Abstract
Small protein B (SmpB) is an essential component of the highly conserved tmRNA–SmpB system that has the dual function of releasing stalled ribosomes from damaged messenger RNAs and targeting incompletely synthesized protein fragments for degradation. Nuclear magnetic resonance (NMR) analysis of SmpB from Aquifex aeolicus revealed an antiparallel β-barrel structure, with three helices packed outside the core of the barrel. While the overall structure of SmpB appears to be unique, the structure does contain an embedded oligonucleotide binding fold; in this respect SmpB has similarity to several other RNA-binding proteins that are known to be associated with translation, including IF1, ribosomal protein S17 and the N-terminal domain of aspartyl tRNA synthetase. Conserved amino acids on the protein surface that are most likely to directly interact with the tmRNA were identified. The presence of widely separated clusters of conserved amino acids suggests that SmpB could function either by stabilizing two distal regions of the tmRNA, or by facilitating an interaction between the tmRNA and another component of the translational apparatus.
Keywords: NMR/protein structure/SmpB/SsrA/tmRNA
Introduction
Small protein B (SmpB) is a component of a system used by prokaryotes for releasing stalled ribosomes from damaged messenger RNAs and targeting incompletely synthesized protein fragments for degradation (Karzai et al., 1999, 2000; Wower et al., 2001). Another com ponent of this system is tmRNA (also known as SsrA RNA or 10Sa RNA), to which the SmpB binds with high affinity. The tmRNA has the remarkable ability to function as both a transfer RNA and a messenger RNA, since it can be charged by alanine tRNA synthetase (Komine et al., 1994; Ushida et al., 1994) and it contains within its primary sequence the nucleotides that code for (typically) 10 amino acids that are added to the C-terminus of the incompletely synthesized proteins (Tu et al., 1995). This C-terminal peptide tag is believed to target the protein fragments for degradation (Keiler et al., 1996; Gottesman et al., 1998). The tmRNA–SmpB system thus serves two important biological functions: first, it releases ribosomes that are stalled on damaged mRNAs that lack an in-frame stop codon; and secondly, it provides a general quality control system that promotes the degradation of incomplete protein fragments that otherwise might have inappropriate cellular activities due to improper folding or lack of appropriate control. SmpB has been shown to be required for the function of the tmRNA (Karzai et al., 1999; Wiegert and Schumann, 2001; Barends et al., 2001), and has no known functions that are independent of its role within the tmRNA–SmpB system for peptide tagging and ribosome rescue.
The tmRNA contains ∼360 nucleotides, and its secondary structure has been analyzed extensively (Williams and Bartel, 1998; Knudsen et al., 2001). A tRNA-like domain is the site at which charging with alanine occurs, and has been proposed to be the site of SmpB binding (Karzai et al., 1999; Barends et al., 2002). The nucleotides that confer the mRNA function are located within another region of the tmRNA, and the molecule is also believed to contain four pseudoknot structures (Felden et al., 1996; Williams and Bartel, 1998; Knudsen et al., 2001). In addition to binding SmpB, tmRNA has been shown to form a ternary complex with elongation factor Tu (EF-Tu) and GTP (Rudinger-Thirion et al., 1999; Barends et al., 2000, 2001; Zvereva et al., 2001), and has also been shown to interact with ribosomal protein S1 (Wower et al., 2000) and alanine tRNA synthetase (Komine et al., 1994; Ushida et al., 1994). It has recently been proposed that the tmRNA and SmpB are parts of a larger ribonucleoprotein complex containing the additional components phosphoribosyl pyrophosphate synthase, RNase R and a protein of unknown function encoded by the yfbG gene (Karzai and Sauer, 2001). Previous studies have shown that tmRNA-defective strains of Escherichia coli have subtle phenotypes such as temperature-sensitive growth, reduced motility, inability to support growth of λimmP22 hybrid phage, induction of Alp protease activity and enhanced activity of several repressor proteins (Kirby et al., 1994; Komine et al., 1994; Retallack et al., 1994; Retallack and Friedman, 1995).
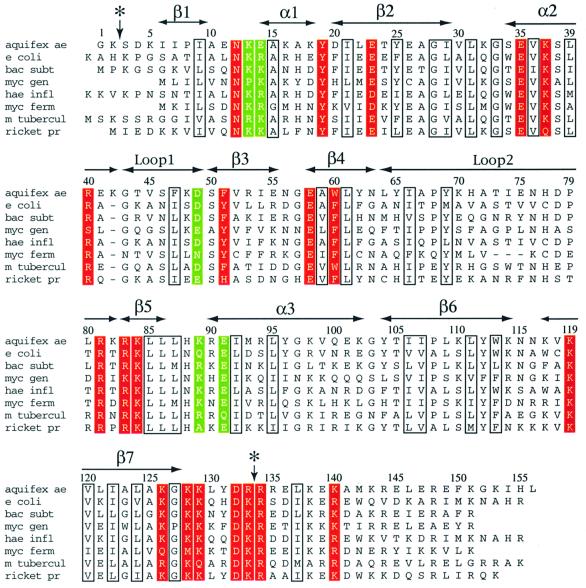
Like the tmRNA, the SmpB protein is well conserved among prokaryotic species, and a gene encoding an SmpB protein with similar mass and sequence has been identified in all prokaryotic genomes that have been sequenced (Keiler et al., 2000). The appearance of SmpB in the organism with the smallest number of genes, Mycoplasma genitalium, provides additional evidence suggesting its importance. The degree of amino acid conservation in SmpB is quite high (Figure 1), ensuring that the proteins from the various prokaryotes are of similar three-dimensional structure. The subject of the present work is the ∼18 kDa SmpB from the thermophilic prokaryote Aquifex aeolicus, which was selected in the hope that a thermophilic version of the protein would be most suitable for structural analysis.
