Abstract
Recent studies of mRNA export factors have provided additional evidence for a mechanistic link between mRNA 3′-end formation and nuclear export. Here, we identify Nab2p as a nuclear poly(A)-binding protein required for both poly(A) tail length control and nuclear export of mRNA. Loss of NAB2 expression leads to hyperadenylation and nuclear accumulation of poly(A)+ RNA but, in contrast to mRNA export mutants, these defects can be uncoupled in a nab2 mutant strain. Previous studies have implicated the cytoplasmic poly(A) tail-binding protein Pab1p in poly(A) tail length control during polyadenylation. Although cells are viable in the absence of NAB2 expression when PAB1 is overexpressed, Pab1p fails to resolve the nab2Δ hyperadenylation defect even when Pab1p is tagged with a nuclear localization sequence and targeted to the nucleus. These results indicate that Nab2p is essential for poly(A) tail length control in vivo, and we demonstrate that Nab2p activates polyadenylation, while inhibiting hyperadenylation, in the absence of Pab1p in vitro. We propose that Nab2p provides an important link between the termination of mRNA polyadenylation and nuclear export.
Keywords: hnRNP/mRNA export/nuclear poly(A)-binding protein/polyadenylation
Introduction
The majority of RNA polymerase II transcripts are modified by 3′-end endonucleolytic cleavage and polyadenylation (for reviews see Minvielle-Sebastia and Keller, 1999; Zhao et al., 1999). In mammals, poly(A) tails reach a maximum of 200–300 nucleotides, and the nuclear poly(A) tail-binding protein PABPN1 (formerly PABP2) is essential for regulating the efficiency and extent of polyadenylation. The PABPN1 protein binds to nascent oligo(A) tails during pre-mRNA 3′-end processing and interacts with the multisubunit cleavage and polyadenylation specificity factor (CPSF) and poly(A) polymerase to promote rapid formation of the extended poly(A) tail (Wahle, 1991, 1995). When the tail reaches 200–300 nucleotides, the processive phase of the polyadenylation reaction terminates (Wahle, 1995; Keller et al., 2000).
In Saccharomyces cerevisiae, poly(A) tails are considerably shorter (70–90 nucleotides), and a PABPN1 ortholog does not exist. Instead, the cytoplasmic poly(A) tail-binding protein Pab1p (PABP1 in metazoans) has been proposed to coat the poly(A) tail during polyadenylation to restrict tail length in a manner analogous to PABPN1 (Zhao et al., 1999). Indeed, human PABP1 shuttles between the nucleus and cytoplasm (Afonina et al., 1998), suggesting that this predominantly cytoplasmic protein also functions in the nucleus. The Pab1 protein is associated with cleavage factor IA (CF IA), which is involved in both the cleavage and polyadenylation steps, and Pab1p is required for poly(A) tail length regulation in vitro (Amrani et al., 1997; Minvielle-Sebastia et al., 1997). An alternative to this model is that long tails may be synthesized initially, and then shortened by a poly(A) nuclease (PAN) activity that is recruited by Pab1p bound to poly(A) (Brown and Sachs, 1998). In support of this latter possibility, poly(A) tails >200 nucleotides are added to a cytochrome c (CYC1) precursor RNA in vitro using extracts from pan mutant strains. However, several observations argue against the proposal that Pab1p is the sole nuclear poly(A) tail-binding protein that regulates tail length by coating the poly(A) tail. First, the steady-state concentration of Pab1p in the nucleus is low so it must compete with more abundant nuclear RNA-binding proteins for poly(A) tail binding (Anderson et al., 1993a). Secondly, the very long tails produced in vitro in the absence of Pab1p are not detectable in either pab1 or pan mutant strains in vivo, which show tails only 3–20 nucleotides longer (Sachs and Davis, 1989; Brown and Sachs, 1998). Why this difference in maximum tail length in vitro and in vivo? Tail length may be restricted in the nucleus by competition between different transcripts for 3′-end processing factors, or termination of polyadenylation might be induced by ongoing nuclear export (Brown and Sachs, 1998). Alternatively, a tail length regulatory factor might be lost, or inactivated, during preparation of the extracts used for mRNA 3′-end formation in vitro.
Following transcript maturation, nuclear mRNA– hnRNP complexes are exported into the cytoplasm through the nuclear pore complex (NPC) (for reviews see Cole, 2000; Cullen, 2000). Specialized interactions between mRNA-binding proteins and NPC-associated factors are critical for mRNA export (Rodrigues et al., 2001). For example, the yeast protein Yra1p binds preferentially to mature mRNA in the nucleus (Sträßer and Hurt, 2000). In turn, Yra1p is recognized by the hetero dimer Mex67p–Mtr2p, which also associates with nucleoporins to facilitate nuclear mRNA export (Santos-Rosa et al., 1998; Hurt et al., 2000; Sträßer et al., 2000). During, and/or following, mRNA export through the NPC, the major class of nuclear pre-mRNA/mRNA-binding proteins, the hnRNPs, dissociate from the mRNA and are recycled back into the nucleus by members of the importin/karyopherin β family.
Previous work has suggested that polyadenylation is also important for mRNA export (Eckner et al., 1991; Huang and Carmichael, 1996). Indeed, recent studies on one class of mRNA export factor support the possibility that these two processes are mechanistically coupled (Hilleren and Parker, 2001; Hilleren et al., 2001; Jensen et al., 2001). What is the link between poly(A) tail addition and export? One model suggests that some mutations in mRNA export factors impair the release of nascent mRNA from its transcriptional site and lead to prolongation of the polyadenylation phase, possibly by depleting a 3′-end processing release/termination factor. In this report, we provide evidence that the yeast hnRNP Nab2p plays dual roles in the termination of mRNA polyadenylation and nuclear export.
Results
Hyperadenylated mRNAs accumulate in the nucleus following Nab2p depletion
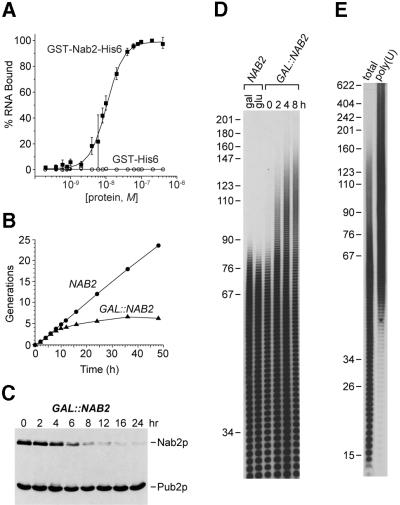
Current models for the regulation of poly(A) tail length during mRNA 3′-end formation suggest that Pab1p coats the poly(A) tail during polyadenylation (Zhao et al., 1999). Because Pab1p is not a major protein in the nucleus, we speculated that additional and more abundant nuclear poly(A)-binding proteins might function in polyadenylation. A candidate nuclear poly(A) tail-binding protein is Nab2p, which binds poly(A) in vitro and also appears to play a role in mRNA export (Anderson et al., 1993b; Wilson et al., 1994; Green et al., 2002). To determine if Nab2p and Pab1p possessed similar binding affinities for poly(A), we employed a standard nitrocellulose filter binding assay. Previous work has shown that Pab1p occupies a binding site of ∼12 nucleotides and human PABP1 binds tightly to (rA)25 (Kd = 7 nM) (Görlach et al., 1994). Interestingly, yeast Nab2p showed a similar (rA)25 binding affinity (Figure 1A, Kd = 10.5 nM).
