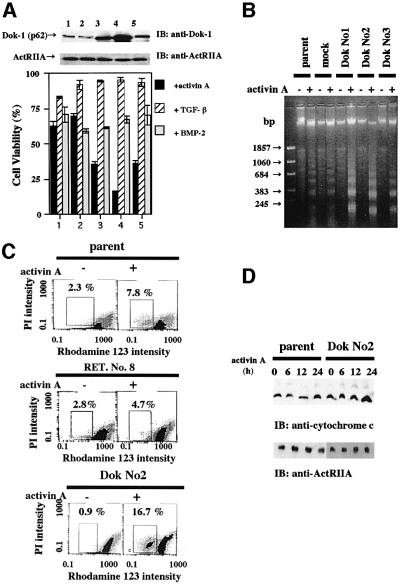
Since Dok-1 is likely to be an indispensable gene for activin-induced apoptosis, we next wished to establish an HS72 cell overexpressing Dok-1. Three independent Dok-1-overexpressing cell lines, Dok Nos 1, 2 and 3, were established and used for characterization. As shown in Figure 2A, augmentation of activin-induced growth inhibition was observed in all three Dok-1-overexpressing HS72 cell lines. In contrast, growth inhibition induced either by TGF-β or by BMP-2 in Dok-1-overexpressing HS72 cell lines was indistinguishable from that observed in parental cells, which also suggested the specific roles of Dok-1 in activin signaling. To assess whether growth inhibition is due to activin-induced apoptosis, we next analyzed fragmentation of genomic DNA (Figure 2B), mitochondrial membrane potential (Figure 2C) and cytochrome c release into cytoplasm (Figure 2D). In Dok-1-overexpressing and activin A-induced HS72 cells, we observed augmentation of genomic DNA fragmentation, reduction of the mitochondrial membrane potential (increase of the population with low rhodamine 123 intensity) and an increase of cytochrome c in cytoplasm, compared with activin A-treated parental and mock-infected cells. These findings clearly show that Dok-1 overexpression enhances activin-induced apoptosis.
Fig. 2. Acceleration of activin A-induced apoptosis in HS72 cells overexpressing Dok-1. (A) Comparison of cell viability of parental, mock-transfected and Dok-1-overexpressing HS72 cells. Cells were incubated with activin A (25 ng/ml), TGF-β (5 ng/ml) or BMP-2 (25 ng/ml) for 24 h. Cell viability was determined by MTT assay (lower panel). Upper panel: expression of Dok-1 protein or ActRIIA was shown by immunoblotting (upper panel). Lane 1, parental cells; lane 2, mock-transfected cells; lane 3, Dok No. 1; lane 4, Dok No. 2; lane 5, Dok No. 3. (B) DNA fragmentation analysis. Cells were incubated with or without activin A for 24 h. Genomic DNAs were extracted from each cell line, electrophoresed in 1.5% agarose gel and visualized. (C) Mitochondrial membrane potential assay. Cells were cultured with or without activin A (25 ng/ml) for 12 h, then labeled with rhodamine 123 and analyzed by flow cytometry (Epics Elite). PI, propidium iodide. (D) Cytochrome c release from mitochondria to cytoplasm. Cells were cultured in the presence of activin A (25 ng/ml) and, at appropriate culture periods, cytoplasmic proteins were extracted. Cytochrome c released was detected by immunoblotting following SDS–PAGE.
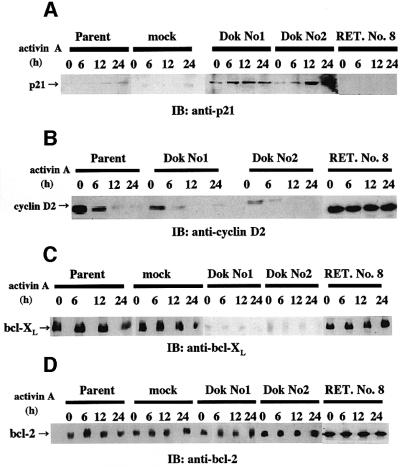
We next assessed the levels of p21, cyclin D2, bcl-XL and bcl-2 in Dok-1-overexpressing HS72 cells, since activin A-induced apoptosis of HS72 cells involves regulation of p21, cyclin D2 and bcl-XL without any change of bcl-2 (Yamato et al., 1997; Koseki et al., 1998). In Dok No. 1 and No. 2 cells, basal p21 expression was slightly increased and, after activin A treatment, p21 expression was significantly increased, compared with that of parental or mock-transfected cells (Figure 3A). Consistent with the previous report (Yamato et al., 1997), expression of cyclin D2 was almost completely suppressed by activin treatment of parental cells (Figure 3B). In Dok-1-overexpressing HS72 cells, the decrease in the cyclin D2 level by activin treatment was augmented (Figure 3B). Furthermore, we observed down-regulation of bcl-XL by activin treatment for 24 h in parental cells and a significant decrease of basal bcl-XL expression in Dok No. 1 and No. 2 cells (Figure 3C). On the other hand, the expression of bcl-2 was not altered either by activin treatment or by Dok-1 overexpression (Figure 3D). These results demonstrate that Dok-1 overexpression up-regulates the activin signaling pathway and down-regulates the anti-apoptosis gene in HS72 cells.
Fig. 3. Expression of p21, cyclin D2, bcl-XL and bcl-2 in HS72 cells overexpressing Dok-1. Expression levels of p21 (A), cyclin D2 (B), bcl-XL (C) and bcl-2 (D) were analyzed by immunoblotting. HS72 cells were treated with 25 ng/ml of activin A for the indicated times. Lysates were prepared, and probed with anti-p21, anti-cyclin D2, anti-bcl-XL or anti-bcl-2 antibodies.
Inhibition of the ras pathway accelerated activin-induced apoptosis
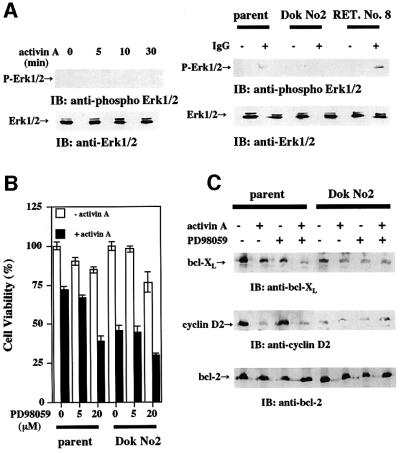
We next wished to determine whether the inhibition of the ras pathway has a role in activin-induced apoptosis, because it was demonstrated that Dok-1 negatively regulated the ras/Erk pathway (Yamanashi et al., 2000). We did not observe phosphorylation of Erk by activin stimulation (Figure 4A, left panel). Next, we examined phosphorylation of Erk by stimulating the cells with anti-mouse IgM (rabbit IgG), which co-cross-links BCR and FcγRIIB to activate mitogen-activated protein (MAP) kinase (Yamanashi et al., 2000). Erk was activated by IgG treatment in parental cells, but not in Dok-1-overexpressing cells (Figure 4A, right panel). This result indicates that the ras/Erk pathway is inhibited in Dok-1-overexpressing cells. To confirm this finding, we tested the effect of the MAP kinase inhibitor, PD98059, on activin-induced apoptosis. In the presence of PD98059, activin-induced apoptosis was augmented (Figure 4B). Suppression of the expression of bcl-XL by activin was also augmented in the presence of PD98059 (Figure 4C). Interestingly, the expression of cyclin D2 was not altered significantly in the presence of PD98059. From these results, we concluded that the inhibition of the ras/Erk pathway by Dok-1 has an important implication in activin-induced apoptosis.