Fig. 1. Sequence alignment for SmpB. Amino acid sequences of SmpB from eight species of prokaryotes are shown: A.aeolicus, E.coli, Bacillus subtilis, M.genitalium, Haemophilus influenzae, Mycoplasma fermentans, Mycobacterium tuberculosis and Rickettsia prowazekii. The positions of conserved amino acids and elements of secondary structure are shown. Well conserved hydrophobic, glycine and proline residues are boxed; these are the residues most likely to be essential for structural purposes. Residues that are accessible on the protein surface, and are similar or identical in >90% of the SmpB sequences, are shaded in red. These residues are most likely to be involved in interactions with the tmRNA or other components of the translational apparatus. Other surface residues that are relatively well conserved (in 80–90% of the sequences) are highlighted in green. It should be noted that a significantly larger set of sequences than the eight shown was used in determining which residues are the most conserved among diverse species. A vertical arrow and asterisk indicate the sites near the N-terminus and C-terminus of the protein that are sensitive to cleavage by trypsin.
A search among structures in the Protein Data Bank (PDB) reveals that SmpB has no significant primary sequence homology to any protein of known structure, a result that is becoming less common as the number of solved protein structures has greatly increased in the last few years. A BLAST search (Altschul et al., 1997) of the non-redundant protein sequence data available at the National Center for Biotechnology Information (NCBI) web site shows that SmpB is not significantly homologous to any proteins other than SmpB in different species.
The present work describes the results of nuclear magnetic resonance (NMR) experiments used to characterize the structure of SmpB from A.aeolicus. The availability of this structural model will be essential in ultimately determining the mechanisms by which tmRNA, SmpB, EF-Tu and other components work in concert to rescue stalled ribosomes from damaged messenger RNAs and tag incompletely synthesized proteins for degradation.
Results
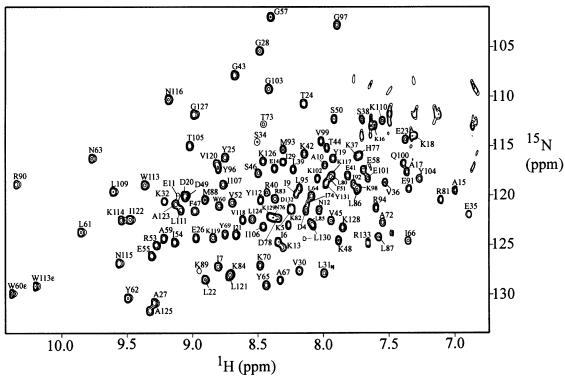
The purified A.aeolicus SmpB protein was found to be quite soluble and stable, and generally well suited for structural characterization by NMR methods (Figure 2). Preliminary two-dimensional NMR data indicated that the great majority of the resonances were well dispersed, as is typical for a folded protein. A fraction of the resonances, however, had chemical shifts that were overlapping and near the values typical of random coil, with relatively narrow line widths, as is typical of flexible or disordered structure. This observation led us to perform controlled proteolysis experiments, which were successful in identifying a trypsin-resistant core fragment of SmpB. Limited proteolysis with trypsin resulted in a fragment consisting of residues 1–133 of the full-length 156-residue SmpB, as confirmed by N-terminal sequencing and mass spectrometry. A fragment consisting of residues 3–133 could also be obtained by trypsin digestion; this fragment and the 1–133 fragment were separable using cation exchange chromatography. The NMR spectra of the trypsin-resistant fragments were found to be nearly identical to those of the full-length SmpB, with the only major difference being that the very sharp resonances with random-coil-like chemical shifts were absent. These results provide evidence that SmpB contains a stable core of well defined structure, with the last 23 residues of the full-length protein most likely being flexible or disordered. Triple-resonance and multi-dimensional NMR methods were used to characterize the trypsin-cleaved fragment consisting of residues 1–133 and the full-length protein.
Fig. 2. 15N-1H correlated HSQC spectrum of SmpB. The spectrum was obtained from a sample of A.aeolicus SmpB, residues 1–133, at 30°C in 10 mM phosphate plus 50 mM NaCl at pH 6. Resonance assignments for 122 of the most clearly resolved cross peaks are labeled.
