Abstract
Enhancers have been defined as cis-acting DNA sequences that stimulate transcription from a linked promoter in a distance- and orientation-independent manner. How enhancers activate gene transcription over vast chromosomal distances within metazoan genomes remains poorly understood. Here, we show that the transcription factor GAGA can stimulate transcription by linking an enhancer to its cognate promoter. Strikingly, in addition to facilitating activation by a remote enhancer in cis, GAGA can direct activation of a promoter by an enhancer located on a separate DNA molecule. Enhancer function in trans is critically dependent on POZ domain-mediated GAGA oligomerization, enabling GAGA to bind two DNA molecules simultaneously. Transcriptional activation by an enhancer functioning in trans was observed both in transfected cells and in reconstituted transcription reactions. We propose that GAGA facilitates long-range activation by providing a protein bridge that mediates enhancer–promoter communication.
Keywords: enhancer–promoter interaction/GAGA/POZ domain/transcription
Introduction
Enhancers are cis-regulatory DNA elements that can activate gene transcription irrespective of their orientation or distance relative to the transcription start site (Serfling et al., 1985; Ptashne, 1986; Blackwood and Kadonaga, 1998; Dillon and Sabbattini, 2000). A fundamental question in transcriptional regulation is how enhancers find and control their target promoters over long distances. In the large genomes of metazoans, regulatory elements that determine the patterns of gene expression frequently act over thousands of base pairs (bp). The principal models proposed to explain distal enhancer function invoke either protein–protein interactions resulting in the formation of DNA loops, sliding of proteins along the DNA, transfer of superhelical torsion or the generation of large chromatin domains that facilitate or prohibit gene activation (Ptashne, 1986; Martin et al., 1996; Blackwood and Kadonaga, 1998; Dorsett, 1999; Dillon and Sabbattini, 2000; Engel and Tanimoto, 2000). The key feature of the ‘looping model’ is that enhancer–promoter interactions occur via contacts between DNA bound transcription factors. The other models all involve some form of protein scanning or transmission of a signal along the DNA between the enhancer and promoter. Therefore, whereas the cis configuration of enhancer and promoter is an essential prerequisite for the ‘scanning model’, the looping model allows for activation to occur in trans.
The need for dynamic long-distance associations between enhancers and promoters is particularly evident during metazoan development, which requires a precise orchestration of temporal and spatial gene expression (Fraser and Grosveld, 1998; Dorsett, 1999; Farkas et al., 2000). For example, the many enhancers and silencers that regulate the Drosophila homeotic Ultrabithorax (Ubx) gene are spread out over >100 kilobases (kb) (Farkas et al., 2000). One key regulatory sequence is a Polycomb response element (PRE), located ∼25 kb upstream of the Ubx transcription start site (Pirrotta, 1997; Lyko and Paro, 1999; Farkas et al., 2000; Mahmoudi and Verrijzer, 2001). We were intrigued by the preponderance of binding sites for the transcription factor GAGA within this PRE as well as in the Ubx promoter itself.
GAGA is encoded by the essential Trithorax-like (Trl) gene and is a versatile protein that performs a multitude of chromosomal tasks (Farkas et al., 1994, 2000; Henikoff and Vermaak, 2000; Mahmoudi and Verrijzer, 2001). GAGA is required for the normal expression of a wide range of different genes, including the homeotic genes such as Ubx, other pattern formation genes, heat shock genes and histone genes. Rather than activating naked DNA templates, it is believed that GAGA activates RNA polymerase II transcription, largely by relief of chromatin repression (Croston et al., 1991; Tsukiyama et al., 1994; Okada and Hirose, 1998). Trl mutations enhance position effect variegation, suggesting that GAGA also counteracts heterochromatin-mediated silencing (Farkas et al., 1994). Finally, there is evidence that GAGA plays a role in chromosome condensation and segregation (Bhat et al., 1996; Platero et al., 1998; Henikoff and Vermaak, 2000). GAGA contains two major structural domains: a zinc finger DNA binding domain (DBD) and an N-terminal POZ (poxvirus and zinc finger; also called BTB, broad complex tramtrack bric-a-brac) protein–protein interaction domain (Figure 1A). We and others have previously shown that the POZ domain mediates multimerization into higher order GAGA oligomers that bind co-operatively to multiple sites present in its natural target promoters (Espinas et al., 1999; Katsani et al., 1999). Furthermore, our electron microscopy experiments provided the first hint that GAGA oligomers may bind two DNA molecules simultaneously, and that this property of GAGA was critically dependent on POZ domain-mediated oligomerization (Katsani et al., 1999).