Fig. 4. Involvement of the ras/Erk pathway in activin-induced apoptosis. (A) Detection of activated Erk in HS72 cells. HS72 cells were treated with activin A (25 ng/ml, left panel) or rabbit IgG to mouse IgM (30 µg/ml, right panel) for the indicated times. Lysates were prepared, and probed with anti-Erk or anti-phospho-Erk antibodies. (B and C) Effect of the ras pathway inhibitor (PD98059) on activin-induced apoptosis. The cell viability of HS72 cells treated with activin A (25 ng/ml) in the absence or presence of PD98059 (5 or 20 µM) was determined by MTT assay (B). HS72 cells were incubated with activin A (25 ng/ml) in the absence or presence of PD98059 (20 µM) for 24 h. Lysates were prepared, and probed with anti-bcl-XL, anti-cyclin D2 or anti-bcl-2 antibodies (C).
Up-regulation of p21 and SBD promoter activity by Dok-1
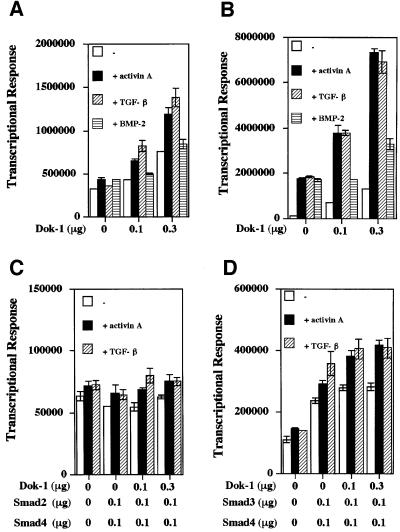
Up-regulation of the cell cycle inhibitor p21WAF1/Cip1 by activin A is common to both HS72 cells and hepatoma HepG2 cells: in both cell types, activin A induces growth arrest (Yamato et al., 1997; Zauberman et al., 1997). Thus, we next examined the effects of Dok-1 expression on ligand-dependent and Smad-dependent induction of p21 promoter and Smad-dependent promoter (SBD) activities in HepG2 cells (Figure 5). Dok-1 expression augmented basal promoter activity as well as activin- and TGF-β-induced promoter activity in HepG2 cells (Figure 5A and B). BMP-2-induced promoter activity was not significantly affected by Dok-1 expression. Furthermore, we tested the effect of Dok-1 expression on Smad-dependent promoter (SBD) activity. Dok-1 expression up-regulated Smad3-dependent promoter activity, but not Smad2-dependent promoter activity (Figure 5C and D). This is consistent with the result obtained in Dok-1-overexpressing HS72 cells, in which the basal level of p21 protein expression is augmented and is up-regulated further by activin treatment (Figure 3).
Fig. 5. Effects of Dok-1 on activin A-induced activation of the p21 promoter or the SBD promoter. Up-regulation of p21 (A) or SBD (B) promoter activity by Dok-1 cDNA expression in HepG2 cells. HepG2 cells were transfected with p21-lux (0.1 µg) or SBD-lux (0.1 µg), CMV-β-gal (0.1 µg) and the indicated amounts of Dok-1 cDNA, and incubated with activin A (50 ng/ml), TGF-β (5 ng/ml) or BMP-2 (50 ng/ml) for 24 h. The luciferase activity of cell lysates was measured and normalized to the β-galactosidase activity. The values represent the means ± SE of triplicate determinations. Effects of Dok-1 on Smad2 + Smad4-induced (C) and Smad3 + Smad4-induced p21 promoter activity (D). HepG2 cells were transfected with p21-lux (0.1 µg), CMV-β-gal (0.1 µg), Dok-1 and Smad2 (0.1 µg) and Smad4 (0.1 µg) (C), or Smad3 (0.1 µg) and Smad4 (0.1 µg) (D) and treated with or without 50 ng/ml of activin A for 24 h. The luciferase activity of the cell lysates was measured and normalized to β-galactosidase activity. The values represent the means ± SE of triplicate determinations.
Association of Dok-1 with Smad proteins and activin receptors
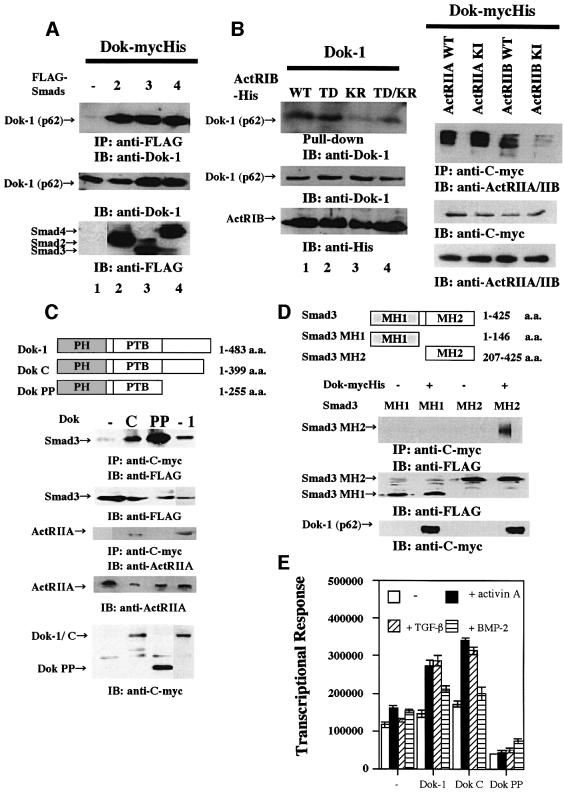
Smads are phosphorylated by the type I receptor and translocated to the nucleus. In the nucleus, Smads bind to many different transcription factors and mediate the various biological activities of the TGF-β superfamily dependent on the particular cell type (Miyazono et al., 2001). We assayed whether Dok-1 interacts with Smads. In COS-7 cells that co-expressed Dok-mycHis and FLAG-Smads, specific interactions of Dok-1 with Smad2, 3 or 4 were observed (Figure 6A). We next investigated whether Dok-1 binds to activin receptors. Dok-1 interacted more strongly with the wild-type and the constitutively active activin type IB receptor (ActRIB), compared with the kinase-inactive receptors (Figure 6B, left panel). Furthermore, Dok-1 also interacted with the wild-type or kinase-inactive (KI) activin type IIA receptor (ActRIIA). In contrast, we observed only a very weak interaction of Dok-1 with the ActRIIB receptor (Figure 6B, right panel).