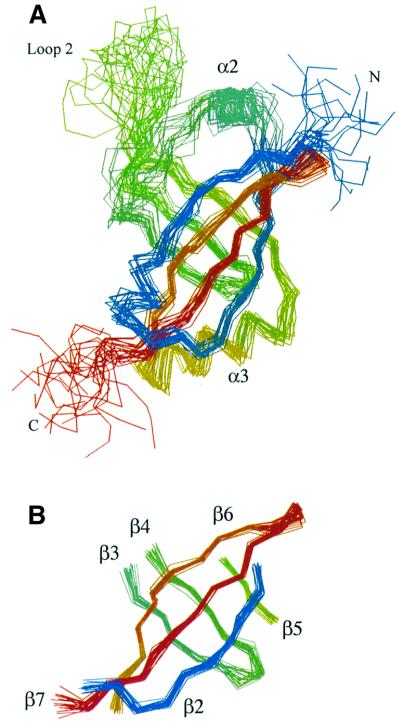
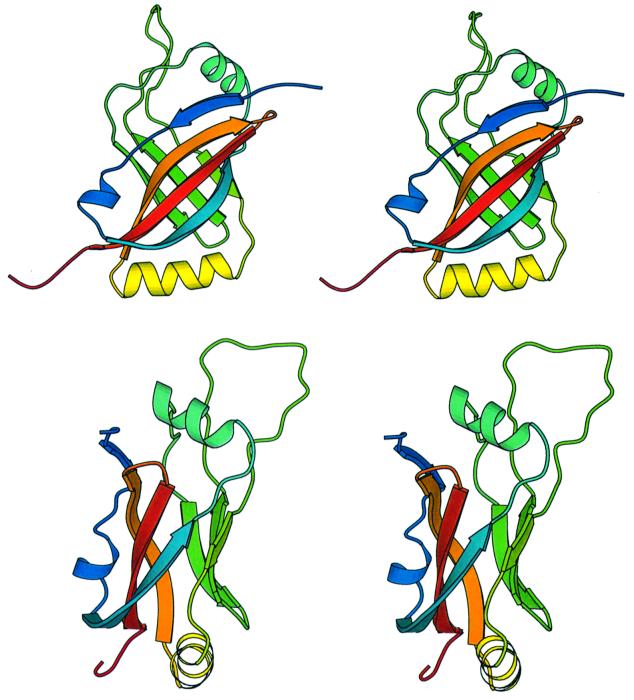
The structure of the trypsin-resistant core (residues 1–133) of SmpB was determined from constraints derived from NMR data, specifically distance constraints derived from observed NOE intensities, and torsion angle and hydrogen bond constraints for the regions identified as having regular β-sheet or helical secondary structure. The structure of the domain was quite well defined by the NMR data, with >2000 unambiguous distance constraints (559 of which were long range) being derived from the nuclear Overhauser effect (NOE) spectra. Nearly complete chemical shift assignments were obtained for the 1H, 13C and 15N nuclei. Of particular significance, complete resonance assignments were obtained for all of the leucine, isoleucine, valine, proline and alanine side chains (among others); assignment of NOE cross peaks derived from these side chains were critical in accurately defining the hydrophobic core of the protein. A superposition of a set of structures that are equally consistent with the NMR-derived constraints is shown in Figure 3. These structures are a fair representation of the full range that is consistent with the NMR data, and are all quite similar (Figure 4). Structural statistics for residues 1–133 of SmpB are summarized in Table I. Coordinates for residues 1–133 of the SmpB structure have been deposited in the PDB and assigned identification number 1K8H.
Fig. 3. Superpositions of the backbones of 20 low energy structures of SmpB. Structures shown are equally consistent with the NMR data, color-ramped from blue near the N-terminal to red near the C-terminus of the protein. The 20 models are a fair representation of the full range of structures that are consistent with the NMR-derived constraints and reasonable molecular geometry. (A) Superposition of the 20 models was performed by minimizing the differences in the coordinates of the backbone atoms of residues 6–128. Residues 1–133 of the protein are shown. (B) The 20 structures were superimposed by minimizing the differences in the coordinates of the backbone atoms within the six strands of the β-barrel; only the residues of these six β-strands are shown.
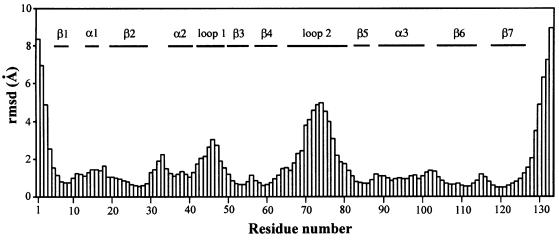
Fig. 4. Root mean square deviation (r.m.s.d.) for the backbone coordinates of SmpB. R.m.s.d. for the backbone heavy atoms versus residue number is shown for a set of 20 structures that satisfy the NMR-derived structural constraints and have a near-minimum value for the CNS energy function. R.m.s.d. values were calculated using a set of 20 structures that were superimposed by minimizing the differences in the coordinates of the backbone atoms of residues 6–128. The figure shows that the β-strands are quite well defined by the NMR data (r.m.s.d. ∼1 Å), the helices and short loops connecting the strands of the β-barrel are moderately well defined (r.m.s.d. ∼1–2 Å), while the positions of the residues in the longer loops and the few residues nearest the N- and C-termini are not as well determined (r.m.s.d. 3–9 Å). Since all parts of the structure are not equally well determined, the r.m.s.d. statistics can vary significantly depending on which parts of the structure are selected to do the superposition. If the structures are superimposed by minimizing the difference in the coordinates of only the residues of the β-strands, the r.m.s.d. for the residues in the six-stranded β-barrel is ∼0.5 Å (see Table I and Figure 3B).
Table I. Summary of refinement and structural statistics for the A.aeolicus SmpBa.
| Intraresidue NOEs | 832 |
| Sequential NOEs (residue i to i + 1) | 519 |
| Medium-range NOEs (residue i to i + 2, 3, 4) | 170 |
| Long-range NOEs | 559 |
| Dihedral angle restrains (phi and psi) | 142 |
| Hydrogen bond restrains | 43 |
| Total structural restraints | 2265 |
| Number of unique starting structures for simulated annealing | 10 |
| Number of simulated annealing runs, differing in initial trajectories | 100 |
| R.m.s.d. for backbone atoms, residues 1–133b | 2.27 Å |
| R.m.s.d. for side chains, residues 1–133 | 3.46 Å |
| R.m.s.d. for backbone atoms, residues 6–128b | 1.63 Å |
| R.m.s.d. for side chains, residues 6–128 | 2.76 Å |
| R.m.s.d. for backbone atoms, β barrel onlyc | 0.50 Å |
| R.m.s.d. for side chains, β barrel only | 1.54 Å |
| Average number of NOE violations >0.2 Å (per structure) | 7.0 |
| Average number of NOE violations >0.5 Å (per structure) | 0.15 |
| Number of NOE violations >0.6 Å | 0 |
| Residues in most favored regions of Ramachandran plot | 70.0% |
| Residues in additional allowed regions of Ramachandran plot | 20.9% |
| Residues in generously allowed regions of Ramachandran plot | 5.5% |
| Residues in disallowed regions of Ramachandran plotd | 3.6% |
| R.m.s.d. for covalent bonds | 0.0019 Å |
| R.m.s.d. for covalent angles | 0.38° |
| R.m.s.d. for improper angles | 0.29° |
aStatistics are derived from a set of 20 low-energy structures, a set that is representative of the range of structures that are consistent with the structural constraints. The values of the CNS energy function, averaged for the 20 low-energy structural models are Etotal = 184.4, Ebond = 8.5, Eangle = 91.7, Eimproper = 14.7, Evdw = 43.1, Ecdih = 2.4, and Enoe = 23.9.
bSuperimposition was carried out using residues 6–128.
cSuperimposition was carried out using the β-barrel residues only.
dThe four residues in disallowed regions of Ramachandran plot are A10, K32, L80 and N115; these residues are in loop or turn regions where the uncertainty in the structure may account for their presence in disallowed regions.