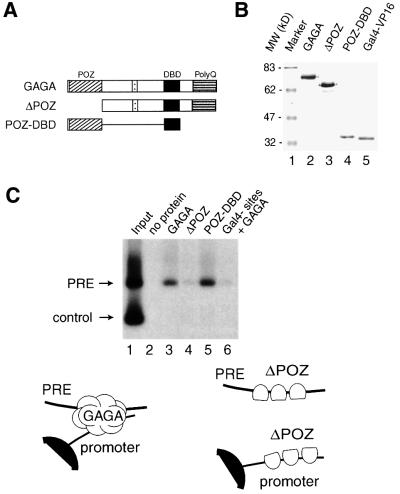
Fig. 1. GAGA is a bridging factor that can bind two DNA molecules simultaneously. (A) Schematic representation of full-length GAGA (519 residues), ΔPOZ (a deletion mutant comprising amino acids 120–519 lacking the POZ domain) and POZ-DBD [a polypeptide comprising the POZ domain (amino acids 1–126) fused to the DNA binding domain (amino acids 298–378)]. All polypeptides contained an N-terminal HA tag. (B) Expression and purification of recombinant proteins used in this study. Full-length GAGA, ΔPOZ and POZ-DBD were expressed in insect Sf9 cells using a baculovirus expression system. FLAG-tagged Gal4-VP16 was expressed in bacteria using a T7 expression system. The recombinant proteins were immunopurified from cell extracts using either the FLAG or HA tag, essentially as described previously (Katsani et al., 1999). Eluted proteins were analyzed by SDS–PAGE followed by Coomassie Blue staining. The molecular weights (kDa) of the protein standards are indicated. (C) GAGA can link the Ubx PRE to biotinylated GAGA sites immobilized on streptavidin beads. Radiolabeled input DNA comprised a 400 bp PRE fragment containing nine GAGA sites and a 140 bp control DNA fragment lacking GAGA sites. The input DNA was incubated with the immobilized GAGA sites in either the presence or absence of the indicated proteins. Following a 45 min incubation, the beads were washed, and bound DNA was recovered and analyzed by agarose gel electrophoresis followed by autoradiography.
In this study, we address the mechanism by which remote enhancers promote the initiation of transcription by RNA polymerase II. Since binding sites for the transcription factor GAGA are common components of Drosophila promoters as well as distal regulatory elements, we wondered whether GAGA might be specifically tailored to facilitate long-range interactions. Our results suggest that GAGA can provide a protein bridge, permitting activation by a remote enhancer or even an enhancer located on a separate DNA molecule. We were able to demonstrate enhancer function in trans both in reconstituted in vitro transcription reactions and in transfected cells. Collectively, these results suggest that a continuous DNA path is not strictly required for enhancer function. The implications of these findings for models to explain long-range gene activation are discussed.
Results
GAGA can form a protein link between separate DNA fragments
To examine GAGA’s ability to bring distal DNA elements together, hemagglutinin (HA) epitope-tagged full-length GAGA, a deletion mutant lacking the POZ domain (ΔPOZ) and a minimal construct comprising the DBD and POZ domains (POZ-DBD) were immunopurified from extracts of insect Sf9 cells infected with recombinant baculoviruses (Figure 1A and B). While all these GAGA polypeptides bind DNA efficiently, only GAGA and POZ-DBD are capable of oligomerization and co-operative binding to multiple sites. To test whether GAGA was able to act as a protein link between separate DNA fragments, we performed DNA pull-down experiments. Biotinylated oligonucleotides containing GAGA sites were coupled to a streptavidin resin. These beads were incubated with a radiolabeled Ubx PRE fragment containing nine GAGA sites and an unrelated control fragment in either the presence or absence of GAGA protein. After extensive washing, bound DNA was recovered and analyzed by agarose gel electrophoresis followed by autoradiography (Figure 1C). Whereas the immobilized GAGA sites alone were unable to retain the labeled PRE DNA (lane 2), specific association of the fragment was observed in the presence of GAGA protein (lane 3). Binding was dependent on the GAGA sites, since only the PRE and not the control DNA was recovered. Moreover, when Gal4 sites were coupled to the beads instead of GAGA sites, the PRE fragment was no longer retained in the presence of GAGA (lane 6). In contrast to full-length GAGA, ΔPOZ, which still binds DNA but can no longer multimerize, failed to link the PRE fragment to the beads (lane 4). In the presence of POZ-DBD, however, the PRE fragment was retained (lane 5), establishing that the DNA binding and multimerization domains of GAGA are both necessary and sufficient for simultaneous binding to two DNA molecules.
