Abstract
The human single-stranded DNA-binding protein, replication protein A (RPA) binds DNA in at least two different modes: initial [8–10 nucleotides (nt)] and stable (∼30 nt). Switching from 8 to 30 nt mode is associated with a large conformational change. Here we report the 2.8 Å structure of the RPA trimerization core comprising the C-terminal DNA-binding domain of subunit RPA70 (DBD-C), the central DNA-binding domain of subunit RPA32 (DBD-D) and the entire RPA14 subunit. All three domains are built around a central oligonucleotide/oligosaccharide binding (OB)-fold and flanked by a helix at the C-terminus. Trimerization is mediated by three C-terminal helices arranged in parallel. The OB-fold of DBD-C possesses unique structural features; embedded zinc ribbon and helix–turn–helix motifs. Using time-resolved proteolysis with trypsin, we demonstrate that the trimerization core does not contribute to the binding with substrates of 10 nt, but interacts with oligonucleotides of 24 nt. Taken together, our data indicate that switching from 8–10 to 30 nt mode is mediated by DNA binding with the trimerization core.
Keywords: DNA-binding/OB-fold/replication protein A/single-stranded DNA/subunit interaction
Introduction
The eukaryotic single-stranded (ss) DNA-binding protein, replication protein A (RPA), is a central player in replication, recombination and repair. This heterotrimeric protein binds to ssDNA generated during these processes. Through specific protein–protein interactions, RPA is also thought to contribute to the assembly of the recombination, repair and replication machines. The human homolog of RPA is a heterotrimer with subunits of ∼70, 32 and 14 kDa (RPA70, RPA32 and RPA14, respectively) (Wold, 1997; Iftode et al., 1999).
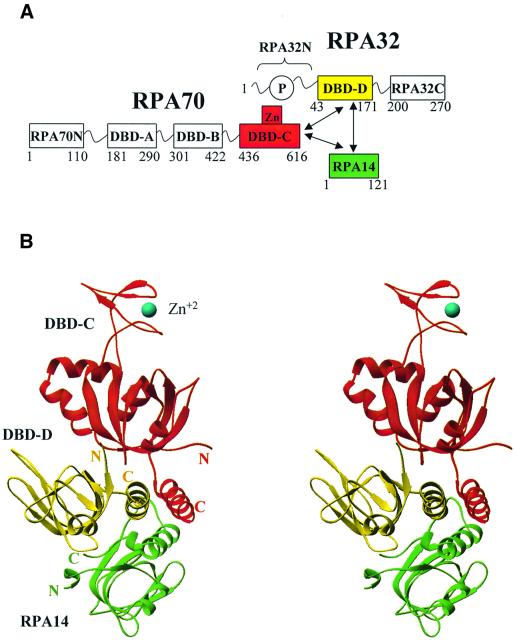
RPA has a modular structure (Figure 1A). The RPA70 subunit has four domains; the N-terminal protein–protein interaction domain (RPA70N; amino acids 1–110; Jacobs et al., 1999) and three ssDNA-binding domains (DBDs) arranged in tandem, DBD-A (amino acids 181–290), DBD-B (amino acids 301–422) and DBD-C (amino acids 436–616) (Philipova et al., 1996; Bochkarev et al., 1997; Brill and Bastin-Shanower, 1998; Bochkareva et al., 2000). DBD-C contains a conserved Cys4-type zinc-binding motif (Wold, 1997); the bound zinc modulates DNA-binding (Bochkareva et al., 2000). This region has been suggested to be involved in binding to damaged DNA, but to date, the role of this motif is not clear. The RPA32 subunit consists of three domains. The central ssDNA-binding domain, DBD-D (amino acids 43–171; Bochkareva et al., 1998) is flanked at the N-terminus by a small, unstructured domain, which carries all known human RPA phosphorylation sites (RPA32N; 1–40; Gomes et al., 1996; Zernik-Kobak et al., 1997), and at the C-terminus by a protein–protein interaction module (RPA32C; 200–270; Mer et al., 2000). The entire small subunit, RPA14, is folded in one structural domain. The C-terminal part of this subunit is important for trimerization. Other functions of RPA14 are currently not clear. Trimerization is mediated by three domains: DBD-C (in RPA70), DBD-D (in RPA32) and RPA14, which together form the trimerization core (Bochkareva et al., 2000).
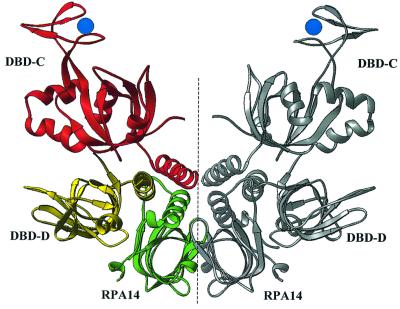
Fig. 1. Structural organization of the DBD-C/-D/RPA14 trimer. (A) Schematic showing the RPA domain structure. Domains are presented as boxes, their borders are indicated. Zn, zinc ribbon; P, unstructured, phosphorylated N-terminus of subunit RPA32. Domains comprising the trimerization core are colored in red, yellow and green (for DBD-C, DBD-D and RPA14, respectively). The regions of subunit interaction are indicated by arrows. (B) Stereo ribbon presentation of DBD-C, DBD-D and RPA14 viewed along the axis of the three-helical bundle (colors as in A). The coordinated zinc atom is shown in blue as a sphere. The N- and C-termini of each domain are indicated. The drawing was generated using RIBBONS (Carson, 1997).