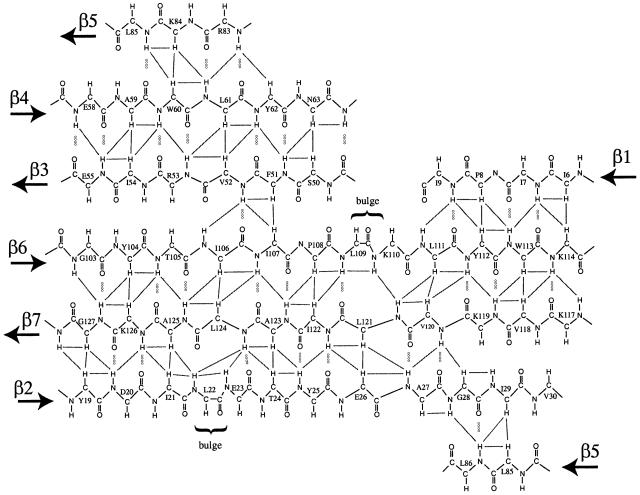
The NMR results show that the core of the SmpB protein consists of a closed barrel of six antiparallel β strands, designated β2–β7 in Figure 5. Strand β1 is antiparallel to strand β6 but is not an integral component of the closed β-barrel (Figure 5). The β-strands within SmpB differ significantly in length. Strands β2, β6 and β7 each consist of ∼11 residues and are connected by an extensive network of hydrogen bonds to form a large sheet; strands β1, β3, β4 and β5 are relatively short, consisting of four to six residues each. The regular networks of hydrogen bonds in strands β2 and β6 are interrupted by single-residue bulges at residues Leu22 and Leu109. These bulges are well defined by the NMR data, and contribute to the overall twist of the β-sheet. Interestingly, the residues of strands β2 and β6 near each bulge are well conserved among the various species, suggesting that the bulges are a conserved structural feature (Figure 1). The SmpB structure contains three regular α helices: (i) a short helix (α1) links strands β1 and β2; (ii) a two-and-a-half turn helix (α2) links strands β2 and β3; and (iii) and a three-and-a-half turn helix (α3) links strands β5 and β6. Each of these three helices has a distinct (and conserved) hydrophobic face that contacts conserved hydrophobic residues on the external surface of the β-barrel. The most extensive region of non-regular secondary structure is residues 64–82 (designated loop 2), which link strands β4 and β5. Ribbon diagrams depicting the fold of SmpB are shown in Figure 6.
Fig. 5. β-sheet structure of the SmpB protein. Note that strand β5 appears twice in the figure, to show its anti-parallel arrangement relative to strands β2 and β4, completing the closed β-barrel structure. Some of the pairs of protons for which NOE cross peaks are observed are indicated by lines. Inter-strand hydrogen bonds are indicated by dotted lines.
Fig. 6. Stereoview ribbon diagrams of SmpB. The two views differ by a 90° rotation. The protein is color-ramped from blue at the N-terminus to red at the C-terminus. The coordinates for SmpB have been submitted to the Protein Data Bank and have been assigned PDB code 1K8H. The diagram was created using MOLSCRIPT (Kraulis, 1991).
Amino acids are conserved among similar proteins from widely divergent species for reasons related to either structure or function; residues within a protein core are often conserved for structural reasons, while conserved residues on a protein surface are often important for function. When the amino acid sequences of SmpB from several species are aligned (as in Figure 1) and this alignment is compared with the structural model, it becomes possible to identify and distinguish those amino acids that are most likely to be essential for either structural or functional purposes. Hydrophobic residues within the core of the protein, as well as conserved residues in turns, are likely to have essential structural roles; these residues are boxed in Figure 1. Well conserved amino acids on the surface of the protein were also identified; these residues are most likely to make essential contacts with the tmRNA or other RNA or protein components of the translational apparatus, as part of the mechanism of ribosome rescue and tagging of peptides for degradation.