GAGA facilitates activation by a remote enhancer
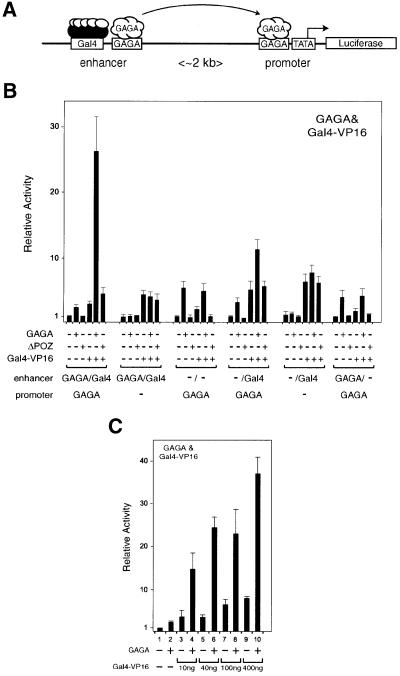
An interesting corollary of these results is that GAGA may facilitate transcriptional activation by bringing a remote enhancer into close proximity of its cognate promoter. To test this idea, we constructed a template containing an enhancer comprising Gal4 binding sites and GAGA elements separated by >2 kb (positioned 2494 bp upstream and 2004 bp downstream of the transcription start site) of intervening DNA from a promoter. The promoter contains GAGA elements (positioned at –24) proximal to a TATA box directing expression of the luciferase gene (Figure 2A). Thus, the enhancer can be bound by both Gal4-VP16 and GAGA, whereas the promoter can only be recognized by GAGA. It should be noted that, similar to the constructs used in this study, natural GAGA response elements typically contain four or more GAGA binding sites recognized by a GAGA oligomer (Espinas et al., 1999; Katsani et al., 1999). Human 911 cells, which lack endogenous GAGA, were co-transfected with the reporter plasmid in the absence or presence of expression vectors for GAGA, ΔPOZ or Gal4-VP16 (Figure 2B). Expression of GAGA alone only weakly activates transcription and, likewise, Gal4-VP16 acting through the distal enhancer induces a modest stimulation of transcription. However, concomitant expression of GAGA and Gal4-VP16 activates transcription to a level that is more than the product of the activation by GAGA and Gal4-VP16 separately. The GAGA mutant ΔPOZ, which is unable to oligomerize, does not co-operate with Gal4-VP16. Primer extension experiments confirmed that transcripts initiated at the appropriate transcription start site within the promoter, either in the presence or absence of enhancer-binding activators (data not shown). Taken together, these results are consistent with the idea that the observed co-operation between GAGA and Gal4-VP16 is the consequence of GAGA’s ability to bring the enhancer and promoter into close proximity of each other, thus facilitating activation by the enhancer-bound Gal4-VP16.
Fig. 2. GAGA promotes activation from a remote enhancer. (A) Structure of the reporter [pGL3-prom(GAGA)Enh(GAGA/Gal4)] used to examine GAGA’s effect on long-range activation via a distal cis-acting enhancer. The basic reporter plasmid containing five GAGA sites proximal to a minimal TATA box-containing core promoter, separated by >2 kb of intervening DNA from a distal enhancer comprising five GAGA sites adjacent to five Gal4 sites. Mutant versions were generated that lack the promoter proximal GAGA sites [pGL3-prom-Enh(GAGA/Gal4)], the GAGA sites in the enhancer [pGL3-prom(GAGA)Enh(Gal4)] or the Gal4 sites in the enhancer [pGL3-prom(GAGA)Enh(GAGA)]. (B) The distinct reporter plasmids were co-transfected in the presence of various combinations of expression vectors for GAGA, ΔPOZ or Gal4-VP16, as indicated. Complete reporter [pGL3-prom(GAGA)Enh(GAGA/Gal4)], reporter lacking the proximal GAGA sites [pGL3-prom-Enh(GAGA/Gal4)], reporter lacking distal enhancer sites [pGL3-prom(GAGA)], reporter lacking the distal GAGA sites [pGL3-prom(GAGA)Enh(Gal4)], reporter lacking both proximal and distal GAGA sites [pGL3-prom-Enh(Gal4)] and reporter lacking the Gal4 enhancer sites [pGL3-prom(GAGA)Enh(GAGA)]. (C) The complete reporter plasmid, pGL3-prom(GAGA)Enh(GAGA/Gal4), was co-transfected in the presence of 50 ng expression vector lacking (lanes 1, 3, 5, 7 and 9) or containing (lanes 2, 4, 6, 8 and 10) GAGA, and increasing amounts of the expression vector for Gal4-VP16 as indicated (lanes 3 and 4, 10 ng; lanes 5 and 6, 40 ng; lanes 7 and 8, 100 ng; and lanes 9 and 10, 400 ng).
To test this idea further, we constructed a series of distinct reporter templates in which the proximal or distal GAGA elements or the Gal4 binding sites were removed. When the opportunity for GAGA to bridge between the enhancer and the promoter was blocked by removal of the promoter proximal GAGA sites, it no longer mediated co-operative activation with Gal4-VP16. Moreover, this experiment reveals that unlike Gal4-VP16, GAGA is unable to activate from a remote enhancer position. As expected, when both GAGA and Gal4 sites were removed from the enhancer, only activation by GAGA through the promoter sites was observed. On a template containing promoter proximal GAGA sites and an enhancer comprising only Gal4 binding sites, both GAGA and Gal4-VP16 could activate. However, in the presence of both factors the level of activation was approximately additive and there was no strong synergy. In the absence of all GAGA sites there was no activation by GAGA, and GAGA failed to co-operate with Gal4-VP16. Finally, in the absence of Gal4 sites, Gal4-VP16 could not stimulate transcription, irrespective of the presence or absence of GAGA.
We tested the ability of a fixed amount of GAGA to co-operate with a wide concentration range of Gal4-VP16. As shown in Figure 2C, the amount of synergic transcriptional activation mediated by GAGA was largely independent of the amount of Gal4-VP16 expressing plasmid transfected into the cells. This result again supports the notion that the synergy between GAGA and Gal4-VP16 results from facilitated enhancer promoter communication, rather than, for instance, facilitated binding of Gal4-VP16 by GAGA.