Fig. 6. Interaction of Dok-1 with Smads and activin receptors. (A) Interactions of Dok-1 with Smad2, 3 or 4. Lysates of COS-7 cells transfected with FLAG-Smad2 and Dok-mycHis (lane 2), FLAG-Smad3 and Dok-mycHis (lane 3), or FLAG-Smad4 and Dok-mycHis (lane 4) were immunoprecipitated with anti-FLAG antibody and probed with anti-Dok-1 polyclonal antibody. In lane 1, a lysate of cells transfected with Dok-mycHis alone was analyzed. (B) Interaction of Dok-1 with ActRIB and ActRIIs. Left panel: lysates of COS-7 cells transfected with His6-tagged wild-type ActRIB (ActRIB-His-WT) and Dok-1 (lane 1), His6-tagged constitutively active ActRIB (ActRIB-His-TD) and Dok-1 (lane 2), His6-tagged kinase-negative ActRIB (ActRIB-His-KR) and Dok-1 (lane 3), or ActRIB-His-TD/KR and Dok-1 (lane 4) were pulled-down with Probond™ resin, and probed with anti-Dok-1 monoclonal antibody. Right panel: lysates of cells that co-expressed Dok-mycHis and activin receptors were immunoprecipitated with anti-C-myc antibody and blotted with anti-ActRIIA/IIB antibody. (C) Mapping of domains of Dok-1 that are required for Smad or ActRIIA interaction. Upper panel: diagram of Dok-1 mutants. Lower panel: lysates of COS-7 cells that co-expressed mycHis-tagged Dok-1 deletion mutants and FLAG-tagged Smad3 or ActRIIA were immunoprecipitated with anti-C-myc antibody and blotted with anti-FLAG or anti-ActRIIA antibody. In the leftmost lane, a lysate of cells transfected with Dok-mycHis cDNA alone was analyzed. (D) Mapping of domains of Smad that are required for Dok-1 interaction. Upper panel: diagram of Smad3 mutants. Lower panel: lysates of COS-7 cells that co-expressed Dok-mycHis and FLAG-tagged Smad3 mutants were immunoprecipitated with anti-C-myc antibody and blotted with anti-FLAG antibody. (E) Effect of Dok-1 deletion mutants on p21 promoter activity. HepG2 cells were transfected with p21-lux (0.1 µg), CMV-β-gal (0.1 µg) and Dok deletion mutants (0.3 µg) and treated or not with activin A (50 ng/ml), TGF-β (5 ng/ml) or BMP-2 (50 ng/ml) for 24 h. The luciferase activity of the cell lysates was measured and normalized to β-galactosidase activity. The values represent the means ± SE of triplicate determinations.
To determine the domains of Dok-1 required for the interaction with Smads and activin receptors, a series of Dok-1 deletion mutants (Dok C and Dok PP) was generated (Figure 6C, upper panel). Dok C (amino acids 1–399) contained the PH and PTB domains and lacked the short C-terminal region of tyrosine phosphorylation sites, whereas Dok PP (amino acids 1–255) completely lacked all tyrosine phosphorylation sites. Smad3 interacted with both Dok C and Dok PP, whereas ActRIIA interacted with Dok C, but not with Dok PP (Figure 6C, lower panel). Next, we generated Smad3 deletion mutants, MH1 (1–146 amino acids) and MH2 (amino acids 207–425), to determine the domain of Smad3 required for the interaction between Smad3 and Dok-1. The MH2 domain but not the MH1 domain of Smad3 interacted with Dok-1 (Figure 6D). Additionally, we tested the effect of Dok-1 deletion mutants on ligand-induced p21 promoter activity. Dok C up-regulated activin A- and TGF-β-induced p21 promoter activity, whereas Dok PP down-regulated it (Figure 6E).
Activin A-dependent association and dissociation of Dok-1 with Smad proteins and activin receptors
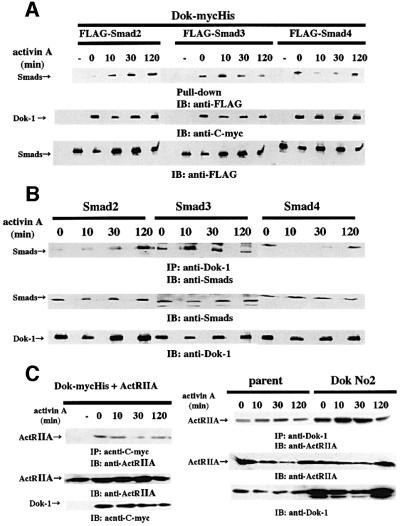
We tested whether interaction of Dok-1 with Smads or activin receptors is altered by ligand stimulation. As shown in Figure 7A, Dok-mycHis interacted with FLAG-Smad2 or FLAG-Smad3, and these interactions were accelerated significantly after ligand stimulation. The interaction of Dok-1 with Smad2 was gradually enhanced after ligand stimulation, whereas the interaction of Dok-1 with Smad3 reached a maximum after 10 min stimulation and weakened thereafter. Dok-1 was also found to bind to Smad4 in the absence of ligand stimulation (Figure 7A). The extent of interaction was markedly attenuated after 10 min incubation with activin A, then was augmented and returned to the basal level after 120 min of stimulation (Figure 7A). In HS72 cells, we tested the effect of activin treatment on the interaction between endogeneous Smads and Dok-1. We observed the time-dependent association and dissociation of Smads with Dok-1, which are similar to those observed in transfected 293 cells (Figure 7B). The interaction of Dok-1 with ActRIIA or ActRIB was not changed by ligand stimulation either in 293 cells or in HS72 cells (Figure 7C and data not shown).
Fig. 7. Effect of activin A on interactions of Dok-1 and Smads or activin receptors. (A) 293 cells that expressed Dok-mycHis plus FLAG-Smad2, FLAG-Smad3 or FLAG-Smad4 were incubated with 50 ng/ml of activin A for the indicated times. Lysates were purified with Probond™ resin, and probed with anti-FLAG antibody. In the leftmost lanes of each transfection, lysates of cells transfected with FLAG-Smad alone were analyzed. In the leftmost lane, a lysate of non-transfected cells was analyzed. (B) Endogenous interaction of Dok-1 with Smads in HS72 cells. HS72 cells were incubated with or without 25 ng/ml of activin A for the indicated times. Lysates were immunoprecipitated with anti-Dok-1 monoclonal or polyclonal antibodies and probed with anti-Smad2 or anti-Smad3 polyclonal antibodies, or anti-Smad4 monoclonal antibody. (C) Left panel: 293 cells that co-expressed Dok-mycHis and ActRIIA were incubated with 50 ng/ml of activin A for the indicated times. Lysates were immunoprecipitated with anti-C-myc antibody and probed with anti-ActRIIA antibody. In the leftmost lane, a lysate of cells transfected with ActRIIA cDNA alone was analyzed. Right panel: HS72 cells were incubated with 25 ng/ml of activin A for the indicated times. Lysates were immunoprecipitated with anti-Dok-1 monoclonal antibody and probed with anti-ActRIIA antibody.