RPA binding to ssDNA occurs via a multistep pathway and is associated with a significant conformational change as demonstrated by biochemical methods, electron microscopy and crystallography (reviewed by Iftode et al., 1999). First, globular RPA binds to 8–10 nucleotides (nt) in an unstable manner (Blackwell and Borowiec, 1994). Current evidence associates this binding with DBD-A and DBD-B, the two major DNA-binding domains that harbor most of the binding activity of the full trimer (Pfuetzner et al., 1997; Walther et al., 1999). Cross-linking experiments indicate that DBD-D does not interact with ssDNA in this mode (Mass et al., 1998; Lavrik et al., 1999; Bastin-Shanower and Brill, 2001). The second step is associated with a significant conformational change to a mode with an occluded binding size of ∼30 nt per trimer (Kim et al., 1992; Blackwell et al., 1996; Gomes et al., 1996). The latter mode is thought to be facilitated by two minor DNA-binding domains, DBD-C and DBD-D (Philipova et al., 1996; Bochkareva et al., 1998, 2000; Brill and Bastin-Shanower, 1998; Lavrik et al., 1998). In this mode, DBD-A, -B, -C and -D, together directly contact between 23 and 27 nt (Bastin-Shanower and Brill, 2001). Several more nucleotides bridge the space between trimers bringing the occluded size up to ∼30 nt. An intermediate 13–14 nt (Lavrik et al., 1999) or 12–22 nt (Bastin-Shanower and Brill, 2001) binding mode has recently been characterized. In this mode, RPA binds to ssDNA with higher affinity, as compared with the 8–10 nt mode, and does not contact ssDNA with DBD-D, as compared with the 30 nt mode.
To understand the mechanism of trimerization and obtain insight into the high-order ssDNA-binding modes, we have resolved the structure of the RPA trimerization core containing DBD-C, DBD-D and RPA14. DBD-C shares a significant similarity with other DBDs, and also carries some unique structural features such as a zinc ribbon and helix–turn–helix motif. Proteolytic analysis indicates that DBD-C and DBD-D do not contribute to the initial 8 nt binding mode, but stabilize the high-order binding modes. Analysis of intermolecular contacts also suggests a model for the intact RPA trimer.
Results and discussion
General description of the structure
The RPA trimerization core comprising DBD-C (RPA70436–616), DBD-D (RPA3243–171) and intact RPA14 (DBD-C/-D/RPA14) was characterized and its crystals grown as described previously (Bochkareva et al., 2000). Experimental data were collected from a selenomethionine (Se-Met)-containing crystal and the structure was resolved using a combination of the molecular replacement (MR) and multiwave anomalous dispersion (MAD) methods. The structure has been refined to a working R-factor of 23.6% (Rfree 28.3%) using all data with F >2σ between 20 and 2.8 Å. The unit cell contains two structurally similar independent trimers, I and II, related by a non-crystallographic two-fold symmetry. Additional details are provided in the Materials and methods section and Table I.
Table I. Data collection and refinement statistics.
| Data processinga (Space group P3121, unit cell: a = b = 88.5, c = 341.1Å) | |||||
|---|---|---|---|---|---|
| Se-Met MAD |
High resolution |
||||
| Data set | Peak | Inflection | High energy | Low energy | Low energy |
| Energy (KeV) | 12.662 | 12.660 | 12.95 | 12.30 | 12.30 |
| Anomalous or isomorphous | ano | ano | ano | ano | iso |
| Resolution (Å) | 2.8 | 2.8 | 2.8 | 2.8 | 2.7 |
| No. of reflections measured | 349 842 | 350 610 | 353 398 | 342 411 | 233 991 |
| No. of independent reflections | 73 451 | 73 434 | 73 628 | 72 533 | 43 573 |
| R-factor (%) (overall/outer shell) | 5.9/79.5 | 5.8/78.3 | 5.8/88.1 | 5.2/74.8 | 6.2/53.2 |
| I/σ (overall/outer shell) | 18.7/1.4 | 19.2/1.4 | 17.3/1.3 | 20.5/1.3 | 23.1/2.7 |
| Completeness (%) |
99.7/98.1 |
99.7/98.1 |
99.9/99.7 |
98.4/85.3 |
99.2/99.7 |
| Experimental MAD phase calculationb (20–2.8Å) | |||||
| No. of anomalous scatterers (Se/Zn) | 30/2 | ||||
| FOM (34240 acentric reflections) | 0.60 | ||||
| FOM (5088 centric reflections) |
0.53 |
|
|
|
|
| Structure refinementc (20–2.8Å) | |||||
| Sigma cutoff | (F >2σ )d | (F >0 ) | |||
| No. of reflections (total/test set) | 36 244/3681 | 38 232/3868 | |||
| Completeness (total/test set) (%) | 92.3/9.4 | 97.4/9.9 | |||
| R/Rfree (%) | 23.6/28.3 | 24.5/28.9 | |||
| No. of non-hydrogen atoms |
6571 (two Zn; no water) |
|
|
|
|
| Model r.m.s.d. from ideality | |||||
| Bonds/angles (Å/degrees) | 0.019/2.2 | ||||
The parameters definition and statistics from aDenzo/Scalepack, bSHARP and cCNS.
dSigma cutoff applied during the refinement.
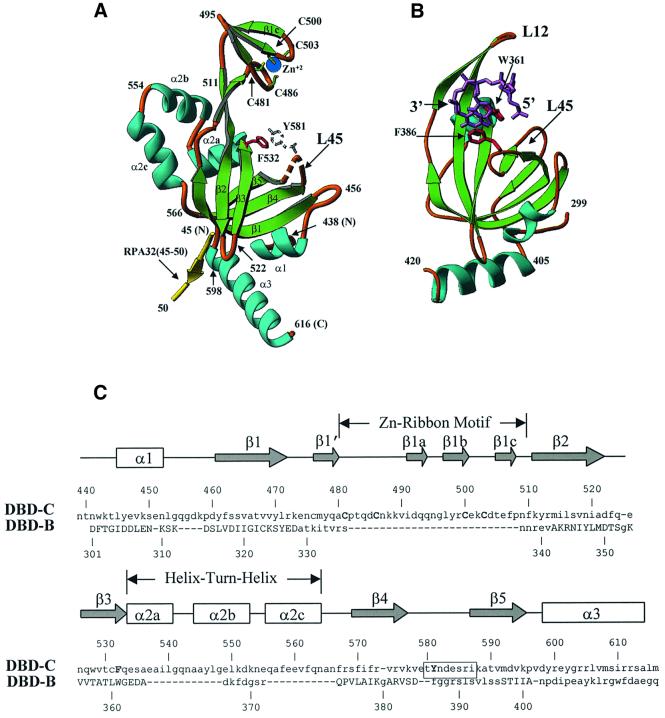
The structure of the trimerization core is shown in Figure 1B. All three domains, DBD-C, DBD-D and RPA14, are structurally similar; that is, each domain is built around a central oligonucleotide/oligosaccharide binding (OB)-fold (Murzin, 1993) and flanked by an α-helix at the C-terminus (amino acids 598–616 in DBD-C; Figure 2A). This similarity extends to the DBD-B domain, the structure of which was resolved in a complex with ssDNA (Figure 2B). Although the C-terminal helix is conserved in DBD-B, -C, -D and RPA14, its orientation with respect to the OB-fold differs significantly. Except for a missing C-terminal α-helix, RPA70N and DBD-A also share the same fold (Bochkarev et al., 1997; Daughdrill et al., 2001).