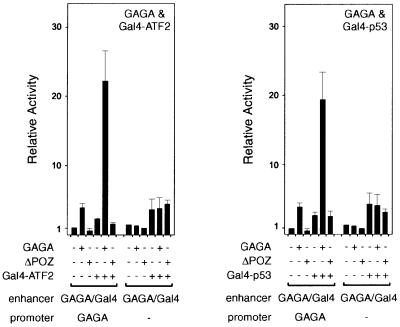
Finally, we wished to ensure that the ability of GAGA to promote long-range enhancer function is independent of the nature of the enhancer-bound activator. Therefore, we tested the co-operation between GAGA and fusions of the Gal4 DBD to either the activator ATF2 or p53 (Figure 3). In both cases, we observed a clear synergic activation of enhancer-mediated transcription in the presence of GAGA. Deletion of the promoter proximal GAGA sites, preventing GAGA from linking the enhancer to the promoter, resulted in a loss of co-operation between GAGA and the enhancer-bound activator. Taken together, our results suggest that GAGA promotes enhancer function by physically linking it to its cognate promoter.
Fig. 3. The ability of GAGA to promote long-range enhancer function is activator independent. Cells were co-transfected with either the complete reporter pGL3-prom(GAGA)Enh(GAGA/Gal4) (lanes 1–6) or the reporter lacking the proximal GAGA sites [pGL3-prom-Enh(GAGA/Gal4)] (lanes 7–12) in the presence of various combinations of expression vectors for GAGA, ΔPOZ, Gal4-ATF2 or Gal4-p53, as indicated.
GAGA is a bridging factor that can mediate enhancer function in trans
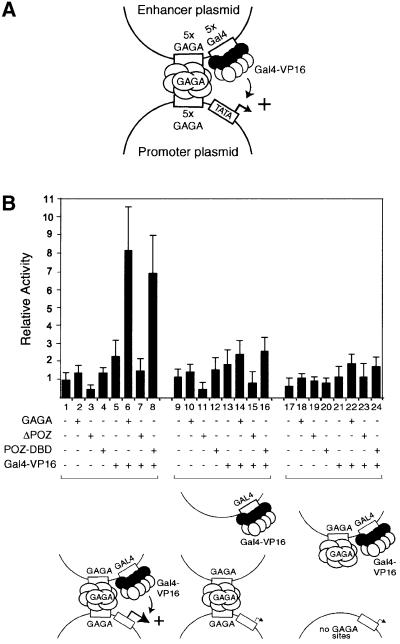
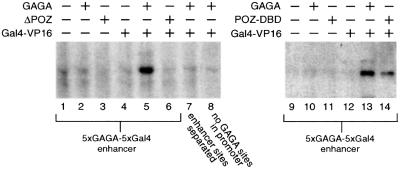
The extreme example of long-range regulation would be the activation of a promoter by an enhancer located on a separate DNA molecule. To determine whether GAGA is able to mediate enhancer–promoter interactions in trans, we designed the experiment illustrated in Figure 4A. We constructed an enhancer plasmid containing Gal4 binding sites adjacent to GAGA elements, and a separate promoter plasmid with GAGA sites proximal to a core promoter directing expression of the luciferase gene. Cells were co-transfected with these promoter and enhancer plasmids in either the absence or presence of expression vectors for GAGA, ΔPOZ, POZ-DBD or Gal4-VP16 (Figure 4B). No appreciable activation was observed by either GAGA, ΔPOZ or POZ-DBD alone (lanes 1–4). As expected, since the promoter plasmid lacks Gal4 binding sites, there was no significant activation by Gal4-VP16 alone (lane 5). Strikingly, co-expression of GAGA and Gal4-VP16 resulted in a clear stimulation of transcription, suggesting that GAGA brings the enhancer and promoter together to allow activation by the enhancer-bound Gal4-VP16 (lane 6). Indeed, activation was dependent on GAGA oligomerization, since deletion of the POZ domain (ΔPOZ) abrogated transcriptional stimulation by Gal4-VP16 (lane 7). Moreover, the DBD and multimerization domains comprising POZ-DBD were sufficient to mediate activation (lane 8). When the Gal4 and GAGA binding sites in the enhancer were separated and placed in distinct plasmids, transactivation was lost (lanes 9–16). Finally, activation by the enhancer was dependent on the presence of GAGA sites in the reporter plasmid since their removal abolished activation (lanes 17–24). Taken together, these results strongly suggest that GAGA can control transcription by providing a protein link between enhancer and promoter.
Fig. 4. GAGA mediates enhancer function in trans in transiently transfected cells. (A) Hypothetical model for GAGA function. Two distinct plasmids were constructed: an enhancer plasmid containing Gal4 binding sites adjacent to GAGA elements, and a promoter plasmid with GAGA sites proximal to a core promoter in front of the luciferase gene. We speculated that GAGA might direct the enhancer plasmid to the reporter plasmid to allow activation by Gal4-VP16. This model was tested in transiently transfected cells. (B) Human 911 cells were co-transfected with the promoter and enhancer plasmids in the absence or presence of expression vectors for GAGA, ΔPOZ, POZ-DBD or Gal4-VP16. The reporter construct pGL35-tk-GAGA (lanes 1–16) contains five GAGA elements upstream of the thymidine kinase promoter fused to the luciferase gene, whereas pGL3-tk (lanes 17–24) lacks GAGA elements. These reporter constructs were co-transfected with either the intact enhancer plasmid 5×GAGA-5×Gal4-pBluescript (lanes 1–8 and 17–24) or two ‘separated’ enhancer plasmids (lanes 9–16): one containing only the GAGA sites (5×GAGA-pBluescript) and another containing only the Gal4 binding sites (5×Gal4-pBluescript). These reporters were co-transfected with the indicated combinations of expression vectors for no protein, GAGA, ΔPOZ, POZ-DBD or Gal4-VP16 expression vector.