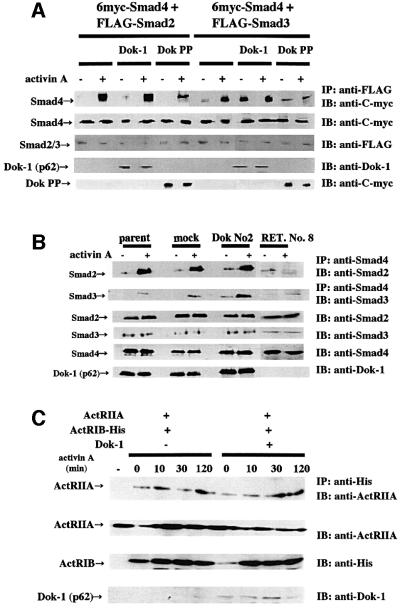
We next examined the effect of Dok-1 on the association of Smad4 with Smad2 or Smad3 in 293 cells and HS72 cells. No change in association of Smad2 with Smad4 was observed when co-transfected with Dok-1 (Figure 8A, left panel). In contrast, the association of Smad3 with Smad4 was strikingly enhanced when co-transfected with Dok-1 (Figure 8A, right panel). The expression of Dok PP inhibited the association of Smad3 and Smad4 (Figure 8A). This may explain why the expression of Dok PP inhibited the up-regulation of p21 promoter activity by activin A and TGF-β (Figure 6C). In HS72 cells, the overexpression of Dok-1 strengthened the activin-dependent association of Smad3 with Smad4, but did not affect the interaction between Smad2 and Smad4. In RET. No. 8 cells, the association of Smad3 with Smad4 decreased (Figure 8B). The interaction of ActRIB and ActRIIA did not change even in the presence of Dok-1 (Figure 8C). These results strongly suggest that Dok-1 accelerates hetero-oligomerization of Smad3 and Smad4, and affects the activin signaling pathway.
Fig. 8. Effect of Dok-1 on Smad hetero-oligomerization and complex formation with activin receptors. (A) 293 cells that co-expressed FLAG-Smad2 and 6myc-Smad4 (left panel) or FLAG-Smad3 and 6myc-Smad4 (right panel) with or without Dok-1 or Dok PP were cultured with or without 50 ng/ml of activin A for 2 h. The cell lysates were immunoprecipitated with anti-FLAG antibody and probed with anti-C-myc antibody. (B) HS72 cells were incubated with 25 ng/ml of activin A for the indicated times. Lysates were immunoprecipitated with anti-Smad4 antibody and probed with anti-Smad2 or anti-Smad3 antibodies. (C) 293 cells that co-expressed ActRIB-His and ActRIIA with or without Dok-1 were cultured with or without 50 ng/ml of activin A for the indicated times. The cell lysates were immunoprecipitated with anti-His antibody and probed with anti-ActRIIA antibody. In the leftmost lane, a lysate of cells transfected with ActRIIA cDNA alone was analyzed.
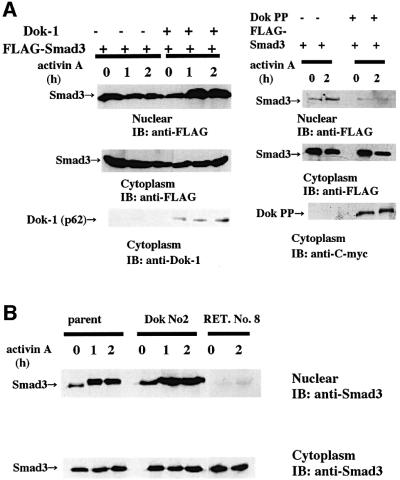
Since the Smad3/4 complex translocates to the nucleus upon ligand stimulation, we examined the effect of Dok-1 on nuclear translocation of Smad3. In Dok-1-transfected 293 cells (Figure 9A, left panel) and HS72 cells (Figure 9B), we readily detected an increase of nuclear Smad3 after ligand stimulation, indicating that Dok-1 is a positive regulator of Smad3/4 complex formation and translocation of Smad3 to the nucleus. In the presence of Dok PP or mutated Dok-1 (RET. No. 8), activin failed to induce Smad3 nuclear translocation (Figure 9A, right panel, and B).
Fig. 9. Augmentation of Smad3 nuclear translocation by Dok-1. (A) 293 cells that expressed FLAG-Smad3 in the absence or presence of Dok-1 (left panel) or mycHis-tagged Dok PP (right panel) were incubated with or without 50 ng/ml of activin A for the indicated times. Nuclear and cytoplasmic lysates were extracted and blotted with anti-FLAG antibody. Cytoplasmic lysates were also probed with anti-Dok-1 antibody or anti-C-myc antibody (bottom panel). (B) HS72 cells were incubated with 25 ng/ml of activin A for the indicated times. Nuclear and cytoplasmic lysates were extracted and blotted with anti-Smad3 antibody.
Activin signaling does not induce Dok-1 tyrosine phosphorylation
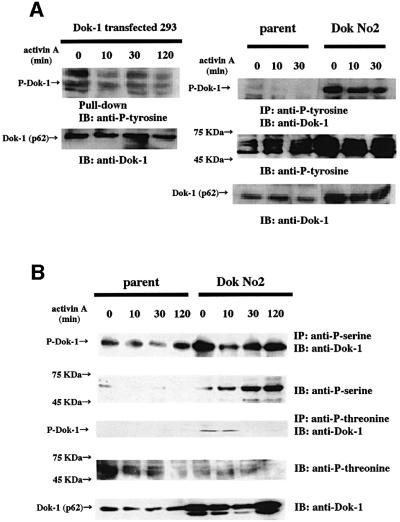
Since Dok-1 is a target of protein tyrosine kinase, like src and abl (Songyang et al., 2001), we tested whether Dok-1 is tyrosine phosphorylated in response to activin signaling. We did not detect any increase of tyrosine phosphorylation by activin stimulation for up to 120 min in Dok-1-transfected 293 cells (Figure 10A, left panel) or HS72 cells (Figure 10A, right panel). In Dok-1-overexpressing HS72 cells, the basal level of tyrosine phosphorylation increased, presumably because of the overexpressed Dok-1 protein. Since Dok-1 has many serine (8.5%) and threonine (5.5%) residues, and interacts with activin receptors, which are serine/threonine kinases, we examined the effect of activin A on serine and threonine phosphorylation of Dok-1. Activin treatment did not appreciably augment the level of serine phosphorylation of Dok-1, whereas it increased the overall level of serine phosphorylation (Figure 10B, upper two panels). In contrast, threonine phosphorylation of Dok-1 decreased after 30 min of ligand stimulation (Figure 10B, lower panels). These results support the model that Dok-1 acts as an adaptor protein and regulator for Smads and receptors rather than as a substrate for receptor serine/threonine kinase.