Fig. 2. Similarities and differences between DBD-C and DBD-B. (A) A detailed view of DBD-C colored in green (β-strands), sky-blue (α-helices) and gold (loops); a fragment of RPA32 (amino acids 45–50) is shown in yellow. Amino acid numbers are shown in black. A disordered loop is indicated by the dashed line. Conserved aromatic amino acids (the position of Y581 is putative) and the four cysteines that coordinate zinc ion are shown in red, colourless, green-and-yellow and blue, respectively. (B) The structure of DBD-B and the mode of its interaction with ssDNA. DBD-B is colored as DBD-C in (A) and has an orientation approximately the same as DBD-C. ssDNA is colored magenta (Protein Data Bank entry 1jmc). (C) Structure-based sequence alignment of DBD-C and DBD-B. The β-strands are indicated by arrows and the α-helices by boxes. Secondary structure elements are referred to as in agreement with the nomenclature of the OB-fold. The positions of the zinc ribbon and helix–turn–helix are outlined. The sequences of DBD-C and DBD-B were aligned using program O based on the three-dimensional structure; the residues that are capitalized in DBD-B indicate those that were included in the alignment (r.m.s.d. is 1.82 Å for 69 Cα atoms). Functionally important amino acids of DBD-C, four cysteines and two conserved aromatics, are capitalized. The boxed residues are those that are disordered in the structure.
The trimerization interface is mediated by three C-terminal α-helices arranged in parallel. This hydrophobic interaction is stabilized by Tyr599, Tyr602, Leu606, Val607, Ile610 and Ala614 of RPA70; Met152, Phe155, Ile159, Leu160, Ile163 and Met167 of RPA32; and Leu98, Ala102, Ile105, Phe109 and Phe112 of RPA14. Three OB-folds are packed in tandem and run about half-a-turn around the three-helical bundle (clockwise in order RPA14, DBD-D, DBD-C). Outside the three-helical bundle, DBD-C has only one contact with the extreme N-terminus of DBD-D and no contacts with RPA14.
The structures of RPA14 and DBD-D, taken individually and as a dimeric substructure, are virtually identical to that in the crystals of the RPA14/RPA32 dimer core that has been described previously (Bochkarev et al., 1999) and will not be discussed here in detail.
Structural features of DBD-C
The structure of DBD-C (Figure 2A) resembles that of the other DNA-binding domains in RPA in that it is built around an OB-fold. A structure-based sequence alignment of DBD-B and DBD-C was generated using the program O. Best-fit superposition of 69 Cα atoms gives an atomic root mean square deviation (r.m.s.d.) of 1.82 Å (Figure 2C); that between DBD-A and DBD-C was 1.4 Å with 63 Cα’s. For comparison, the r.m.s.d. between DBD-A and DBD-B was 1.5 Å with 74 Cα included.
Several lines of evidence strongly suggest that the DNA-binding site in DBD-C (and therefore, in all four DBDs) is conserved. First is the structural conservation of the common protein motif, the OB-fold. Secondly, there is functional conservation of key aromatic amino acids, Phe532 and Tyr581, analogs of which have been found in all four DBDs (Phe238 and Phe269 in DBD-A, Trp361 and Phe386 in DBD-B, and Trp107 and Phe135 in DBD-D). These have been shown to stack with DNA bases in DBD-A and DBD-B (Bochkarev et al., 1997, 1999). The functional importance of these aromatic amino acids has been confirmed by mutational analysis both in vivo and in vitro (Philipova et al., 1996; Brill and Bastin-Shanower, 1998; Bastin-Shanower and Brill, 2001). Thirdly, in the absence of DNA, the loop connecting β-strands 4 and 5 (L45 loop) in DBD-A and DBD-B is flexible. Upon DNA binding, this loop clamps down on DNA and stabilizes the interaction via the stacking of conserved aromatics with DNA bases (Figure 2B) (Bochkarev et al., 1997; Bochkareva et al., 2001). In DBD-C, the structure of which was resolved without DNA, the L45 loop (amino acids 580–586) is disordered (i.e. flexible).
DBD-C contains several significant features, distinct from the other DBDs, the most striking being a zinc ribbon motif (amino acids 480–510) embedded in the OB-fold between β-strands 1 and 2 (Figure 2A and C). This motif is formed by three antiparallel β-strands (β1a, β1b and β1c). The zinc ion is tetrahedrally coordinated by four cysteines, 481, 486, 500 and 503, provided by two extended loops. The zinc ribbon is found in several transcription factors (Qian et al., 1993), ribosomal proteins (Hard et al., 2000) and the NAD-dependent deacetylase SIR2 (Min et al., 2001). Although zinc ribbon motifs have been reported in several structures, the functional role of this motif is poorly understood. Here, the position and orientation of the zinc ribbon with respect to the rest of the protein suggests its possible function. The central part of the zinc motif does not seem to contact ssDNA directly. However, a part of the loop between β1′ and β1a, in particular, amino acids Asn487, Lys488 and Lys489, which flank Cys486, are located in close proximity to the putative ssDNA-binding cleft and therefore might interact with the phosphate backbone of bound ssDNA (discussed below). Thus any destabilization of the zinc ribbon structure could have a negative impact on ssDNA-binding. This could explain the known influence of this motif on DNA-binding (Lin et al., 1998; Lao et al., 1999; Bochkareva et al., 2000). Interestingly, another ssDNA-protein, gp32 from bacteriophage T4, also contains a zinc-binding motif in a similar position, but in gp32, this is a zinc finger rather than a zinc ribbon (Shamoo et al., 1995). This finding suggests a conservation of function between the two motifs. In gp32, the zinc finger is important for cooperative binding (Nadler et al., 1990). Thus, the zinc ribbon in RPA may contribute to cooperativity (as known for gp32), DNA-binding (as predicted for zinc ribbon motifs of some transcription factors; Awrey et al., 1998) or regulation of ssDNA-binding by redox effects (Park et al., 1999; You et al., 2000).