Following an experimental design similar to that used in the transfection experiments (Figure 4B), we tested the ability of GAGA to mediate enhancer trans-activation in reconstituted transcription assays. Reactions contained both promoter and enhancer plasmids and were conducted in the presence or absence of the purified recombinant transcriptional regulators shown in Figure 1B. Human nuclear extract lacking GAGA was used as a source of general transcription factors and transcription was monitored by primer extension (Figure 5). While neither GAGA nor Gal4-VP16 alone supported activation (lanes 2, 4, 10 and 12), they strongly stimulated transcription when added together (lanes 5 and 13). Removal of the POZ domain abolished activation (lane 6), while POZ-DBD was sufficient to mediate enhancer function in trans (lane 14), reinforcing the notion that GAGA oligomerization is essential for trans-activation. Importantly, co-operation between GAGA and Gal4-VP16 requires an intact enhancer, since separation of the Gal4 and GAGA binding sites abolished activation (lane 7). Likewise, removal of the GAGA sites in the promoter plasmid reduced transcription to basal levels (lane 8). In agreement with our transfection studies, these results demonstrate the ability of GAGA to function as a bridging factor, allowing promoter activation by an enhancer located on a separate DNA molecule.
Fig. 5. Enhancer stimulated trans-activation in reconstituted transcription reactions. In vitro transcription reactions containing both promoter and enhancer plasmids were performed using a human nuclear extract that lacks GAGA, as a source of basal transcription factors. Transcription reactions containing 50 ng of the promoter plasmid were conducted in the absence or presence of recombinant GAGA, ΔPOZ, POZ-DBD and Gal4-VP16, as indicated. The promoter plasmid contained five GAGA sites upstream of a minimal core promoter, with the exception of lane 8, where the promoter lacked GAGA sites. Reactions also contained a 6-fold excess of 5×GAGA-5×Gal4-pBluescript enhancer plasmid, with the exception of lane 7, which contained equal amounts of 5×GAGA-pBluescript and 5×Gal4-pBluescript plasmids. Transcription products were detected by primer extension, separated on a 7% denaturing polyacrylamide gel and subjected to autoradiography. The results shown were quantified by phosphoimager analysis and revealed a minor reduction in transcription levels in the presence of the activators alone (∼0.8- to 1-fold basal levels), a 6- to 7-fold activation in the presence of both GAGA and Gal4-VP16, and 5-fold activation by POZ-DBD and Gal4-VP16.
Discussion
Here, we have addressed the mechanism by which a remote enhancer can regulate its cognate promoter over long distances. Our results show that GAGA can form a protein link between separate DNA elements. This bridging function of GAGA is critically dependent on oligomerization via its POZ domain. We show that in cells, as well as in reconstituted transcription experiments, GAGA can facilitate gene activation by acting as an anchor that links the enhancer to a promoter. GAGA can tether an enhancer to a promoter to allow activation of transcription even when they are located on separate DNA molecules. Collectively, these results demonstrate that a protein link provided by GAGA is sufficient to mediate productive enhancer–promoter communication.
Together with earlier studies in which the DNA molecules harboring enhancer and promoter were joined together either by a streptavidin–biotin bond (Mueller-Storm et al., 1989) or as intertwined plasmid catenanes (Dunaway and Droge, 1989), our results using a natural transcription regulator establish that a continuous DNA path is not strictly required for enhancer function. Consequently, in these experiments trans-activation occurs in the absence of protein scanning along the DNA or the transfer of superhelical tension. Rather, bringing the enhancer into close physical proximity to the promoter appears sufficient to allow transcriptional activation. Collectively, these findings argue in favor of the notion that promoter–enhancer contacts that may occur by random collision are stabilized by protein–protein interactions. When located in cis, such promoter–enhancer interactions would cause looping of the intervening DNA, as has been observed during transcription, DNA replication and recombination (Su et al., 1991; Schleif, 1992; Ringrose et al., 1999). Recently, it was shown that telomere looping permits activation of an inserted gene by a downstream enhancer in yeast cells, where long-range activation does not normally occur (De Bruin et al., 2001). Moreover, a number of in vivo studies in metazoans have also argued in favor of a DNA looping model for gene control by distal elements (Dillon et al., 1997; Fraser and Grosveld, 1998; Engel and Tanimoto, 2000).
Clearly, such a looping mechanism does not rule out a role for the intervening DNA, which may be bound by factors that either constrain or facilitate enhancer– promoter interactions. For example, the packaging of DNA into chromatin can augment long-range activation, possibly by increasing DNA flexibility or reducing the distance between enhancer and promoter (Kamakaka et al., 1993; Barton and Emerson, 1994; Wong et al., 1997; Ringrose et al., 1999). Other proteins such as HMG1/Y appear to play a role in facilitating long-distance interactions through an architectural mechanism (Bagga et al., 2000). Interestingly, genetic screens in Drosophila have identified Chip and Nipped-B as potential facilitators of long-range activation that function through specific sequences located between enhancers and promoters (Dorsett, 1999).