Fig. 10. Phosphorylation of Dok-1. (A) Left panel: 293 cells that expressed Dok-mycHis were incubated with or without 50 ng/ml of activin A for the indicated times. The cell lysates were mixed with Probond™ resin and probed with anti-phosphotyrosine antibody. Right panel: HS72 cells were incubated with or without 25 ng/ml of activin A for the indicated times. The cell lysates were immunoprecipitated with anti-phosphotyrosine antibody and probed with anti-Dok-1 polyclonal antibody. (B) HS72 cells were incubated with or without 25 ng/ml of activin A for the indicated times. The cell lysates were immunoprecipitated with anti-phosphoserine or anti-phosphothreonine antibodies and probed with anti-Dok-1 polyclonal antibody.
Discussion
The response of mouse B-cell hybridoma HS72 cells to activin has been characterized in earlier studies: activin A up-regulates p21 expression, suppresses cyclin D2 expression and induces accumulation of the hypophosphorylated form of retinoblastoma protein (Yamato et al., 1997). Similar molecular changes have also been reported in hepatoma cells that are growth arrested by activin A (Zauberman et al., 1997). Apoptosis induced by activin A is suppressed by bcl-2 (Koseki et al., 1995) and also by the inhibitory Smad, Smad7 (Ishisaki et al., 1998), indicating the involvement of the conventional apoptotic pathway as well as the activin/TGF-β-specific signal transduction pathway in activin A-induced B-cell apoptosis. However, the precise molecular mechanisms of activin-induced apoptosis remain to be determined.
In order to identify essential genes responsible for activin A-induced apoptosis in mouse B-cell hybridoma HS72 cells, we performed retrovirus-mediated gene trap screening. We have used the modified poly(A) trap retrovirus vector system, called the RET system, which was invented originally by Ishida and Leder (1999). By using the RET system, we identified the rasGAP-binding protein, Dok-1, as an indispensable molecule in activin A-induced B-cell apoptosis (Figures 1 and 2). In cells in which Dok-1 is deleted by gene trapping, activin A no longer induces apoptosis (Figure 1). Conversely, in Dok-1-overexpressing B-cell lines, apoptosis induced by activin A was augmented (Figures 2 and 3). In contrast, apoptosis induced by BMP-2 was not greatly affected in either Dok-1-deleted cells or Dok-1-overexpressing cells. Since TGF-β-induced apoptosis in HS72 cells is very weak, we could not determine the effects of Dok-1 on TGF-β in this cell. However, Dok-1 cDNA expression augmented basal and activin A- and TGF-β-induced p21 and SBD promoter activity, as well as Smad3- and Smad4-induced p21 promoter activity in HepG2 cells (Figure 5). Thus, Dok-1 and Smad have synergistic activity on p21 promoter stimulation. This synergistic effect is evoked by a direct interaction of Dok-1 with Smad (Figure 6). The N-terminal PH and PTB domains of Dok-1 are involved in the association with Smads (Figure 6C). Both Smad2 and Smad3 associate with Dok-1 in a ligand-dependent manner (Figure 7). Interestingly, Dok-1 also associates with ActRIIs, ActRIB and TGF-βRI (Figures 6 and 7, and data not shown) and stimulates the association of Smad3 with Smad4, in parallel with the dissociation of Dok-1 from Smad4 (Figures 7 and 8). Since the Smad3/4 complex has the ability to bind directly to DNA (Miyazono et al., 2001), it is likely that Dok-1 plays a role as a docking/adaptor protein to link receptors with the Smad proteins and to regulate the tight association of Smad3 with Smad4, resulting in the augmentation of activin signaling. Interestingly, in the absence of ligand, Dok-1 associates strongly with Smad4 whereas, upon ligand stimulation, Dok-1 dissociates easily from Smad4 (Figure 7). Thus, we propose a model in which Dok-1 inhibits the non-specific interaction between Smad3 and Smad4 in the absence of ligand, thus preventing leaky signaling. On ligand challenge, Dok-1 dissociates from Smad4, allowing the association of Smad3 with Smad4 to propagate the signaling.
The role of Dok-1 in activin A-induced B-cell apoptosis is reminiscent of the role of Disabled-2 (Dab2) in TGF-β-sensitive human fibrosarcoma HT-1080 cells. Dab2 works as an essential adaptor protein linking receptors with Smads, aiding in transmission of signaling from TGF-β receptors to Smads (Hocevar et al., 2001). Structurally, Dab2 is composed of an N-terminal PTB domain followed by a proline-rich domain, and the PTB domain is involved in Smad association. Dok-1 and Dab2 possess similar roles in the interaction between Smads and receptors. However, unlike Dok-1, Dab2 does not associate with Smad4 (Hocevar et al., 2001), suggesting distinct roles for Dok-1 and Dab2 in activin/TGF-β signaling. Since Dab2 is localized in clathrin-coated vesicles (Morris and Cooper, 2001), there is a possibility that these adaptor proteins are involved in the association of activin/TGF-β receptors with the endocytic machinery and endosomes.
In B-lineage cells, Dok-1 has been characterized as a tyrosine-phosphorylated protein acting downstream of a variety of protein tyrosine kinase pathways: oncoprotein Bcr–Abl activation (Yamanashi and Baltimore, 1997), antigenic activation of the B-cell receptor (Yamanashi et al., 2000) and c-Kit activation through stem cell factor (Van Dijk et al., 2000). Intriguingly, activin signaling did not induce tyrosine phosphorylation, but prevented threonine phosphorylation of Dok-1 (Figure 10). The balance between phosphorylation and dephosphorylation is important in TGF-β/activin signal transduction. Protein phosphatase-2A(PP2A)-Bα associates with the TGF-β receptor and dephosphorylates and inactivates p70 S6 kinase (Petritsch et al., 2000). PP2A-Bα enhances the growth-inhibitory activity of TGF-β (Griswold-Prenner et al., 1998). Therefore, activin-induced dephosphorylation of Dok-1 may also result from interaction between signaling molecules for activin receptors and protein phosphatase. Dok-1 acts as an adaptor protein downstream of receptor serine/threonine kinase as well as receptor tyrosine kinase. Dok-1 consists of PH domain, a putative PTB domain and a C-terminal potential tyrosine phosphorylation site (Carpino et al., 1997; Yamanashi and Baltimore, 1997). The PH domain is likely to be involved in phospholipid binding and to be necessary for targeting Dok-1 to the membrane. The PTB domain is also found in insulin receptor substrate-1, Shc and the Disabled family (Kavanaugh and Williams, 1994; Sakai et al., 1994; Morris and Cooper, 2001), and recognizes phosphotyrosine-containing sequences (Kavanaugh and Williams, 1994). In addition, structural studies indicate that the PTB domain can recognize broader types of sequences, including unphosphorylated sequences (Kuriyan, 1997). The multiple tyrosine residues in the Dok-1 C-terminal region are phosphorylation sites for tyrosine kinases. Since Dok-1 is not tyrosine phosphorylated by activin stimulation (Figure 10), the functions of the C-terminal tyrosine residues in activin signaling are currently unknown. We observed that a Dok-1 mutant, Dok C, lacking several C-terminal tyrosine phosphorylation sites, augments Smad activity more significantly than wild type, whereas another mutant, Dok PP, lacking all tyrosine phosphorylation sites, acts as a dominant-negative mutant (Figure 6C). This finding suggests a regulatory role for the tyrosine phosphorylation sites in signal trasnduction.