Fig. 1. Nab2p depletion leads to hyperadenylation of mRNA. (A) High affinity binding of Nab2p to poly(A). Nitrocellulose filter binding assays were performed using (rA)25 and either GST–Nab2-His6 (closed squares) or GST–His6 (open circles). (B) Generation plot showing growth inhibition of YJA517-1C (GAL::NAB2) following glucose shift (0 h). (C) Immunoblot analysis using mAb 3F2 (anti-Nab2p) and mAb 3B1 (anti-Pub2p). YJA517-1C cells, grown in galactose, were shifted into glucose for 0–24 h. (D) Poly(A) tail analysis of NAB2 versus GAL::NAB2 cells grown in either galactose (gal) or glucose (glu) for 0–8 h. Each lane represents equal amounts of labeled RNA. The indicated size markers are pBR322 MspI fragments. (E) Selection of hyperadenylated RNAs following Nab2p depletion in vivo. Poly(A) tail lengths were determined for total RNA isolated from YJA517-1C cells grown in glucose for 16 h (total) or following selection of hyperadenylated RNA from this total pool by poly(U)–Sephadex chromatography [poly(U)].
Since Nab2p and Pab1p bound to poly(A) with similar affinities in vitro, we investigated whether Nab2p was required for poly(A) tail length control in vivo. The NAB2 gene was cloned downstream of the GAL1 promoter on a low-copy plasmid to create pGAL::NAB2. The YJA517-1C haploid strain (see Table I), which carries both a nab2Δ chromosomal deletion and the pGAL::NAB2 plasmid, was viable on galactose but failed to grow on glucose (Figure 1B). Immunoblot analysis demonstrated that Nab2p depletion occurred slowly following repression of NAB2 expression, and that growth was compromised following ∼80% loss of Nab2p at 8 h (Figure 1C). Although no significant defects in either pre-mRNA splicing or pre-rRNA processing were detectable during this time period (data not shown), a striking effect on polyadenylation was observed. As early as 2 h following glucose-induced repression, when intracellular Nab2p levels had declined <2-fold, maximum poly(A) tail length rose dramatically, and by 8 h maximum tail length had increased from ∼80 to >150 nucleotides (Figure 1D). This increase of >70 nucleotides was not due simply to alteration of the carbon source since cells that expressed wild-type Nab2p levels possessed normal poly(A) tail lengths when cultured in either glucose or galactose. Because non-mRNA RNA polymerase II transcripts are also polyadenylated under certain conditions (Chapon et al., 1997; Abou Elela and Ares, 1998; van Hoof et al., 2000), hyperpolyadenylated RNAs produced following Nab2p depletion were isolated selectively by poly(U)– Sephadex chromatography at an elevated formamide concentration (Figure 1E). These RNAs were identified by cDNA library construction followed by DNA sequencing of 100 cDNA clones. Whereas a variety of mRNAs were present in this long poly(A) tail pool, no snRNAs, snoRNAs or telomerase RNA were identified (our unpublished data). Therefore, Nab2p depletion leads to selective hyperadenylation of mRNA.
Table I. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| YJA512 | MATa/α leu2Δ2/leu2Δ2 ura3-52/ura3-52 NAB2/nab2::LEU2 pNAB2.15 | Anderson et al. (1993b) |
| YJA513 | MATα leu2Δ2 ura3-52 nab2Δ::LEU2 pNAB2.15 | this study |
| YJA515 | MATα leu2Δ2 ura3-52 nab2Δ::LEU2 pNAB2.19 | this study |
| YJA517 | MATa/MATα leu2Δ2/leu2Δ2 ura3-52/ura3-52 NAB2/nab2Δ::LEU2 pGAL::NAB2 | this study |
| YJA517-1C | MATa leu2Δ2 ura3-52 nab2Δ::LEU2 pGAL::NAB2 | this study |
| YJA517-1D | MATα leu2Δ2 ura3-52 NAB2 | this study |
| YJA221 | MATα leu2Δ2 his3Δ2 trp1-189 ura3-52 nab2Δ::LEU2 pnab2-21 | this study |
| YJA223 | MATα leu2Δ2 his3Δ200 trp1-189 ura3-52 nab2Δ::LEU2 TRP1::nab2-21 | this study |
| L4717 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Hong et al. (1997) |
| YKN105 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pPAB1.6 | this study |
| YKN106 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pPAB1.7 | this study |
| YRH201C | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pGAL::NAB2 | this study |
| YRH202 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pGAL::NAB2 YEp13 | this study |
| YRH204 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pPAB1 | this study |
| YHCS220 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab2Δ::HIS3 pGAL::NAB2 pHCS220 | this study |
| YAS394 | MATa pab1::HIS3 RPL46::LEU2 ade2 his3 leu2 trp1 ura3 can1 | Sachs and Deardorff (1992) |
| BMA64 | MATa ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 | Baudin-Baillieu et al. (1997) |
| YSD10 | MATa TAP::FIP1-TRP1-Kl ade2-1 leu2-3,112 ura3-1 trp1Δhis3-11,15 can1-100 | S.Dheur and L.Minvielle-Sebastia (in preparation) |
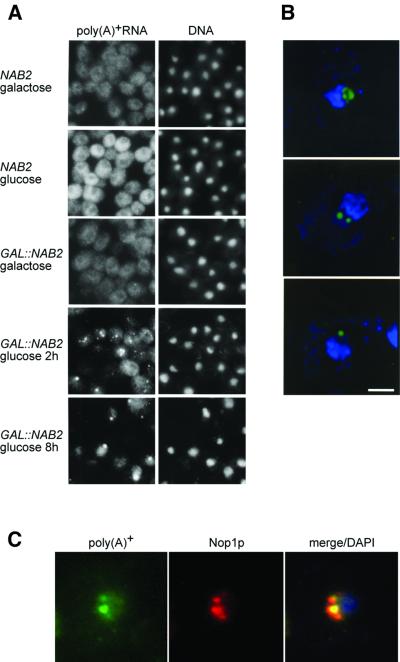
Some temperature-sensitive (ts) strains deficient in mRNA export at restrictive growth temperatures also accumulate hyperadenylated mRNAs when nuclear export is blocked (Kadowaki et al., 1994; Tseng et al., 1998; Hilleren and Parker, 2001; Jensen et al., 2001). Thus, we examined the effect of depleting Nab2p on the intracellular distribution of poly(A)+ RNAs using fluorescence in situ hybridization (FISH). Cells expressing the wild-type NAB2 gene during growth in either glucose or galactose possessed a disperse cytoplasmic poly(A)+ RNA staining pattern (Figure 2A). Although YJA517-1C (GAL::NAB2) cells also showed a normal poly(A)+ RNA distribution in galactose, when these cells were shifted to glucose, intense subnuclear regions, or foci, appeared within 2 h with a concomitant loss of cytoplasmic fluorescence signal (Figure 2A and B). Similarly to the mRNA transport-defective strain mtr2-1 (Kadowaki et al., 1994), these poly(A)+ RNA-rich foci co-localized with the nucleolar protein Nop1p (Figure 2C). These results supported the hypothesis that Nab2p was required for mRNA export and, similarly to other characterized mRNA export mutants, the hyperadenylated phenotype was due to an export block. To test this hypothesis, we created a conditional lethal nab2 allele and examined whether the poly(A) tail and export phenotypes were linked obligatorily in this mutant.