In this study, we have addressed one aspect of the mechanisms of action of GAGA, a versatile protein that performs a multitude of chromosomal tasks (Farkas et al., 2000; Henikoff and Vermaak, 2000). Our proposal that GAGA acts as a bridging factor that promotes enhancer– promoter communication may explain a number of in vivo observations on GAGA function. First, the promoter proximal GAGA sites in the Ubx gene have been implicated in regulation by the ABX and BXD distal regulatory elements (Laney and Biggin, 1992). Secondly, a study specifically designed to identify promoter elements essential for enhancer–promoter interactions revealed that the GAGA elements in the engrailed (en) promoter are essential for enhancer-dependent activation (Orihara et al., 1999). Thirdly, the eve promoter possesses intrinsic insulator properties that were found to be critically dependent on GAGA function (Ohtsuki and Levine, 1998). These results led to the proposal by Ohtsuki and Levine (1998) that GAGA might confer insulator function via trapping of distal enhancers. The results presented in this paper now provide a molecular mechanism by which GAGA may capture enhancers by acting as a protein anchor. Similar mechanisms may occur during natural examples of activation in trans such as transvection, where an enhancer on one chromosome activates an allelic promoter on another paired chromosome (Henikoff and Comai, 1998; Dorsett, 1999). Furthermore, GAGA may contribute to the non-random integration of P-elements harboring the en promoter near the chromosomal en locus (Hama et al., 1990), or of the preferential integration of P-elements harboring a PRE near chromosomal PREs (Fauvarque and Dura, 1993). In conclusion, our experiments demonstrate that GAGA can mediate trans-activation by providing a protein bridge that directs enhancer– promoter association. We propose that these findings reflect a more general mechanism for communication between enhancers and promoters.
Materials and methods
Plasmid constructs
Details on plasmid construction will be provided upon request. To construct the TATA pGL3 reporter Luciferase construct, the SmaI–XhoI fragment of TATA-CAT (Jonat et al., 1990) was inserted into the BamHI (end-filled)–XhoI restricted pGL3-Basic vector. The reporter plasmids used in the cis-activation experiments were subsequently derived from the TATA pGL3 reporter by inserting, as indicated, five GAGA sites into the BamHI–SalI sites immediately downstream from the TATA box at position –24, five GAGA sites, five Gal4 sites or five GAGA plus five Gal4 sites into the KpnI–SacI sites positioned at –2494 or +2004 to generate the following constructs: complete reporter, pGL3-prom (GAGA)Enh(GAGA/Gal4); reporter lacking the promoter proximal GAGA sites, pGL3-prom-Enh(GAGA/Gal4); reporter lacking enhancer sites, pGL3-prom(GAGA); reporter lacking the distal GAGA sites, pGL3-prom(GAGA)Enh(Gal4); reporter lacking both proximal and distal GAGA sites, pGL3-prom-Enh(Gal4); reporter lacking the Gal4 enhancer sites, pGL3-prom(GAGA)Enh(GAGA).
The reporter plasmid tk-pGL3 has been described previously (van Dam et al., 1998). 5×GAGA-tk-pGL3 was constructed by cloning five GAGA sites into the KpnI–SacI sites of tk-pGL3.
The mammalian expression vector Gal4-VP16 contains the 78 C-terminal amino acids of VP16 fused to the first N-terminal 147 amino acids of Gal4 under the control of an SV40 promoter and has been described previously (Sadowski et al., 1988). The SV40-driven mammalian expression vectors Gal4-p53 (codons 1–73) and Gal4-ATF2 (codons 1–112) have been described previously (Fields et al., 1990; Duyndam et al., 1996).
Vectors expressing GAGA, ΔPOZ or POZ-DBD were generated by inserting DNA fragments encoding full-length GAGA, the ΔPOZ mutant (amino acids 120–519) or the POZ-DBD mutant (amino acids 1–120 and 321–379) into pcDNA3.1+ (Invitrogen).
DNA pull-down assays
Immobilized GAGA sites were generated by coupling biotinylated double-stranded oligonucleotides harboring five GAGA sites to streptavidin beads. These beads were pre-incubated with binding buffer [80 mM KCl, 10% glycerol, 25 mM HEPES pH 7.6, 5 mM MgCl2, 0.1% NP-40, 10 µM ZnCl2, 0.05 µg/µl bovine serum albumin and 100 µg/ml poly(dIdC)–(dIdC)] for 1 h at room temperature (RT). After removal of excess buffer, radiolabeled input DNA (∼10 ng) comprising a 400 bp PRE fragment containing nine GAGA sites and a 140 bp control DNA fragment lacking GAGA sites were added in either the presence or absence of recombinant purified GAGA, ΔPOZ or POZ-DBD (∼50 ng) in 50 µl binding buffer. Following a 45 min incubation at RT on a rotating wheel, beads were washed with binding buffer. Bound DNA was recovered and subjected to 1.8% agarose gel electrophoresis followed by autoradiography. HA epitope-tagged full-length GAGA, a deletion mutant lacking the POZ domain (ΔPOZ) and a minimal construct comprising the DBD and POZ domain (POZ-DBD) (see Figure 1) were immunopurified from extracts of insect Sf9 cells infected with recombinant baculoviruses as described previously (Katsani et al., 1999).