The Dok protein family is composed of at least five members (Grimm et al., 2001). Dok-1, -2 and -3 are expressed mainly in hematopoietic tissues. Dok-1 and -2 associate with rasGAP and inhibit MAP kinase signaling to inhibit cell growth (Yamanashi and Baltimore, 1997; Tamir et al., 2000). In contrast, Dok-4 and -5 do not associate with rasGAP, but enhance c-RET-dependent activation of MAP kinase in neuronal differentiation (Grimm et al., 2001). Activin/TGF-β signaling activates the MAP kinase pathway depending on the cell type (Massagué and Chen, 2000). Thus differential expression of distinct members of the Dok family and their association with the receptor serine/threonine kinases and Smads may explain the complexity and specificity of signal transduction for the TGF-β superfamily including activins. In this study, we showed that the inhibition of the ras/Erk pathway by either Dok-1 overexpression or PD98059 treatment had a significant implication for activin-induced apoptosis in B cells. Similarly, Hognason et al. (2001) reported that epidermal growth factor (EGF)-induced apoptosis was potentiated by inhibition of the ras pathway. We found that Dok-1 regulated activin-induced apoptosis by suppressing MAP kinase activation, inhibiting the expression of the anti-apoptosis gene, bcl-XL, and accelerating the formation of the Smad3/4 hetero-oligomer. Down-regulation of bcl-XL is also reported in TGF-β-induced B-cell apoptosis (Saltzman et al., 1998). Dok-1 must play physiologically important roles in B-cell apoptosis induced by activin.
An emerging theme in cell signaling research is the regulation of membrane receptors by scaffolding, docking and adaptor proteins. Membrane receptors, which are expressed at low levels on the cell surface, commonly use adaptor/docking proteins to amplify signaling following receptor activation (Pawson and Scott, 1997). A number of proteins have been reported to associate with receptors for the TGF-β superfamily or with the Smad proteins (Miyazono et al., 2001). However, reports of adaptor proteins that associate both with receptors and with Smad proteins are few. In a previous study, we identified an activin receptor-interacting PDZ protein, called ARIP1, as a scaffolding protein that unites the activin receptors with Smad and thereby regulates activin-mediated signaling in neuronal cells (Shoji et al., 2000). In addition, the FYVE domain-containing proteins, SARA and Hgs/Hrs, are implicated as molecules that recruit Smad proteins to receptors to initiate activin/TGF-β signaling (Tsukazaki et al., 1998; Miura et al., 2000). Dab2 is also reported to link TGF-β receptors with Smad proteins (Hocevar et al., 2001). In addition, TGF-β receptor-associated protein 1 (TRAP1) interacts with TGF-β receptors and Smad4 in a ligand-dependent fashion and diminishes the interaction of Smad4 with Smad2 (Wurthner et al., 2001). Our characterization of Dok-1 as an essential molecule linking the activin receptors with Smad proteins in activin-induced apoptosis in B cells suggests a novel mode of regulation of activin signaling by the Dok family of adaptor/docking proteins.
Materials and methods
Cell lines and antibodies
HS72 cells (a gift from Dr T.Nishihara at Kyushu Dental College) were maintained in Iscove’s modified Dulbecco’s medium (IMDM, Gibco-BRL) supplemented with 5% fetal calf serum (FCS). HepG2 cells, HEK293 cells and COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Sigma) supplemented with 10% FCS. The anti-p21 (C-19), anti-cyclin D2 (34B1-3), anti-cytochrome c (H-104), anti-bcl-2 (C-2), anti-bcl-XL (L-19), anti-Smad2/3 (E-20), anti-Smad4 (B-8) and anti-Dok-1 monoclonal (A-3) and polyclonal (M-276) antibodies were purchased from Santa Cruz; anti-Erk1/2 and anti-phospho-Erk1/2 (E10) antibodies from Cell Signaling; anti-FLAG (M2) and anti-phosphoserine (PSR-45) antibodies from Sigma; anti-phosphothreonine antibody (PTR-8) from BioMaker; anti-C-myc antibody (9E11) from NeoMarkers; anti-Penta His antibody from Qiagen; anti-Smad2 and anti-Smad3 antibodies from Zymed; anti-phosphotyrosine antibody (PY20) from Transduction Laboratories; anti-mouse IgM (rabbit IgG) from MBL; anti-activin receptor IIA/IIB antibody (MAB3391) from R&D Systems; and horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG from Biosource.
Gene trapping by retrovirus infection
The pRET vector (a gift from Dr Y.Ishida at Kyoto University) (Ishida and Leder, 1999) was transfected into BOSC23 cells by liposome reagent Fugene6 (Roche Diagnostics). Retroviruses were harvested 2 days after transfection and used to infect 1 × 107 HS72 cells. Two days after infection, cells were cultured in media containing 1 mg/ml G418 for 1 week and RET-integrated cells were selected. After G418 selection, GFP-expressing cells were collected by the use of a cell sorter (Epics Elite, Beckman Coulter). GFP-positive cells were treated with 25 ng/ml of activin A for 2 weeks. Medium containing activin A was changed every 4 days. Proliferating cells were subjected to subcloning by limiting dilution. RNA was extracted from cells with Trizol reagent (Gibco-BRL). First strand cDNA was synthesized by SuperScriptII (Gibco-BRL). Trapped genes were determined by 3′-RACE and nucleotide sequencing (Ishida and Leder, 1999).
MTT assay
HS72 cells were cultured at a density of 2 × 104 cells/well in a 96-well plate with or without activin A for 24 h (Yamato et al., 1997). Cell viability was determined by colorimetric MTT assay. Optical density at 570–630 nm was measured with a plate reader.
Mitochondrial membrane potential assay
HS72 cells (1 × 106) were cultured with or without 25 ng/ml of activin A for 12 h. Rhodamine 123 (10 µM), a lipophilic cation taken up by mitochondria in proportion to the mitochondrial membrane potential (Johnson et al., 1981), was added and the cells were incubated for 15 min at 37°C. Cells were collected by centrifugation, suspended in phosphate buffered-saline (PBS) containing 10 µM propidium iodide (PI), a reagent for detection of dead cells, and analyzed by flow cytometry (Epics Elite) (Shimizu et al., 1996).