Fig. 2. Accumulation of poly(A)+ RNA in the nucleus and nucleolus following loss of Nab2p in vivo. (A) FISH analysis of poly(A)+ RNA distribution using conventional immunofluoresence microscopy. DNA was detected with DAPI. (B) FISH localization of poly(A)+ RNA using deconvolution microscopy. The distribution of poly(A)+ RNA (green) is shown for YJA517-1C (GAL::NAB2) cells shifted to glucose for 2 h. The nucleolus is adjacent to the bulk chromosomal DNA staining (blue). Three nucleolar distributions are displayed (top panel, overall nucleolar; middle panel, two intra-nucleolar foci; bottom panel, a single focus at the nucleolar periphery). Bar = 2 µm. (C) Co-localization of poly(A)+ RNA (green) and Nop1p (red) in GAL::NAB2 cells shifted to glucose for 2 h. Merged poly(A)+ RNA, Nop1p and DNA signals are shown in the right panel.
Uncoupling of mRNA polyadenylation and export defects in a nab2 mutant
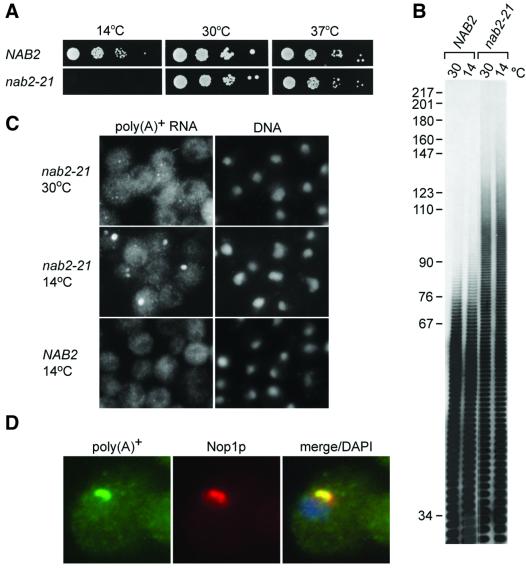
A number of mRNA export mutants have been identified by screening randomly generated ts strains for accumulation of poly(A)+ RNA in the nucleus (Amberg et al., 1992; Kadowaki et al., 1994). These screens failed to identify nab2 ts alleles. As an alternative, the cold-sensitive nab2-21 allele was constructed by eliminating the NAB2 sequence encoding amino acids 424–445. This 21 amino acid deletion includes part of the sixth, and all of the seventh, C3H repeat that is required for binding to poly(A) in vitro (Anderson et al., 1993b). When tested for viability at different temperatures, the nab2-21 strain grew similarly to wild-type strains at both 30 and 37°C, but was growth inhibited at 14°C (Figure 3A). Surprisingly, long poly(A) tails (maximum tail length ∼130 nucleotides) were detected at both permissive (30°C) and restrictive (14°C) growth temperatures, demonstrating that hyperadenylation alone was not growth inhibitory (Figure 3B). In contrast, FISH analysis showed accumulation of poly(A)+ RNA in 1 or 2 foci per nucleus only at 14°C (Figure 3C). In agreement with the Nab2p depletion studies, poly(A)+ RNA accumulated in the nucleus and co-localized with Nop1p (Figure 3D). Since the poly(A) tail length and FISH analyses may not be equally sensitive, these observations did not eliminate the possibility that tail lengths increased at 14°C, or that mRNA export was slightly impaired at 30°C, in nab2-21. Nevertheless, our results were consistent with the hypothesis that Nab2p was required for poly(A) tail length regulation, but long poly(A) tails were not growth inhibitory. To examine this proposal further, we identified genes whose overexpression compensated for loss of NAB2 function.
Fig. 3. Constitutive long poly(A) tails and inducible nuclear accumulation of poly(A)+ RNA in nab2-21. (A) Spot growth analysis of NAB2 and nab2-21 strains. Serial 10-fold dilutions of cells were plated on YPD plates. (B) Poly(A) tail analysis of NAB2 and nab2-21 at 30 and 14°C. (C) FISH analysis of poly(A)+ RNA distribution. (D) Poly(A)+ RNA accumulates in the nucleolus. The localization of poly(A)+ RNA (green) in nab2-21 at 14°C is compared with the nucleolar protein Nop1p (red) and DNA (blue).
PAB1 is a high-copy suppressor of nab2 mutant alleles
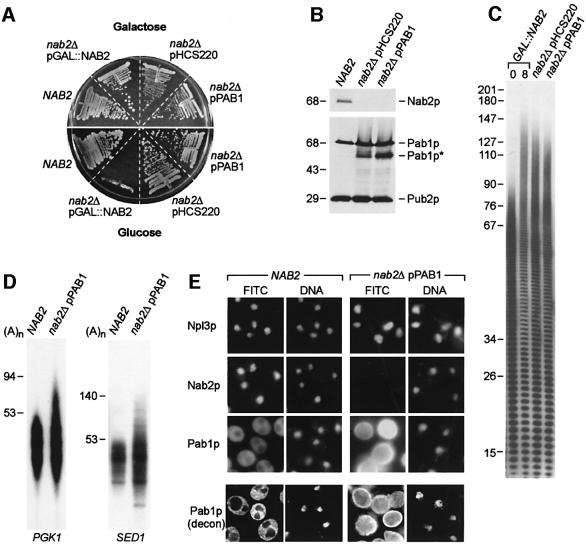
For the high copy suppressor screen, GAL::NAB2 cells (YRH201C) were grown in galactose, transformed with a yeast genomic DNA library cloned into a multicopy plasmid and subsequently transferred to growth on glucose to repress NAB2 transcription. Out of ∼106 transformants, 20 colonies grew in glucose following restreaking, and 19 of these were identified as carrying NAB2 by PCR screening. The remaining strain (nab2Δ pHCS220) did not carry a plasmid-borne NAB2 copy but grew well on glucose (Figure 4A). Sequencing of the pHCS220 plasmid revealed that it contained a 10 kb genomic fragment that included the CHD1, PAB1 and DNF1 genes. All three genes were subcloned individually and tested for suppression. Only the nab2Δ pPAB1 strain grew in glucose, suggesting that Pab1p overexpression compensated for loss of Nab2p (Figure 4A), and this was confirmed by immunoblot analysis (Figure 4B). The intracellular level of the 70 kDa full-length Pab1p increased ∼2-fold, while a 53 kDa Pab1p fragment (Figure 4B, Pab1p*) was not detectable in wild-type NAB2 strains but was present in both nab2Δ pHCS220 and nab2Δ pPAB1 strains. The Pab1p* polypeptide was generated by proteolysis within the C-terminus of Pab1p since it was detectable with monoclonal antibody (mAb) 1G1, which recognizes an N-terminal epitope (our unpublished data). Surprisingly, long poly(A) tails were still detectable in nab2Δ pHCS220 and nab2Δ pPAB1 cells (Figure 4C). To confirm that long poly(A) tails were added to mRNAs in nab2Δ pPAB1 cells, poly(A) tail lengths were determined for several transcripts using the PCR-based poly(A) tail length (PAT) assay (Salles et al., 1999). All of the tested cDNAs that were identified originally in the hyperadenylated cDNA library (see Figure 1E) showed long poly(A) tails in this assay, including PGK1 and SED1 transcripts (Figure 4D). Therefore, mRNAs with long poly(A) tails were present in both nab2-21 and nab2Δ pPAB1 cells under conditions permissive for growth.