Transient transfections
Human 911 cells were transfected on 3 cm dishes using the calcium phosphate precipitation method. After a 6 h incubation, cells were washed with HEPES-buffered saline and culture medium was added. After 15 h, cells were harvested and lysed in 250 µl lysis buffer (25 mM Tris–phosphate pH 7.8, 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′N′ tetraacetic acid, 10% glycerol, 1% Triton X-100). Luciferase activity was measured according to the manufacturer’s protocol (Promega).
For the long-distance cis-activation experiments (Figure 2), cells were co-transfected with 1 µg of the indicated reporter constructs, 50 ng of pcDNA3.1+ derived vectors expressing either no protein, GAGA or ΔPOZ, either in the presence or absence of 5 ng of the Gal4-VP16, 100 ng of Gal4-ATF2 or 150 ng of Gal4-p53 expression vectors, as indicated. pBluescript plasmid was added as carrier DNA to bring the total DNA content of each reaction to 5 µg. For the trans-activation experiments, cells were co-transfected with 350 ng of the indicated reporter construct (5×GAGA-tk-pGL3 or tk-pGL3), 50 ng of pcDNA3.1+-derived vectors expressing either no protein, GAGA, ΔPOZ or POZ-DBD, as indicated, in the presence or absence of 10 ng of the Gal4-VP16 expression vector. All transfections contained either 1.5 µg enhancer plasmid (5×GAGA-5×Gal4-pBluescript) or both enhancer plasmids 5×GAGA-pBluescript and 5×Gal4-pBluescript (1.5 µg each). pBluescript was added as carrier DNA to bring the total DNA content of each reaction to 5 µg.
In vitro transcription
Transcription reactions were performed essentially as described previously (Wang et al., 1997). Transcription reactions (25 µl) containing the promoter plasmid (50 ng) and enhancer plasmid (300 ng 5×GAGA/5×Gal4-pBluescript and 300 ng empty pBluescript vector, or 300 ng each of 5×GAGA-pBluescript and 5×Gal4-pBluescript) were pre-incubated in the presence or absence of recombinant purified proteins (∼100 ng GAGA, ΔPOZ or POZ-DBD and ∼20 ng Gal4-VP16). Forty-five minutes of incubation was permitted prior to addition of 50 µg HeLa nuclear extract, which lacks GAGA, as a source of basal transcription factors. After a further 20 min incubation at RT, transcription was initiated by the addition of NTPs (0.5 mM). Transcription products were detected by primer extension, separated on a 7% denaturing polyacrylamide gel and subjected to autoradiography. The FLAG-tagged Gal4-VP16 protein was expressed in bacteria and immunopurified using an anti-FLAG affinity column essentially as described previously (Chiang and Roeder, 1993).
Acknowledgments
Acknowledgements
We thank J.Svejstrup, L.Comai, L.Fradkin, E.Kalkhoven, R.Vries and J.van der Knaap for comments on the manuscript.
References
- Bagga R., Michalowski,S., Sabnis,R., Griffith,J.D. and Emerson,B.M. (2000) HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res., 28, 2541–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.C. and Emerson,B.M. (1994) Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev., 8, 2453–2465. [DOI] [PubMed] [Google Scholar]
- Bhat K.M., Farkas,G., Karch,F., Gyurkovics,H., Gauz,J. and Schedl,P. (1996) The GAGA factors is required in the early Drosophila embryo not only for transcription regulation but also for nuclear division. Development, 122, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M. and Kadonaga,J.T. (1998) Going the distance: a current view of enhancer action. Science, 281, 61–63. [DOI] [PubMed] [Google Scholar]
- Chiang C.M. and Roeder,R.G. (1993) Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Res., 6, 62–64. [PubMed] [Google Scholar]
- Croston G.E., Kerrigan,L.A., Lira,L.M., Marshak,D.R. and Kadonaga,J.T. (1991) Sequence speicifc antirepression of histone HI-mediated inhibition of basal RNA polymerase II transcription. Science, 251, 643–649. [DOI] [PubMed] [Google Scholar]
- DeBruin D., Zaman,Z., Liberatore,R.A. and Ptashne,M. (2001) Telomere looping permits gene activation by a downstream UAS in yeast. Nature, 409, 109–113. [DOI] [PubMed] [Google Scholar]
- Dillon N. and Sabbattini,P. (2000) Functional gene expression domains: defining functional unit of eukaryotic gene regulation. BioEssays, 22, 657–665. [DOI] [PubMed] [Google Scholar]
- Dillon N., Trimborn,T., Strouboulis,J., Fraser,P. and Grosveld,F. (1997) The effect of distance on long-range chromatin interactions. Mol. Cell, 1, 131–139. [DOI] [PubMed] [Google Scholar]
- Dorsett D. (1999) Distant liaisons: long-range enhancer–promoter interactions in Drosophila. Curr. Opin. Genet. Dev., 9, 505–514. [DOI] [PubMed] [Google Scholar]
- Dunaway M. and Droge,P. (1989) Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature, 341, 657–659. [DOI] [PubMed] [Google Scholar]
- Duyndam M.C., van Dam,H., van der Eb,A.J. and Zantema,A. (1996) The CR1 and CR3 domains of the adenoviros type 5 E1A proteins can independently mediate activation of ATF-2. J. Virol., 70, 5852–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J.D. and Tanimoto,K. (2000) Looping, linking and chromatin activity: new insights into β-globin locus regulation. Cell, 100, 499–502. [DOI] [PubMed] [Google Scholar]