Cytochrome c release assay
HS72 cells (1 × 106) were cultured with or without 25 ng/ml of activin A for various times. Cells were collected by centrifugation and lysed in lysis buffer [1 mM NaH2PO4, 8 mM Na2HPO4, 75 mM NaCl, 250 mM sucrose, 190 mg/ml digitonin and protease inhibitor cocktail (Sigma)] (Heibein et al., 1999). After incubation for 10 min on ice, lysates were centrifuged. The protein concentration of the supernatants was determined by Coomassie Protein Assay Reagent (Pierce), and protein (50 µg) was separated by 15% SDS–PAGE and then electroblotted onto a PVDF membrane (Millipore). Immunodetection was performed by an ECL Plus Western blotting detection system (Amersham).
DNA construction
Murine Dok-1 cDNA was a gift from Dr Y.Yamanashi (Yamanashi and Baltimore, 1997). The Dok-1 coding sequence was amplified by PCR and then subcloned into the XhoI site of pcDNA3.1/Myc-His(+) A (Invitrogen). The resultant DNA was designated as Dok-mycHis. Dok C was generated by digestion and religating using the StuI site of Dok-mycHis. Dok PP was prepared by digestion and religating using the EcoRV site. Smad3 MH1 (amino acids 1–146 of Smad3) and Smad3 MH2 (amino acids 207–425 of Smad3) were amplified by PCR and subcloned into the EcoRI–XhoI site of pcDNA3FLAG.
Generation of Dok-1-overexpressing cell
Dok-1 cDNA cloned into the retroviral vector MSCV puro (a gift from Dr Y.Yamanashi) (Yamanashi and Baltimore, 1997) was transfected into BOSC23 cells using Fugene6. Retroviruses were used to infect HS72 cells. One day later, Dok-1-integrated HS72 cells were selected in medium containing 7 µg/ml of puromycin (Sigma). The medium was changed every 4 days. After 2 weeks of selection, subcloning was performed by limiting dilution. Dok-1 expression of each clone was confirmed by immunoblotting.
p21 and SBD promoter assay
The p21 promoter in the pGL2-basic luciferase reporter vector was a gift from Dr X.-F.Wang of Duke University Medical Center (Datto et al., 1995). SBD-lux was from Dr C.-H.Heldin. HepG2 cells plated at a density of 5 × 104 cells/well in 12-well plates were transfected by using TransFast liposome reagent (Promega) according to the manufacturer’s protocol. Luciferase activity was measured by a luciferase assay system (Promega), and was normalized to β-galactosidase activity (Tsuchida et al., 1995).
Immunoprecipitation, pull-down and western blotting
COS-7 cells or 293 cells were maintained at a density of 5 × 105 cells/6 cm dish. Transfection was performed by using TransFast liposome reagent (Promega). HS72 cells were maintained at a density of 1 × 106 cells/6 cm dish. After 2 days, 293 cells or HS72 cells were incubated with or without 50 ng/ml of activin A for various times, and lysed. Protein concentrations of supernatants were determined by Coomassie Protein Assay Reagent (Pierce). Protein G–Sepharose-pre-adsorbed lysates (200 µg) were incubated with primary antibody by rotating at 4°C overnight. Immunoprecipitated proteins were washed three times. For the pull-down assay, proteins were incubated with Probond™ resin (Invitrogen) at 4°C for 2 h. The precipitated proteins were separated by SDS–PAGE, and transferred to PVDF membranes. The membranes were blocked in blocking buffer (20 mM Tris–HCl pH 7.5, 0.15 M NaCl, 5% skim milk and 0.05% Tween-20) or phosphorylation blocking buffer (20 mM Tris–HCl pH 7.5, 0.15 M NaCl, 1% gelatin and 0.05% Tween-20). Immunodetection was performed by an ECL Plus Western blotting detection system (Amersham).
Nuclear extracts were prepared by NE-PER Nuclear and Cytoplasmic Extraction reagent (Pierce) according to the manufacturer’s protocol. Nuclear extracts were separated by SDS–PAGE and transferred to PVDF membranes.
Acknowledgments
Acknowledgements
We thank Dr T.Nishihara for the suggestion of apoptosis assays in HS72 cells and helpful discussions, Dr Y.Ishida for introducing us to RET vectors and the retrovirus-mediated gene trap method, Dr Y.Yamanashi for p62 (dok-1) cDNA and helpful discussions, Dr X.-F.Wang for the p21 luciferase construct, Dr K.Miyazono for Smad cDNAs, Dr Y.Eto for recombinant human activin A, Dr Y.Hasegawa for bovine activins, and the Genetics Institute for BMP. This research was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.T. and H.S.), and was also supported by a grant from The Inamori Foundation for Research to K.T.
References
- Asashima M., Nakano,H., Shimada,K., Kinoshita,K., Ishii,K., Shibai,H. and Ueno,N. (1990) Activin A (erythroid differentiation factor) induction in mesodermal early amphibian embyos. Roux’s Arch. Dev. Biol., 198, 330–335. [DOI] [PubMed] [Google Scholar]
- Attisano L., Wrana,J.L., Cheifetz,S. and Massagué,J. (1992) Novel activin receptors distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell, 68, 97–108. [DOI] [PubMed] [Google Scholar]
- Carpino N., Wisniewski,D., Strife,A., Marshak,D., Kobayashi,R., Stillman,B. and Clarkson,B. (1997) p62dok: a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell, 88, 197–204. [DOI] [PubMed] [Google Scholar]
- Datto M.B., Yu,Y. and Wang,X.-F. (1995) Functional analysis of the transforming growth factor β responsive elements in the WAF1/p21 promoter. J. Biol. Chem., 270, 28623–28628. [DOI] [PubMed] [Google Scholar]
- Eto Y., Tsuji,T., Takezawa,M., Takano,S., Yokogawa,Y. and Shibai,H. (1987) Purification and characterization of erythroid differentiation factor (EDF) isolated from human leukemia cell line THP-1. Biochem. Biophys. Res. Commun., 142, 1095–1103. [DOI] [PubMed] [Google Scholar]
- Grimm J., Sachs,M., Britsch,S., Di Cesare,S., Schwarz-Romond,T, Alitalo,K. and Birchmeier,W. (2001) Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J. Cell Biol., 154, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold-Prenner I., Kamibayashi,C., Maruoka,E.M., Mumby,M.C. and Derynck,R. (1998) Physical and functional interactions between type I transfoming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol. Cell. Biol., 11, 6595–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibein J.A., Barry,M., Motyka,B. and Bleackley,R.C. (1999) Granzyme B-induced loss of mitochondrial inner membrane potential (ΔΨm) and cytochrome c release are caspase independent. J. Immunol., 163, 4683–4693. [PubMed] [Google Scholar]
- Hocevar B.A., Smine,A., Xu,X.X. and Howe,P.H. (2001) The adaptor molecule Disabled-2 links the transforming growth factor β receptors to the Smad pathway. EMBO J., 20, 2789–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hognason T., Chatterjee,S., Vartanian,T., Ratan,R.R., Ernewein,K.M. and Habib,A.A. (2001) Epidermal growth factor receptor induced apoptosis: potentiation by inhibition of ras signaling. FEBS Lett., 491, 9–15. [DOI] [PubMed] [Google Scholar]