Fig. 4. PAB1 is a high-copy suppressor of nab2Δ. (A) Growth (24°C) of wild-type NAB2 and nab2Δ strains carrying pHCS220 or pPAB1. (B) Immunoblot analysis of Nab2p, Pab1p and Pab1p* expression in NAB2, nab2Δ pHCS220 and nab2Δ pPAB1 strains. Equal protein loads are shown by immunoblotting with mAb 2B1 (anti-Pub2p). Sizes are indicated in kilodaltons. (C) Poly(A) tail length analyses of nab2Δ pHCS220 and nab2Δ pPAB1 strains are compared with hyperadenylated RNAs seen following shift of YJA517-1C (GAL::NAB2) cells to glucose for 8 h. (D) Hyperadenylated PGK1 and SED1 mRNAs in nab2Δ pPAB1 cells. Poly(A) tail lengths were determined using LM-PAT. Poly(A) tail lengths are indicated and were determined by comparison with a 100 nucleotide DNA ladder. (E) Pab1p appears in the nucleus when overexpressed in the nab2Δ pPAB1 strain. Panels in the top three rows show localization of Npl3p, Nab2p and Pab1p using conventional immunofluorescence and mAbs 1E4, 3F2 and 1G1, respectively, in NAB2 and nab2Δ pPAB1 cells at 24°C. The panels in the bottom row are deconvolution images (Pab1p, decon) showing Pab1p distribution in wild-type NAB2 versus nab2Δ pPAB1 cells. Pab1p localization in vacuoles is probably due to increased Pab1p turnover in nab2Δ pPAB1.
An important question that emerged from these studies was the mechanism of PAB1 suppression. PAB1 is also a dose-dependent suppressor of rna15-2 and, since Pab1p and Rna15p are CF I subunits, a previous study proposed that elevated nuclear levels of Pab1p compensate directly for loss of Rna15p activity in mRNA 3′-end formation (Amrani et al., 1997). Since it was possible that overexpressed Pab1p also accumulated in the nucleus of nab2Δ cells and compensated for Nab2p loss, we examined the subcellular distribution of Pab1p in nab2Δ pPAB1 cells. In agreement with a previous study (Anderson et al., 1993a), Pab1p was localized predominantly in the cytoplasm in wild-type cells while Npl3p and Nab2p were distributed in the nucleus (Figure 4E). Although the distribution of Npl3p was identical in the nab2Δ pHCS220 and nab2Δ pPAB1 strains that were devoid of Nab2p, overexpressed Pab1p was detectable throughout these cells. Decon volution microscopy demonstrated that Pab1p was not detectable in the nuclei or vacuoles of NAB2 wild-type cells, but was present in both of these compartments in nab2Δ pPAB1 cells (Figure 4E). Thus, PAB1 overexpression in a nab2Δ background led to the appearance of Pab1p in the nucleus, and correlated with resolution of the nab2Δ growth defect. However, Pab1p levels were elevated throughout the cell so it was possible that an increase in the cytoplasmic level of Pab1p was responsible for viability of the nab2Δ pPAB1 strain. To determine if PAB1 suppression required nuclear import of Pab1p, nab2Δ suppression analysis was performed using a modi fied PAB1 allele on a low-copy plasmid that expressed a nuclear-targeted form of Pab1p.
Nuclear targeting of Pab1p suppresses the nab2Δ growth defect
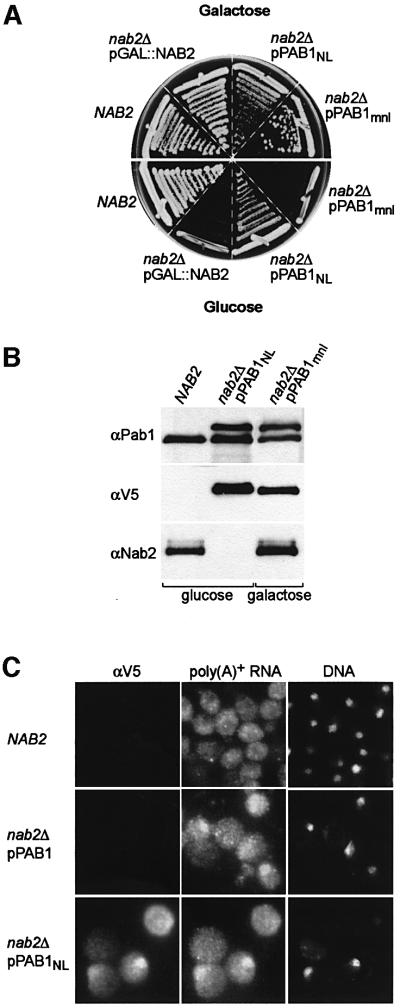
We used plasmid shuffling to replace the multicopy pPAB1 plasmid in the nab2Δ pPAB1 strain with a CEN plasmid (pRS315) carrying a modified PAB1 allele, which encoded Pab1p tagged with the SV40 nuclear localization (NL) sequence (PKKKRKV) at the N-terminus and a V5 epitope at the C-terminus. The nab2Δ pPAB1NL strain was viable at 24°C (Figure 5A), and the Pab1 and Pab1NL proteins were expressed at a similar level in nab2Δ pPAB1NL cells (Figure 5B). Low-copy PAB1 suppression of nab2Δ required nuclear targeting of Pab1p since nab2Δ PAB1mnl, a strain carrying a PAB1 allele with a non-functional (PKTKRKV) SV40 mutant NL (mnl) sequence (nab2Δ pPAB1mnl), was not viable in glucose (Figure 5A) although the corresponding protein, Pab1mnl, was expressed in galactose in the presence of pGAL::NAB2 (Figure 5B). As opposed to wild-type Pab1p, the Pab1NL protein, visualized by cell immunofluorescence using the anti-V5 antibody, accumulated in the nucleus although this protein was also detectable in the cytoplasm (Figure 5C). Significantly, poly(A)+ RNA was detectable in both the nuclear and cytoplasmic compartments in the majority of cells expressing either pPAB1 or pPAB1NL, suggesting that cell viability was due to partial resolution of the nab2Δ mRNA export block by PAB1 overexpression in these strains (compare Figures 5C and 2A). However, nab2Δ pPAB1NL cells with the highest nuclear concentration of Pab1NL protein also showed the most intense nuclear poly(A)+ RNA signals, which suggested that very high levels of Pab1p in the nucleus might impair mRNA export. In contrast to the effects on mRNA export and cell viability, pPAB1NL expression exacerbated the nab2Δ hyperadenylation defect and maximum poly(A) tail lengths increased to ∼10 nucleotides longer in nab2Δ pPAB1NL as compared with nab2Δ pPAB1 cells (data not shown). On the basis of this latter observation, we speculated that Nab2p might be the primary factor that regulates poly(A) tail length in vivo by acting either as a polyadenylation termination, or a poly(A) tail trimming, factor. Loss of termination/trimming activity might delay mRNA entry into the export pool. To look at this possibility, we examined whether Nab2p was directly required for poly(A) tail length control during 3′-end processing in vitro.