Another interesting feature of DBD-C is a three-helical cap at the back of the domain (amino acids 533–566), which carries characteristic hallmarks of a homeodomain (helix–turn–helix) motif (Branden and Tooze, 1991). The structural similarity suggests possible involvement of this domain in binding to double-stranded (ds) DNA. Indeed, interaction with dsDNA has been mapped to or around this area (Lao et al., 1999). Unlike DBD-C, the similar position in other DBDs is occupied either by a single helix, (DBD-A and B) or by a flexible loop (DBD-D).
Outside the three-helix trimerization interface, DBD-C has only one contact with DBD-D. Ile46 at the extreme N-terminus of DBD-D is only 3 Å away from Asp522 at the tip of the loop between β-strands 2 and 3 (L23) of DBD-C (Figure 2A). However, this contact may be of functional importance. This structure-based notion is supported by experimental data. Deletion of 50 amino acids from the extreme C-terminus of RPA70 removes the C-terminal helical interface of DBD-C, but not the L23 loop. This deletion completely abolishes the interaction of RPA70 with RPA14, but not with RPA32. In contrast, deletion of 110 amino acids, which removes the L23 loop, also abolishes an interaction between RPA70 and RPA32 (Kim et al., 1996). Although RPA32N, the domain containing all known phosphorylation sites of human RPA, is not present within the structure of the trimeric core, the direction of the backbone at amino acid 45 orients RPA32N inside of, or in close proximity to, the putative ssDNA-binding cleft of DBD-C (between Phe532 and Tyr581). This cleft is ∼20 Å away from Ile46. Strikingly, the DNA-binding cleft of DBD-D is also ∼20 Å from Ile46 (Bochkarev et al., 1999). Thus, structural data suggests that RPA32N might interact with DBD-C and/or DBD-D to regulate the DNA binding of the core.
Model for DBD-C bound to ssDNA
The structure has been determined of DBD-AB bound to oligo(dC)8, but as yet there is no understanding of the cooperative involvement of DBD-C in binding longer lengths of DNA. As discussed above, strong evidence exists that the DNA-binding site is conserved in all four DBDs. In the DBD-AB–ssDNA complex structure, each DBD binds three nucleotides (dC1–3 and dC6–8, respectively) in a very similar manner (Bochkarev et al., 1997). The binding is mediated by three types of protein–DNA interaction: (i) stacking between aromatic amino acids and bases, (ii) electrostatic interaction between basic amino acids and the phosphate backbone, and (iii) hydrogen bonding (most of these are non-sequence-specific) between side chains and bases. Only stacking is conserved between the two DBDs. In contrast, the residues that hydrogen bond with DNA do not occupy topologically equivalent positions, and differ in their number.
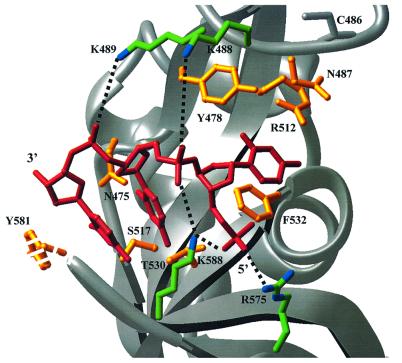
To identify putative residues that may interact with ssDNA, DBD-B and DBD-C were aligned by using homologous structural elements (Figure 2C), and then DBD-C was oriented on the ssDNA fragment of DBD-B (PDB entry 1jmc). Not surprisingly, the ssDNA fragment of DBD-B fitted DBD-C snugly (Figure 3); the DNA bases contacted the inner portion of DBD-C, and the sugar-phosphate backbone was exposed on the outside. Phe532 and Tyr581 are in position to stack with flanking DNA bases. Basic amino acids Lys488, Lys489, Arg575 and Lys588 are oriented to contact the phosphate backbone of DNA. Side chains of Asn475, Tyr478, Asn487, Ser517, Arg513 and Thr530 are (or can be) in close proximity to the putative positions of DNA bases. Interestingly, most (except for Arg513) of these residues predicted to hydrogen bond with bases are capable of serving both as donors and acceptors of hydrogen bonds. This observation suggests how DBD-C can accommodate different DNA bases in a non-sequence-specific manner.
Fig. 3. DBD-C modeled on a ssDNA molecule derived from the DBD-AB/ssDNA co-crystal structure. The DBD-C was aligned with DBD-B as shown in Figure 2C and then docked onto the 3 nt DBD-B binding site (shown in red). Side chains in DBD-C (the position for Y581 is putative) that are orientated to interact with the sugar-phosphate backbone are colored in green and blue (for carbon and nitrogen atoms, respectively), and those in position to hydrogen bond, or stack with, DNA bases are yellow. Putative hydrogen bonds with the phosphate groups are shown as dashed lines. The position of conserved C486 is shown as a reference.
Homology analysis suggests that the mechanism of binding may be conserved from yeast to human. Of 12 residues predicted to contact ssDNA in human DBD-C, 10 are identical in its yeast (Saccharomyces cerevisiae) homolog, and two more residues are functionally similar (Lys588 is substituted with Arg, and Ser517 with Thr). For comparison, the percentage of identity and similarity between the RPA70 subunits from human and yeast was 32.5% and 44.5%, respectively (Iftode et al., 1999).