- Ishida Y. and Leder,P. (1999) RET: a poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res., 27, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishisaki A., Yamato,K., Nakao,A., Nonaka,K., Ohguchi,M., Ten Dijke,P. and Nishihara,T. (1998) Smad7 is an activin-inducible inhibitor of activin-induced growth arrest and apoptosis in mouse B cells. J. Biol. Chem., 273, 24293–24296. [DOI] [PubMed] [Google Scholar]
- Johnson L.V., Walsh,M.L., Bochus,B.H. and Chen,L.B. (1981) Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J. Cell Biol., 88, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh W.M. and Williams,L.T. (1994) An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science, 266, 1862–1865. [DOI] [PubMed] [Google Scholar]
- Koseki T., Yamato,K., Krajewski,S., Reed,J.C., Tsujimoto,Y. and Nishihara,T. (1995) Activin A-induced apoptosis is suppressed by BCL-2. FEBS Lett., 376, 247–250. [DOI] [PubMed] [Google Scholar]
- Koseki T., Yamato,K., Ishisaki,A., Hashimoto,O., Sugino,H. and Nishihara,T. (1998) Correlation between bcl-X expression and B-cell hybridoma apoptosis induced by activin A. Cell Signal., 10, 517–521. [DOI] [PubMed] [Google Scholar]
- Kuriyan J. (1997) Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biomol. Struct., 26, 259–288. [DOI] [PubMed] [Google Scholar]
- Ling N., Ying,S.Y., Ueno,N., Shimasaki,S., Esch,F., Hotta,M. and Guillemin,R. (1986) Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature, 321, 779–782. [DOI] [PubMed] [Google Scholar]
- Massagué J. and Chen,Y.-G. (2000) Controlling TGF-β signaling. Genes Dev., 14, 627–644. [PubMed] [Google Scholar]
- Mathews L.S. and Vale,W. (1991) Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell, 65, 973–982. [DOI] [PubMed] [Google Scholar]
- Miura S. et al. (2000) Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol. Cell. Biol., 20, 9346–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., Kusanagi,K. and Inoue,H. (2001) Divergence and convergence of TGF-β/BMP signaling. J. Cell Physiol., 187, 265–276. [DOI] [PubMed] [Google Scholar]
- Morris S.M and Cooper,J.A. (2001) Disabled-2 colocalizes with the LDLR in clathrin-coatd pits and interacts with AP-2. Traffic, 2, 111–123. [DOI] [PubMed] [Google Scholar]
- Nishihara T., Okahashi,N. and Ueda,N. (1993) Activin A induces apoptotic cell death. Biochem. Biophys. Res. Commun., 197, 985–991. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Scott,J.D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Petritsch C., Beug,H., Balmain,A. and Oft,M. (2000) TGF-β inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev., 14, 3093–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R., Iwamatsu,A., Hirano,N., Ogawa,S., Tanaka,T., Mano,H. Yazaki,Y. and Hirai,H. (1994) A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J., 13, 3748–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A., Munro,R., Searfoss,G., Franks,C., Jaye,M. and Ivashchenko,Y. (1998) Transfoming growth factor-β-mediated apoptosis in the Ramos B-lymphoma cell line is accompanied by caspase activation and bcl-XL downregulation. Exp. Cell Res., 242, 244–254. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Eguchi,Y., Kamiike,W., Waguri,S., Uchiyama,Y., Matsuda,H. and Tsujimoto,Y. (1996) Bcl-2 blocks loss of mitochondrial membrane potential while ICE inhibitors act at a different step during inhibition of death induced by respiratory chain inhibitors. Oncogene, 13, 21–29. [PubMed] [Google Scholar]
- Shoji H., Tsuchida,K., Kishi,H., Yamakawa,N., Matsuzaki,T., Nakamura,T. and Sugino,H. (2000) Identification and characterization of a PDZ protein that interacts with activin type II receptors. J. Biol. Chem., 275, 5485–5492. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Yamanashi,Y., Liu,D. and Baltimore,D. (2001) Domain-dependent function of the rasGAP-binding protein p62Dok in cell signaling. J. Biol. Chem., 276, 2459–2465. [DOI] [PubMed] [Google Scholar]
- Tamir I., Stolpa,J.C., Helgason,C.D., Nakamura,K., Bruhns,P., Daeron,M. and Cambier,J.C. (2000) The rasGAP-binding protein p62dok is a mediator of inhibitory FcγRIIB signals in B cells. Immunity, 12, 347–358. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Vaughan,J.M., Wiater,E, Gaddy-Kurten,D. and Vale,W. (1995) Inactivation of activin-dependent transcription by kinase-deficient activin receptors. Endocrinology, 136, 5493–5503. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T., Chiang,T.A., Davison,A.F., Attisano,L. and Wrana,J.L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell, 95, 779–791. [DOI] [PubMed] [Google Scholar]
- Vale W., Rivier,J., Vaughan,J., McClintock,R., Corrigan,A., Woo,W., Karr,D. and Spiess,J. (1986) Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature, 321, 776–779. [DOI] [PubMed] [Google Scholar]
- Van Dijk T.B., van Den Akker,E., Amelsvoort,M.P., Mano,H., Lowenberg,B. and von Lindern,M. (2000) Stem cell factor induces phosphatidylinositol 3′-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells. Blood, 96, 3406–3413. [PubMed] [Google Scholar]
- Wurthner J.U., Frank,D.B., Felici,A., Green,H.M., Cao,Z., Schneider,M.D., McNally,J.G., Lechleider,R.J. and Roberts,A.B. (2001) Transfoming growth factor-β receptor-associated protein 1 is a Smad4 chaperone. J. Biol. Chem., 276, 19495–19502. [DOI] [PubMed] [Google Scholar]
- Yamanashi Y. and Baltimore,D. (1997) Identification of the Abl-and rasGAP-associated 62 kDa as a docking protein, Dok. Cell, 88, 205–211. [DOI] [PubMed] [Google Scholar]
- Yamanashi Y., Tamura,T., Kanamori,T., Yamane,H., Nariuchi,H., Yamamoto,T. and Baltimore,D. (2000) Role of the rasGAP-associated docking protein p62dok in negative regulation of B cell receptor-mediated signaling. Genes Dev., 14, 11–16. [PMC free article] [PubMed] [Google Scholar]
- Yamato K., Koseki,T., Ohguchi,M., Kizaki,M., Ikeda,Y. and Nishihara,T. (1997) Activin A induction of cell-cycle arrest involves modulation of cyclin D2 and p21CIP1/WAF1 in plasmacytic cells. Mol. Endocrinol., 11, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Zauberman A., Oren,M. and Zipori,D. (1997) Involvement of p21WAF1/Cip1, CDK4 and Rb in activin A mediated signaling leading to hepatome cell growth inhibition. Oncogene, 15, 1705–1711. [DOI] [PubMed] [Google Scholar]