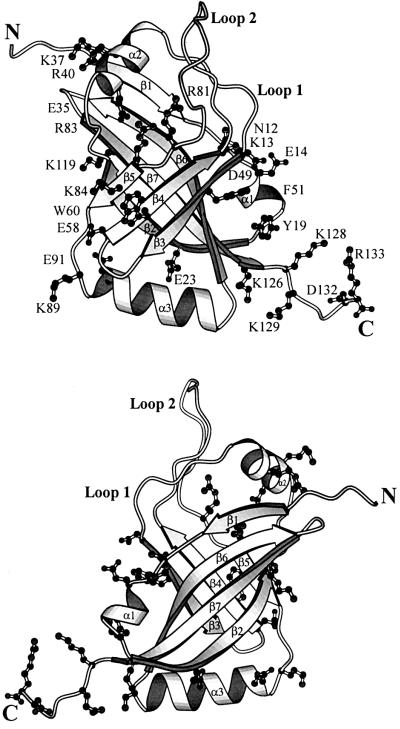
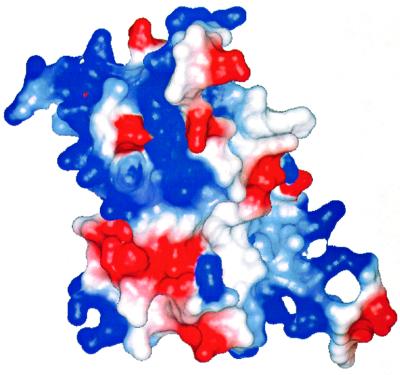
Conserved residues cover a substantial fraction of the surface of SmpB. This is particularly apparent when the protein is viewed facing strands β3, β4 and β5 of the β-barrel (Figure 7). The conserved surface residues are clustered into two regions of the protein surface. One of these regions consists of a broad surface centered on the solvent-exposed side of the strands β4 and β5, and contains residues Glu35, Lys37, Arg40, Glu58, Trp60, Arg81, Arg83, Lys84, Lys89, Glu91 and Lys119. These residue types are typical of those that populate RNA binding sites in other RNA-associated proteins, where lysine and arginine can have favorable electrostatic interactions with RNA, glutamate can serve as a hydrogen bond acceptor, and tryptophan can interact with RNA via ring stacking. This surface is therefore a likely region of contact with RNA. A second conserved surface contains residues Asn12, Lys13, Glu14, Tyr19, Glu23, Asp49, Phe51, Lys126, Lys128, Lys129, Asp132 and Arg133, and represents a second likely RNA-binding site (Figure 7). A plot of the electrostatic potential shows that the most conserved regions of the protein surface are, in the main, positively charged (Figure 8), which would facilitate interactions with the RNA backbone. The total charge of SmpB is clearly positive; the calculated isoelectric pH of the protein is 9.9. Interestingly, SmpB contains a surface region centered on the solvent-exposed side of strands β2, β6 and β7, which is mostly hydrophilic but lacking in well conserved residues (Figure 7), and therefore is likely to be solvent exposed when SmpB is bound in complex with the tmRNA.
Fig. 7. Positions of conserved residues on the surface of SmpB. A view facing strands β3, β4 and β5 of the β-barrel (top panel) shows that conserved residues cover a large fraction of the protein surface. Conserved amino acids are clustered into two regions of the protein surface, separated by a rather large distance and angle, suggesting that SmpB could bring together and stabilize distal regions of the tmRNA, or perhaps facilitate an interaction with another component of the translational apparatus while binding to the tmRNA. In contrast, the face of the β-barrel centered on strands β2, β6 and β7 (bottom panel) is centered on a surface that lacks conserved hydrophilic residues; this latter surface is most likely to be solvent exposed when SmpB is in its functional complex with the tmRNA.
Fig. 8. Electrostatic surface potential plot for SmpB. The view faces strands β3, β4 and β5, where the majority of the conserved surface amino acids are visible. The surface charge is predominantly positive (blue) on the surfaces containing the highest density of conserved residues (upper right and lower left), indicating regions of likely interaction with RNA. It is noted, however, that negatively charged amino acids (red) can also make specific interacts with RNA bases by serving as hydrogen bond acceptors. The figure was prepared using the program MOLMOL (Koradi et al., 1996).
NMR data provide evidence that the structure of the trypsin-resistant core of SmpB (residues 1–133) is uninfluenced by the hydrophilic C-terminal ‘tail’ residues 134–156. Triple resonance spectra were acquired for the full-length and truncated versions of the protein, and used to compare the chemical shifts of the 1H, 13C and 15N nuclei. For the region of the molecule where the resonance assignments are essentially complete (residues 3–131), no significant chemical shift changes could be attributed to the removal of the C-terminal tail. The resonances of the nuclei within the C-terminal tail were identified by comparing the spectra of the full-length and truncated versions of the protein; however, we were unable to make sequence-specific assignments for the majority of these tail residues due to the severe overlap of their chemical shifts near the random coil values. Overall, the NMR data support a model where the globular trypsin-resistant core of SmpB is structurally independent of the C-terminal tail, which most likely has a random coil structure, at least when the purified SmpB is isolated in solution.
Lack of structure in the C-terminal tail does not necessarily imply that it is functionally unimportant; evidence to the contrary is provided by the presence of well conserved, positively charged residues Arg134 and Lys140, which have the potential to specifically interact with RNA. In this sense, SmpB is reminiscent of several of the proteins within the large and small ribosomal subunits, such as L15, L21e and L37e (Ban et al., 2000), and S4 (Sayers et al., 2000), which contain substantial charged tails at the N- and/or C-terminal of an otherwise globular domain. In the context of the structure of the complete ribosomal subunits, these charged protein tails were found to penetrate to the interior of the ribosome and make specific contacts with the ribosomal RNA (Ban et al., 2000; Wimberly et al., 2000). By analogy, we suggest that the C-terminal tail of SmpB may become structured and have specific interactions within the context of the complete and functional tmRNA–SmpB ribonucleoprotein complex, perhaps penetrating into the interior of the tmRNA. Aquifex aeolicus SmpB that is missing the C-terminal tail residues 134–156 was found to have a significantly reduced affinity for the tmRNA (J.Wower, unpublished data).