The trimer core does not contribute to the binding with 10 nt, but interacts with substrates of 24 nt
The structural basis of the 8–10 nt binding mode, and how it resolves to the high-order binding modes, remain speculative. Current consensus associates the 8–10 nt mode with DBD-A and -B. To large extent, this model is based on the structural evidence that a tandem of DBD-A and -B interact with 8 nt of ssDNA (Bochkarev et al., 1997). As demonstrated above, DBD-A, -B and -C share similar structure, and most likely, similar binding mechanism. This similarity raises the possibility that the 8–10 nt binding mode can also be mediated by DBD-B and -C. Indeed, this alternative model is consistent with the reported cross-linking experiments. Initial binding is mediated by the RPA70 subunit, and RPA32 does not contribute in this mode (Mass et al., 1998, 2001; Kolpashchikov et al., 1999; Bastin-Shanower and Brill, 2001).
The intact RPA trimer and some of its subcomplexes demonstrate a different pattern of digestion with trypsin in the presence and absence of DNA. In the absence of DNA, RPA70 is quickly degraded. However, the presence of oligo(dT)30 in the proteolytic buffer protects a fragment comprising amino acids ∼169–616 (i.e. ∼DBD-ABC) (Gomes et al., 1996). Similar effect has been observed in a smaller fragment of RPA70 containing only DBD-A and -B (Gomes et al., 1996; E.Bochkareva, unpublished data). In contrast, the trimerization core, and DBD-C in particular, is resistant to trypsin in the absence of DNA. However, DBD-C is susceptible to tryptic digestion in the presence of metal chelating agents, such as EDTA (Bochkareva et al., 2000).
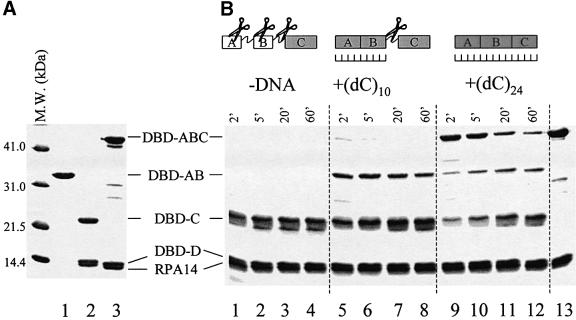
The RPA digestion pattern may not only depend on the presence or absence of DNA, but also on the size of DNA added in the digestion buffer. If the 8–10 nt mode is mediated by DBD-A and B, then a limited proteolysis of DBD-ABC in the presence of the 10 nt oligonucleotide may dissect two fragments: a complex containing DBD-AB (Figure 4A, lane 1) bound to 10 nt, and DBD-C (Figure 4A, lane 2), which is resistant to trypsin. In contrast, if binding to 10 nt is mediated by DBD-BC, we could expect to detect a complex of DBD-BC bound to 10 nt and free (if not degraded) DBD-A, i.e. a fragment under 120–130 amino acids (∼14 kDa or less).
Fig. 4. Time-resolved proteolysis of DBD-ABC/-D/RPA14 with trypsin in the presence of different ssDNA substrates. (A) The major DNA-binding domain of RPA70 (DBD-AB; RPA70 fragment 181–432) (lane 1), trimerization core (DBD-C/-D/RPA14) (lane 2) and trimeric subcomplex that contains all four DBDs (DBD-ABC/-D/RPA14) (lane 3; repeated in B, lane 13) were expressed in bacteria, purified, resolved by gel electrophoresis and visualized by Coomassie Blue staining. Molecular mass markers are on the left. (B) Limited digestion of DBD-ABC/-D/RPA14 with trypsin in the absence of DNA (lanes 1–4), the presence of oligo(dC)10 (lanes 5–8), and the presence of oligo(dC)24 (lanes 9–12). Purified DBD-ABC/-D/RPA14 (10 µg) was pre-incubated with a 3× molar excess of the indicated oligonucleotides (or none) on ice for 30 min and then digested with 10 ng of trypsin. Aliquots were removed at the designated times, resolved by gel electrophoresis and visualized by Coomassie Blue staining.
To evaluate this hypothesis, we generated a trimeric complex containing RPA70 fragment 181–616, RPA32 fragment 43–171 and intact RPA14 (DBD-ABC/-D/RPA14; Figure 4A, lane 3 and B, lane 13), and compared the trypsin digestion pattern of this subcomplex under three conditions: (i) in the absence of DNA, (ii) in the presence of oligo(dC)10, and (iii) in the presence of oligo(dC)24. In the absence of DNA, in a course of a few minutes, DBD-ABC was truncated to a small proteolytically resistant fragment (Figure 4B, lanes 1–4), which has previously been characterized to comprise amino acids 432–616 (i.e. ∼DBD-C; Bochkareva et al., 2000). In agreement with the previous report, this fragment was further degraded by trypsin, when EDTA was added to the digestion buffer (10 mM EDTA tested, data not shown).
One new band appeared on the gel in the presence of oligo(dC)10 (Figure 4B, lanes 5–8), as compared with the proteolysis in the absence of DNA. The presence of EDTA in the digestion buffer did not noticeably affect susceptibility of this fragment to trypsin, i.e. this fragment does not include DBD-C (data not shown). This band co-migrated on the gel with RPA70181–432. We concluded that an interaction with (dC)10 stabilizes DBD-AB, but does not stabilize the linker between DBD-B and C; it remains susceptible to proteolysis. Finally, in agreement with previous reports, an interaction with oligo(dC)24 structurally stabilized the entire DBD-ABC fragment making it resistant to trypsin (Figure 4B, lanes 9–12).
Several lines of evidence now indicate that the 8–10 nt binding mode is mediated by DBD-A and -B. First, our data exclude DBD-B and -C mediating binding with 8–10 nt. Secondly, in the full trimer, DBD-D does not cross-link with 12 and 17 nt substrates (Bastin-Shanower and Brill, 2001). Although we cannot exclude that under the experimental conditions (3-fold molar excess of ssDNA) DBD-C might interact with DNA, the binding affinity of the trimerization core is 10- to 100-fold less than that of DBD-AB (Lao et al., 1999; Bochkareva et al., 2000). This excludes the pair of DBD-C and -D as a potential mediator for this binding mode. Thirdly, DBD-AB directly contacts 8 nt in the crystals and a 10 nt substrate selectively stabilizes a conformation of this fragment in solution in the presence of DBD-C and -D.