Fig. 5. Targeting of Pab1p to the nucleus suppresses the nab2Δ growth defect. (A) Suppression of nab2Δ by low-copy pPAB1NL. Plate growth of NAB2, nab2Δ pGAL::NAB2, nab2Δ pPAB1NL and nab2Δ pPAB1mnl strains on YPGal (galactose) and YPD (glucose) at 24°C is shown. (B) Immunoblot analysis of NAB2, nab2Δ pPAB1NL and nab2Δ pPAB1mnl strains grown on YPD (glucose) and YPGal (galactose). Proteins were detected using anti-V5 (αV5), anti-Pab1p 1G1 (αPab1p) and anti-Nab2p 3F2 (αNab2p) antibodies. (C) SV40 NLS-tagged Pab1p accumulates in the nucleus. The localizations of V5-tagged Pab1p, poly(A)+ RNA and DNA are compared in NAB2, nab2Δ pPAB1 and nab2Δ pPAB1NL strains.
Nab2p stimulates polyadenylation while restricting tail length in vitro
A potential role for Nab2p in mRNA 3′-end formation in vitro was examined using extracts from a pab1Δ strain (YAS394). We used these extracts because preliminary studies using wild-type extracts indicated that Nab2p was not essential for CYC1 precursor RNA poly(A) tail length regulation (our unpublished data). However, the discovery that Pab1p is required for poly(A) tail length control in vitro suggested that the presence of Pab1p in these cell extracts might mask any role for Nab2p in polyadenylation (Amrani et al., 1997; Minvielle-Sebastia et al., 1997). In addition, Kap104p and Nab2p form a complex and Kap104p might bind to free Nab2p in the extract and prohibit its RNA-binding activity (Lee and Aitchison, 1999). In support of this possibility, we determined that Kap104p was present in the cell extracts used for polyadenylation, and the majority of Nab2p in these extracts could be immunodepleted with an anti-Kap104p mAb (data not shown). Therefore, in vitro 3′-end processing assays were performed with extracts from a pab1Δ strain in the absence or presence of recombinant Nab2p.
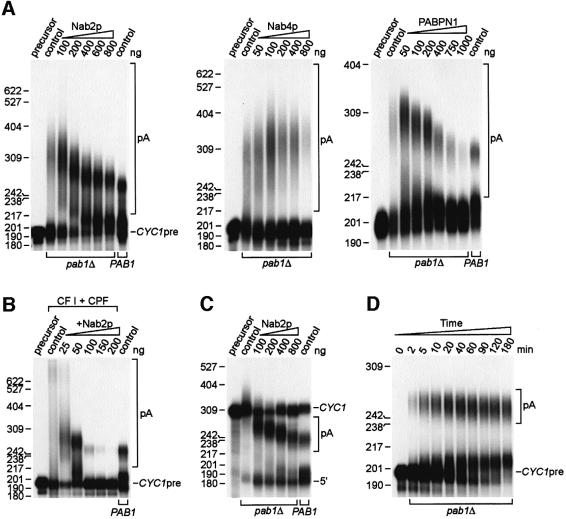
Using pre-cleaved precursor CYC1 RNA (CYC1pre), previous in vitro polyadenylation studies have shown that poly(A) tail lengths are restricted to 50–70 nucleotides using wild-type PAB1 extracts while pab1Δ extracts hyperadenylate CYC1pre (Amrani et al., 1997; Minvielle-Sebastia et al., 1997) (Figure 6A, compare pab1Δ and PAB1 control lanes). Addition of bacterially expressed Pab1p to pab1Δ extracts results in shortening of the poly(A) tail to within the normal 50–70 nucleotide range. A similar effect on polyadenylation was seen when recombinant Nab2p was added to pab1Δ extracts with maximum poly(A) tail length restricted to 60–80 nucleotides following addition of ≥400 ng of Nab2p (Figure 6A). Recombinant Nab2p also stimulated polyadenylation 2- to 3-fold. Since poly(A) tails were ∼10 nucleotides shorter when a PAB1 wild-type extract was used for polyadenylation (Figure 6A, last lane), Pab1p might recruit PAN activity to shorten tails further. The effect of Nab2p on poly(A) tail length control was specific since the addition of another yeast hnRNP, Nab4p, to pab1Δ extracts led to enhanced hyperadenylation. Hyperadenylation was also inhibited in the presence of the bovine nuclear poly(A) tail-binding protein PABPN1. However, polyadenylation was also strongly impaired at ≥400 ng of PABPN1, possibly because PABPN1 bound to the poly(A) tail was unable to interact productively with the yeast 3′-end processing machinery. Hyperadenylation was also observed when the purified cleavage and polyadenylation factors CF I and CPF were used for CYC1pre-RNA polyadenylation in the absence of Pab1p (Amrani et al., 1997; Minvielle-Sebastia et al., 1997). Addition of low levels (25–50 ng) of Nab2p increased CYC1pre polyadenylation while limiting tail length to 60–80 nucleotides, while ≥100 ng of Nab2p inhibited polyadenylation (Figure 6B). This activity did not require pre-cleaved RNA substrates since Nab2p also enhanced polyadenylation while restricting poly(A) tail lengths during a coupled cleavage–polyadenylation reaction with extracts (Figure 6C), as well as in a reconstituted reaction with purified factors (data not shown).
Fig. 6. Nab2p is required for poly(A) tail length control during polyadenylation in vitro in the absence of Pab1p. (A) Recombinant Nab2p stimulates polyadenylation but inhibits hyperadenylation in vitro. Reactions were performed using pre-cleaved CYC1 precursor RNA (CYC1pre) and either YAS394 (pab1Δ) or wild-type (PAB1) cell extracts. Extracts were either used directly (control lanes) or supplemented with 100–800 ng of GST–Nab2p-His6 (left panel), 50–800 ng of GST–Nab4p (middle panel) or 50–1000 ng of purified bovine PABPN1 (right panel). Polyadenylated RNAs are bracketed (pA). The indicated size markers are pBR322 MspI fragments. (B) Hyperadenylation is inhibited by Nab2p during polyadenylation in vitro using purified cleavage and polyadenylation factors. Reactions were performed using CYC1pre, CF I + CPF, 10 ng of GST–Nab4p and 25–200 ng of GST–Nab2p-His6. The last lane is a control reaction using wild-type PAB1 extract. (C) Nab2p controls poly(A) tail length during coupled cleavage–polyadenylation in vitro. Reactions were performed using CYC1 RNA instead of pre-cleaved precursor. The positions of the CYC1 RNA (CYC1), polyadenylated CYC1 (pA) and the 5′ cleavage fragment (5′) are indicated. (D) Time course analysis showing that Nab2p restricts poly(A) tail length to 60–80 nucleotides during polyadenylation and does not promote tail trimming.
To discriminate between potential roles for Nab2p in limiting tail length during polyadenylation versus poly(A) tail trimming following synthesis, we examined the time course of polyadenylation in pab1Δ extracts following addition of 400 ng of GST–Nab2-His6. Poly(A) tails in the range of 60–80 nucleotides appeared as soon as 2 min following the initiation of synthesis and, although the conversion of CYC1pre continued up to 180 min, tail length distribution did not change significantly during this period (Figure 6D). We conclude that Nab2p is the primary factor that controls poly(A) tail length during mRNA 3′-end formation, which is consistent with its predominantly nuclear localization.