Discussion
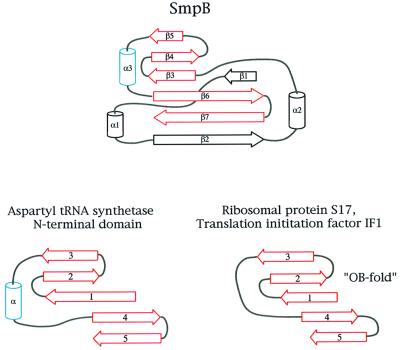
Primary sequence comparisons attest to the uniqueness of SmpB; however, the structural results provided by the present work provide an additional means for comparing SmpB with other proteins. Searches using the programs DALI (Holm and Sander, 1997) and VAST (available at the NCBI web site) indicated that there are no structures within the PDB that have significant similarity to SmpB, based on comparison of the coordinates of backbone atoms. However, a more qualitative inspection of the overall topology of SmpB reveals the presence of an oligonucleotide binding (OB) fold (Murzin, 1993) within the structure of its β-sheet. In this respect, SmpB is similar to several other RNA-binding proteins that are known to be associated with translation (Draper and Reynaldo, 1999), including ribosomal protein S17 and the prokaryotic translation initiation factor IF1 (Figure 9). Like SmpB, proteins IF1 and S17 each contain substantial RNA binding surfaces. IF1 binds to the 16S RNA in a transient manner at the ribosomal A-site (Carter et al., 2001) while S17 is a more permanent part of the ribosomal small subunit, with extensive contacts to the 16S RNA (Wimberly et al., 2000). The similarity between SmpB and the N-terminal domain of aspartyl tRNA synthetase (DRS) is even more notable due to the presence of a helix that is in a position analogous to helix α3 in SmpB (Figure 9). This tRNA synthetase domain directly contacts the anticodon loop when in complex with the tRNA (Cavarelli et al., 1993). These apparent subtle similarities between the structures of IF1, S17, DRS and SmpB, combined with their close association with the RNA components of the translational apparatus, suggest that they may share a common ancestor or be linked by an evolutionary relationship. Functionally, SmpB may be more like a ribosomal protein than a transient RNA-binding protein, such as a tRNA synthetase or initiation factor, since the available evidence suggests it is an integral part of the tmRNA ribonucleoprotein complex (Karzai and Sauer, 2001).
Fig. 9. Relationship of SmpB to other RNA-binding proteins. The structure of SmpB contains an embedded oligonucleotide binding (OB-fold); in this respect, SmpB is related to several other proteins associated with the translational apparatus. Ribosomal protein S17 and translation initiation factor IF1 are other members of the OB-fold family of proteins, and contain a five-stranded antiparallel β-sheet (shown in red), with the strands connected in a Greek-key topology. The N-terminal domain of aspartyl tRNA synthetase also contains the OB-fold within its structure, as does SmpB. The secondary structures of the aspartyl tRNA synthetase, S17, IF1 and SmpB are derived from PDB entries 1ASZ, 1RIP, 1AH9 and 1K8H, respectively.
In addition to SmpB, the structures of other proteins that contact RNA and are associated with the process of translation have been found to contain six-stranded closed β-barrels; these include domain III of EF-Tu (Nissen et al., 1995) and domains within translation initiation factor IF2 (Meunier et al., 2000; Roll-Mecak et al., 2000). However, the connectivity of the strands within the β-barrels of the EF-Tu and the IF2 domains differs from that in SmpB, providing evidence against a close evolutionary relationship between these proteins.
How and where might SmpB bind to the tmRNA? The tmRNA, with ∼360 nucleotides, is substantially larger than SmpB alone. The secondary structure of the RNA contains a tRNA-like domain, a region with an mRNA-like function, and four pseudoknots (PK1–PK4), the first of which (PK1) is located between the mRNA- and tRNA-like domains within the tmRNA primary sequence (Karzai et al., 2000; Zwieb et al., 2001). Previously it has been reported that modification of the PK2, PK3 and PK4 pseudoknots does not remove the peptide tagging function of the tmRNA–SmpB complex (Nameki et al., 2000), which suggests that SmpB binds to the mRNA- or tRNA-like regions or PK1. SmpB does not prohibit binding of EF-Tu to the tRNA-like domain or block the interaction of the pseudoknots and mRNA-like region with ribosomal protein S1 (Wower et al., 2000), and actually stimulates the function of alanine tRNA synthetase as it charges the tRNA-like domain with alanine (Karzai et al., 2000; Barends et al., 2001). This information serves to restrict even further where the SmpB–tmRNA interactions may occur. Recently, Barends et al. (2001) used a combination of in vitro kinetic, gel-shift and enzymatic protection assays to provide additional evidence that SmpB and EF-Tu simultaneously bind to the amino acid acceptor stem region of Ala-tmRNA. Further insight into the nature of the SmpB–tmRNA interaction is provided by the results of the present work, which show that conserved residues of SmpB are distributed over a large region of the protein surface, and on opposite sides of the protein structure (Figure 7), suggesting that SmpB contains more than one interaction surface and makes more than just a single simple contact with the tmRNA. The evidence that is now available suggests that SmpB may serve to bring together and stabilize a particular conformation and relative orientation of the mRNA- and tRNA-like regions of the tmRNA, in a role analogous to that of some of the ribosomal proteins that contact distal regions of the ribosomal RNA. In another (alternative) role, SmpB, via its substantial and widely spaced RNA-binding surfaces, could serve as a mediator of the interaction between the tmRNA and the stalled ribosome as part of the mechanism of ribosome rescue by transference of the decoding function from the messenger RNA to the mRNA-like region of the tmRNA; perhaps a surface of SmpB can interact with the 16S ribosomal RNA at or near the sites usually occupied by tRNA. In support of this latter scenario, it has been reported that SmpB is required for the association of tmRNA with 70S ribosomes (Karzai et al., 1999). Clearly, however, a detailed and definitive answer as to exactly how SmpB binds to the tmRNA and enables the functions of ribosome rescue and peptide tagging must await an NMR or crystallographic analysis of SmpB protein–RNA complexes. Biophysical studies designed to shed more light on the details of the SmpB–tmRNA complex and its interactions are currently in progress.