Analysis of intermolecular contacts suggests a model for the intact RPA trimer
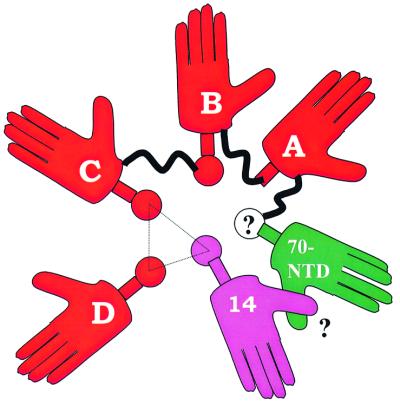
In the crystals, two independent molecules of the trimer core are packed in such a way that three helices of one molecule complement the three-helix interface of the other, forming a six-helix packing arrangement; six OB-folds run around this multi-helical bundle (Figure 5). This packing arrangement suggested that other parts of RPA, not present in the core structure, might contribute helices that interact with the trimerization interface. This would be in accord with electron microscope (EM) images, which show that individual structural modules of apo RPA have a globular arrangement with ∼6-fold symmetry (Alani et al., 1992). The intact RPA trimer contains six OB-folds; RPA70N, four DNA binding domains (A, B, C and D) and RPA14, structures of which are now available. One obvious candidate to contribute a helix to the pool is DBD-B. It is reasonable to postulate that DBD-B occupies the place clockwise from DBD-C and provides its helix to the pool (Figure 6). DBD-A does not seem to provide one, since neither in the presence nor in the absence of ssDNA does this domain have a helix at the C-terminus (Bochkareva et al., 2001). The C-terminal part of RPA70N contains a ‘transient’ helix comprising amino acids 105–110 (Jacobs et al., 1999) that might be stabilized upon binding (Uesugi et al., 1997; Uesugi and Verdine, 1999; Mer et al., 2000). We hypothesize that RPA70N deposits the induced, C-terminal helix to the helical pool. To be consistent with the six-fold internal symmetry of RPA and with the multi-helical bundle model, RPA70N must be positioned clockwise from DBD-A, between DBD-A and RPA14. Thus, RPA14 might also contribute to stabilization of RPA70N, thus providing a structural role for RPA14.
Fig. 5. The six-helix bundle maintains the crystallographic dimer of DBD-C/-D/RPA14 trimers (dimer-of-trimers). A ribbon representation of the dimer-of-trimers, with one trimer colored as in Figure 1A and the other in silver. The crystallographic dimer-of-trimer is viewed along an axis perpendicular to the non-crystallographic axis of symmetry (dashed line).
Fig. 6. Suggested model of the apo RPA trimer. Cartoon representing RPA as a hexamer of OB-folds centered around a multi-helical bundle. Helices are represented by circles, and the six OB-folds by palms, which are labeled as 14, 70-NTD (N-terminal domain), A, B, C and D (for RPA14, RPA70N, DBD-A, -B, -C and -D, respectively). The trimerization core is outlined with a triangle. Putative elements, an interaction between RPA70N and RPA14, and the transient helix of RPA70N, are shown with question mark. See text for more details.
Implication for the multistep ssDNA-binding mechanism
A clear picture now emerges as to how the four DBDs cooperatively bind ssDNA. We propose that the globular conformation of apo RPA is stabilized by a multi-helical pool, with five of the six OB-folds (except for DBD-A) contributing helices to this pool together with a helix induced in RPA70N.
It has been demonstrated experimentally that the initial 8–10 nt binding mode is mediated by DBD-A and -B. An interaction with oligo(dC)8 aligns DBD-A and -B in tandem in the crystals, and perhaps it does so in the full RPA trimer. The functional role for this initial binding could be three-fold: (i) to anchor RPA to ssDNA, (ii) to properly orient the trimer with respect to a defined polarity of DNA, and perhaps, (iii) to recruit other proteins of replication, recombination or repair systems.
In the next step, which involves a significant conformational transition, RPA switches to a higher-order binding mode; it elongates on ssDNA in a 5′ to 3′ direction to involve DBD-C and DBD-D (i.e. the trimerization core) (de Laat et al., 1998; Iftode and Borowiec, 2000; Bastin-Shanower and Brill, 2001). This interaction has been hypothesized to position DBD-C and then DBD-D in tandem after DBD-B, (sequential model of binding) (Bochkareva et al., 2001). Our new data are in agreement with the sequential model, in that DBD-C could reorientate and contact the 3′ protruding end of ssDNA in tandem after DBD-B. Upon binding, the linker that connects DBD-B and DBD-C becomes resistant to proteolysis. DBD-A, -B and -C (without DBD-D) contact between 13–14 nt (Kolpashchikov et al., 1999) and 22 nt (Bastin-Shanower and Brill, 2001). It is not known whether this 13–14 nt (intermediate) binding mode is a stable and functionally important one or represents a transitional state on the way from the 8 to 30 nt mode.
The 30 nt mode is a cooperative effect mediated by all four DBDs. This notion is supported by a number of experimental facts. First, an interaction of the trimerization core with oligo(dC)24 stabilizes a conformation of the linker between DBD-B and -C. Secondly, substrates of 23 nt and longer cross-link with DBD-D. Thirdly, mutation of conserved aromatic amino acids in DBD-C and -D compromise binding of the intact RPA trimer to these substrates in vitro, and is lethal for yeast (Philipova et al., 1996; Bastin-Shanower and Brill, 2001). Our new structural and proteolytic data are consistent with DBD-A, -B, -C and -D contacting DNA sequentially. However, the trimerization core does not seem to provide enough room for DBD-D to align with DBD-ABC. Instead, upon exit from DBD-C, ssDNA may wrap around the trimerization core to reach the DBD-D. This may explain the extra nucleotides (23–27 nt) required to contact all four DBDs, as compared with the 18–20 nt predicted from the sequential model. Analysis of high magnification EM data supports this hypothesis. RPA/ssDNA filaments appear to be more irregular and kinked as compared with the smoothly contoured gp32/ssDNA and DBD-AB/ssDNA filaments (Treuner et al., 1996; Eckerich et al., 2001).