Discussion
Nab2p is required for poly(A) tail length control during mRNA 3′-end formation
In this report, we demonstrate that the yeast hnRNP Nab2p is required for mRNA poly(A) tail length control in vivo and in vitro. Evidence that Nab2p plays an essential and direct role in limiting tail length during polyadenylation includes: (i) Nab2p depletion in vivo leads to the appearance of hyperadenylated mRNA; (ii) unlike mutations in mRNA export factors, long poly(A) tails are not the indirect result of a block to mRNA export since hyperadenylated mRNA is present in nab2-21 with a normal subcellular distribution of poly(A)+ RNA; (iii) either PAB1 overexpression, or expression of nuclear-targeted Pab1p, fails to correct the hyperadenylation phenotype of nab2 mutant strains; (iv) Nab2p stimulates polyadenylation while inhibiting hyperadenylation in vitro in the absence of Pab1p; and (v) in contrast to Pab1p, Nab2p does not show poly(A) tail trimming activity in vitro. Why have previous biochemical fractionation studies failed to uncover Nab2p as a factor that regulates polyadenylation in vitro? First, these studies employed whole-cell extracts that contain relatively high Pab1p levels, and we show here that Pab1p masks the polyadenylation function of Nab2p in vitro. Secondly, Nab2p in these extracts is bound to Kap104p and this association probably interferes with Nab2p–RNA interactions (Lee and Aitchison, 1999).
Previous studies have suggested that Pab1p is an important 3′-end processing factor in vivo since it is required for polyadenylation in vitro, co-purifies with CF I and interacts directly with Rna15p, which is an essential factor for polyadenylation (Amrani et al., 1997; Minvielle-Sebastia et al., 1997). Interestingly, PAB1 is also a multicopy suppressor of rna15-2 (Amrani et al., 1997). While NAB2 expression is essential for normal polyadenylation in vivo, our results do not exclude a role for Pab1p in mRNA 3′-end formation or the possibility that Nab2p and Pab1p are both associated with the poly(A) tail in the nucleus. An alternative possibility is that Nab2p binds to A-rich tracts within specific pre-mRNAs and the extent of polyadenylation is regulated by Nab2p interactions with Pab1p bound to the poly(A) tail.
Based on previous studies and the work reported here, we propose that poly(A) tails are synthesized to a default length in the presence of Nab2p, and that Pab1p recruits PAN activity for message-specific trimming of the poly(A) tail. According to this proposal, Nab2p is recruited initially to the mRNA 3′-end, possibly by the growing poly(A) tail. What is the evidence that the poly(A) tail is important for Nab2p function? This hnRNP contains an unusual RNA-binding motif consisting of seven C3H repeats, which is required for poly(A)-binding activity in vitro (Anderson et al., 1993b). Deletion of two of these repeats in the nab2-21 allele results in mRNA hyperadenylation, suggesting that poly(A) binding is compromised in this mutant. In addition, Pab1p masks the hyperadenylation inhibitory function of Nab2p in vitro. Since these two proteins are not structurally related, but both bind to poly(A), it is likely that Pab1p blocks Nab2p activity in vitro by sequestration of the poly(A) tail. This is consistent with our observation that another unrelated poly(A)-binding protein, PABPN1, also inhibits polyadenylation in vitro. The presence of excess Pab1p in cell extracts might limit accessibility to the poly(A) tail and also prevent hyperadenylation by simultaneous recruitment of PAN activity. Following tail binding, we suggest that Nab2p interacts with additional 3′-end processing factor(s), possibly Nab4p, since these two hnRNPs interact in the two-hybrid system (Uetz et al., 2000), resulting in the termination of polyadenylation.
Potential roles for Nab2p, Pab1p and the poly(A) tail in mRNA export
Considerable evidence supports the proposal that mRNA 3′-end processing and nuclear export are tightly coupled in vivo (Brodsky and Silver, 2000; Hilleren et al., 2001; Jensen et al., 2001). When temperature-sensitive dbp5/rat7, gle1/brr3, mex67, mtr2, nup159/rat8p and rip1 strains are shifted to restrictive growth temperatures, mRNA export is blocked and hyperadenylated mRNA accumulates in the nucleus. Since long poly(A) tails are not detectable at permissive growth temperatures in these export mutants, inhibition of mRNA export may lead to the titration of a factor that is required for the termination of polyadenylation (Hilleren and Parker, 2001; Jensen et al., 2001).
Although we have provided evidence that Nab2p is a polyadenylation termination factor, perhaps the most surprising result of our studies is that hyperadenylation can occur in the absence of a detectable mRNA export defect in nab2-21 cells. Of course, polyadenylation control might be impaired, but not completely compromised, in nab2-21 at 30°C, and thus transcripts are released at rates compatible with normal mRNA export. An important caveat is that the poly(A) tail length and poly(A)+ RNA localization assays are probably not equally sensitive so there may be an undetectable export problem in nab2-21 at 30°C. Another possibility is that Nap2p is required independently for the regulation of mRNA polyadenylation and nuclear export. For mRNA export, Nab2p might associate with transcripts during 3′-end formation and interact subsequently with an NPC-associated factor to form a complex that promotes mRNP targeting to the NPC, similar to the function of the Mex67p–Mtr2p complex for transcribed regions of the mRNA (for reviews see Cole, 2000; Cullen, 2000). In support of this idea, Nab2p shuttles between the nucleus and cytoplasm and interacts in vivo with the NPC-associated protein, Gfd1p (Hodge et al., 1999; Strahm et al., 1999; Duncan et al., 2000). An alternative model is that Nab2p association with the nascent transcript antagonizes the activity of the nuclear exosome, which monitors mRNA 3′-end processing and retains incorrectly processed transcripts at the transcription site (Hilleren et al., 2001). It is not clear why poly(A)+ RNA accumulated in a nucleolar region in GAL::NAB2 and nab2-21 cells, but it is interesting to note that in rat7-1 rrp6Δ mutants, in which both mRNA export and nuclear exosome activities are inhibited, HSP104 transcripts also accumulate in a subnuclear region that stains poorly with 4′,6-diamidino-2-phenylindole (DAPI), possibly the nucleolus (Hilleren et al., 2001). According to this model, increased nuclear Pab1p levels restore cell growth in nab2Δ pPAB1 or nab2Δ pPAB1NL cells by partially restoring mRNA export, perhaps by inhibiting nuclear exosome activity. The goal of future studies is to distinguish between these potential roles of Nab2p in nuclear mRNA export.
Materials and methods
Yeast strains and plasmids
Table I lists the yeast strains used in this study. The pGAL::NAB2 plasmid was constructed by subcloning the BglII–SalI fragment from pNAB2-3B6 into pRD53 (BamHI–SalI) (Wilson et al., 1994). The pNAB2-3B6 plasmid contains the NAB2 coding sequence, generated by PCR using primers MSS47 (5′-CCCGGATCCCGGTACAGCGGATAGCGC-3′) and MSS48 (5′-CCCGAATTCGATCATAAGGAAGTGGAAG-3′), inserted into pSP72 (Promega Biotech, Madison, WI) at EcoRI–BamHI sites (Anderson et al., 1993b). To isolate a strain expressing an inducible/repressible NAB2 gene, pGAL::NAB2 was used to transform YJA511 to Ura+ to generate YJA517. YJA517 was sporulated, and tetrads dissected to allow for identification of a strain carrying a NAB2 chromosomal deletion. To study the effect of depleting Nab2p in vivo, YJA517-1C (GAL::NAB2) cells were first grown in YPGal to mid-log phase, and then diluted into YPD and grown at 30°C for 0–30 h.