Materials and methods
Protein cloning, expression and purification
The gene encoding the SmpB protein was amplified by PCR from A.aeolicus genomic DNA, kindly provided by Dr Robert Huber. PCR primers were designed to incorporate unique NdeI and BamHI restriction sites, permitting the insertion of the amplified smpb gene into the pET-9a vector (Novagen) for expression in E.coli. DNA sequencing confirmed the correct sequence of the full-length smpb gene. Cells were grown at 37°C in Luria broth supplemented with kanamycin, and protein expression was induced when the cells reached an OD600 of 0.6 with 0.4 mM isopropyl-β-thiogalactopyranoside (IPTG). Four hours after the addition of IPTG, the cells were harvested by centrifugation and stored at –80°C. Thawed cells were lysed by the addition of lysozyme followed by sonication. Cellular nucleic acids were precipitated by adding polyethylenimine (PEI) to the cell lysate, up to 1% volume (v/v). After centrifugation to remove the nucleic acids and cell debris, cellular proteins (including SmpB) were precipitated from the supernatant by adding ammonium sulfate, up to 65% saturation. The precipitated proteins were separated by centrifugation, and the protein pellet was dialyzed against a 50 mM Tris–Cl buffer at pH 7.5. The dialyzed proteins were passed through a quaternary amine anion exchange column (UNOsphere Q, Bio-Rad), which bound many cellular proteins, but not the SmpB. Proteins that did not bind to the anion exchange column were then loaded to a sulfopropyl agarose cation exchange column (UNOsphere S, Bio-Rad), and eluted with a linear 0.1–0.4 M gradient of NaCl at a flow rate 2 ml/min. Fractions containing nearly pure SmpB were identified by SDS–PAGE and combined. The buffer was changed to 10 mM sodium phosphate plus 50 mM NaCl at pH 6 and samples were concentrated for NMR analysis. The identity of the SmpB protein was confirmed by N-terminal sequencing. Samples of SmpB enriched in 15N and/or 13C were prepared as above, but with M9 minimal media containing 1 g/l 15N ammonium chloride and/or 1 g/l 13C glucose (Cambridge Isotope Laboratories) as the source of nitrogen and/or carbon.
Protease-cleaved fragments of SmpB were obtained by adding trypsin to purified SmpB, so that the trypsin concentration was 0.1 mg/ml. This limited proteolysis was carried out at 37°C for 15 min, using an SmpB concentration of typically 5–10 mg/ml in buffer containing 100 mM NaCl and 50 mM Tris–Cl pH 7.5. The trypsin cleavage was stopped by the addition of 10 µg/ml PMSF. Mass spectrometry showed that these cleavage conditions resulted in two fragments of SmpB, consisting of residues 1–133 (the major product) and 3–133, with numbering as in Figure 1. The two fragments were easily separated using cation exchange chromatography (UNOsphere S, Bio-Rad), where the larger fragment eluted at higher NaCl concentration than the smaller fragment. Small and large SmpB tryptic fragments eluted at approximately 0.1 and 0.2 M NaCl concentration, respectively, in a 0–0.5 M NaCl gradient.
NMR spectroscopy
NMR spectra were recorded at temperatures between 30 and 40°C using a 500 MHz Varian Inova spectrometer equipped with a triple-resonance probe and z-axis pulsed-field gradient. NMR samples typically contained 1 to 2 mM of the protein SmpB and 10 mM sodium phosphate plus 50 mM NaCl in 90% H2O/10% D2O or 100% D2O at pH 6. Backbone resonance assignments were obtained using three-dimensional HNCA, HNCO, HCACO, HNCACB and HN(CO)CACB spectra that correlate the backbone protons to the N, Cα, Cο and Cβ signals of the same and adjacent amino acid residues. Side chain 1H resonance assignments were obtained using two-dimensional 2QF-COSY and TOCSY spectra, and three-dimensional 15N-1H-1H HMQC-TOCSY and 1H-1H-13C HCCH-TOCSY spectra. NOE cross peaks were identified in two-dimensional 1H-1H NOE spectra, a three-dimensional 15N-1H-1H HMQC-NOE spectrum, and a three-dimensional 13C-edited 1H-1H HMQC-NOE spectrum. The 13C-edited 1H-1H NOE spectrum was collected in 90% H2O/10%D2O solvent, so that NOE peaks between amide and side-chain protons could be resolved by the chemical shift of a side-chain 13C nucleus.
Structure determination
The structure of the SmpB protein was determined using the hybrid distance geometry-simulated annealing and energy minimization protocols within the CNS program suite (Brünger et al., 1998), with the goal of identifying the full range of structures that are consistent with the distance and angle constraints derived from the NMR data, while having reasonable molecular geometry, consistent with a minimum value of the CNS energy function. Distance constraints were derived from the intensities of cross peaks within the multi-dimensional NOE spectra. Whenever possible, NOE cross peaks were identified in spectra with relatively short (60 ms) mixing times, to minimize the effects of spin diffusion on the structure calculation. Peaks were placed into one of four categories, ranging from strong to very weak, and assigned to interproton distance bounds as follows: strong (<3.2 Å), medium (<3.6 Å), weak (<4.2 Å) and very weak (<4.5 Å). NOE cross peaks identified in the 15N-edited or 13C-edited three-dimensional NOE spectra obtained with longer mixing times (120 ms), or in two-dimensional homonuclear spectra with mixing times of up to 160 ms, were assigned to interproton distance bounds as follows: strong and medium peaks (<5.5 Å), weak and very weak peaks (<7.0 Å), with these more generous distance ranges being used so that errors would not be introduced due to the influence of spin diffusion on peak intensities. Pseudoatom corrections to distance restraints were included as follows: when NOEs involving methyl protons of valine and leucine were not stereospecifically assigned, distances were measured from the center of the two methyl groups, and 2 Å was added to the interproton distance. For NOEs involving other methyl protons, distances were measured from the center of the methyl group and 1 Å was added to the interproton distance. For NOEs involving methylene protons with no stereospecific assignment, distances were measured from the center of the methylene group and 0.7 Å was added to the interproton distance. For NOEs involving delta and epsilon protons on tyrosine and phenylalanine rings that were not uniquely assigned, distances were measured from the central point between the two delta (or epsilon) protons, and 2.5 Å were added to the interproton distance. Backbone torsion angle restraints for phi and psi were only included for regions of regular β-strand or α-helix structure that were clearly identified by characteristic NOE cross peak patterns, 13C chemical shifts and slowly exchanging amide protons. In these cases, phi and psi were restricted to –120° ± 25 and 150° ± 25, respectively, for β-strands, and –60° ± 25 and –60° ± 25, respectively, for α-helices. Hydrogen bond restraints were imposed using distance bounds, and were only included for regions of regular β-strand or α-helix structure, and where the amide proton was substantially protected from solvent exchange.