Materials and methods
Proteins
The RPA trimerization core (RPA70436–616/3243–171/14; also DBD-C/-D/RPA14) was characterized, expressed and crystallized as described previously (Bochkareva et al., 2000). RPA70181–616/3243–171/14 (DBD-ABC/-D/RPA14) was cloned, expressed and purified using similar protocol.
Structure solution
Four-wavelength 2.8 Å MAD experiment and high resolution data (2.7 Å) were collected from a single Se-Met-modified 3-core crystal on the 19ID beamline at the Structural Biology Center, Argonne National Laboratory. Data were integrated and scaled with Denzo/Scalepack (Otwinoski and Minor, 1997; Table I). The MR program AmoRe (Navaza, 1994) found positions for two independent RPA14/RPA3243–171 dimers (Bochkarev et al., 1999). Using MR phases, a 4 Å anomalous difference map was calculated in PHASES (Furey and Swaminathan, 1997) and revealed coordinates of most of the 30 Se and two zinc atoms. The 2.8Å MAD experimental map was generated using SHARP (de La Fortelle and Bricogne, 1997). In the experimental map, continuous density was observed for both crystallographically independent trimers, except for amino acids 112–116 in both RPA32, and amino acids 580–586 in both RPA70 subunits. The model was built with O (Jones et al., 1991) and refined against 2.8 Å resolution data using the CNS (Brünger et al., 1998) program. The final model contained 6571 non-hydrogen atoms (822 residues, two zinc ions, and no ordered water), had a working R-factor of 23.6% and a Rfree value of 28.3% for 36 244 reflections with F >2σ between 20.0 and 2.8 Å. The crystals contained an unusually high (71%) content of solvent. Seventy-eight percent of amino acids were in most favorable areas with no outliers. Data collection and refinement statistics is summarized in Table I. The atomic coordinates have been deposited at the Protein Data Bank (ID code 1L1O).
Proteolysis
Purified DBD-ABC/-D/RPA14 (10 µg) was treated with 10 ng of trypsin in 25 µl solution containing 20 mM HEPES pH 7.5, 150 mM NaCl and 5 mM dithiothreitol (DTT) and 10 µM ZnCl2. Where indicated, the protein was preincubated with a 3-fold molar excess of oligo(dC)10 or oligo(dC)24 (both 5′-phosphorylated) for 30 min on ice. Fractions were resolved by SDS gel electrophoresis and visualized by Coomassie Blue staining.
Acknowledgments
Acknowledgements
We thank Sanjay Bidichandani and Gillian Air for critical reading of the manuscript, members of the Structural Biology Center (SBC) 19-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory (ANL) for support. This work was supported by NIH grant GM61192–01 (to A.B.) and CIHR grant MOP13939 (to S.P.L.M.). Use of the ANL SBC beamlines at APS was supported by the US Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-ENG-38.
References
- Alani E., Thresher,R., Griffith,J.D. and Kolodner,R.D. (1992) Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol., 227, 54–71. [DOI] [PubMed] [Google Scholar]
- Awrey D.E. et al. (1998) Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J. Biol. Chem., 273, 22595–22605. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower S.A. and Brill,S.J. (2001) Functional analysis of the four DNA binding domains of replication protein A: the role of RPA2 in ssDNA binding. J. Biol. Chem., 276, 36446–36453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell L.J. and Borowiec,J.A. (1994) Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol., 14, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell L.J., Borowiec,J.A. and Masrangelo,I.A. (1996) Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol., 16, 4798–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- Bochkarev A., Bochkareva,E., Frappier,L. and Edwards,A.M. (1999) The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J., 18, 4498–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E., Frappier,L., Edwards,A.M. and Bochkarev,A. (1998) The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem., 273, 3932–3936. [DOI] [PubMed] [Google Scholar]
- Bochkareva E., Korolev,S. and Bochkarev,A. (2000) The role for zinc in replication protein A. J. Biol. Chem., 275, 27332–27338. [DOI] [PubMed] [Google Scholar]
- Bochkareva E., Belegu,V., Korolev,S. and Bochkarev,A. (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J., 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C. and Tooze,J. (1991) Introduction to Protein Structure. Garland Publishing, Inc., NY.
- Brill S.J. and Bastin-Shanower,S. (1998) Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol., 18, 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. In Carter,C.W.Jr and Sweet,R.M. (eds), Methods in Enzymology: Macromolecular Crystallography, Part B. Vol. 277. Academic Press, Orlando, FL, pp. 493–505.