The nab2-21 allele was created by PCR-mediated deletion of nucleotides 1269–1323 in the NAB2 coding region. The 5′ and 3′ ends of NAB2 were amplified from pNAB2.19 in separate reactions using MSS128 (5′-AGTACTCTGCCTGGAGGGCCAAATAAACAATCAATTC-3′) and a T3 polymerase promoter primer (5′-ATTAACCCTCACTAAAG-3′) or MSS129 (5′-CCTCCAGGCAGAGTACT-3′) and a T7 promoter primer (5′-TAATACGACTCACTATAGGGAGA-3′). The two PCR products were purified and combined in a reaction with T7 and T3 primers. The product corresponding to the correct size was gel purified and cloned into pRS314 (EcoRI–SstI) to generate pnab2-21.
The nab2Δ pPAB1NL strain was constructed by amplification of the PAB1 open reading frame using MSS1261 (5′-GCGCTCGAGCCC GGGCCAAAGAAGAAGCGTAAGGTTGATATTACTGATAAGAC AGCTGAACAATTG-3′) and MSS1259 (5′-GCGGGGTGACCAGCTTGCTCAGTTTGTTGTTCTTGC-3′) followed by insertion into XhoI–BstEII-cut pYES2/SPB4 (Invitrogen, Carlsbad, CA) to generate an NLS-PAB1-V5 gene fusion. The mutant NLS-PAB1-V5 was created using primers MSS1262 (5′-GCGCTCGAGCCCGGGATGGCTCC AAAGACTAAGCGTAAGGTTGATATTACTGATAAGACAGCTGA ACAATTG-3′) and MSS1259. The PAB1 5′- and 3′-untranslated regions (UTRs) were amplified using MSS1287 (5′-GCGGTCGACGAGGTCATACTGTATGAAGCCACAAAG-3′)/MSS1263 (5′-GCGCCCGGGTTTATTTTTATTGGTTTTTTAGTTTTTTTTGG-3′) and MSS1268 (5′-GCGTCTAGATGCTCTATGTAATCACCTACTTCCC-3′)/MSS1288 (5′-GCGGAGCTCCTGTTACGGTTGCTTTCCCTTGCTC-3′) primer pairs, respectively. The 5′-UTR was inserted into SalI–SmaI, and the 3′-UTR into XbaI–SacI, of pRS315 to generate pPAB1.5. Wild-type and mutant NLS-PAB1-V5 fragments were excised with SmaI and XbaI and inserted into SmaI–XbaI-digested pPAB1.5 to generate the pPAB1.6 (wt NLS-PAB1-V5) and pPAB1.7 (mutant NLS-PAB1-V5) plasmids, which were transformed into YRH201C to create YKN105 and YKN106 strains, respectively. Transformed cells initially were plated on SG-Leu and then on YPD to test for nab2Δ suppression. For YKN105, cells were streaked to 5-fluoro-orotic acid plates to select for the loss of pGAL::NAB2.
Filter binding assay
Dissociation constants were determined using a standard filter binding assay and (rA)25 (Dharmacon Research, Lafayette, CO). Binding reactions were performed at 30°C for 30 min in 20 mM Tris–HCl pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.5 U/µl Superase-In (Ambion, Austin, TX) using 5 pM 32P end-labeled A25 RNA and increasing amounts of purified GST–Nab2-His6 or GST-His6 fusion proteins. The resulting protein–RNA complexes were filtered as described (Wong and Lohman, 1993), and bound/unbound RNA fractions quantified using a Storm phosphorImager (Molecular Dynamics, Sunnyvale, CA). Each point on the curve represents the mean of three independent experiments.
Poly(A) tail length determination, isolation of hyperadenylated RNAs and cDNA library construction
Total RNA was prepared from whole cells, and poly(A) tail lengths were determined as described (Minvielle-Sebastia et al., 1998). To isolate hyperpolyadenylated RNAs, total RNA was incubated with 1.0 g of poly(U)–Sephadex (Gibco-BRL, Rockville, MD) in 20 ml of BB70 buffer (10 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.5% SDS, 0.5 M LiCl, 70% formamide) at room temperature for 60 min. The poly(U)–Sephadex was washed three times with 10 ml of BB70 and three times with 10 ml of BB (BB minus formamide), and RNAs were eluted by 10 washes with 4 ml of EB (10 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.05% SDS) for 5 min each at room temperature. Two 20 ml fractions were collected, and the eluted RNA was converted into a cDNA library (Stratagene, La Jolla, CA).
Ligation-mediated RT–PCR (LM-RT–PCR) was performed as described (Salles et al., 1999). Reaction products were fractionated on 8% denaturing polyacrylamide gels and visualized by autoradiography.
In situ hybridization, cellular immunofluorescence and immunoblotting
The subcellular distribution of poly(A)+ RNA was examined by in situ hybridization using a published procedure (Amberg et al., 1992). Cell immunofluoresence was performed as described (Wilson et al., 1994) using the following antibodies (dilutions): anti-Pab1p 1G1 (1:20 000), anti-Nab2p 3F2 (1:500), anti-Nop1p A66 (1:5000), anti-digoxigenin (1:50) (Roche Molecular Biochemicals, Indianapolis, IN), anti-V5 (1:500) (Invitrogen, Carlsbad, CA), Alexa Fluor 488 goat anti-mouse IgG1 and Alexa Fluor 555 goat anti-mouse IgG2a (Molecular Probes, Eugene, OR). Images were obtained using either a Zeiss Axioplan 2 microscope equipped with a ×100 fluorescence/differential contrast objective or a deconvolution microscope (Applied Precision, Issaquah, WA).
Immunoblotting was performed as described (Anderson et al., 1993b) using the following antibodies and dilutions: anti-Nab2p 3F2 (1:500), anti-Pub1p 2B1 (1:5000), anti-Pab1p 1G1 (1:5000), anti-V5 (1:500) and horseradish peroxidase-conjugated sheep anti-mouse secondary (1:5000) (Amersham Pharmacia Biotech, Piscataway, NJ).
High-copy suppressor screen
YRH201C cells, grown in YPGal, were transformed with a YEp13 genomic library (ATCC, Manassas, VA) as described (Anderson et al., 1993b), and then plated onto SD-Ura-Leu at 24°C. To determine the transformation efficiency, 1% of the transformation was plated onto SGal-Ura-Leu. This procedure yielded ∼106 Leu+ transformants, of which 20 were able to grow on SD-Ura-Leu. These colonies were screened by whole-cell PCR to eliminate library plasmids carrying NAB2. The pHSN220 plasmid was rescued and sequenced using primers MSS737 (5′-AGTCCTGCTCGCTTCGC-3′) and MSS738 (5′-ATGTCGGCGATATAGGC-3′). PAB1 was PCR amplified from L4717 genomic DNA using primers MSS744 (5′-GGGTCTAGAAGGTCATACTGTATGAAGCC-3′) and MSS745 (5′-GGGGGATCCCTGTTACGGTTGCTTTCCC-3′), and the resulting fragment was cloned into YEp13 (XbaI–BamHI) to generate pPAB1. YRH201C cells transformed with pPAB1 were plated onto SGal-Ura-Leu. Leu+ transformants were replated to SD-Ura-Leu to confirm that PAB1 was a nab2Δ high-copy suppressor.