Ten diverse starting structures were generated by subjecting a random coil model to the CNS simulated annealing protocol using only the dihedral angle constraints. These 10 structures were then used as starting models for 100 runs of the simulated annealing protocol. Most of the simulated annealing runs resulted in similar structures with similar energies. From this final set of refined models, a set of 20 structures were selected that satisfy the following criteria: (i) their CNS energy term was at or very near the minimum value obtained; (ii) there were no interproton distance constraint violations of >0.6 Å; and (iii) no torsion angle constraint violations exceeded 2.5°. These 20 models are a fair representation of the full range of structures that satisfy the NMR-derived restraints while having reasonable molecular geometry, as defined by the CNS energy function. Structural statistics (Table I) were calculated with the assistance of the program PROCHECK-NMR (Laskowski et al., 1996).
Acknowledgments
Acknowledgements
This work was supported by a grant from the Robert A. Welch Foundation (F-1353) and a grant from the American Cancer Society (GMC-89306).
References
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Barends S., Wower,J. and Kraal,B. (2000) Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry, 39, 2652–2658. [DOI] [PubMed] [Google Scholar]
- Barends S., Karzai,A.W., Sauer,R.T., Wower,J. and Kraal,B. (2001) Simultaneous and functional binding of SmpB and EF-Tu.GTP to the alanyl acceptor arm of tmRNA. J. Mol. Biol., 314, 9–21. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons,W.H., Broderson,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.C. and Moras,D. (1993) Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Draper D.E. and Reynaldo,L.P. (1999) RNA binding strategies of ribosomal proteins. Nucleic Acids Res., 27, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B., Himeno,H., Muto,A., Atkins,J.F. and Gestleland,R.F. (1996) Structural organization of Escherichia coli tmRNA. Biochimie, 78, 979–983. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Roche,E., Zhou,Y. and Sauer,R.T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev., 12, 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1997) DALI/FSSP classification of three-dimensional protein folds. Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A.W. and Sauer,R.T. (2001) Protein factors associated with the SsrA–SmpB tagging and ribosome rescue complex. Proc. Natl Acad. Sci. USA, 98, 3040–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA–SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- Keiler K.C., Shapiro,L. and Williams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial gemiomes: a two piece tmRNA functions in Caulobacter. Proc. Natl Acad. Sci. USA, 97, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J.E., Trempy,J.E. and Gottesman,S. (1994) Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol., 176, 2068–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B., Wower,J., Zwieb,C. and Gorodkin J. (2001) tmRDB (tmRNA database). Nucleic Acids Res., 29, 171–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Kitabatake,M., Yokogawa,T., Nishikawa,K. and Inokuchi,H. (1994) A tRNA like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi P., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann,J.A., MacArthur,M.W., Kaptein,R. and Thorton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Meunier S., Spurio,R., Czisch,M., Wechselberger,R., Guenneugues,M., Gualerzi,C.O. and Boelens,R. (2000) Structure of the fMet-tRNAfMet-binding domain of B. stearothermophilus initiation factor IF2. EMBO J., 19, 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki N., Tadaki,T., Himeno,H. and Muto A. (2000) Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett., 470, 345–349. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Tirup,S., Polekhina,G., Reshatnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Retallack D.M. and Friedman,D.I. (1995) A role for small stable RNA in modulating the activity of DNA-binding proteins. Cell, 83, 227–235. [DOI] [PubMed] [Google Scholar]
- Retallack D.M., Johnson,L.L. and Friedman,D.I. (1994) Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J. Bacteriol., 176, 2082–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., Cao,C., Dever,T.E. and Burley,S.K. (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes GDP and GTP binding. Cell, 103, 781–792. [DOI] [PubMed] [Google Scholar]
- Rudinger-Thirion J., Giege,R. and Felden,B. (1999) Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA, 5, 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Gerstner,R.B., Draper,D.E. and Torchia,D.A. (2000) Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry, 39, 13602–13613. [DOI] [PubMed] [Google Scholar]
- Tu G.F., Reid,G.E., Zhang,J.G., Moritz,R.L. and Simpson,R.J. (1995) C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem., 270, 9322–9326. [DOI] [PubMed] [Google Scholar]
- Ushida C., Himeno,H., Watanabe,T. and Muto,A. (1994) tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res., 22, 3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert T. and Schumann,W. (2001) SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol., 183, 3885–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.P. and Bartel,D.P. (1998) The tmRNA website. Nucleic Acids Res., 26, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Wower I.K., Zwieb,C.W., Guven,S.A. and Wower,J. (2000) Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J., 19, 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J., Wower,I.K., Kraal,B. and Zwieb,C.W. (2001) Quality control of the elongation step of protein synthesis by tmRNP. J. Nutr., 131, 2978S–2982S. [DOI] [PubMed] [Google Scholar]
- Zvereva M.I., Ivanov,P.V., Teraoka,Y., Topilina,N.I., Dontsova,O.A., Bogdanov,A.A., Kalkum,M., Nierhaus,K.H. and Shpanchenko,O.V. (2001) Complex of tmRNA and EF-Tu: complex modes of interaction. J. Biol. Chem., 276, 47702–47708. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Guven,S.A., Wower,I. and Wower,J. (2001) Three-dimensional folding of the tRNA-like domain of Escherichia coli tmRNA. Biochemistry, 40, 9587–9595. [DOI] [PubMed] [Google Scholar]