- Espinas M.L., Jimenez-Garcia,E., Vaquero,A., Canudas,S., Bernues,J. and Azorin,F. (1999) The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem., 274, 16461–16469. [DOI] [PubMed] [Google Scholar]
- Farkas G., Gausz,J., Galloni,M., Reuter,G., Gyurkovics,H. and Karch,F. (1994) The Trithorax-like gene encodes the Drosophila GAGA factor. Nature, 371, 806–808. [DOI] [PubMed] [Google Scholar]
- Farkas G., Leibovitch,B.A. and Elgin,S.C. (2000) Chromatin organization and transcriptional control of gene expression in Drosophila. Gene, 253, 117–136. [DOI] [PubMed] [Google Scholar]
- Fauvarque M. and Dura,J. (1993) Polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev., 7, 1508–1520. [DOI] [PubMed] [Google Scholar]
- Fields S. and Jang,S. (1990) Presence of a potent transcription activating sequence in the p53 protein. Science, 249, 1046–1049. [DOI] [PubMed] [Google Scholar]
- Fraser P. and Grosveld,F. (1998) Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol., 10, 361–365. [DOI] [PubMed] [Google Scholar]
- Hama C., Ali,Z. and Kornberg,T.B. (1990) Region-specific recombin ation and expression are directed by portions of the Drosophila engrailed promotor. Genes Dev., 4, 1079–1093. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Comai,L. (1998) Trans-sensing effects: the ups and downs of being together. Cell, 93, 329–332. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Vermaak,D. (2000) Bugs on drugs go GAGAA. Cell, 103, 695–698. [DOI] [PubMed] [Google Scholar]
- Jonat C., Rahmsdorf,H.J., Park,K.K., Cato,A.C., Gebel,S., Ponta,H. and Herrlich,P. (1990) Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell, 62, 1189–1204. [DOI] [PubMed] [Google Scholar]
- Kamakaka R.T., Bulger,M. and Kadonaga,J.T. (1993) Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev., 7, 1779–1795. [DOI] [PubMed] [Google Scholar]
- Katsani K.R., Hajibagheri,M.A. and Verrijzer,C.P. (1999) Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J., 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney J.D. and Biggin,M.D. (1992) Zeste, a nonessential gene, potentially activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev., 6, 1531–1541. [DOI] [PubMed] [Google Scholar]
- Lyko F. and Paro,R. (1999) Chromosomal elements conferring epigenetic inheritance. BioEssays, 21, 824–832. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T. and Verrijzer,C.P. (2001) Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene, 20, 3055–3066. [DOI] [PubMed] [Google Scholar]
- Martin D.I., Fiering,S. and Groudine,M. (1996). Regulation of β-globin gene expression: straightening out the locus. Curr. Opin. Genet. Dev., 6, 488–495. [DOI] [PubMed] [Google Scholar]
- Mueller-Storm H.P., Sogo,J.M. and Schaffner,W. (1989) An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell, 58, 767–777. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S. and Levine,M. (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev., 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M. and Hirose,S. (1998) Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activated fushi tarazu gene transcription in vitro. Mol. Cell. Biol., 18, 2455–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihara M., Hosono,C., Kojima,T. and Saigo,K. (1999) Identification of engrailed promoter elements essential for interactions with a stripe enhancer in Drosophila embryos. Genes Cells, 4, 205–218. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1997) Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet., 13, 314–318. [DOI] [PubMed] [Google Scholar]
- Platero J.S., Csink,A.K., Quintanilla,A. and Henikoff,S. (1998) Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol., 140, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. (1986) Gene regulation by proteins acting nearby and at a distance. Nature, 322, 697–701. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Chabanis,S., Angrand,P.O., Woodroofe,C. and Stewart,A.F. (1999) Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J., 18, 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma,J., Treizenberg,S. and Ptashne,M. (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature, 335, 563–564. [DOI] [PubMed] [Google Scholar]
- Schleif R. (1992) DNA looping. Annu. Rev. Biochem., 61, 199–223. [DOI] [PubMed] [Google Scholar]
- Serfling E., Jasin,M. and Schaffner,W. (1985) Enhancers and eukaryotic gene transcription. Trends Genet., 1, 224–230. [Google Scholar]
- Su W., Jackson,S., Tjian,R. and Echols,H. (1991) DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev., 5, 820–826. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Becker,P.B. and Wu,C. (1994) ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature, 367, 525–532. [DOI] [PubMed] [Google Scholar]
- vanDam H., Huguier,S., Kooistra,K., Baguet,J., Vial,E., van der Eb,A.J., Herrlich,P., Angel,P. and Castellazi,M. (1998) Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev., 12, 1227–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.H., Zou,S. and Tjian,R. (1997) TAFII250-dependent transcrip tion of cyclin A is directed by ATF activator proteins. Genes Dev., 11, 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Shi,Y.B. and Wolffe,A.P. (1997) Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J., 16, 3158–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]