In vitro polyadenylation and recombinant protein preparation
Standard 3′-end processing assays were performed for 60 min at 30°C essentially as described (Minvielle-Sebastia et al., 1997) with 2 µl (∼20 µg of protein) of YAS394 (pab1Δ) or BMA64 (PAB1) cell extracts. For reconstitution assays with purified factors, reactions contained 0.5 µl of partially purified CF I (Mono Q fraction from CF II purification; Minvielle-Sebastia et al., 1998) and 0.5 µl of CPF (affinity purified from strain YSD10, which expresses a TAP-tagged Fip1p fusion protein; unpublished data). Where specified, the reactions were supplemented with either 25–800 ng of GST–Nab2p-His6, 50–800 ng of GST–Nab4p (Minvielle-Sebastia et al., 1998) or 50–1000 ng of purified bovine PABPN1 (kind gift of Elmar Wahle, University of Halle-Wittenberg, Halle Saale, Germany). For the polyadenylation time course, a 12-fold reaction mixture containing YAS394 extract was supplemented with 9.6 µg of GST–Nab2p-His6 and pre-incubated at 30°C for 15 min in the absence of the substrate RNA. Polyadenylation was initiated by addition of labeled transcript, aliquots of 20 µl were withdrawn at the times indicated, and reactions were stopped and analyzed on 40 cm long 6% polyacrylamide–8.3 M urea gels.
For preparation of full-length recombinant Nab2p, a His6 tag was cloned in-frame at the 3′ end of the NAB2 open reading frame. The NAB2 gene was amplified from L4717 genomic DNA using primers MSS1131 (5′-GCGGAATTCATGTCTCAAGAACAGTACACAGAAAAC-3′) and MSS1132 (5′-GCGGTCGACTCAGCCGCTGCTGTGATGATGATGATGATGGCTGCTGCCGTTCATTTCCGTATCTTGTTCTTG-3′), and the PCR product cloned into pGEX-4T-1 (Amersham Pharmacia Biotech) between the EcoRI and SalI sites. The resulting GST–Nab2p-His6 protein was expressed in BL21-CodonPlus (DE3) cells (Stratagene) and purified using His-Bind (Novagen, Madison, WI) and glutathione–Sepharose (Amersham Pharmacia Biotech) chromatography.
Acknowledgments
Acknowledgements
We thank J.Aris, A.de Bruyn Kops, S.Butler, T.-H.Chang, E.Hurt, C.Guthrie, A.Sachs, K.Sträßer and E.Wahle for strains, reagents and protocols. We also thank M.Paddy for help with deconvolution microscopy, and J.Aris, S.Butler and A.Lewin for comments on the manuscript. This work was supported by NIH grant GM46272 (M.S.S.), a pre-doctoral trainee award to R.E.H. (NIH T32-AI-07110), and by the CNRS, the Ministère de la Recherche Scientifique, La Fondation pour la Recherche Médicale/Fondation BNP-Paribas and La Ligue Nationale contre le Cancer (L.M.-S). S.D. was the recipient of a post-doctoral fellowship from La Ligue Nationale contre le Cancer.
References
- AbouElela S. and Ares,M.,Jr (1998) Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J., 17, 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonina E., Stauber,R. and Pavlakis,G.N. (1998) The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem., 273, 13015–13021. [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein,A.L. and Cole,C.N. (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6, 1173–1189. [DOI] [PubMed] [Google Scholar]
- Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.T., Paddy,M.R. and Swanson,M.S. (1993a) PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 6102–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.T., Wilson,S.M., Datar,K.V. and Swanson,M.S. (1993b) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol., 13, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A., Guillemet,E., Cullin,C. and LaCroute,F. (1997) Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast, 13, 353–356. [DOI] [PubMed] [Google Scholar]
- Brodsky A.S. and Silver,P.A. (2000) Pre-mRNA processing factors are required for nuclear export. RNA, 6, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E. and Sachs,A.B. (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol., 18, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C., Cech,T.R. and Zaug,A.J. (1997) Polyadenylation of telomerase RNA in budding yeast. RNA, 3, 1337–1351. [PMC free article] [PubMed] [Google Scholar]
- Cole C.N. (2000) mRNA export: the long and winding road. Nature Cell Biol., 2, E55–E58. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. (2000) Connections between the processing and nuclear export of mRNA: evidence for an export license? Proc. Natl Acad. Sci. USA, 97, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Umen,J.G. and Guthrie,C. (2000) A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol., 10, 687–696. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ellmeier,W. and Birnstiel,M.L. (1991) Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J., 10, 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach M., Burd,C.G. and Dreyfuss,G. (1994) The mRNA poly(A)-binding protein: localization, abundance and RNA-binding specificity. Exp. Cell Res., 211, 400–407. [DOI] [PubMed] [Google Scholar]
- Green D.M., Marfatia,K.A., Crafton,E.B., Zhang,X., Cheng,X. and Corbett,A.H. (2002) Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem., 277, 7752–7760. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (2001) Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA, 7, 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. McCarthy,T., Rosbash,M., Parker,R. and Jensen,T.H. (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature, 413, 538–542. [DOI] [PubMed] [Google Scholar]
- Hodge C.A., Colot,H.V., Stafford,P. and Cole,C.N. (1999) Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J., 18, 5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B., Brockenbrough,S., Wu,P. and Aris,J.P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Carmichael,G.G. (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol., 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Sträßer,K., Segref,A., Bailer,S., Schlaich,N., Presutti,C., Tollervey,D. and Jansen,R. (2000) Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem., 275, 8361–8368. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Patricio,K., McCarthy,T. and Rosbash,M. (2001) A block to mRNA nuclear export in S.cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell, 7, 887–898. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi,M., Chen,S. and Tartakoff,A.M. (1994) Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell, 5, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R.W., Kühn,U., Aragón,M., Bornikova,L., Wahle,E. and Bear,D.G. (2000) The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol., 297, 569–583. [DOI] [PubMed] [Google Scholar]
- Lee D.C. and Aitchison,J.D. (1999) Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem., 274, 29031–29037. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L. and Keller W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J., Wiederkehr,T., Strahm,Y. and Keller,W. (1997) The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA, 94, 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B.J., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B. and Davis,R.W. (1989) The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell, 58, 857–867. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. and Deardorff,J.A. (1992) Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell, 70, 961–973. [DOI] [PubMed] [Google Scholar]
- Salles F.J., Richards,W.G. and Strickland,S. (1999) Assaying the polyadenylation state of mRNAs. Methods, 17, 38–45. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Pante,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog,B., Zenklusen,D., Rychner,E., Kantor,J., Rosbash,M. and Stutz,F. (1999) The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J., 18, 5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. and Hurt,E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K., Bassler,J. and Hurt,E. (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol., 150, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.S., Weaver,P.L., Liu,Y., Hitomi,M., Tartakoff,A.M. and Chang,T.H. (1998) Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J., 17, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P. et ak. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- Wahle E. (1995) Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem., 270, 2800–2808. [DOI] [PubMed] [Google Scholar]
- Wilson S.M., Datar,K.V., Paddy,M.R., Swedlow,J.R. and Swanson,M.S. (1994) Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol., 127, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I. and Lohman,T.M. (1993) A double-filter method for nitrocellulose-filter binding: Application to protein–nucleic acid interactions. Proc. Natl Acad. Sci. USA, 90, 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]