- Daughdrill G.W., Ackerman,J., Isern,N.G., Botuyan,M.V., Arrowsmith,C., Wold,M.S. and Lowry,D.F. (2001) The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res., 29, 3270–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Sugasawa,K., Weterings,E., Jaspers,N.G. and Hoeijmakers,J.H. (1998) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev., 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In Carter,C.W.Jr and Sweet,R.M. (eds), Methods in Enzymology: Macromolecular Crystallography, Part A. Vol. 276. Academic Press, Orlando, FL, pp. 472–494. [DOI] [PubMed]
- Eckerich C., Fackelmayer,F.O. and Knippers,R. (2001) Zinc affects the conformation of nucleoprotein filaments formed by replication protein A (RPA) and long natural DNA molecules. Biochim. Biophys. Acta, 1538, 67–75. [DOI] [PubMed] [Google Scholar]
- Furey W. and Swaminathan,S. (1997) PHASES-95: a program package for the processing and analysis of diffraction data from macromolecules. In Carter,C.W.Jr and Sweet,R.M. (eds), Methods in Enzymology: Macromolecular Crystallography, Part B. Vol. 277. Academic Press, Orlando, FL, pp. 590–620. [DOI] [PubMed]
- Gomes X.V., Henricksen,L.A. and Wold,M.S. (1996) Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry, 35, 5586–5595. [DOI] [PubMed] [Google Scholar]
- Hard T., Rak,A., Allard,P., Kloo,L. and Garber,M. (2000) The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold. J. Mol. Biol., 296, 169–180. [DOI] [PubMed] [Google Scholar]
- Iftode C. and Borowiec,J.A. (2000) 5′ → 3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry, 39, 11970–11981. [DOI] [PubMed] [Google Scholar]
- Iftode C., Daniely,Y., and Borowiec,J.A. (1999) Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol., 34, 141–180. [DOI] [PubMed] [Google Scholar]
- Jacobs D.M., Lipton,A.S., Isern,N.G., Daughdrill,G.W., Lowry,D.F., Gomes,X. and Wold,M.S. (1999) Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR, 14, 321–331. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim C., Snyder,R.O. and Wold,M.S. (1992) Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol., 12, 3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.K., Stigger,E. and Lee,S.H. (1996) Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J. Biol. Chem., 271, 15124–15129. [DOI] [PubMed] [Google Scholar]
- Kolpashchikov D.M., Weisshart,K., Nasheuer,H.P., Khodyreva,S.N., Fanning,E., Favre,A. and Lavrik,O.I. (1999) Interaction of the p70 subunit of RPA with a DNA template directs p32 to the 3′-end of nascent DNA. FEBS Lett., 450, 131–134. [DOI] [PubMed] [Google Scholar]
- Lao Y., Lee,C.G. and Wold,M.S. (1999) Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry, 38, 3974–3984. [DOI] [PubMed] [Google Scholar]
- Lavrik O.I., Kolpashchikov,D.M., Nasheuer,H.P., Weisshart,K. and Favre,A. (1998) Alternative conformations of human replication protein A are detected by crosslinks with primers carrying a photoreactive group at the 3′-end. FEBS Lett., 441, 186–190. [DOI] [PubMed] [Google Scholar]
- Lavrik O.I., Kolpashchikov,D.M., Weisshart,K., Nasheuer,H.P., Khodyreva,S.N. and Favre,A. (1999) RPA subunit arrangement near the 3′-end of the primer is modulated by the length of the template strand and cooperative protein interactions. Nucleic Acids Res., 27, 4235–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.L., Shivji,M.K., Chen,C., Kolodner,R., Wood,R.D. and Dutta,A. (1998) The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem., 273, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Mass G., Nethanel,T. and Kaufmann,G. (1998) The middle subunit of replication protein A contacts growing RNA–DNA primers in replicating simian virus 40 chromosomes. Mol. Cell. Biol., 18, 6399–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass G., Nethanel,T., Lavrik,O.I., Wold,M.S. and Kaufmann,G. (2001) Replication protein A modulates its interface with the primed DNA template during RNA–DNA primer elongation in replicating SV40 chromosomes. Nucleic Acids Res., 29, 3892–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G., Bochkarev,A., Gupta,R., Bochkareva,E., Frappier,L., Ingles, C.J., Edwards,A.M. and Chazin,W.J. (2000) Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell, 103, 449–456. [DOI] [PubMed] [Google Scholar]
- Min J., Landry,J., Sternglanz,R. and Xu,R.M. (2001) Crystal structure of a SIR2 homolog–NAD complex. Cell, 105, 269–279. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler S.G., Roberts,W.J., Shamoo,Y. and Williams,K.R. (1990) A novel function for zinc(II) in a nucleic acid-binding protein. Contribution of zinc(II) toward the cooperativity of bacteriophage T4 gene 32 protein binding. J. Biol. Chem., 265, 10389–10394. [PubMed] [Google Scholar]
- Navaza J. (1994) An automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Otwinoski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. In Carter,C.W.Jr and Sweet,R.M. (eds), Methods in Enzymology: Macromolecular Crystallography, Part A. Vol. 276. Academic Press, Orlando, FL, pp. 307–326. [DOI] [PubMed]
- Park J.S., Wang,M., Park,S.J. and Lee,S.H. (1999) Zinc finger of replication protein A, a non-DNA binding element, regulates its DNA binding activity through redox. J. Biol. Chem., 274, 29075–29080. [DOI] [PubMed] [Google Scholar]
- Pfuetzner R.A., Bochkarev,A., Frappier,L. and Edwards,A.M. (1997) Replication protein A. Characterization and crystallization of the DNA binding domain. J. Biol. Chem., 272, 430–434. [DOI] [PubMed] [Google Scholar]
- Philipova D., Mullen,J.R., Maniar,H.S., Lu,J., Gu,C. and Brill,S.J. (1996) A hierarchy of SSB protomers in replication protein A. Genes Dev., 10, 2222–2233. [DOI] [PubMed] [Google Scholar]
- Qian X., Jeon,C., Yoon,H., Agarwal,K. and Weiss,M.A. (1993) Structure of a new nucleic-acid-binding motif in eukaryotic transcriptional elongation factor TFIIS [published erratum appears in Nature (1995) 376, 279]. Nature, 365, 277–279. [DOI] [PubMed] [Google Scholar]
- Shamoo Y., Friedman,A.M., Parsons,M.R., Konigsberg,W.H. and Steitz,T.A. (1995) Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA [published erratum appears in Nature (1995) 376, 616]. Nature, 376, 362–366. [DOI] [PubMed] [Google Scholar]
- Treuner K., Ramsperger,U. and Knippers,R. (1996) Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol., 259, 104–112. [DOI] [PubMed] [Google Scholar]
- Uesugi M. and Verdine,G.L. (1999) The α-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl Acad. Sci. USA, 96, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M., Nyanguile,O., Lu,H., Levine,A.J. and Verdine,G.L. (1997) Induced α helix in the VP16 activation domain upon binding to a human TAF. Science, 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- Walther A.P., Gomes,X.V., Lao,Y., Lee,C.G. and Wold,M.S. (1999) Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry, 38, 3963–3973. [DOI] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- You J.S., Wang,M. and Lee,S.H. (2000) Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry, 39, 12953–12958. [DOI] [PubMed] [Google Scholar]
- Zernik-Kobak M., Vasunia,K., Connelly,M., Anderson,C.W. and Dixon,K. (1997) Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem., 272, 23896–23904. [DOI] [PubMed] [Google Scholar]